Abstract
In vitro human cell line models have been widely used for cancer pharmacogenomic studies to predict clinical response, to help generate pharmacogenomic hypothesis for further testing, and to help identify novel mechanisms associated with variation in drug response. Among cell line model systems, immortalized cell lines such as Epstein-Barr virus (EBV)-transformed lymphoblastoid cell lines (LCLs) have been used most often to test the effect of germline genetic variation on drug efficacy and toxicity. Another model, especially in cancer research, uses cancer cell lines such as the NCI-60 panel. These models have been used mainly to determine the effect of somatic alterations on response to anticancer therapy. Even though these cell line model systems are very useful for initial screening, results from integrated analyses of multiple omics data and drug response phenotypes using cell line model systems still need to be confirmed by functional validation and mechanistic studies, as well as validation studies using clinical samples. Future models might include the use of patient-specific inducible pluripotent stem cells and the incorporation of 3D culture which could further optimize in vitro cell line models to improve their predictive validity.
Keywords: anticancer therapy, cancer cell line collections, drug response, in vitro human cell line models, lymphoblastoid cell lines, NCI-60 panel, pharmacogenomics
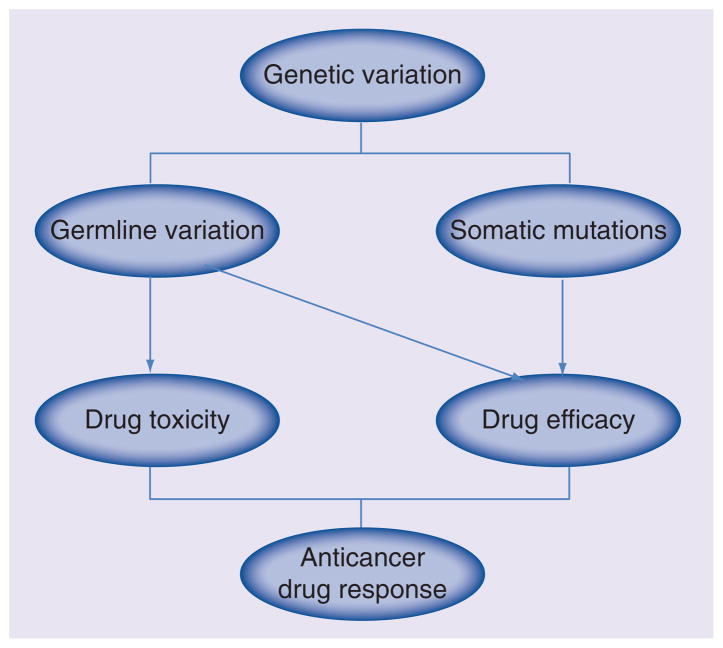
Patient response to anticancer treatment varies widely, and one major factor contributing to this variation is host genetic background – including both germline and somatic genetic variation (Figure 1). Pharmaco genomics is the study of the role of inherited and acquired genetic variation in drug response [1]. Preclinical models such as cell line model systems may be particularly useful to help predict anticancer drug response and to help further our understanding of mechanisms of drug action in cases when there is limited access to clinical samples and/or the cost to obtain clinical samples to study drug response is too great [2]. Since both germline genetic variants and tumor somatic alterations can contribute to variable drug response, cell lines focused on germline DNA as well as on somatic alterations are both important in pharmacogenomic research. Currently, there are two common types of in vitro human cell line models. One involves immortalized cell line model systems such as Epstein-Barr virus (EBV)-transformed lymphoblastoid cell lines (LCLs) which can be used to study the effect of germline variation on both drug efficacy and adverse events [3–26], while the other one involves cancer cell line model systems such as the NCI-60 cancer cell panel [27], the Cancer Cell Line Encyclopedia (CCLE) [28], and the Cancer Genome Project (CGP) [29], all of which can be used to investigate the effect on drug efficacy of somatic mutations in addition to germline genetic variation.
Figure 1.
Cancer pharmacogenomics.
The application of in vitro human cell line models to study variation in drug response has many advantages. The cell lines represent a renewable resource and, for many of these cell line systems, extensive multiomic data (such as genomics, epigenomics, transcriptomics, proteomics and metabolomics) are available or is being made available through public databases. Additional results from novel high throughput assays could be continuously accumulated for these cells in a relatively short time period. In general, cell lines are well-controlled systems and many phenotypes (such as cytotoxicity, growth rate, gene expression change, intracellular metabolites) could be measured by various high-throughput assays for any given drug or combinations of drugs with fewer confounding factors than are found for clinical sample. Finally, as mentioned earlier, a great deal of molecular data are publicly available, which makes these models extremely valuable for laboratories around the world. However, as for any model system, there are also limitations associated with these cell lines. The microenvironment and drug pharmacokinetic effects on clinical response can’t be assessed [2]. Gene expression profiles in cell line models are not identical with those for primary tissues [30]. Cell culture might also introduce new mutations and change the cell line characteristics. Therefore, further functional validation and clinical confirmation of biomarkers discovered using in vitro cell line models will be required.
Since both immortalized cell line models and cancer cell line models have both contributed to a series of advances in cancer pharmacogenomics, in subsequent paragraphs, we have reviewed some of the discoveries made with EBV-transformed LCL and cancer cell line models. Finally, we will also describe the future possibility of generating patient-specific inducible pluripotent stem (iPS) cell systems as well as incorporating 3D culture to improve the clinical predictive validity of data obtained with in vitro cell line models.
EBV-transformed lymphoblastoid cell line models
EBV-transformed LCLs are immortalized cell lines derived from human peripheral B lymphocytes. These cells contain normal diploid karyotypes and represent germline variation of the donor [31]. Whole exome and genome sequencing have showed 99% concordance in DNA sequence between LCLs and peripheral blood cells from the same individual [32,33]. However, the analysis of DNA methylation revealed similar methylation patterns only in promoter regions [34]. Furthermore, the regulation of gene expression is tissue specific [35,36]. EBV transformation and cell culture processes might also introduce chromosomal instability and cellular changes (such as cell growth rate, baseline ATP levels) in addition to nongenetic factors that could influence drug response cytotoxicity assays performed with LCLs [37,38]. Therefore, functional validation in additional appropriate cell lines and/or clinical samples is required, and the use of LCLs at early passage is recommended. We understand that biomarkers identified through LCLs might not be validated by functional assay or clinical study and this could be partly due to the confounding factors. Finally, LCL models might only be appropriate for the study of genes influencing the mechanism of drug action (i.e., pharmacodynamics) because many drug metabolizing enzymes and drug transporters are not highly expressed in these cell lines [39].
Many large LCL collections from different human populations are commercially available from the Coriell Institute [40], which is an important cell line repository funded by the NIH [41]. Specifically, National Institute of General Medical Science (NIGMS) and National Human Genome Research Institute (NHGRI) collections such as the Human Variation Panel and LCLs from the International HapMap Project [3–26] have been used extensively for cancer pharmacogenomic studies. Recently all of the LCLs from the 1000 Genomes Project have also been deposited into the NHGRI collection, cell lines from 1092 individuals in 14 populations with diverse ancestry, and each cell line contains publicly available Next-Generation sequencing information for common, low-frequency and rare variants [42]. In addition, biobank sample is another important source for large LCL collections from population-based cohorts. Many Institutions across the world have established various Biobanks, such as the National Biobank of Korea (NBK) [43], most of which have tight links to the Electronic Medical Records for future research. A method has been reported to successfully generate LCLs from small volumes of cryopreserved whole blood samples, which might make large scale generation of LCLs from biobank samples possible [44]. Since most studies published thus far are studies using the Human Variation Panel and HapMap LCLs, in this review we will focus mainly on those two models.
