Abstract
Influenza hemagglutinin (HA) is the major surface glycoprotein on influenza viruses and mediates viral attachment and subsequent fusion with host cells. The HA is the major target of the immune response, but due to its high level of variability, as evidenced by substantial antigenic diversity, it had been historically considered to elicit only a narrow, strain-specific antibody response. However, a recent explosion in the discovery of broadly neutralizing antibodies (bnAbs) to influenza virus has identified two major supersites of vulnerability on the HA through structural characterization of HA-antibody complexes. These commonly targeted epitopes are involved with receptor binding as well as the fusion machinery and, hence, are functionally conserved and less prone to mutation. These bnAbs can neutralize viruses by blocking infection or the spread of infection by preventing progeny release. Structural analyses of these bnAbs show they exhibit striking similarities and trends in recognition of the HA and use recurring recognition motifs, despite substantial differences in their germline genes. This information can be utilized in design of novel therapeutics as well as in immunogens for improved vaccines with greater breadth and efficacy.
1 Introduction
Influenza viruses cause major respiratory disease each year, commonly known as the flu, and are a significant health challenge and economic burden worldwide (Molinari et al. 2007). Several countermeasures are available to combat the flu such as inhibitors against the surface glycoprotein neuraminidase or the M2 proton channel; however, the effectiveness of these antivirals has become severely diminished as viruses evolve to become drug-resistant (Bright et al. 2006; de Jong et al. 2005; Kiso et al. 2004). Vaccinations against the flu, which were first administered in the 1940s, remains the best means of flu control and prevention. However, unlike other infectious diseases with available vaccine regimens, such as for smallpox, polio, and measles, there is currently no “magic bullet” to abolish future flu infections due to the high diversity and ever-changing antigenicity of the influenza viruses. Therefore, design and development of a universal or more long-term flu vaccine would be highly desirable for the elicitation of antibody responses that can accommodate for the enormous diversity and continual changes in influenza viruses and which target the highly conserved functional epitopes.
Fortunately, only a few subtypes of influenza viruses have caused human pandemics and these are type A H1N1, H2N2, and H3N2. In fact, H1N1 and H3N2 have dominated the human type A viruses for nearly a century (1918-present) with a brief interlude by H2N2 viruses (1957-1968). Human type B viruses have two lineages but these viruses do not lead to the same mortality rates associated with human A viruses. All of these viruses are under constant surveillance and are closely monitored to follow influenza activity such as illnesses, severity, and to determine what the dominant circulating virus will be in any given year (Salzberg 2008). Vaccines are, therefore, predictions of candidate strains that may circulate in the upcoming season. Currently, two influenza A strains (H1N1 and H3N2) and one or two influenza B strains (Victoria and/or Yamagata lineages) are included in the annual vaccine, as these viruses currently circulate in humans on an annual basis. The vaccines are administered by injection of inactivated virus (the “flu shot”) or by an intranasal spray of live, attenuated virus. However, the effectiveness of the vaccine is highly dependent on the match between the strains in its formulation and the dominant circulating virus. This selection process is further complicated by the high mutability rate of influenza viruses and, thus, the vaccine formulations need to be updated accordingly almost every year.
In addition to seasonal flu, unpredictable outbreaks from other HA subtypes can sporadically infect humans and cause severe disease such as H5N1, H7N7, H9N2, as well as the recent H7N9 and H10N8 viruses (Chen et al. 2014; Gao et al. 2013). These viruses have been associated with an devastatingly high mortality rate, which can reach up to ~60%, compared to ~0.01% for seasonal viruses (CDC 2010). Fortunately, none of these deadly viruses have been able to spread by sustainable human-to-human transmission. Nonetheless, the unpredictability and pandemic potential of these divergent viruses underscores the need for broader spectrum therapy and pandemic preparedness.
Hemagglutinin (HA) is the major surface glycoprotein on influenza viruses and is the primary target for the humoral immune response to influenza virus. The HA currently has been classified into 18 distinct subtypes (Tong et al. 2013), based upon their reactivity to polysera for type A viruses (designated H1–H18), and two lineages for type B viruses (Victoria and Yamagata). Type A HAs can be further classified into two phylogenetic groups; group 1: H1, H2, H5, H6, H8, H9, H11, H12, H13, H16, H17, and H18; and group 2: H3, H4, H7, H10, H14, and H15.
The influenza HA glycoprotein is a class I viral fusion protein and mediates viral entry into host cells. It is synthesized as a precursor polypeptide, termed HA0, and assembles into pre-fusion homotrimers (Wilson et al. 1981). The HA0 trimers are subsequently matured by host proteases, which generate disulfide-linked HA1 and HA2 subunits. In general terms, the HA can structurally be considered to be composed of “head” and “stem” domains (Fig. 1). The globular membrane-distal HA head is composed entirely of HA1 residues, which form the receptor binding pocket (three per HA trimer) that mediate interactions between the virus and host cell sialosides (Weis et al. 1988). Directly below the HA head, the membrane-proximal HA stem contains the fusion machinery. The stem is largely α-helical and is composed primarily of HA2 residues and some ascending and descending HA1 residues. The mature HA (HA1/HA2) is similar in structure to the HA0 precursor except at the cleavage site where the fusion peptide, located at the N-terminus of the HA2 subunit, now inserts deeply into a hydrophobic pocket around the trimer axis. In the HA0 form, the uncleaved fusion peptide forms a loop that protrudes at the HA surface (Chen et al. 1998; Stevens et al. 2004). The pre-fusion mature HA conformation is pH-dependent and metastable, and acidification triggers major conformational changes among the central HA2 helices that lead to the post-fusion state (Bullough et al. 1994; Skehel et al. 1982). Despite the diversity of the HAs, they almost all share the conserved function of host cell binding and membrane fusion, with the exception of the recently identified H17 and H18 HAs from bat influenza viruses (Sun et al. 2013; Tong et al. 2013; Zhu et al. 2013b). Therefore, blocking receptor binding or inhibiting the progression of HA maturation and membrane fusion are effective means of preventing or ameliorating HA-mediated virus infection (Brandenburg et al. 2013).
