Schizophrenia is a complex neuropsychiatric disorder with high heritability (80–90%) and phenotypic variability common to other psychiatric spectrum disorders (International Schizophrenia et al., 2009). In 2011, the Schizophrenia Psychiatric GWAS Consortium (PGC) published the largest schizophrenia GWAS to date, with an initial discovery sample of 21 856 individuals and a replication sample of 29 839 individuals (Schizophrenia Psychiatric Genome-Wide Association Study, 2011). Of the seven genome-wide significant hits, the strongest finding was in an intron of the primary transcript for human microRNA 137 (miR-137). Since then, multiple studies have identified associations between miR-137 and brain function in patients with schizophrenia strongly supporting miR-137 involvement in the disease etiology (Green et al., 2013; Whalley et al., 2012).
MicroRNAs (miRNAs), small non-coding RNA molecules predominantly negatively regulating gene expression, are involved in normal neurodevelopmental and physiological processes, and are implicated in the pathogenesis of several neuropsychiatric disorders (Amiel et al., 2012). MiR-137 is a brain-enriched miRNA shown to modulate proliferation and differentiation of adult neurons and affect neuronal maturation, dendritic spine development and morphogenesis (Schizophrenia Psychiatric Genome-Wide Association Study, 2011). In addition to miR-137, four of the seven genome-wide significant genes from PGC were predicted and experimentally validated as miR-137 gene targets (Kwon et al., 2011). These results warrant further investigation into a potential mechanism of susceptibility to schizophrenia mediated by dysregulation of miR-137 and its downstream targets.
Variable Nucleotide Tandem Repeats (VNTRs) have been previously associated with neuropsychiatric disorders such as attention deficit hyperactivity disorder, addiction, and schizophrenia (McGeary, 2009; Mill et al., 2005; Prata et al., 2009); however, the molecular effects of VNTRs on disease development have been largely understudied and their role in the pathogenesis of neuropsychiatric disorders remains unclear. A 15bp VNTR at the 5’ end of the miR-137 precursor was first discovered in a melanoma cell line in 2008 (Bemis et al., 2008), and was identified in our postmortem brain sample of 54 schizophrenia cases and 54 controls donated by the Mount Sinai School of Medicine using Sanger sequencing. In this sample the VNTR consisted of 9 alleles (3–10 and 13 repeats) with varying frequency; the major VNTR allele is 3 (72% frequency), followed by 4 (9%), then 9 (6%) and all others were under 5%. We hypothesize that this VNTR is involved in disease susceptibility by affecting the expression of a schizophrenia-associated miRNA and its schizophrenia-associated downstream targets and we tested this hypothesis using in vitro approaches.
Due to low allele frequency of the VNTR alleles we were not able to reliably estimate their effects on miR-137 expression in the postmortem sample (data not shown). Therefore, we attempted to assess the effect of the VNTR alleles individually on miR-137 expression in HEK293 cells. These cells were selected based on their low endogenous miR-137 expression, high transfection efficiency and gene expression profile similar to that of neurons (Shaw et al., 2002). Cells were transfected with expression vectors carrying each VNTR allele (3–13 repeats) in the precursor of miR-137 and cells carrying the 9 and 13 VNTR alleles exhibited significantly reduced miR-137 expression as measured by qPCR (Figure 1a; Bonferroni-corrected p<0.038 and p<0.025, respectively).
Figure 1.