Human Variation Panel
Taking advantage of the Coriell Institute NIGMS Human Variation Panel collection, our laboratory created a LCL model system containing 96 African–American (AA), 96 Caucasian–American (CA) and 96 Han Chinese–American (HCA) LCLs (sample sets HD100AA, HD100CAU, HD100CHI) derived from unrelated healthy individuals to study cancer pharmacogenomics. For each cell line, extensive multiomics data have been generated, including 1.3 million genotyped SNPs plus 5.4 million imputed SNPs, 54,613 mRNA expression probe sets, 485,577 DNA methylation probes, 733 miRNA expression probe sets, as well as cytotoxicity phenotype for 23 anticancer drugs and radiation therapy [10,11,13–16,18,19,23,45,46]. As mentioned earlier, one advantage of using cell line models is that, as technology develops, more assays, such as metabolomics or proteomics can be added to existing data sets to make the models more comprehensive. In addition to data accumulation, novel integrative analysis methods have also been developed for biomarker discovery [47–51].
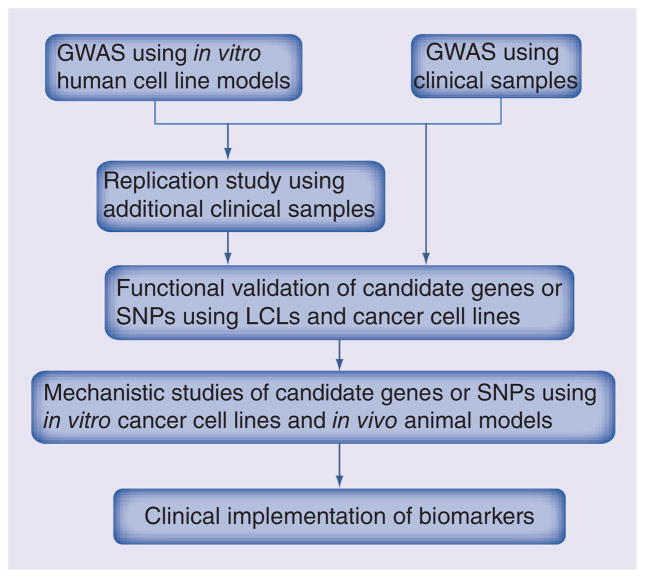
One major application of cell line models is to generate pharmacogenomic hypotheses that might help us to identify novel mechanisms of drug action and/or help to design better treatment regimens (Figure 2). Through a genome-wide association study (GWAS) using the Human Variation Panel with enriched molecular features, we discovered that basal mRNA expression of the FKBP5 gene was significantly associated with gemcitabine cytotoxicity (IC50) with a Bonferroni corrected p-value of 0.0001 [13]. Functional validation using siRNA showed that knockdown of FKBP5 altered tumor cell sensitivity to gemcitabine in pancreatic and breast cancer cell lines [13]. Mechanistic study revealed that FKBP5 acted as a scaffolding protein for Akt and PHLPP, and influenced cancer cell death following genotoxic stress by promoting dephosphorylation of Akt at amino acid S473 [52]. In vivo pancreatic cancer cell line xenograft mouse studies showed that downregulation of FKBP5 promoted tumor growth and resistance to gemcitabine, while combination treatment with gemcitabine and an Akt inhibitor reversed the effect [53]. Mulholland et al. subsequently confirmed that downregulation of FKBP5 increased Akt activity by reducing PHLPP-Akt interaction [54]. FKBP5 pharmacogenomic studies using normal and tumor DNA samples from pancreatic cancer patients treated with gemcitabine discovered 404 SNPs using Next-Generation DNA sequencing, which included 326 SNPs with minor allele frequencies (MAF) greater than 1%. SNP rs73748206 in Intron 2 of FKBP5 was associated with patient overall survival and affected interaction with the glucocorticoid receptor as well as level of FKBP5 gene expression [55]. Mitra et al. reported that another FKBP5 intronic SNP, rs3798346, was significantly associated with event-free survival and overall survival in pediatric acute myeloid leukemia (AML) patients treated with cytarabine [56]. This is one among many examples for which our group and others have used LCLs to characterize biomarkers at both functional and mechanistic levels [10–12,14–16,18,19,21–24].
Figure 2. Outline of using in vitro human cell line models to predict clinical response to anticancer therapy.
This flow diagram outlines the strategy of how to use an in vitro human cell line model to predict clinical response.
GWAS: Genome-wide association study;
LCL: Lymphoblastoid cell line.
In addition to the discovery of novel germline biomarkers that might contribute to anticancer drug response, these cell lines can also be used to functionally characterize biomarkers associated with clinical response phenotypes during GWAS performed using patient DNA samples (Figure 2). Those studies make possible the use of additional functional genomic approaches that can help us to determine how genetic variation affects clinical drug response. Many of these studies were designed in an attempt to identify pharmacogenomic biomarkers associated with the endocrine therapy (selective estrogen receptor modulators [SERMs] or aromatase inhibitors [AIs]) in the treatment of ER+ breast cancer using samples from large clinical trials. One of the first studies was designed to identify pharmacogenomic biomarkers associated with AI-induced musculoskeletal adverse events (MS-AE). A signal close to the TCL1A gene on chromosome 14 was observed for 293 cases and 585 controls using germline patient DNA samples from the MA.27 Phase III clinical trial that was designed to compare the efficacy of two AIs, anastrozole and exemestane. Four SNPs close to this gene were associated with MS-AE with p-values of 2.23 × 10−6 to 6.67 × 10−6. It should be pointed out that these p-values were not genome-wide significant. However, functional studies using Human Variation Panel LCLs selected on the basis of genotypes for the GWAS SNP signals indicated that SNP rs11849538 regulated the estrogen-dependent expression of TCL1A and – downstream – of IL17RA through the creation of a new estrogen response element (ERE) [9]. Mechanistic studies indicated that estradiol (E2) induced SNP-dependent TCL1A expression which also influenced the expression of IL-17, IL-12, IL-12RB2, IL-1R2, as well as NF-κB transcriptional activity [57]. These mechanistic finding could have implications for other disease areas beyond AI-induced MS-AE, specifically, rheumatologic diseases such as rheumatoid arthritis (RA). In a similar fashion, a clinical GWAS using DNA samples from 772 postmenopausal women with estrogen receptor (ER)-positive breast cancer prior to the initiation of AI therapy identified 18 SNPs in or near the TSPYL5 gene on chromosome 8 – all in high linkage disequilibrium (LD) – that were strongly associated with basal plasma estradiol (E2) concentration with a p-value for the top SNP that was less than 3.5 × 10−8. In this case, the SNP p-value was genome-wide significant. Using LCLs with different genotypes for the imputed rs2583506 SNP demonstrated that that SNP created a functional ERE and regulated E2-dependent TSPYL5 expression which, in turn, influenced E2 biosynthesis by inducing CYP19A1 expression [17]. Another clinical GWAS using 592 cases and 1171 matched control DNA samples from subjects enrolled in the NSABP P-1 and P-2 SERM breast cancer prevention trial identified SNPs in or near the ZNF423 and CTSO genes that were associated with breast cancer risk during SERM therapy [8]. Functional studies using LCLs with known genotypes for these SNPs revealed that both ZNF423 and CTSO were involved in the SNP-dependent and estrogen-dependent induction of BRCA1 expression. ZNF423 appears to be an estrogen-inducible BRCA1 transcription factor [8]. These novel findings may help us to move toward more highly personalized SERM prevention therapy and, as a result, to change the current treatment paradigm, in other words, treating 50 women to prevent one case of breast cancer [8].
The common theme running through these studies is that LCLs are a powerful model system to functionally and mechanistically characterize common germline genetic variation associated with clinical response. This type of approach adds significantly to pharmacogenomic studies for which clinical or population-based replication is difficult to do and for which the power might be limited. Therefore, to be able to functionally characterize clinical pharmacogenomic GWAS signals serves as a useful alternative approach to replication studies while at the same time providing novel insights into the underlying biology associated with identified pharmacogenomic biomarkers.