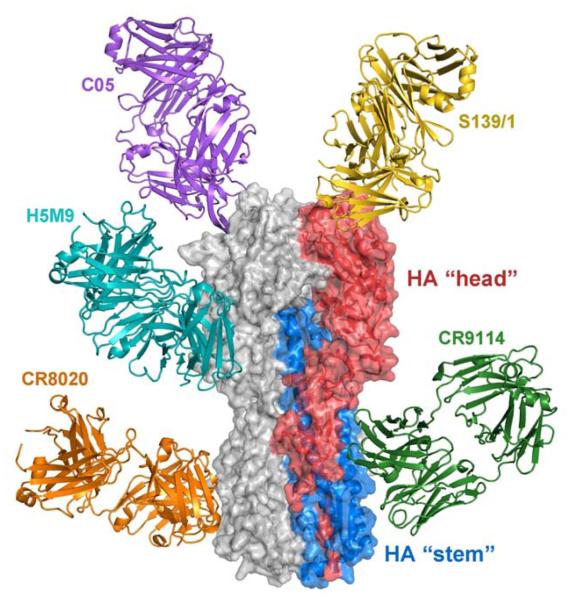
Fig. 1.
Structure of the influenza hemagglutinin (HA) and broadly neutralizing antibody (bnAb) binding sites. A single protomer of the trimeric HA is colored with HA1 and HA2 residues in red and blue, respectively. The membrane-distal HA “head” contains the receptor binding site (three per HA) and is comprised of HA1 residues. The membrane-proximal HA “stem” contains the fusion machinery and is primarily comprised of HA2 residues. All antibodies (C05, PDB 4FP8; S139/1, PDB 4GMS; H5M9, PDB 4MHH; CR8020, PDB 3SDY; CR9114, PDB 4FQI) have been modeled onto the A/Hong Kong/1/1968 (H3N2) HA (PDB 4FNK).
There has been a recent explosion in the discovery of broadly neutralizing antibodies (bnAbs) to human influenza viruses. These bnAbs target conserved sites on the HA head and stem and block influenza infection or its progression at critical HA junctures. These bnAbs have varying levels of cross-reactivity against divergent strains within and across subtypes. Moreover, structural studies by x-ray crystallography and electron microscopy (EM) studies have revealed supersites of vulnerability on the HA that are recognized by bnAbs [see recent reviews (Corti and Lanzavecchia 2013; Ekiert and Wilson 2012; Julien et al. 2012)]. Altogether, analysis of the growing arsenal of these bnAb-HA structures have revealed recurring modes of recognition that are now the main source of inspiration for design of vaccines and antibody-based therapeutics in the form of small molecules, peptides, and designed proteins against the HA.
2 HA Head-Reactive Antibodies
The HA head has long been considered to be able to elicit only a narrow, strain-specific antibody response as it undergoes rapid mutation to change its surface antigenicity by sequence variation and glycan incorporation to evade and escape recognition from our immune system. Accordingly, classical antigenic sites on the HA head have been mapped by tracking patterns of natural amino acid variation, as well as laboratory escape mutants, and these sites have been long considered as hot spots for antigenic drift (Caton et al. 1982; Wiley et al. 1981). These antigenic sites are prone to higher sequence variability and, thus, the cross-reactivity of antibodies to strains other than to the immunizing strain is severely restricted. For example, antibody 2D1, which binds near the top of the HA, recognizes only HAs from the pandemic 1918 and 2009 H1N1 viruses, which are highly antigenically similar at the 2D1 epitope despite being separated by nearly a century (Xu et al. 2010).
However, the notion of strain specificity has been challenged by a recent surge in the isolation and characterization of a number of broader-spectrum antibodies against the HA head. Depending on the sequence conservation of the epitope, the antibodies can have broader reactivity. The epitopes of some of these antibodies are located at various locations on the globular HA head (Fig. 1) (Cho et al. 2014; Dreyfus et al. 2012; Fleury et al. 1999; Fleury et al. 2000; Zhu et al. 2013a). For instance, antibody H5M9 contacts a nearly invariant epitope among H5 HAs at the vestigial esterase domain distant from the receptor binding site and somewhat closer to the fusion domain (Zhu et al. 2013a). In addition, antibody CR8071 recognizes an epitope that is highly conserved among nearly all flu type B strains also on the side of the HA head (Dreyfus et al. 2012). Antibodies HC45 and BH151 also bind a similar region on H3 HAs, but their breadth has not been determined (Fleury et al. 1999; Fleury et al. 2000). Antibodies GC0757 and GC0857 also target the side of the HA head, but bind a different epitope on the opposite face of the HA (Cho et al. 2014). Yet, the vast majority of the broader-spectrum antibodies against the HA head target the receptor binding site (Barbey-Martin et al. 2002; Ekiert et al. 2012; Fleury et al. 1998; Hong et al. 2013; Lee et al. 2012; Schmidt et al. 2013; Tsibane et al. 2012; Whittle et al. 2011; Xu et al. 2013). Since the receptor binding site is functionally conserved for receptor binding, it has restricted sequence variation compared to the rest of the HA head (Martin et al. 1998), and now appears to be a fascinating and amenable target of bnAbs.
2.1 Receptor Binding Site-Targeted Antibodies
The HA receptor binding site is a broad, shallow pocket located at the apex of the globular HA head. The framework of the receptor binding pocket is formed by the 130 loop, 150 loop, 220 loop, and 190 helix, which are designated by their numbering in the HA sequence (Fig. 2a). To date, 11 receptor binding site-targeted antibodies have been structurally characterized. All the receptor binding site-targeted antibodies commonly insert a complementarity determining region (CDR) loop into the binding pocket and thereby directly block HA from interacting with host-cell sialosides (Fig. 2b). However, due to the small footprint of the receptor binding site, most antibodies can only insert a single CDR loop, typically using heavy chain CDR loop 3 (HCDR3) and occasionally HCDR2. Since the receptor binding site is located near the top of the HA, there is substantial conformational freedom of the antibody approach angle to this site (Fig. 2c), which plays a role in their breadth of recognition. The antibodies can be loosely classified as having either greatly expanded individual subtype recognition or an increased level of heterosubtypic recognition, and thus these different bnAbs have quite varying degrees of breadth and potency. Nonetheless, the convergence of interactions at the receptor binding site substantiates the concept that this epitope is a supersite of vulnerability akin to an extensive glycan-dependent site found on the HIV-1 Env trimer (Kong et al. 2013).
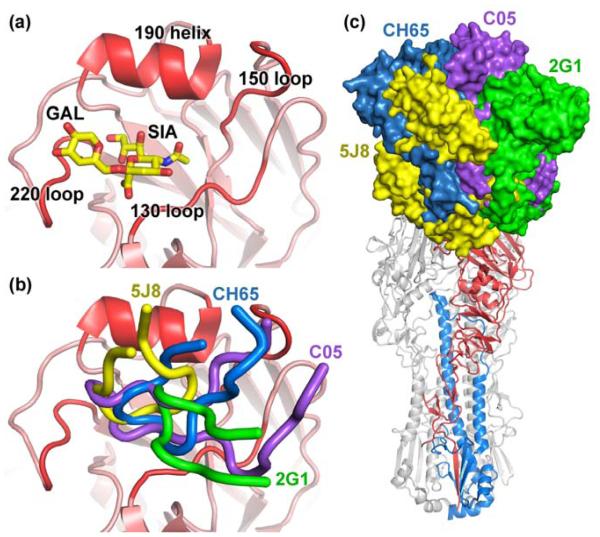
Fig. 2.