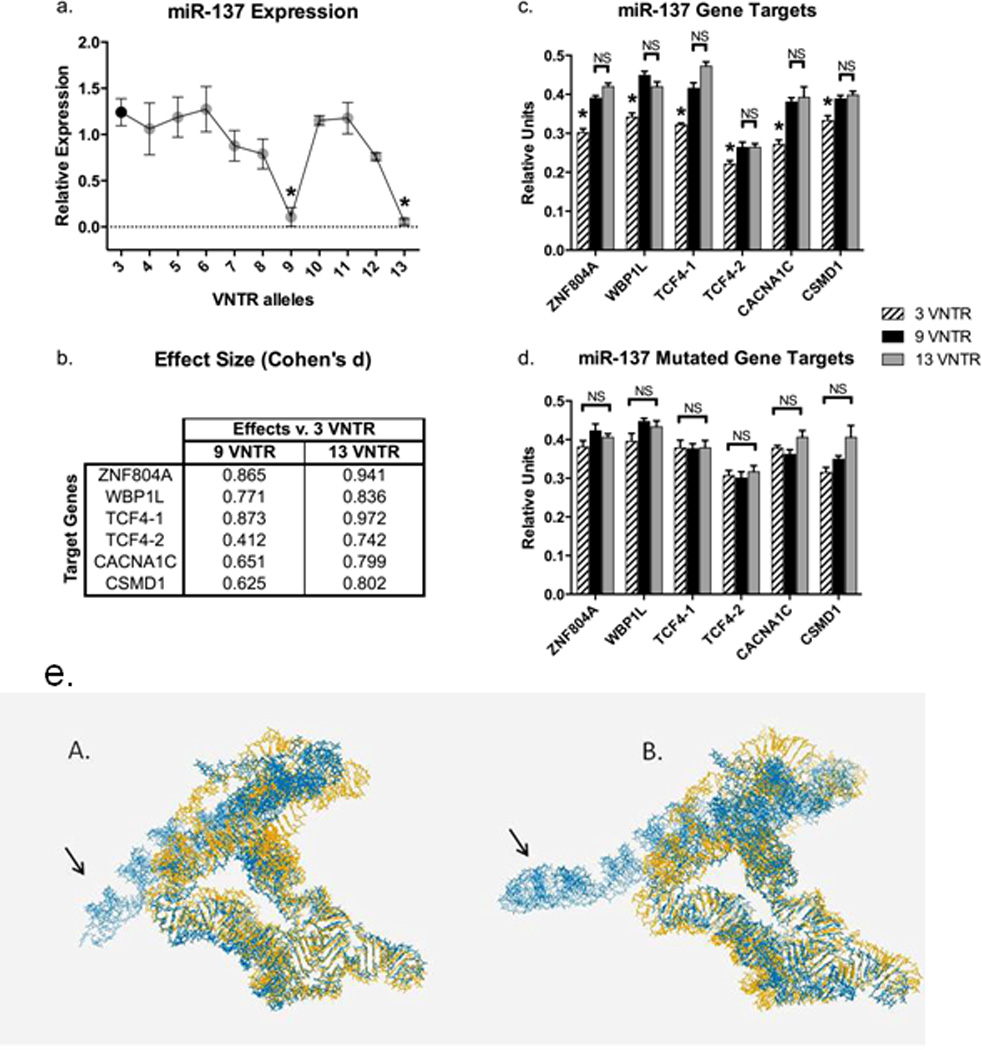
(a) MiRNA expression was assessed by qPCR and relative expression values calculated using 2−ΔΔCt. Data were analyzed using the Kruskal-Wallis test, followed by Dunn’s post-hoc test correcting for multiple testing (GraphPad Prism 6). Two VNTR alleles, 9 and 13, exhibited significantly reduced expression of miR-137, while 7, 8, and 12 showed a trend of reduction in miR-137 expression.
Asterisks indicate significant differences. All experiments were performed in triplicate and results shown are means ± SEM.
(b) Effect sizes of luciferase activity differences (Cohen’s d) between 9 and 13 VNTR alleles compared to the 3 VNTR allele.
(c) Luciferase assay in HEK293 cells; co-expression of miR-137-VNTR alleles with wild-type gene target sequences of schizophrenia candidate genes: ZNF804A, WBP1L, TCF4-1, TCF4-2, CACNA1C, and CSMD1. 9 and 13 VNTR alleles exhibited significantly higher luciferase activity compared to 3 VNTR allele for all target genes. (d) Once gene target sequences were mutated no difference between 9 and 13 compared to 3 was observed. The data were analyzed using Mann-Whitney test followed by Bonferroni correction for multiple testing. Asterisks indicate significant differences. NS indicates no significant differences. All experiments were performed in triplicate and results shown are means ± SEM.
(e) The impact of the VNTR on the 3D structure of the miR-137 precursor was analyzed using RNAcomposer (http://rnacomposer.cs.put.poznan.pl/). The total length of the miR-137 precursor sequences varied from 350 bases (3 VNTR allele) to 500 bases (13 VNTR allele). In this composite figure, the 3D structure of miR-137 for the 3 VNTR allele is colored in yellow, whereas the 9 (panel A) and 13 (panel B) VNTR alleles are overlaid in blue. Although, the 3D structures share similarity, the local substructures containing the 9 and 13 VNTR alleles deviate significantly from the folding pattern of the 3 VNTR allele (noted by the arrows).