HapMap LCLs
In addition to the Human Variation Panel, another widely applied model system uses HapMap LCLs. This system consists of 270 cell lines from three ethnic groups, including 30 CEPH trios, 30 Yoruban (YRI) trios, as well as 45 unrelated Japanese and 45 unrelated Han Chinese. For each sample, about 7 million common genetic variants (MAF >5%) have been genotyped by the International HapMap Project [58]. Recently additional information for less common and rare variants has been generated by the 1000 Genome Project using Next-Generation Sequencing [42]. This model has also been extensively characterized at the molecular level, including DNA CNV, DNA methylation, mRNA and miRNA expression, protein expression as well as cytotoxicity data for various anticancer drugs [3–7,12,20–22,24–26,59]. These data have also been used to develop statistical methodology related to the integrated analysis of different omic data to help identify pharmacogenomic biomarkers [6,12]. In addition to the analysis of SNP, expression and drug cytotoxicity data, a comprehensive GWAS for CNVs identified drug-related CNVs that were independent of SNP signatures [4] and the analysis of protein expression for 441 human signaling proteins and transcription factors in 68 HapMap YRI LCLs found enrichment of protein quantitative trait loci (pQTLs) for cisplatin and paclitaxel response. Functional studies indicated that knockdown of SMC1A or ZNF569 altered cisplatin or paclitaxel response [22].
Similar to application of the Human Variation Panel LCLs, the HapMap cell lines have also been used for many cancer pharmacogenomic studies designed to identify and understand biomarkers for the clinical prediction of drug response. One GWAS using HapMap LCLs reported that the rs1649942 SNP was associated with carboplatin sensitivity by influencing gene expression, and further phase 1 validation indicated that this SNP was correlated with decreased progression-free survival (PFS) and overall survival (OS) in Australian Ovarian Cancer Study (AOCS) patients, but not in the phase 2 validation [7]. A meta-analysis using HapMap and other LCLs from different ethnic groups, together with a genotyping study performed with clinical DNA samples, found that the rs1203633 SNP was associated with cytarabine-induced cytotoxicity and overall survival in AML patients [60]. In addition to their use to discover biomarkers related to drug efficacy, the HapMap LCLs have also been used to identify biomarkers for drug-induced toxicity. Combination analysis of GWAS using HapMap LCLs and breast cancer patient samples from patients treated with paclitaxel found that the rs7254081 SNP in the intron of RFX2 correlated with paclitaxel response in LCLs was also associated with paclitaxel-induced sensory peripheral neuropathy through transregulation of expression of 18 genes. Functional studies showed knockdown of RFX2 affected nerve cell response to paclitaxel [24]. Even though LCLs have limitations, taken together, all of these examples clearly demonstrate the utility of these cell lines in identifying functional genomic biomarkers that can help predict clinical drug efficacy or drug-induced toxicity.
Besides these publically available LCLs, patient-derived LCLs have also been generated from different disease populations, including LCLs generated from participants in the Cholesterol and Pharmacogenetics (CAP) clinical trial and LCLs from patients enrolled in the Childhood Asthma Management Program (CAMP) clinical trial [61,62]. The 516 LCLs from Caucasian patients in the CAP trial have been used for cancer pharmacogenomic studies, and the rs531572 SNP in the MGMT gene was found to be associated with temozolomide response through the cis-regulation of gene expression [63]. Recently, another large genome-wide association study (GWAS) also utilized these 516 LCLs to screen 29 chemotherapeutic agents and identified 22 loci associated with drug response through single drug analysis as well as association analysis for drugs in the same class [64].
Cancer cell line models
Both germline DNA and tumor DNA can contribute to efficacy and/or toxicity related to anticancer treatment. Therefore, we can’t overemphasize the importance of tumor cell lines to study cancer pharmacogenomics. Cancer cell lines serve as extremely useful in vitro models for a specific tumor type to study cancer biology and treatment response (Table 1). Early collections of cancer cell lines included the NCI-60 cancer cell line panel, probably the most widely used model for studying cancer biology and the role of somatic alteration in drug response [27], as well as cancer-specific collections such as the breast cancer cell line panel and colorectal cancer (CRC) cell line panel. Many recent publications also included additional large-scale collections of various cancer cell lines with extensive genomic characterization and drug cytotoxicity profiles [28,29]. In addition, isogenic cancer cell line with knockout/knock in of specific gene/allele has also been used for cancer pharmacogenomics [65–67].
Table 1.
Examples of in vitro human cell line model systems that have been used in cancer pharmacogenomic studies.
| In vitro human cell line model | Genetic variation | |
|---|---|---|
| Immortalized cell lines | ||
| EBV-transformed LCLs | Human Variation Panel | Germline |
| EBV-transformed LCLs | HapMap LCLs | Germline |
|
| ||
| Cancer cell lines | ||
| Heterogeneous panels | NCI-60 panel | Germline and somatic |
| CCLE | Germline and somatic | |
| CGP | Germline and somatic | |
| GSK cancer cell line panel | Germline and somatic | |
| Homogeneous panels | Breast cancer cell line panel | Germline and somatic |
| Colorectal cancer cell line panel | Germline and somatic | |
| Isogenic cancer cell lines | Germline and somatic | |
CCLE: Cancer cell line encyclopedia; CGP: Cancer genome project; EBV: Epstein-Barr virus; LCL: Lymphoblastoid cell line.
NCI-60 cancer cell line panel
The National Cancer Institute (NCI) assembled the NCI-60 cell line model system in the late 1980s. This panel contains a set of 60 human cancer cell lines from nine common types of cancer and has been used extensively for cancer pharmacogenomic studies [27,68]. This cell line panel also contains extensive omics data at the DNA/RNA/Protein levels, including karyotype, runs of homozygosity (ROH) patterns, SNP data, copy number variation (CNV), DNA mutations, DNA methylation, DNA fingerprinting, mRNA and miRNA expression, protein levels, endogenous estrogen and estrogen metabolite levels, metabolomic data, metabolite levels in the culture media, and drug response data [69–84]. Recently, all coding variants in these cell lines have been identified using whole exome sequencing [85]. All of these public data make the NCI-60 panel a very useful tool for investigators to explore from various perspectives. For example, computational scientists have used this panel to develop novel systematic approaches to integrate multiomics data [86,87]. The gene expression profiles and common genomic alterations showed high concordance between cancer cell lines and corresponding primary tumor tissues [28,88]. However, recent studies have also shown that cancer cells are not tumor tissue, and gene expression patterns in the NCI-60 panel were found to differ from those in primary tumor tissue [30]. Therefore, further clinical validation of identified biomarkers is also required for data gathered with this model system.
John Weinstein’s group used NCI-60 panel to explore the relationship between molecular genetic markers and drug response phenotype. For example, their analyses of CNV, gene expression and drug response data suggested that ASNS might be a biomarker for L-asparaginase (L-ASP) treatment response in ovarian cancer [89,90]. Further analysis of ASNS protein level using a large panel of ovarian cancer cell lines supported a negative correlation between ASNS protein level and response [91], and functional studies indicated that knockdown of ASNS sensitized ovarian cancer cell lines to L-ASP treatment [92]. In another example, the analysis of DNA mutations in 24 known cancer genes helped make it possible to identify a BRAF mutation, V600E, that was associated with phenothiazine (an antipsychotic drug) response in melanoma [93,94]. Finally, an analysis of proteomic signatures found 27 proteins were correlated to paclitaxel response [95]. Moreover, they also developed an online database, CellMiner, to integrate all of the omics data from the NCI-60 cell line model system [96,97].
Many pharmacogenomic studies from other groups were also performed with the NCI-60 cell lines by taking advantages of the public availability of the rich data for these cell lines. For example, SLFN11 gene expression was correlated with response to DNA-damaging agents while activation of the PI3K-Akt signaling pathway and basal level of cleaved PARP1 were associated with response to PI3K inhibitors [98,99]. Integrated analyses of pharmacogenomic data using both NCI-60 panel data and clinical data showed that genetic variants in genes encoding drug-metabolizing enzymes and transporters were associated with gemcitabine response in pancreatic cancer patients. In addition, an eight-gene signature for predicting response to antimicrotubule agents in lung and breast cancer patients was developed [100,101]. Another analysis of the consumption and release (CORE) profiles of 219 metabolites performed with NCI-60 cancer cell culture media, together with mRNA expression data, showed that glycine consumption and expression by the mitochondrial glycine biosynthetic pathway was significantly correlated with cancer cell proliferation rate as well as mortality for breast cancer [73].
In the early 1990s, Takao Yamori’s group in Japan established a JFCR39 panel that was similar to the NCI-60 panel. This panel contained 30 cancer cells that are part of the NCI-60, together with another 3 breast cancer cell lines and 6 stomach cancer cell lines that were chosen for inclusion because of the high incidence of gastric cancer in Japan [102]. Multiple layers of molecular features and drug response data for 130 chemicals have been generated using this panel [103], which will be also helpful for cancer pharmacogenomics, especially for tumors with high prevalence in Japan.