Recognition of the HA receptor binding site. a The receptor binding site of HA is framed by the 130 loop, 150 loop, 190 helix, and the 220 loop (labeled and highlighted in red), which binds sialoglycans (PDB 2YP4). b Receptor binding site-targeted antibodies 5J8 (yellow, PDB 4M5Z), CH65 (blue, PDB 3SM5), 2G1 (green, PDB 4HG4), and C05 (purple, PDB 4FP8) insert CDR loops into the binding pocket to compete with the sialoglycan receptor. c Superimposition of the receptor binding site-targeted antibodies reveal that they bind the HA using different angles of approach.
2.1.1 Receptor Binding Site Recognition of a Single Subtype
Antibodies CH65 and CH67, which derive from the same B-cell clonal lineage, have been characterized to bind and neutralize seasonal H1 strains that have infected humans since 1986 (Schmidt et al. 2013; Whittle et al. 2011). However, these antibodies have weak to no activity against the 2009 pandemic H1 strain. In contrast, antibody 5J8, which was isolated from a human donor using hybridoma technology, has reactivity against HAs from the 1918 and 2009 pandemic H1N1 viruses as well as other seasonal isolates (Hong et al. 2013; Krause et al. 2011). Crystal structures of CH65, CH67, and 5J8 in complex with the HA reveal that they all recognize an epitope in and around the receptor binding site and all insert their HCDR3 loop into the binding pocket. The pandemic HA strains incorporate an amino-acid insertion at position 133a (between residues 133 and 134 in the 130 loop), which changes the local conformation of the loop and is a binding determinant for these antibodies. CH65 and CH67 cannot accommodate the 133a insertion (typically a lysine residue) and are sterically blocked from binding. In contrast, 5J8 accommodates the insertion by using a different angle of approach on the HA, compared with CH65 and CH67, and thus avoids the main-chain bulge due to 133a insertion. In addition, 5J8 makes favorable electrostatic interactions with the side chain of the 133a residue. As such, these antibodies converge onto the receptor binding site of human H1 isolates, yet use different sets of interactions and angles of approach, and complement one another for coverage of human H1 isolates that have circulated since H1N1 viruses reemerged in humans in 1977.
H2N2 viruses have circulated in humans starting from 1957 but persisted only for 10 years. However, there has been recent concern that H2N2 viruses may reemerge in humans as immunity has dropped in the population (Nabel et al. 2011). Antibodies 2G1, 8F8, and 8M2 were isolated from human donors by the hybridoma technology and were found to recognize and neutralize human H2N2 viruses that spanned from 1957-1963 (Krause et al. 2012; Xu et al. 2013). Just as for the anti-H1 receptor binding site-targeted antibodies, crystal structures these three antibodies in complex with the HA reveal that these antibodies all reach into the binding pocket. Antibody 8F8 primarily inserts HCDR3, whereas 8M2 and 2G1 use the hydrophobic HCDR2 encoded by the VH1-69 germline gene. The 8M2 and 2G1 antibodies adopt different orientations (~180° rotation around VH/VL) on the HA to center their HCDR2 in the receptor binding site while their HCDR3s interact on the periphery on opposite ends of the receptor binding site. These studies also isolated escape mutants in and around the receptor binding site. Interestingly, these mutants come at a detrimental fitness cost to the virus such that the receptor binding properties of the HA have diminished. This finding substantiates the notion of limited mutability of the receptor binding site due to the conservation of function.
2.1.2 Heterosubtypic Recognition of the Receptor Binding Site
While the anti-H1 or anti-H2 antibodies discussed above are limited to a single subtype, antibodies C05 and S139/1 remarkably have achieved some level of heterosubtypic reactivity and can bind multiple HA strains from both phylogenetic groups, including the H1, H2, H3, and other divergent subtypes (Ekiert et al. 2012; Lee et al. 2012). Antibody S139/1 was selected from H3N2 immunized mice, and was the first reported heterosubtypic antibody that mapped to the receptor binding site, possessing reactivity against H1, H2, H3, H5, H9, and H13 subtypes (Yoshida et al. 2009). The crystal structure of S139/1 in complex with the HA reveals that the antibody reaches into the receptor binding site using HCDR2 (Lee et al. 2012). Binding and neutralization studies confirmed that S139/1 does indeed have heterosubtypic activity, albeit with narrow specificity within each subtype. Nonetheless, these results suggest that divergent isolates across subtypes and groups can share a similar receptor binding site epitope.
Antibody C05 was isolated from phage libraries constructed from the immune repertoires of individuals who had been infected with seasonal H1N1 influenza viruses (Ekiert et al. 2012). C05 has potent neutralizing activity against H1, H2, H3, and H9 viruses and has a broader breadth of recognition within these subtypes as compared to S139/1. Unlike all other receptor binding site-targeted antibodies described thus far, C05 binds the HA exclusively through the heavy chain. The majority of the interactions are mediated through an exceptionally long HCDR3, which is inserted into the receptor binding site. Since the antibody essentially uses a single antibody loop, the C05 epitope on HA is very compact. As such, C05 makes minimal contacts with variable residues around the receptor binding site, which affords the antibody its exceptional reactivity spectrum.
2.2 Enhanced Affinity Through Avidity
Due to the large footprints of antibodies compared with sialosides on the HA, antibodies to the HA head inevitably contact variable residues outside of the receptor binding pocket that limits their overall breadth of recognition. One way that these receptor binding site-targeted antibodies appear to accommodate this limitation is by enhanced affinity through avidity. Although monovalent Fab (fragment antigen binding) bind with reasonable affinity to some strains, the enhanced avidity of bivalent IgG (immunoglobulin G) increases the breadth of recognition to additional strains and subtypes where Fab binding is relatively weak. This feature has been seen for a number of the receptor binding site-targeted antibodies such as S139/1, where the monovalent Fab binds weakly to non-H3 isolates but the bivalent IgG substantially increases its affinity, positively correlating with neutralizing activity (Lee et al. 2012). Therefore, antibodies making relatively low affinity Fab interactions with the receptor binding site can have significant antiviral activity when enhanced by avidity, thereby extending the breadth of neutralization to highly divergent influenza virus strains and subtypes. It appears that avidity is a general feature for other receptor binding site-targeted antibodies as observed for C05, 5J8, and CH65 (Ekiert et al. 2012; Hong et al. 2013). Thus, to gain broad recognition against the receptor binding site, it may be favorable for the Fab portion of the antibody to have intermediate specificity for a number of HA strains to account for the variability of the residues in and around the receptor binding site, which can then be rescued by bivalent binding of the IgG.