To further clarify the mechanism by which the VNTR alleles impact miR-137 function, we performed a series of luciferase assays to measure the strength of interaction between miR-137 and its previously validated gene targets: TCF4, CACNA1C, WBP1L (formerly: C10orf26), CSMD1 and ZNF804A (Kim et al., 2012; Schizophrenia Psychiatric Genome-Wide Association Study, 2011). Considering that only 9 and 13 VNTR alleles exhibited a significant impact on miR-137 expression, we assessed those alleles’ effects on miR-137 function compared to the major VNTR allele, 3 (as the control). Cells transfected with 9 and 13 VNTR alleles showed elevated luciferase activity compared to 3 VNTR allele and these effects were not observed with the mutated target sequences, confirming that these interactions are miRNA- and gene target-specific (Figures 1b, 1c and 1d).
In conclusion, we report a novel mechanism by which a schizophrenia-implicated miRNA’s function is controlled by a VNTR that modulates miR-137 expression and its schizophrenia-associated target genes. Given that the VNTR is present within the miR-137 precursor sequence we hypothesized that 9 and 13 VNTR alleles affect mature miR-137 maturation, expression and function by altering the precursor’s secondary and tertiary structures. Using three dimensional (3D) RNA folding algorithms we observed differences in the tertiary structures caused by the 9 and 13 VNTRs, which we hypothesize is disrupting the mature miRNA’s processing from its precursor sequence (Figure 1e). Although additional studies are needed to further clarify the mechanisms underlying the VNTR’s effects on miR-137 function, our results offer intriguing insights into a potentially novel mechanism of susceptibility to schizophrenia that aligns major GWAS findings within a unified genetic and molecular mechanism in order to provide greater understanding of otherwise disparate findings.
References
- Amiel J, de Pontual L, Henrion-Caude A. miRNA, development and disease. Advances in genetics. 2012;80:1–36. doi: 10.1016/B978-0-12-404742-6.00001-6. [DOI] [PubMed] [Google Scholar]
- Bemis LT, Chen R, Amato CM, Classen EH, Robinson SE, Coffey DG, Erickson PF, Shellman YG, Robinson WA. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer research. 2008;68(5):1362–1368. doi: 10.1158/0008-5472.CAN-07-2912. [DOI] [PubMed] [Google Scholar]
- Green MJ, Cairns MJ, Wu J, Dragovic M, Jablensky A, Tooney PA, Scott RJ, Carr VJ. Genome-wide supported variant MIR137 and severe negative symptoms predict membership of an impaired cognitive subtype of schizophrenia. Molecular psychiatry. 2013;18(7):774–780. doi: 10.1038/mp.2012.84. [DOI] [PubMed] [Google Scholar]
- International Schizophrenia C. Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AH, Parker EK, Williamson V, McMichael GO, Fanous AH, Vladimirov VI. Experimental validation of candidate schizophrenia gene ZNF804A as target for hsa-miR-137. Schizophrenia research. 2012;141(1):60–64. doi: 10.1016/j.schres.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeary J. The DRD4 exon 3 VNTR polymorphism and addiction-related phenotypes: a review. Pharmacology, biochemistry, and behavior. 2009;93(3):222–229. doi: 10.1016/j.pbb.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill J, Asherson P, Craig I, D'Souza UM. Transient expression analysis of allelic variants of a VNTR in the dopamine transporter gene (DAT1) BMC genetics. 2005;6:3. doi: 10.1186/1471-2156-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prata DP, Mechelli A, Fu CH, Picchioni M, Toulopoulou T, Bramon E, Walshe M, Murray RM, Collier DA, McGuire P. Epistasis between the DAT 3' UTR VNTR and the COMT Val158Met SNP on cortical function in healthy subjects and patients with schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(32):13600–13605. doi: 10.1073/pnas.0903007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Psychiatric Genome-Wide Association Study C. Genome-wide association study identifies five new schizophrenia loci. Nature genetics. 2011;43(10):969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G, Morse S, Ararat M, Graham FL. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2002;16(8):869–871. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- Whalley HC, Papmeyer M, Romaniuk L, Sprooten E, Johnstone EC, Hall J, Lawrie SM, Evans KL, Blumberg HP, Sussmann JE, McIntosh AM. Impact of a microRNA MIR137 susceptibility variant on brain function in people at high genetic risk of schizophrenia or bipolar disorder. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2012;37(12):2720–2729. doi: 10.1038/npp.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]