Breast cancer cell line panel & colorectal cancer cell line panel
Because of lineage confounding factors and the limited number of cell lines for each type of cancer in the NCI-60 panel, homogeneous cancer cell line models might be more informative and robust for the prediction of clinical response in particular cancer types and/or subtypes. For example, a collection of breast cancer cell lines as well as a panel of CRC cell lines have been well characterized and widely used for biomarker discovery.
This breast cancer panel contains approximately 51 cell lines and extensive omics data generated by Joe Gray’s group, including CNV data, DNA methylation data, transcriptional profiles, protein levels and phosphorylation levels for selected genes in commonly deregulated signaling pathways [104–107]. Most of these cell lines are available from ATCC. This model includes representatives of different subtypes of breast cancer with unique molecular features that are observed in primary tumors. Based on CNV and gene-expression profiles which displayed the same heterogeneity as that seen in primary tumors, these cell lines have been divided into five subtypes, luminal A, luminal B, ERBB2-associated, basal-like and normal-like. These different subtypes of breast cancer cell lines showed variable response to therapeutic agents [104,108].
This breast cancer cell line panel is a particularly useful tool to help discover pharmacogenomic biomarkers related to specific breast cancer subtypes. For example, basal-like breast cancer cell lines were more sensitive to polyamine analogues, and a 13-gene signature was associated with response to these drugs [109]. Cell lines with low copy number of BRCA1 were sensitive to PARP inhibitors, and a 7-gene signature for DNA repair pathways was associated with drug response in basal subtype tumors [110]. Because multiple omic data are available for these cell lines, more integrated or network analyses have been performed to identify biomarkers associated with different therapies, especially targeted therapies [108,111–113]. Integrated analysis of drug response data and multiomics data indicated that one third of drugs showed subtype-specific response. For example, ERBB2-amplified cell lines were sensitive to ERBB2-targeted agents, and basal subtype cell lines were sensitive to platinum salts [108]. Recently the analysis of drug response data for 90 anticancer agents in 70 breast cancer cell lines showed that combinations of additional omic features with transcriptional signatures substantially improved the prediction of drug response [113].
A collection of CRC cell lines has also been used for cancer pharmacogenomics. It includes 70 CRC cell lines, many of which are available from ATCC. The analysis of whole exome mutation and CNV data indicated that CRC cell line panel was a great representative for primary CRC at the genomic level, and the difference between paired cell lines and tumor tissue from which the cell lines are derived might represent the heterogeneity of primary tumor [114,115]. This panel of CRC cell lines was used to investigate the differential pharmacological response of anti-ERBB1 monoclonal antibody (cetuximab), and results showed that the direct growth inhibition was associated with mutation status in KRAS, BRAF and PIK3CA in exon 20, while the immune killing was correlated with ERBB1 expression level [116]. Furthermore, Schlicker et al. classified CRC tumors as five subtypes using gene expression feature, and different subtypes of CRC cell lines displayed various response to targeted inhibitors [116,117].
Large cancer cell line collections
Because of the limited power for biomarker discovery using only 60 cancer cell lines from the NCI-60 panel, recently, several pharmacogenomic studies were performed using large collections of cancer cell lines for which extensive multiomic data, sequencing data, as well as drug response phenotypes for a series of cytotoxic and targeted agents were obtained.
One study used the Cancer Cell Line Encyclopedia (CCLE) which contains 947 widely used human cancer cells from 36 tumor types, and 24 anticancer drugs were screened in approximately 500 cell lines. Plasma cell lineage was found to be correlated with response to IGF1 receptor inhibitors; AHR expression was associated with response to MEK inhibitor in NRAS-mutant line; and SLFN11 expression was correlated with sensitivity to topoisomerase inhibitors [28]. Another study, the Cancer Genome Project (CGP), used 639 human cancer cell lines and 130 anticancer drugs were screened. EWS-FLI1 rearrangement was found to be associated with response to a PARP inhibitor (olaparib) in sarcoma [29]. The GSK cancer cell line panel contained 311 cancer cell lines and 19 compounds were tested. Integrated analysis suggested that primary oncogenic targets were critical for predicting drug sensitivity [118]. These cell lines were used successfully to derive models to predict several different chemotherapeutic outcomes in different cancers, as demonstrated by Paul Geeleher, Nancy J Cox and R Stephanie Huang [119].
However, lack of consistency of drug response phenotypes between the CCLE and CGP sets generated by the two groups were reported subsequently [120,121]. Between the two largest cancer cell line panels, CCLE and CGP, overlap information is available for mutation data in 64 genes, expression of 12153 genes and drug response data for 15 compounds in 471 cell lines. Although the expression and mutation data were highly reproducible, the analysis showed poor concordance between studies, which might help explain significant differences in the drug response data [120,121]. Since currently no single parameter can best represent drug response, it might be best to consider multiple parameters for analysis, including the slope of the dose-response curve, the area under the curve, the maximum effect and potency (IC50) [122]. These studies further emphasize the importance of standardizing the tools and methods for the characterization of drug response phenotypes and also the importance of functional validation and clinical replication of biomarkers identified using cell lines or other model systems.
Isogenic cancer cell lines
In addition to the cancer cell lines derived from primary tumor tissues, isogenic cancer cell lines with knockout/knockin of specific genes/alleles have also been used for cancer pharmacogenomics, especially functional pharmacogenomics [65,67]. With parental cells as an appropriate study control, isogenic cancer cell lines have been applied for drug screening to discover gene/genotype selective compounds. For example, a novel cytidine nucleoside analog was found to inhibit the growth of isogenic KRAS mutant cell lines [65], and stressed isogenic TP53 mutant cancer cells was sensitive to PLK1 inhibitors [123]. Di Nicolantonio et al. established isogenic nontransformed cell lines with knockin of EGFR, KRAS, BRAF and PIK3CA mutations, and the study of 90 drugs on isogenic breast epithelial cells indicated that EGFR mutant cells were sensitive to EGFR tyrosine kinase inhibitors, erlotinib and gefitinib, but not KRAS or BRAF mutant cells [66,67].
Future perspective for optimizing in vitro human cell line models to improve their clinical predictive validity
Due to the difficulty of access to clinical samples, in vitro human cell line models will remain an important resource for cancer pharmacogenomic research in spite of their limitations. Therefore, it will be necessary to find strategies to overcome the limitations of in vitro cell line models and to enhance their potential for identifying functional biomarkers to increase the prediction value of these biomarkers in clinical settings. Included among novel approaches that might help to achieve these goals is the application of iPS cell technology, as well as the incorporation of 3D culture to better simulate the in vivo microenvironment.
iPS cells
iPS cells were first reported by Yamanaka’s group in 2006 [124]. These cells possess the capability of unlimited replication and have the potential to differentiate into all types of somatic cells. Currently, the most widely studied and most promising cell sources for the generation of patient-specific iPS cells are skin fibroblasts and adult human adipose stem cells (hASCs) [125]. Recently, Rajesh et al. also reported that iPS cells could be generated from LCLs, a potential tool for generating individual-specific iPS cells [126]. Previous studies suggested that iPS cell-derived hepatocytes showed characteristic phenotypic abnormalities that were similar to primary hepatocytes from the same patients, and that they could be used for drug toxicity testing [127]. Therefore, with further improvement in derivation technologies, characterization methods, as well as cultivation and differentiation protocols [125], in vitro human cell line models derived from patient-specific iPS cells have great potential to make it possible to study the host genetic effect on drug response in a more tissue or cell type specific manner. In the future, paired cancer and tissue-specific cells differentiated from iPS cells from the same individual with the same genetic background [128] may be informative in vitro tools to identify biomarkers contributing to drug related efficacy and toxicity.