2.3 Receptor Mimicry by Antibodies
The HA recognizes the chemical features of the sialic acid receptor (e.g. the acetamide, carboxylate, and glycerol groups) using highly conserved residues in different regions of the binding pocket. In accordance, the receptor binding site-targeted antibodies take advantage of these highly conserved HA residues and have found ways of mimicking portions of the glycan receptor using amino acid residues. Striking trends in recognition of the receptor binding site are now becoming apparent as more structural details of these antibodies are uncovered.
The portion of the pocket that is occupied by the acetamide group of sialic acid is almost absolutely conserved across all HAs and is formed by the hydrophobic residues Trp153 and Leu194 (some strains use a similarly placed hydrophobic Ile194). As such, nearly all receptor binding site-targeted antibodies commonly insert a hydrophobic amino acid in this acetamide-binding pocket. There appears to be some freedom in the nature of the hydrophobic amino acid that can be placed here, as some antibodies use large aromatic side chains such as a phenylalanine (the anti-H2 antibodies 2G1 and 8M2), tyrosine (anti-H2 antibody 8F8 and anti-H3 antibody HC19), or tryptophan (heterosubtypic antibody C05) (Fig. 3a) whereas other antibodies insert smaller hydrophobic side chains such as valine or proline (anti-H1 antibodies CH65 and CH67 or 5J8, respectively). These structures reveal a compelling conserved strategy used by these antibodies to insert amino acids with similar properties into a hydrophobic pocket in the HA receptor binding site.
Fig. 3.
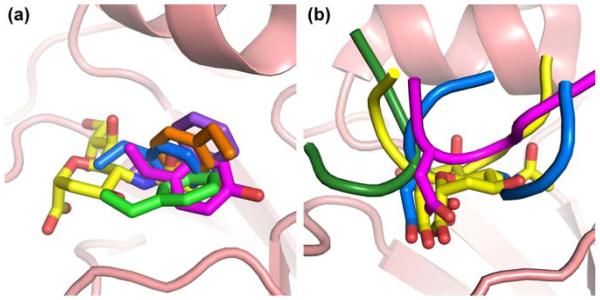
Common trends in HA receptor binding site-targeted antibodies. a Antibodies HC19 (magenta, PDB 2VIR), C05 (purple, PDB 4FP8), 8F8 (blue, PDB 4HF5), 8M2 (orange, PDB 4HFU), and 2G1 (green, PDB 4HG4) use hydrophobic residues to target the receptor binding site that would be occupied by the acetamide moiety of sialic acid. b Antibodies HC19 (magenta, PDB 2VIR), HC63 (dark green, PDB 1KEN), CH65 (blue, PDB 3SM5), and 5J8 (yellow, PDB 4M5Z) use an Asp residue to insert a carboxylate in the receptor binding site that would be occupied by the carboxylate of sialic acid.
In another region of the receptor binding site, sialic acid inserts its carboxylate into a polar pocket that is adjacent to the pocket for the acetamide moiety. The carboxylate forms specific hydrogen bonds with the main-chain backbone of the 130 loop as well as the side chain of a highly conserved Ser136 or Thr136. Antibodies HC19, HC63, CH65, CH67, and 5J8 mimic this interaction by insertion of an aspartic acid carboxylate into the pocket, which closely overlaps with and utilizes the same hydrogen bonding interactions as the sialic acid receptor (Fig. 3b) (Barbey-Martin et al. 2002; Fleury et al. 1998; Hong et al. 2013; Schmidt et al. 2013; Whittle et al. 2011). However, only a few antibodies insert an aspartic acid into this pocket; other antibodies use an isoleucine, such as S139/1 (Lee et al. 2012), or even the HCDR3 backbone, such as C05 (Ekiert et al. 2012), instead.
Although these receptor binding site-targeted antibodies mimic the acetamide and carboxylate moieties of the receptor, they do not directly contact the region of the binding pocket occupied by the glycerol moiety of sialic acid. As there is only room typically for a single antibody loop to enter into the binding groove, the extent of receptor mimicry therefore may have some spatial limitations. Only antibodies 1F1, 8M2, and C05 approach the pocket for the glycerol moiety of sialic acid (Ekiert et al. 2012; Tsibane et al. 2012; Xu et al. 2013), but do not have the same binding mode and hydrogen bonding interactions to the conserved HA residues as the glycerol moiety.
In summary, the recent identification and structural characterization of a variety of neutralizing antibodies to influenza virus have surprisingly revealed that the receptor binding site can indeed be considered a viable target on the HA head for bnAbs. The small footprint of the receptor binding site for a glycan makes it much more difficult to target considering the larger footprint of an antibody. Notwithstanding, it is remarkable that these antibodies have now been found to share similar motifs for recognition of the receptor binding site despite deriving from different mouse or human germline genes as well as possessing differing specificities for influenza strains and subtypes. The combination of the recurrent binding motifs of these receptor binding site bnAbs is likely of value for design of small molecule drugs and therapeutics.
3 HA Stem-Reactive Antibodies
Traditional antigenic sites were only mapped to the head and, therefore, it was a popular belief that the membrane-proximal HA stem was not targeted by the humoral immune response. Antibody C179, which was isolated from mice immunized with an H2N2 virus, was the first reported discovery of a putative stem-targeted in 1993 that had heterosubtypic neutralizing activity against group 1 H1, H2, H5, and H6 HAs (Okuno et al. 1993; Smirnov et al. 1999). Escape mutants were isolated along the HA stem and away from the HA head and, unlike the HA head-targeted antibodies, C179 did not block receptor binding but rather inhibited the HA fusion activity (Okuno et al. 1993). However, the influenza field largely overlooked this discovery and a structure of this antibody in complex with HA remained elusive for two decades (Fig. 4a) (Dreyfus et al. 2013). It was also unknown whether stem-targeted antibodies similar to C179 circulated in the human immune repertoire and, thus, whether it had any relevance to eliciting human antibodies with similar activity. Thus, while antibody discovery and characterization against the HA head was ongoing, the importance of broadly neutralizing or any neutralizing antibodies to the HA stem would be underappreciated and would not be investigated further for many years.
Fig. 4.