3D culture
Since the tumor microenvironment can influence tumor development, progression, metastasis and response to anticancer agents [129], 3D culture was developed to co-culture tumor cells with stromal cells to mimic the tumor microenvironment in vivo. Many studies have suggested that, compared with cells in 2D culture which only provides the necessary cell culture medium and solid support for cell growth in vitro, cancer cells in 3D culture behave in a fashion more similar to their in vivo behavior. For example, cells in 3D culture represented primary tissues better than cells in 2D culture, and transformed mammary epithelial cells (MEC) in 3D culture regained polarity and their proliferation was inhibited [130]. The 3D culture of breast cell lines showed significant changes in morphology, gene expression profiles, cellular signaling pathways and drug response when compared with 2D culture [131]. Association analysis of molecular features with morphological signatures identified PPARγ as a predictor of the invasive stellate morphological phenotype, which represents triple-negative breast cancer [132]. Many additional examples are or will become available, and this technology may overcome some of the limitations of current cell line models and add significantly to our ability to identify biologically relevant biomarkers associated with drug response.
Conclusion & future perspective
In conclusion, as a result of the limited availability of clinical samples and the high cost of performing clinical trials, in vitro human cell line models have become one of the most widely used preclinical models to study drug response. These cell line models are relatively cheap, easily manipulated and data for these cell lines can be accumulated for use in cancer pharmacogenomics studies. These cell lines have already proven to be extremely valuable to identify pharmacogenomic biomarkers and to help generate and test biological hypotheses. Obviously, any biomarkers identified using preclinical models will need to be further validated in vivo using additional animal models and human samples. However, in spite of scientific developments in the area of cancer pharmacogenomics, the pace of clinical translation of these biomarkers is still slow. A major reason is the scientific evidence required for clinical translation. One way to improve the likelihood of identifying biologically relevant biomarkers is to improve preclinical cell line models. In order to better mimic the primary tissue and surrounding microenvironment, future incorporation of patient-derived tissue specific iPS cell panels and 3D culture methods might improve the clinical translational potential of pharmacogenomic biomarkers and enhance our ability to personalize anticancer drug therapy.
Executive summary.
In vitro human cell line models to predict clinical response to anticancer treatment
In the clinic, patient response to anticancer treatment varies widely.
Pharmacogenomics is the study of the role of inherited and acquired genetic variation in drug response.
Due to limited access and the high cost of clinical samples, in vitro human cell line models which represent germline and/or somatic variation among different individuals or patients have been widely used as a preclinical model to predict clinical drug response in cancer pharmacogenomics.
In vitro human publicly available cell line models are a renewable resource and a reasonably well-controlled system that includes extensive multiomics data.
Further functional validation and clinical confirmation of biomarkers discovered by the use of cell line models is required.
Immortalized cell line models
-
Human Epstein-Barr virus (EBV) transformed lymphoblastoid cells
Immortalized cell line models such as EBV-transformed lymphoblastoid cell lines (LCLs) represent germline variation among different individuals.
Many large LCL collections from NIGMS and NHGRI have been used extensively for cancer pharmacogenomics studies. These collections include the Human Variation Panel and LCLs from the International HapMap Project. Recently all LCLs from the 1000 Genomes Project also became available. In addition, biobank samples are another important source for generating large LCL collections from population-based cohorts.
The Human Variation Panel has been used successfully to discover novel biomarkers contributing to anticancer drug response such as FKPB5, as well as to functionally characterize clinical biomarkers such as TCL1A, TSPYL5, ZNF423 and CTSO that were identified during clinical genome-wide association studies.
HapMap LCLs have also been used to identify novel biomarkers involved in drug efficacy and adverse drug response.
-
Cancer cell line models
Cancer cell line models represent germline and somatic variation among different patients.
The NCI-60 panel was the first widely used cancer cell line panel and contains the most abundant omics data. It was a pioneer effort and represents the most extensively used cell line model in cancer pharmacogenomics.
As homogenous tumor-based cell line models, the breast and colorectal cancer cell lines are informative and robust tools to discover biomarkers for specific breast cancer or colorectal cancer subtypes and corresponding therapies.
Large cancer cell line collections such as the Cancer Cell Line Encyclopedia (CCLE) and the Cancer Genome Project (CGP) have increased our power to identify novel biomarkers involved in variation in drug response.
Isogenic cancer cell line with knockout/knock in of specific genes/alleles could help to discover novel therapies.
Conclusion & future perspective
Due to the limited availability of clinical samples, in vitro human cell line models will remain a critical resource for cancer pharmacogenomics.
Immortalized cell line and cancer cell line models that contain extensive multiomics data have been used successfully to identify genomic biomarkers involved in drug response. Further functional validation and clinical replication will be required to confirm these novel biomarkers.
In order to better mimic the primary tissue, in the future patient-specific inducible pluripotent stem cells and 3D culture could be incorporated into cell line based pharmacogenomic studies to improve their clinical translation efficiency and to better predict clinical response.
Footnotes
Financial & competing interests disclosure
This work was supported by NIH grants R01 CA138461, U19 GM61388 (The Pharmacogenomics Research Network), Mayo Center for Individualized Medicine. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
For reprint orders, please contact: reprints@futuremedicine.com
References
- 1.Wang L, Mcleod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med. 2011;364(12):1144–1153. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voskoglou-Nomikos T, Pater JL, Seymour L. Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clin Cancer Res. 2003;9(11):4227–4239. [PubMed] [Google Scholar]
- 3.Dolan ME, Newbold KG, Nagasubramanian R, et al. Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity. Cancer Res. 2004;64(12):4353–4356. doi: 10.1158/0008-5472.CAN-04-0340. [DOI] [PubMed] [Google Scholar]
- 4.Gamazon ER, Huang RS, Dolan ME, Cox NJ. Copy number polymorphisms and anticancer pharmacogenomics. Genome Biol. 2011;12(5):R46. doi: 10.1186/gb-2011-12-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang RS, Duan S, Kistner EO, Hartford CM, Dolan ME. Genetic variants associated with carboplatin-induced cytotoxicity in cell lines derived from Africans. Mol Cancer Ther. 2008;7(9):3038–3046. doi: 10.1158/1535-7163.MCT-08-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang RS, Duan S, Shukla SJ, et al. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am J Hum Genet. 2007;81(3):427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang RS, Johnatty SE, Gamazon ER, et al. Platinum sensitivity-related germline polymorphism discovered via a cell-based approach and analysis of its association with outcome in ovarian cancer patients. Clin Cancer Res. 2011;17(16):5490–5500. doi: 10.1158/1078-0432.CCR-11-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ingle JN, Liu M, Wickerham DL, et al. Selective estrogen receptor modulators and pharmacogenomic variation in ZNF423 regulation of BRCA1 expression: individualized breast cancer prevention. Cancer Discov. 2013;3(7):812–825. doi: 10.1158/2159-8290.CD-13-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingle JN, Schaid DJ, Goss PE, et al. Genome-wide associations and functional genomic studies of musculoskeletal adverse events in women receiving aromatase inhibitors. J Clin Oncol. 2010;28(31):4674–4682. doi: 10.1200/JCO.2010.28.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang J, Fridley BL, Feng Q, et al. Genome-wide association study for biomarker identification of Rapamycin and Everolimus using a lymphoblastoid cell line system. Front Genet. 2013;4:166. doi: 10.3389/fgene.2013.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalari KR, Hebbring SJ, Chai HS, et al. Copy number variation and cytidine analogue cytotoxicity: a genome-wide association approach. BMC Genomics. 2010;11:357. doi: 10.1186/1471-2164-11-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lacroix B, Gamazon ER, Lenkala D, et al. Integrative analyses of genetic variation, epigenetic regulation, and the transcriptome to elucidate the biology of platinum sensitivity. BMC Genomics. 2014;15(1):292. doi: 10.1186/1471-2164-15-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Fridley B, Kalari K, et al. Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res. 2008;68(17):7050–7058. doi: 10.1158/0008-5472.CAN-08-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Fridley BL, Kalari K, et al. Gemcitabine and arabinosylcytosin pharmacogenomics: genome-wide association and drug response biomarkers. PLoS ONE. 2009;4(11):e7765. doi: 10.1371/journal.pone.0007765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Fridley BL, Kalari K, et al. Discovery of genetic biomarkers contributing to variation in drug response of cytidine analogues using human lymphoblastoid cell lines. BMC Genomics. 2014;15(1):93. doi: 10.1186/1471-2164-15-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Schaid DJ, Fridley BL, et al. Gemcitabine metabolic pathway genetic polymorphisms and response in patients with non-small cell lung cancer. Pharmacogenet Genomics. 2012;22(2):105–116. doi: 10.1097/FPC.0b013e32834dd7e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu M, Ingle JN, Fridley BL, et al. TSPYL5 SNPs: association with plasma estradiol concentrations and aromatase expression. Mol Endocrinol. 2013;27(4):657–670. doi: 10.1210/me.2012-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niu N, Qin Y, Fridley BL, et al. Radiation pharmacogenomics: a genome-wide association approach to identify radiation response biomarkers using human lymphoblastoid cell lines. Genome Res. 2010;20(11):1482–1492. doi: 10.1101/gr.107672.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niu N, Schaid DJ, Abo RP, et al. Genetic association with overall survival of taxane-treated lung cancer patients - a genome-wide association study in human lymphoblastoid cell lines followed by a clinical association study. BMC Cancer. 2012;12:422. doi: 10.1186/1471-2407-12-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’donnell PH, Gamazon E, Zhang W, et al. Population differences in platinum toxicity as a means to identify novel genetic susceptibility variants. Pharmacogenet Genomics. 2010;20(5):327–337. doi: 10.1097/FPC.0b013e3283396c4e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shukla SJ, Duan S, Wu X, Badner JA, Kasza K, Dolan ME. Whole-genome approach implicates CD44 in cellular resistance to carboplatin. Hum Genomics. 2009;3(2):128–142. doi: 10.1186/1479-7364-3-2-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stark AL, Hause RJ, Jr, Gorsic LK, et al. Protein quantitative trait loci identify novel candidates modulating cellular response to chemotherapy. PLoS Genet. 2014;10(4):e1004192. doi: 10.1371/journal.pgen.1004192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan XL, Moyer AM, Fridley BL, et al. Genetic variation predicting cisplatin cytotoxicity associated with overall survival in lung cancer patients receiving platinum-based chemotherapy. Clin Cancer Res. 2011;17(17):5801–5811. doi: 10.1158/1078-0432.CCR-11-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wheeler HE, Gamazon ER, Wing C, et al. Integration of cell line and clinical trial genome-wide analyses supports a polygenic architecture of Paclitaxel-induced sensory peripheral neuropathy. Clin Cancer Res. 2013;19(2):491–499. doi: 10.1158/1078-0432.CCR-12-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W, Duan S, Bleibel WK, et al. Identification of common genetic variants that account for transcript isoform variation between human populations. Hum Genet. 2009;125(1):81–93. doi: 10.1007/s00439-008-0601-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziliak D, O’donnell PH, Im HK, et al. Germline polymorphisms discovered via a cell-based, genome-wide approach predict platinum response in head and neck cancers. Transl Res. 2011;157(5):265–272. doi: 10.1016/j.trsl.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6(10):813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 28.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garnett MJ, Edelman EJ, Heidorn SJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeeberg BR, Kohn KW, Kahn A, et al. Concordance of gene expression and functional correlation patterns across the NCI-60 cell lines and the Cancer Genome Atlas glioblastoma samples. PLoS ONE. 2012;7(7):e40062. doi: 10.1371/journal.pone.0040062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugimoto M, Tahara H, Ide T, Furuichi Y. Steps involved in immortalization and tumorigenesis in human B-lymphoblastoid cell lines transformed by Epstein-Barr virus. Cancer Res. 2004;64(10):3361–3364. doi: 10.1158/0008-5472.CAN-04-0079. [DOI] [PubMed] [Google Scholar]
- 32.Nickles D, Madireddy L, Yang S, et al. In depth comparison of an individual’s DNA and its lymphoblastoid cell line using whole genome sequencing. BMC Genomics. 2012;13:477. doi: 10.1186/1471-2164-13-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Londin ER, Keller MA, D’Andrea MR, et al. Whole-exome sequencing of DNA from peripheral blood mononuclear cells (PBMC) and EBV-transformed lymphocytes from the same donor. BMC Genomics. 2011;12:464. doi: 10.1186/1471-2164-12-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugawara H, Iwamoto K, Bundo M, Ueda J, Ishigooka J, Kato T. Comprehensive DNA methylation analysis of human peripheral blood leukocytes and lymphoblastoid cell lines. Epigenetics. 2011;6(4):508–515. doi: 10.4161/epi.6.4.14876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dimas AS, Deutsch S, Stranger BE, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325(5945):1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwan T, Grundberg E, Koka V, et al. Tissue effect on genetic control of transcript isoform variation. PLoS Genet. 2009;5(8):e1000608. doi: 10.1371/journal.pgen.1000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sie L, Loong S, Tan EK. Utility of lymphoblastoid cell lines. J Neurosci Res. 2009;87(9):1953–1959. doi: 10.1002/jnr.22000. [DOI] [PubMed] [Google Scholar]
- 38.Choy E, Yelensky R, Bonakdar S, et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2008;4(11):e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wheeler HE, Dolan ME. Lymphoblastoid cell lines in pharmacogenomic discovery and clinical translation. Pharmacogenomics. 2012;13(1):55–70. doi: 10.2217/pgs.11.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coriell Biorepository. http://ccr.coriell.org.
- 41.Editor in the field. The Coriell Institute for Medical Research. Nat Genet. 2005;37(1):2. doi: 10.1038/ng0105-2. [DOI] [PubMed] [Google Scholar]
- 42.Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nam HY, Shim SM, Han BG, Jeon JP. Human lymphoblastoid cell lines: a goldmine for the biobankomics era. Pharmacogenomics. 2011;12(6):907–917. doi: 10.2217/pgs.11.24. [DOI] [PubMed] [Google Scholar]
- 44.Amoli MM, Carthy D, Platt H, Ollier WE. EBV Immortalization of human B lymphocytes separated from small volumes of cryopreserved whole blood. Int J Epidemiol. 2008;37(Suppl 1):i41–i45. doi: 10.1093/ije/dym285. [DOI] [PubMed] [Google Scholar]
- 45.Heyn H, Moran S, Hernando-Herraez I, et al. DNA methylation contributes to natural human variation. Genome Res. 2013;23(9):1363–1372. doi: 10.1101/gr.154187.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu Z, Wang L, Hildebrandt MA, Schaid DJ. Testing whether genetic variation explains correlation of quantitative measures of gene expression, and application to genetic network analysis. Stat Med. 2008;27(19):3847–3867. doi: 10.1002/sim.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abo R, Jenkins GD, Wang L, Fridley BL. Identifying the genetic variation of gene expression using gene sets: application of novel gene Set eQTL approach to PharmGKB and KEGG. PLoS ONE. 2012;7(8):e43301. doi: 10.1371/journal.pone.0043301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chalise P, Batzler A, Abo R, Wang L, Fridley BL. Simultaneous analysis of multiple data types in pharmacogenomic studies using weighted sparse canonical correlation analysis. OMICS. 2012;16(7–8):363–373. doi: 10.1089/omi.2011.0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fridley BL, Abo R, Tan XL, et al. Integrative gene set analysis: application to platinum pharmacogenomics. OMICS. 2014;18(1):34–41. doi: 10.1089/omi.2013.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fridley BL, Batzler A, Li L, et al. Gene set analysis of purine and pyrimidine antimetabolites cancer therapies. Pharmacogenet Genomics. 