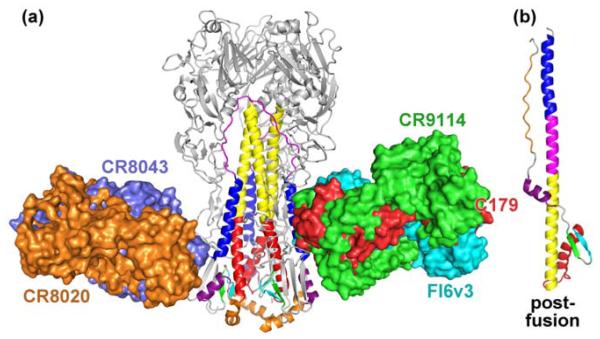
Recognition of the HA stem. a Antibodies CR9114 (green, PDB 4FQI), C179 (red, PDB 4HLZ), and FI6v3 (cyan, PDB 3ZTJ) bind distinct but slightly overlapping epitopes in the HA stem compared with CR8020 (orange, PDB 3SDY) and CR8043 (blue, PDB 4NM8). These stem-targeted antibodies inhibit the postfusion conformation (PDB 1QU1) that is triggered at low pH, shown in (b). [Partially adapted from (Julien et al. 2012).]
3.1 Group 1 HA Recognition
Nearly 15 years after the initial discovery of C179, several stem-targeted antibodies were identified from human combinatorial antibody libraries (Ekiert et al. 2009; Kashyap et al. 2008; Sui et al. 2009; Throsby et al. 2008). Like C179, these antibodies bind antigenically diverse group 1 HAs. Interestingly, the heavy chains of this large panel of antibodies had very high sequence similarity with one another and are encoded by the VH1-69 germline gene. Furthermore, as they undergo very limited somatic hypermutation, such an antibody response could potentially be more readily elicited by vaccination. The structures of two of these antibodies, CR6261 and F10, in complex with HA revealed that these human antibodies bound a highly conserved epitope in the membrane-proximal stem that is shared among group 1 HAs (10 of the 16 known HA subtypes at the time) (Ekiert et al. 2009; Sui et al. 2009). Both antibodies approach at a similar, but not identical, angle to bind the shared HA epitope and the interactions are mediated solely through the antibody heavy chains. In particular, both antibodies insert the hydrophobic tip of their HCDR2 loops, a feature of the VH1-69 germline sequence, into a hydrophobic pocket in the HA stem. The epitope consists of a helical region of the stem composed of HA2 and some HA1 residues and has exceptional sequence conservation as it is comprised of key elements of the fusion machinery. However, the viruses can still bind to host cells, since the HA receptor binding sites are not blocked, and subsequently enter the cell via endosomal compartments. In particular, residues in this stem epitope are involved in the rearrangements from the pre-fusion to post-fusion conformation during the fusion process and, thus, these antibodies block the low pH conformational changes and inhibit the fusion process by locking the HA in its prefusion conformation (Fig. 4). In the endosomes, the viruses are effectively trapped by CR6261 where they can be eventually degraded (Brandenburg et al. 2013). These features are unique to the stem-targeted antibodies, although HC63 is the only example of a head-targeted antibody that can prevent both receptor binding and the low pH conformational change by cross-linking the HA heads of neighboring protomers together (Barbey-Martin et al. 2002).
Two decades after the initial discovery of the mouse monoclonal antibody C179, its crystal structure confirmed that it indeed is a stem-targeted antibody (Dreyfus et al. 2013). C179 binds a similar epitope as CR6261 and F10, but uses a completely different approach angle. Both heavy and light chains mediate interactions with the HA. Similarities can be drawn between C179 and the VH1-69 antibodies, as they both use hydrophobic residues (HCDR3 for C179, HCDR2 for CR6261 and F10) to complement the hydrophobic groove of the HA and block the low pH conformational changes. As such, these structures reveal recurring modes of recognition against a broadly neutralizing and highly conserved epitope at the HA stem.
3.2 Group 2 HA Recognition
The identification of the VH1-69 antibodies that target the HA stem of nearly all group 1 viruses raised hope for the possibility of antibody-based therapy or broader-spectrum influenza vaccine. Yet, these VH1-69 antibodies do not have activity against group 2 HAs, including H3 viruses that currently circulate in humans. In a major advance, two group 2 targeted bnAbs, called CR8020 and CR8043, were isolated by Crucell from memory B cells of healthy donors and have heterosubtypic neutralizing activity in vitro and protect mice from lethal challenge from H3 and H7 viruses (Ekiert et al. 2011; Friesen et al. 2014). The two antibodies were isolated from a single donor and both target a common epitope lower down on the HA stem distinct from the group 1 antibodies (Fig. 4a). Interestingly, CR8020 and CR8043 derive from different germline genes (VH1-18 and VH1-3, respectively) and use different sets of heavy and light chain interactions and alternative angles of approach to target the HA. Moreover, the antibodies have distinct escape mutation sensitivities indicating that there may be versatility in targeting the CR8020 and CR8043 epitopes. Similar to the group 1 targeted antibodies, CR8020 and CR8043 bind key elements of the HA fusion machinery and prevent the conformational changes of the HA that occur low pH. The antibodies also inhibit the maturation process of the HA by blocking the proteolytic cleavage of the HA0 precursor to the mature HA1/HA2 (Ekiert et al. 2011; Friesen et al. 2014). Thus, the identification and structural characterization of these bnAbs have determined a second site of vulnerability on the HA stem.
3.3 Pan-influenza A and Type B Recognition
Structural characterization of the group 1 and the group 2 heterosubtypic stem-targeted antibodies have defined neutralizing epitopes that can be exploited for therapeutics as well as vaccine design. In a further remarkable breakthrough, bnAb FI6v3 was identified and found to cross the group 1 and group 2 barrier (Corti et al. 2011). FI6v3 was isolated from single plasma cells of human donors with activity against nearly all subtypes of influenza type A. It is encoded by the VH3-30 germline gene but surprisingly recognizes a similar epitope as the VH1-69 group 1 specific bnAbs but approaches at a completely different angle to the HA. The FI6v3 binding mode more closely resembles C179 as it uses both heavy and light chains mediate the interactions with the HA and inserts its HCDR3 loop into the hydrophobic groove of the HA stem. The longer HCDR3 has a number of aromatic residues that emulate the binding of the aromatic and non-polar residues of the hydrophobic HCDR2 of the VH1-69 antibodies. A unique feature of FI6v3 is that it can contact the uncleaved fusion peptide in the neighboring HA protomer using its light chain, suggesting that it may also prevent HA maturation (Corti et al. 2011).
However, to have the ultimate universal flu vaccine, an antibody response must be elicited across flu type A as well as both lineages of type B. In another unprecedented discovery, bnAb CR9114 was isolated by Crucell using combinatorial display libraries from human B cells and was found to have activity against all subtypes of influenza type A as well as both lineages of type B viruses (Dreyfus et al. 2012). CR9114 is a VH1-69 encoded antibody and its binding mode highly resembles that of CR6261 and F10, in that it targets a nearly identical epitope in the HA stem solely through its heavy chains and approaches the HA using a similar angle of approach as CR6261. Yet, CR9114 can also recognize group 2 HAs, whereas the CR6261 and F10 VH1-69 antibodies cannot cross the HA group barrier despite the high sequence conservation of this epitope across all HA subtypes. It has been suggested that a glycan at Asn38 conserved among group 2 HAs sterically blocks binding by these antibodies (Ekiert et al. 2009; Sui et al. 2009). CR9114 as well as FI6v3 appear to be able to push the glycan away to enable access to the HA protein surface. It is extraordinary that these pan-influenza antibodies recognize a similar epitope as the group 1 stem-targeted antibodies, yet acquire even greater reactivity, coping with both glycan and some amino acid diversity in and around the epitope. Further investigation of this epitope by other bnAbs may provide additional insight into molecular details of weak spots on the HA.
4 Conclusions and Future Directions
Remarkably, the rapid discovery of bnAbs against the influenza virus did not truly take off until 2008. Since then, an explosion in the discovery and characterization of such bnAbs has advanced the field tremendously and given hope that a “universal” flu vaccine may indeed be possible. Structural characterization of antibody-HA complexes by x-ray crystallography and EM has now defined supersites of vulnerability in the HA stem and in the HA head, particularly in and around the receptor binding site.
The recent identification and elucidation of receptor binding site-targeted antibodies have now proven that the HA head can indeed elicit an antibody response with a broader level of cross-reactivity than previously believed. Although these antibodies can have varying levels of activity, the combination of some of these antibodies can provide nearly complete coverage against particular subtypes. For instance, CH65 and CH67 have activity against many seasonal H1 isolates whereas 5J8 can also bind pandemic H1 strains. By combining these three antibodies, perhaps by linking two Fab arms together to create a bispecific antibody with one Fab arm as CH65 or CH67 and the other arm as 5J8 (Ridgway et al. 1996), it may be possible to generate an IgG with near pan-H1 reactivity.
Other putative receptor binding site-targeted antibodies have broad-spectrum activity but unfortunately no high-resolution structural information have yet been published. For instance, F045-092 reportedly has pan-H3 reactivity (Ohshima et al. 2011) and CR8033 neutralizes both flu B lineages (Dreyfus et al. 2012). Of these antibodies, EM reconstructions of the CR8033-HA complex are available. Higher resolution studies of F045-092 and CR8033 in complex with the HA, in conjunction with the pan-H1 receptor binding site-targeted antibodies, will provide the structural framework for recognition of all subtypes and types of influenza viruses that currently circulate in humans.
The discovery of stem-targeted antibodies with broadly neutralizing activity against group 1 and/or 2 influenza A viruses (Corti et al. 2011; Dreyfus et al. 2013; Ekiert et al. 2009; Ekiert et al. 2011; Friesen et al. 2014; Kashyap et al. 2008; Kashyap et al. 2010; Sui et al. 2009; Throsby et al. 2008) as well as B viruses (Dreyfus et al. 2012) has truly reinvigorated the influenza research community and raised hopes for the development of antibody-based immunotherapy and long-lasting, universal vaccines for influenza. Structures of these antibody-HA complexes have defined two principal, highly conserved epitopes on the HA stem. Rather than by blocking receptor binding, these antibodies can prevent HA maturation as well as the low pH conformational change necessary for fusion of viral and host cell membranes. Some of these antibodies also prevent the release of nascent virions and thus prevent spread of the virus (Brandenburg et al. 2013; Dreyfus et al. 2012). However, despite the detailed efficacy of these bnAbs to neutralize viruses in vitro and in vivo, some concerns have been raised on the accessibility of the membrane-proximal stem epitopes of the densely clustered HA on the surface of viruses. Cryoelectron tomography of viruses with bnAb C179 has now visually confirmed that the HA stem epitope can indeed be accessed by stem-targeted bnAbs (Harris et al. 2013).
The structural characterization of antibody-HA complexes has provided exclusive insight into strategies to harness and employ potent bnAb responses. For instance, vaccination with synthetic peptides that mimic the long α-helix of the HA stem has been reported to provide protection in mice from challenges with H3N2, H1N1, and H5N1 viruses (Wang et al. 2010). An alternative vaccine strategy through the use of chimeric HAs, where different HA heads are linked to the same HA stem, can bias the immune response towards the conserved stem after consecutive immunization with different chimeric HAs (Hai et al. 2012; Krammer et al. 2013). Perhaps this method can also be applied to generate broader-spectrum antibodies that are more selective to the conserved residues in the receptor binding site, while avoiding the variable loops outside of the pocket. Self-assembling nanoparticles have been engineered to display multiple HA spikes and have been shown to elicit bnAb response against the receptor binding site as well as the stem of the HA (Kanekiyo et al. 2013). To focus elicitation against the stem, headless HA constructs have also been engineered that can bind stem-targeted antibodies (Lu et al. 2014). By coupling these headless HAs or similar rational immunogen designs onto self-assembling particles, it may be possible to focus a VH1-69 germline encoded bnAb response solely against the stem, as was similarly performed against the CD4 binding site of the HIV-1 envelope glycoprotein (Jardine et al. 2013).
The structural details of the bnAbs in complex with the HA have also served as a foundation for the computational design of de novo proteins against the stem (Fleishman et al. 2011; Whitehead et al. 2012). Individual residues of the stem-targeted bnAb CR6261 that form favorable interactions along the hydrophobic groove of the HA were grafted onto protein scaffolds. Crystal structures show that the designs do indeed match the computational model even down to the level of side-chain rotamer conformations. Moreover, these designs have antiviral activity and similarly block the low pH conformational change of the HA. In theory, this design method can also extend to design small proteins that target the receptor binding site by combining the recurring themes of recognition, such as receptor mimicry as well as avidity.
In summary, the discovery and structural characterization of receptor binding site and stem-targeted antibodies has shifted the paradigm and outlook for vaccine design against influenza virus. These structures provide a wealth of information that furthers our understanding of how the HA can be targeted and has defined two main supersites of vulnerability and one of two other possible sites (Fig. 1). Recurrent motifs of recognition have been observed both in the receptor binding site and stem epitopes that have directed innovative design of immunogens capable of eliciting broad-spectrum antibodies. The road ahead is certainly bright for a new frontier in the structured-based discovery of broad-spectrum vaccines, immunogens, and therapeutics against influenza viruses.