2011;21(11):701–712. doi: 10.1097/FPC.0b013e32834a48a9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fridley BL, Jenkins G, Schaid DJ, Wang L. A Bayesian hierarchical nonlinear model for assessing the association between genetic variation and drug cytotoxicity. Stat Med. 2009;28(21):2709–2722. doi: 10.1002/sim.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pei H, Li L, Fridley BL, et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16(3):259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hou J, Wang L. FKBP5 as a selection biomarker for gemcitabine and Akt inhibitors in treatment of pancreatic cancer. PLoS ONE. 2012;7(5):e36252. doi: 10.1371/journal.pone.0036252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mulholland DJ, Tran LM, Li Y, et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell. 2011;19(6):792–804. doi: 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ellsworth KA, Eckloff BW, Li L, et al. Contribution of FKBP5 genetic variation to gemcitabine treatment and survival in pancreatic adenocarcinoma. PLoS ONE. 2013;8(8):e70216. doi: 10.1371/journal.pone.0070216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mitra AK, Crews K, Pounds S, et al. Impact of genetic variation in FKBP5 on clinical response in pediatric acute myeloid leukemia patients: a pilot study. Leukemia. 2011;25(8):1354–1356. doi: 10.1038/leu.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu M, Wang L, Bongartz T, et al. Aromatase inhibitors, estrogens and musculoskeletal pain: estrogen-dependent T-cell leukemia 1A (TCL1A) gene-mediated regulation of cytokine expression. Breast Cancer Res. 2012;14(2):R41. doi: 10.1186/bcr3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 59.Stranger BE, Nica AC, Forrest MS, et al. Population genomics of human gene expression. Nat Genet. 2007;39(10):1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gamazon ER, Lamba JK, Pounds S, et al. Comprehensive genetic analysis of cytarabine sensitivity in a cell-based model identifies polymorphisms associated with outcome in AML patients. Blood. 2013;121(21):4366–4376. doi: 10.1182/blood-2012-10-464149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mangravite LM, Engelhardt BE, Medina MW, et al. A statin-dependent QTL for GATM expression is associated with statin-induced myopathy. Nature. 2013;502(7471):377–380. doi: 10.1038/nature12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tantisira KG, Lasky-Su J, Harada M, et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med. 2011;365(13):1173–1183. doi: 10.1056/NEJMoa0911353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown CC, Havener TM, Medina MW, et al. A genome-wide association analysis of temozolomide response using lymphoblastoid cell lines shows a clinically relevant association with MGMT. Pharmacogenet Genomics. 2012;22(11):796–802. doi: 10.1097/FPC.0b013e3283589c50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown CC, Havener TM, Medina MW, et al. Genome-wide association and pharmacological profiling of 29 anticancer agents using lymphoblastoid cell lines. Pharmacogenomics. 2014;15(2):137–146. doi: 10.2217/pgs.13.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Torrance CJ, Agrawal V, Vogelstein B, Kinzler KW. Use of isogenic human cancer cells for high-throughput screening and drug discovery. Nat Biotechnol. 2001;19(10):940–945. doi: 10.1038/nbt1001-940. [DOI] [PubMed] [Google Scholar]
- 66.Di Nicolantonio F, Arena S, Gallicchio M, et al. Replacement of normal with mutant alleles in the genome of normal human cells unveils mutation-specific drug responses. Proc Natl Acad Sci USA. 2008;105(52):20864–20869. doi: 10.1073/pnas.0808757105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Di Nicolantonio F, Arena S, Gallicchio M, Bardelli A. Isogenic mutant human cells: a new tool for personalized cancer medicine. Cell Cycle. 2010;9(1):20–21. doi: 10.4161/cc.9.1.10474. [DOI] [PubMed] [Google Scholar]
- 68.Weinstein JN, Pommier Y. Transcriptomic analysis of the NCI-60 cancer cell lines. C R Biol. 2003;326(10–11):909–920. doi: 10.1016/j.crvi.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 69.Blower PE, Verducci JS, Lin S, et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol Cancer Ther. 2007;6(5):1483–1491. doi: 10.1158/1535-7163.MCT-07-0009. [DOI] [PubMed] [Google Scholar]
- 70.Forbes S, Clements J, Dawson E, et al. Cosmic 2005. Br J Cancer. 2006;94(2):318–322. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436(7047):117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 72.Gaur A, Jewell DA, Liang Y, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67(6):2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 73.Jain M, Nilsson R, Sharma S, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336(6084):1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lorenzi PL, Reinhold WC, Varma S, et al. DNA fingerprinting of the NCI-60 cell line panel. Mol Cancer Ther. 2009;8(4):713–724. doi: 10.1158/1535-7163.MCT-08-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moghaddas Gholami A, Hahne H, Wu Z, et al. Global proteome analysis of the NCI-60 cell line panel. Cell Rep. 2013;4(3):609–620. doi: 10.1016/j.celrep.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 76.Nishizuka S, Charboneau L, Young L, et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci USA. 2003;100(24):14229–14234. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Patnaik SK, Dahlgaard J, Mazin W, et al. Expression of microRNAs in the NCI-60 cancer cell-lines. PLoS ONE. 2012;7(11):e49918. doi: 10.1371/journal.pone.0049918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roschke AV, Tonon G, Gehlhaus KS, et al. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res. 2003;63(24):8634–8647. [PubMed] [Google Scholar]
- 79.Ross DT, Scherf U, Eisen MB, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24(3):227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 80.Ruan X, Kocher JP, Pommier Y, Liu H, Reinhold WC. Mass homozygotes accumulation in the NCI-60 cancer cell lines as compared with HapMap Trios, and relation to fragile site location. PLoS ONE. 2012;7(2):e31628. doi: 10.1371/journal.pone.0031628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shankavaram UT, Reinhold WC, Nishizuka S, et al. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol Cancer Ther. 2007;6(3):820–832. doi: 10.1158/1535-7163.MCT-06-0650. [DOI] [PubMed] [Google Scholar]
- 82.Su G, Burant CF, Beecher CW, Athey BD, Meng F. Integrated metabolome and transcriptome analysis of the NCI60 dataset. BMC Bioinformatics. 2011;12(Suppl 1):S36. doi: 10.1186/1471-2105-12-S1-S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weinstein JN, Myers TG, O’connor PM, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275(5298):343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- 84.Xu X, Veenstra TD. Concentration of endogenous estrogens and estrogen metabolites in the NCI-60 human tumor cell lines. Genome Med. 2012;4(4):31. doi: 10.1186/gm330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Abaan OD, Polley EC, Davis SR, et al. The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res. 2013;73(14):4372–4382. doi: 10.1158/0008-5472.CAN-12-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Federici G, Gao X, Slawek J, et al. Systems analysis of the NCI-60 cancer cell lines by alignment of protein pathway activation modules with “-OMIC” data fields and therapeutic response signatures. Mol Cancer Res. 2013;11(6):676–685. doi: 10.1158/1541-7786.MCR-12-0690. [DOI] [PubMed] [Google Scholar]
- 87.Liu Y, Devescovi V, Chen S, Nardini C. Multilevel omic data integration in cancer cell lines: advanced annotation and emergent properties. BMC Syst Biol. 2013;7:14. doi: 10.1186/1752-0509-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Borrell B. How accurate are cancer cell lines? Nature. 2010;463(7283):858. doi: 10.1038/463858a. [DOI] [PubMed] [Google Scholar]
- 89.Scherf U, Ross DT, Waltham M, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24(3):236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 90.Bussey KJ, Chin K, Lababidi S, et al. Integrating data on DNA copy number with gene expression levels and drug sensitivities in the NCI-60 cell line panel. Mol Cancer Ther. 2006;5(4):853–867. doi: 10.1158/1535-7163.MCT-05-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lorenzi PL, Llamas J, Gunsior M, et al. Asparagine synthetase is a predictive biomarker of L-asparaginase activity in ovarian cancer cell lines. Mol Cancer Ther. 2008;7(10):3123–3128. doi: 10.1158/1535-7163.MCT-08-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lorenzi PL, Reinhold WC, Rudelius M, et al. Asparagine synthetase as a causal, predictive biomarker for L-asparaginase activity in ovarian cancer cells. Mol Cancer Ther. 2006;5(11):2613–2623. doi: 10.1158/1535-7163.MCT-06-0447. [DOI] [PubMed] [Google Scholar]