Abbreviations
- HA
Hemagglutinin
- bnAb
Broadly neutralizing antibody
- EM
Electron microscopy
- CDR
Complementarity determining region
- Fab
Fragment antigen binding
- IgG
Immunoglobulin G
References
- Barbey-Martin C, Gigant B, Bizebard T, Calder LJ, Wharton SA, Skehel JJ, Knossow M. An antibody that prevents the hemagglutinin low pH fusogenic transition. Virology. 2002;294:70–74. doi: 10.1006/viro.2001.1320. [DOI] [PubMed] [Google Scholar]
- Brandenburg B, Koudstaal W, Goudsmit J, Klaren V, Tang C, Bujny MV, Korse HJ, Kwaks T, Otterstrom JJ, Juraszek J, van Oijen AM, Vogels R, Friesen RH. Mechanisms of hemagglutinin targeted influenza virus neutralization. PLoS One. 2013;8:e80034. doi: 10.1371/journal.pone.0080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. JAMA. 2006;295:891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- CDC. Centers for Disease Control and Prevention (CDC) Estimates of deaths associated with seasonal influenza --- United States, 1976-2007. MMWR Morb Mortal Wkly Rep. 2010;59:1057–1062. [PubMed] [Google Scholar]
- Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Fan G, Yang F, Li X, Zhou J, Zou S, Yang L, Chen T, Dong L, Bo H, Zhao X, Zhang Y, Lan Y, Bai T, Dong J, Li Q, Wang S, Zhang Y, Li H, Gong T, Shi Y, Ni X, Li J, Zhou J, Fan J, Wu J, Zhou X, Hu M, Wan J, Yang W, Li D, Wu G, Feng Z, Gao GF, Wang Y, Jin Q, Liu M, Shu Y. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet. 2014 doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- Chen J, Lee KH, Steinhauer DA, Stevens DJ, Skehel JJ, Wiley DC. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–417. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- Cho KJ, Hong KW, Kim SH, Seok JH, Kim S, Lee JH, Saelens X, Kim KH. Insight into highly conserved H1 subtype-specific epitopes in influenza virus hemagglutinin. PLoS One. 2014;9:e89803. doi: 10.1371/journal.pone.0089803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu Rev Immunol. 2013;31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- de Jong MD, Tran TT, Truong HK, Vo MH, Smith GJ, Nguyen VC, Bach VC, Phan TQ, Do QH, Guan Y, Peiris JS, Tran TH, Farrar J. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N Engl J Med. 2005;353:2667–2672. doi: 10.1056/NEJMoa054512. [DOI] [PubMed] [Google Scholar]
- Dreyfus C, Ekiert DC, Wilson IA. Structure of a classical broadly neutralizing stem antibody in complex with a pandemic H2 influenza virus hemagglutinin. J Virol. 2013;87:7149–7154. doi: 10.1128/JVI.02975-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin O, Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RH. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O'Neil RE, Faynboym AM, Horowitz M, Horowitz L, Ward AB, Palese P, Webby R, Lerner RA, Bhatt RR, Wilson IA. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Wilson IA. Broadly neutralizing antibodies against influenza virus and prospects for universal therapies. Curr Opin Virol. 2012;2:134–141. doi: 10.1016/j.coviro.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, Strauch EM, Wilson IA, Baker D. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury D, Barrere B, Bizebard T, Daniels RS, Skehel JJ, Knossow M. A complex of influenza hemagglutinin with a neutralizing antibody that binds outside the virus receptor binding site. Nat Struct Biol. 1999;6:530–534. doi: 10.1038/9299. [DOI] [PubMed] [Google Scholar]
- Fleury D, Daniels RS, Skehel JJ, Knossow M, Bizebard T. Structural evidence for recognition of a single epitope by two distinct antibodies. Proteins. 2000;40:572–578. [PubMed] [Google Scholar]
- Fleury D, Wharton SA, Skehel JJ, Knossow M, Bizebard T. Antigen distortion allows influenza virus to escape neutralization. Nat Struct Biol. 1998;5:119–123. doi: 10.1038/nsb0298-119. [DOI] [PubMed] [Google Scholar]
- Friesen RH, Lee PS, Stoop EJ, Hoffman RM, Ekiert DC, Bhabha G, Yu W, Juraszek J, Koudstaal W, Jongeneelen M, Korse HJ, Ophorst C, Brinkman-van der Linden EC, Throsby M, Kwakkenbos MJ, Bakker AQ, Beaumont T, Spits H, Kwaks T, Vogels R, Ward AB, Goudsmit J, Wilson IA. A common solution to group 2 influenza virus neutralization. Proc Natl Acad Sci U S A. 2014;111:445–450. doi: 10.1073/pnas.1319058110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- Hai R, Krammer F, Tan GS, Pica N, Eggink D, Maamary J, Margine I, Albrecht RA, Palese P. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J Virol. 2012;86:5774–5781. doi: 10.1128/JVI.00137-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AK, Meyerson JR, Matsuoka Y, Kuybeda O, Moran A, Bliss D, Das SR, Yewdell JW, Sapiro G, Subbarao K, Subramaniam S. Structure and accessibility of HA trimers on intact 2009 H1N1 pandemic influenza virus to stem region-specific neutralizing antibodies. Proc Natl Acad Sci U S A. 2013;110:4592–4597. doi: 10.1073/pnas.1214913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Lee PS, Hoffman RM, Zhu X, Krause JC, Laursen NS, Yoon SI, Song L, Tussey L, Crowe JE, Jr., Ward AB, Wilson IA. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J Virol. 2013;87:12471–12480. doi: 10.1128/JVI.01388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, Schief WR. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Lee PS, Wilson IA. Structural insights into key sites of vulnerability on HIV-1 Env and influenza HA. Immunol Rev. 2012;250:180–198. doi: 10.1111/imr.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo M, Wei CJ, Yassine HM, McTamney PM, Boyington JC, Whittle JR, Rao SS, Kong WP, Wang L, Nabel GJ. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature. 2013;499:102–106. doi: 10.1038/nature12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap AK, Steel J, Oner AF, Dillon MA, Swale RE, Wall KM, Perry KJ, Faynboym A, Ilhan M, Horowitz M, Horowitz L, Palese P, Bhatt RR, Lerner RA. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc Natl Acad Sci U S A. 2008;105:5986–5991. doi: 10.1073/pnas.0801367105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap AK, Steel J, Rubrum A, Estelles A, Briante R, Ilyushina NA, Xu L, Swale RE, Faynboym AM, Foreman PK, Horowitz M, Horowitz L, Webby R, Palese P, Lerner RA, Bhatt RR. Protection from the 2009 H1N1 pandemic influenza by an antibody from combinatorial survivor-based libraries. PLoS Pathog. 2010;6:e1000990. doi: 10.1371/journal.ppat.1000990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiso M, Mitamura K, Sakai-Tagawa Y, Shiraishi K, Kawakami C, Kimura K, Hayden FG, Sugaya N, Kawaoka Y. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet. 2004;364:759–765. doi: 10.1016/S0140-6736(04)16934-1. [DOI] [PubMed] [Google Scholar]
- Kong L, Lee JH, Doores KJ, Murin CD, Julien JP, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, Hoffenberg S, Caulfield M, King CR, Hua Y, Le KM, Khayat R, Deller MC, Clayton T, Tien H, Feizi T, Sanders RW, Paulson JC, Moore JP, Stanfield RL, Burton DR, Ward AB, Wilson IA. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol. 2013;20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Pica N, Hai R, Margine I, Palese P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol. 2013;87:6542–6550. doi: 10.1128/JVI.00641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Albrecht R, Blum DL, Ramos I, Fernandez-Sesma A, Edwards KM, Garcia-Sastre A, Basler CF, Crowe JE., Jr. Human monoclonal antibodies to pandemic 1957 H2N2 and pandemic 1968 H3N2 influenza viruses. J Virol. 2012;86:6334–6340. doi: 10.1128/JVI.07158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, Crowe JE., Jr. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol. 2011;85:10905–10908. doi: 10.1128/JVI.00700-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, Yoshida R, Ekiert DC, Sakai N, Suzuki Y, Takada A, Wilson IA. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci U S A. 2012;109:17040–17045. doi: 10.1073/pnas.1212371109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Welsh JP, Swartz JR. Production and stabilization of the trimeric influenza hemagglutinin stem domain for potentially broadly protective influenza vaccines. Proc Natl Acad Sci U S A. 2014;111:125–130. doi: 10.1073/pnas.1308701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J, Wharton SA, Lin YP, Takemoto DK, Skehel JJ, Wiley DC, Steinhauer DA. Studies of the binding properties of influenza hemagglutinin receptor-site mutants. Virology. 1998;241:101–111. doi: 10.1006/viro.1997.8958. [DOI] [PubMed] [Google Scholar]
- Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, Weintraub E, Bridges CB. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine. 2007;25:5086–5096. doi: 10.1016/j.vaccine.2007.03.046. [DOI] [PubMed] [Google Scholar]
- Nabel GJ, Wei CJ, Ledgerwood JE. Vaccinate for the next H2N2 pandemic now. Nature. 2011;471:157–158. doi: 10.1038/471157a. [DOI] [PubMed] [Google Scholar]
- Ohshima N, Iba Y, Kubota-Koketsu R, Asano Y, Okuno Y, Kurosawa Y. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J Virol. 2011;85:11048–11057. doi: 10.1128/JVI.05397-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol. 1993;67:2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway JB, Presta LG, Carter P. 'Knobs-into-holes' engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng. 1996;9:617–621. doi: 10.1093/protein/9.7.617. [DOI] [PubMed] [Google Scholar]
- Salzberg S. The contents of the syringe. Nature. 2008;454:160–161. doi: 10.1038/454160a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AG, Xu H, Khan AR, O'Donnell T, Khurana S, King LR, Manischewitz J, Golding H, Suphaphiphat P, Carfi A, Settembre EC, Dormitzer PR, Kepler TB, Zhang R, Moody MA, Haynes BF, Liao HX, Shaw DE, Harrison SC. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci U S A. 2013;110:264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel JJ, Bayley PM, Brown EB, Martin SR, Waterfield MD, White JM, Wilson IA, Wiley DC. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc Natl Acad Sci U S A. 1982;79:968–972. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov YA, Lipatov AS, Gitelman AK, Okuno Y, Van Beek R, Osterhaus AD, Claas EC. An epitope shared by the hemagglutinins of H1, H2, H5, and H6 subtypes of influenza A virus. Acta Virol. 1999;43:237–244. [PubMed] [Google Scholar]
- Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Shi Y, Lu X, He J, Gao F, Yan J, Qi J, Gao GF. Bat-derived influenza hemagglutinin H17 does not bind canonical avian or human receptors and most likely uses a unique entry mechanism. Cell Rep. 2013;3:769–778. doi: 10.1016/j.celrep.2013.01.025. [DOI] [PubMed] [Google Scholar]
- Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, Cinatl J, ter Meulen J, Lasters I, Carsetti R, Peiris M, de Kruif J, Goudsmit J. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsibane T, Ekiert DC, Krause JC, Martinez O, Crowe JE, Jr., Wilson IA, Basler CF. Influenza human monoclonal antibody 1F1 interacts with three major antigenic sites and residues mediating human receptor specificity in H1N1 viruses. PLoS Pathog. 2012;8:e1003067. doi: 10.1371/journal.ppat.1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TT, Tan GS, Hai R, Pica N, Ngai L, Ekiert DC, Wilson IA, Garcia-Sastre A, Moran TM, Palese P. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc Natl Acad Sci U S A. 2010;107:18979–18984. doi: 10.1073/pnas.1013387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- Whitehead TA, Chevalier A, Song Y, Dreyfus C, Fleishman SJ, De Mattos C, Myers CA, Kamisetty H, Blair P, Wilson IA, Baker D. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat Biotechnol. 2012;30:543–548. doi: 10.1038/nbt.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao HX, Harrison SC. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE, Jr., Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Krause JC, McBride R, Paulson JC, Crowe JE, Jr., Wilson IA. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol. 2013;20:363–370. doi: 10.1038/nsmb.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Igarashi M, Ozaki H, Kishida N, Tomabechi D, Kida H, Ito K, Takada A. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog. 2009;5:e1000350. doi: 10.1371/journal.ppat.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Guo YH, Jiang T, Wang YD, Chan KH, Li XF, Yu W, McBride R, Paulson JC, Yuen KY, Qin CF, Che XY, Wilson IA. A Unique and Conserved Neutralization Epitope in H5N1 Influenza Viruses Identified by an Antibody against the A/Goose/Guangdong/1/96 Hemagglutinin. J Virol. 2013a;87:12619–12635. doi: 10.1128/JVI.01577-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Yu W, McBride R, Li Y, Chen LM, Donis RO, Tong S, Paulson JC, Wilson IA. Hemagglutinin homologue from H17N10 bat influenza virus exhibits divergent receptor-binding and pH-dependent fusion activities. Proc Natl Acad Sci U S A. 2013b;110:1458–1463. doi: 10.1073/pnas.1218509110. [DOI] [PMC free article] [PubMed] [Google Scholar]