- 93.Ikediobi ON, Davies H, Bignell G, et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5(11):2606–2612. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ikediobi ON, Reimers M, Durinck S, et al. In vitro differential sensitivity of melanomas to phenothiazines is based on the presence of codon 600 BRAF mutation. Mol Cancer Ther. 2008;7(6):1337–1346. doi: 10.1158/1535-7163.MCT-07-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Park ES, Rabinovsky R, Carey M, et al. Integrative analysis of proteomic signatures, mutations, and drug responsiveness in the NCI 60 cancer cell line set. Mol Cancer Ther. 2010;9(2):257–267. doi: 10.1158/1535-7163.MCT-09-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shankavaram UT, Varma S, Kane D, et al. CellMiner: a relational database and query tool for the NCI-60 cancer cell lines. BMC Genomics. 2009;10:277. doi: 10.1186/1471-2164-10-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Varma S, Pommier Y, Sunshine M, Weinstein JN, Reinhold WC. High resolution copy number variation data in the NCI-60 cancer cell lines from whole genome microarrays accessible through CellMiner. PLoS ONE. 2014;9(3):e92047. doi: 10.1371/journal.pone.0092047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kwei KA, Baker JB, Pelham RJ. Modulators of sensitivity and resistance to inhibition of PI3K identified in a pharmacogenomic screen of the NCI-60 human tumor cell line collection. PLoS ONE. 2012;7(9):e46518. doi: 10.1371/journal.pone.0046518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zoppoli G, Regairaz M, Leo E, et al. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci USA. 2012;109(37):15030–15035. doi: 10.1073/pnas.1205943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harris M, Bhuvaneshwar K, Natarajan T, et al. Pharmacogenomic characterization of gemcitabine response--a framework for data integration to enable personalized medicine. Pharmacogenet Genomics. 2014;24(2):81–93. doi: 10.1097/FPC.0000000000000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hsu YC, Chen HY, Yuan S, et al. Genome-wide analysis of three-way interplay among gene expression, cancer cell invasion and anti-cancer compound sensitivity. BMC Med. 2013;11:106. doi: 10.1186/1741-7015-11-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kong D, Yamori T. JFCR39, a panel of 39 human cancer cell lines, and its application in the discovery and development of anticancer drugs. Bioorg Med Chem. 2012;20(6):1947–1951. doi: 10.1016/j.bmc.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 103.Nakatsu N, Nakamura T, Yamazaki K, et al. Evaluation of action mechanisms of toxic chemicals using JFCR39, a panel of human cancer cell lines. Mol Pharmacol. 2007;72(5):1171–1180. doi: 10.1124/mol.107.038836. [DOI] [PubMed] [Google Scholar]
- 104.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Loss LA, Sadanandam A, Durinck S, et al. Prediction of epigenetically regulated genes in breast cancer cell lines. BMC Bioinformatics. 2010;11:305. doi: 10.1186/1471-2105-11-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rhee JK, Kim K, Chae H, et al. Integrated analysis of genome-wide DNA methylation and gene expression profiles in molecular subtypes of breast cancer. Nucleic Acids Res. 2013;41(18):8464–8474. doi: 10.1093/nar/gkt643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Iacovides DC, Johnson AB, Wang N, Boddapati S, Korkola J, Gray JW. Identification and quantification of AKT isoforms and phosphoforms in breast cancer using a novel nanofluidic immunoassay. Mol Cell Proteomics. 2013;12(11):3210–3220. doi: 10.1074/mcp.M112.023119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heiser LM, Sadanandam A, Kuo WL, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci USA. 2012;109(8):2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kuo WL, Das D, Ziyad S, et al. A systems analysis of the chemosensitivity of breast cancer cells to the polyamine analogue PG-11047. BMC Med. 2009;7:77. doi: 10.1186/1741-7015-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Daemen A, Wolf DM, Korkola JE, et al. Cross-platform pathway-based analysis identifies markers of response to the PARP inhibitor olaparib. Breast Cancer Res Treat. 2012;135(2):505–517. doi: 10.1007/s10549-012-2188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Heiser LM, Wang NJ, Talcott CL, et al. Integrated analysis of breast cancer cell lines reveals unique signaling pathways. Genome Biol. 2009;10(3):R31. doi: 10.1186/gb-2009-10-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mirzoeva OK, Das D, Heiser LM, et al. Basal subtype and MAPK/ERK kinase (MEK)-phosphoinositide 3-kinase feedback signaling determine susceptibility of breast cancer cells to MEK inhibition. Cancer Res. 2009;69(2):565–572. doi: 10.1158/0008-5472.CAN-08-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Daemen A, Griffith OL, Heiser LM, et al. Modeling precision treatment of breast cancer. Genome Biol. 2013;14(10):R110. doi: 10.1186/gb-2013-14-10-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mouradov D, Sloggett C, Jorissen RN, et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74(12):3238–3247. doi: 10.1158/0008-5472.CAN-14-0013. [DOI] [PubMed] [Google Scholar]
- 115.Douglas EJ, Fiegler H, Rowan A, et al. Array comparative genomic hybridization analysis of colorectal cancer cell lines and primary carcinomas. Cancer Res. 2004;64(14):4817–4825. doi: 10.1158/0008-5472.CAN-04-0328. [DOI] [PubMed] [Google Scholar]
- 116.Ashraf SQ, Nicholls AM, Wilding JL, Ntouroupi TG, Mortensen NJ, Bodmer WF. Direct and immune mediated antibody targeting of ERBB receptors in a colorectal cancer cell-line panel. Proc Natl Acad Sci USA. 2012;109(51):21046–21051. doi: 10.1073/pnas.1218750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schlicker A, Beran G, Chresta CM, et al. Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines. BMC Med Genomics. 2012;5:66. doi: 10.1186/1755-8794-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Greshock J, Bachman KE, Degenhardt YY, et al. Molecular target class is predictive of in vitro response profile. Cancer Res. 2010;70(9):3677–3686. doi: 10.1158/0008-5472.CAN-09-3788. [DOI] [PubMed] [Google Scholar]
- 119.Geeleher P, Cox NJ, Huang RS. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 2014;15(3):R47. doi: 10.1186/gb-2014-15-3-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Haibe-Kains B, El-Hachem N, Birkbak NJ, et al. Inconsistency in large pharmacogenomic studies. Nature. 2013;504(7480):389–393. doi: 10.1038/nature12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Weinstein JN, Lorenzi PL. Cancer: discrepancies in drug sensitivity. Nature. 2013;504(7480):381–383. doi: 10.1038/nature12839. [DOI] [PubMed] [Google Scholar]
- 122.Fallahi-Sichani M, Honarnejad S, Heiser LM, Gray JW, Sorger PK. Metrics other than potency reveal systematic variation in responses to cancer drugs. Nat Chem Biol. 2013;9(11):708–714. doi: 10.1038/nchembio.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sur S, Pagliarini R, Bunz F, et al. A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc Natl Acad Sci USA. 2009;106(10):3964–3969. doi: 10.1073/pnas.0813333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 125.Sun N, Longaker MT, Wu JC. Human iPS cell-based therapy: considerations before clinical applications. Cell Cycle. 2010;9(5):880–885. doi: 10.4161/cc.9.5.10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rajesh D, Dickerson SJ, Yu J, Brown ME, Thomson JA, Seay NJ. Human lymphoblastoid B-cell lines reprogrammed to EBV-free induced pluripotent stem cells. Blood. 2011;118(7):1797–1800. doi: 10.1182/blood-2011-01-332064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Greenbaum LE. From skin cells to hepatocytes: advances in application of iPS cell technology. J Clin Invest. 2010;120(9):3102–3105. doi: 10.1172/JCI44422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Menendez JA, Alarcon T, Corominas-Faja B, et al. Xenopatients 2.0: reprogramming the epigenetic landscapes of patient-derived cancer genomes. Cell Cycle. 2014;13(3):358–370. doi: 10.4161/cc.27770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mcmillin DW, Negri JM, Mitsiades CS. The role of tumour-stromal interactions in modifying drug response: challenges and opportunities. Nat Rev Drug Discov. 2013;12(3):217–228. doi: 10.1038/nrd3870. [DOI] [PubMed] [Google Scholar]
- 130.Birgersdotter A, Sandberg R, Ernberg I. Gene expression perturbation in vitro – a growing case for three-dimensional (3D) culture systems. Semin Cancer Biol. 2005;15(5):405–412. doi: 10.1016/j.semcancer.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 131.Kenny PA, Lee GY, Myers CA, et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1(1):84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Han J, Chang H, Giricz O, et al. Molecular predictors of 3D morphogenesis by breast cancer cell lines in 3D culture. PLoS Comput Biol. 2010;6(2):e1000684. doi: 10.1371/journal.pcbi.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]