Abstract
Enrolling adequate numbers of patients that meet protocol eligibility criteria in a timely manner is critical, yet clinical trial accrual continues to be problematic. One approach to meet these accrual challenges is to utilize technology to automatically screen patients for clinical trial eligibility. This manuscript reports on the evaluation of different automated approaches to determine the metastatic status from unstructured radiology reports using the Clinical Trials Eligibility Database Integrated System (CTED).
The study sample included all patients (N = 5,523) with radiologic diagnostic studies (N = 10,492) completed in a 2 week period. Eight search algorithms (queries) within CTED were developed and applied to radiology reports. The performance of each algorithm was compared to a reference standard which consisted of a physician’s review of the radiology reports. Sensitivity, specificity, positive and negative predicted values were calculated for each algorithm. The number of patients identified by each algorithm varied from 187 to 330 and the number of true positive cases confirmed by physician review ranged from 171 to 199 across the algorithms. The best performing algorithm had sensitivity 94 %, specificity 100%, positive predictive value 90 %, negative predictive value 100 %, and accuracy of 99 %.
Our evaluation process identified the optimal method for rapid identification of patients with metastatic disease through automated screening of unstructured radiology reports. The methods developed using the CTED system could be readily implemented at other institutions to enhance the efficiency of research staff in the clinical trials eligibility screening process.
Keywords: Clinical trials, Eligibility screening, Metastases, Radiology reports, Information extraction, Automation
Introduction
Clinical trials are essential in evaluating new therapies before they become a standard of care. Enrolling adequate numbers of patients that meet protocol eligibility criteria in a timely manner is critical to this process, yet clinical trial accrual continues to be problematic, particularly for cancer studies.1-5 Despite the significant body of literature focusing on barriers to clinical trial accrual6-11, few advances have been made to improve patient recruitment and enrollment.
One approach to meet these accrual challenges is to utilize technology to automatically screen patients for clinical trial eligibility. Successful pre-screening will improve research staff efficiency by reducing the number of ineligible patients requiring manual review, while simultaneously increasing the total number of patients evaluated. The researchers consistently reported doubling the enrollment rates by using electronic screening 12-14, increasing the number of prescreened patients while decreasing the total screening time 12, and significantly increasing the physician referrals.13 Much larger proportions of electronically screened patients were eligible and enrolled in studies compared to conventionally screened patients.15
Automated pre-screening is now feasible because the widespread implementation of Electronic Health Records (EHR). A variety of automated clinical trial screening tools and software that use EHR data have been piloted 16-20, though few are commercially available.21,22 A common limitation of such tools is the inability to utilize unstructured clinical text documents which represent the bulk of clinical information that must be reviewed to determine eligibility. While screening tools based only on discrete data are valuable12-15, accuracy can be improved if information locked in narrative reports is utilized. Although the filed for Information extraction (IE) based on Natural Language Processing (NLP) is growing rapidly, IE use to support research is limited.23
Cancer metastatic status is frequently a key inclusion or exclusion criteria for oncology clinical trials. The current practice is to determine new metastatic disease through manual review of medical records of cancer patients. This approach is highly inefficient due to time required, limited number of patients assessed, and difficulty identifying these patients prior to treatment. Automatic screening can be performed using billing records (ICD-9 diagnosis codes for secondary malignancies). While this is valuable in cancer surveillance and cohort discovery, it is of limited use in clinical trial eligibility screening mainly due to the lag time in billing and the need to identify patients at the time of diagnosis and prior to initiation of treatment. Information to quickly and accurately identify patients with metastatic disease is typically available only in clinical text documents (particularly radiology reports) and has the challenges inherent in extraction through Natural Language Processing due to the complexity of language expression and inconclusive text to express uncertain or negative conditions. Because of these challenges, we evaluated different approaches to determine the metastatic status from unstructured text in radiology reports in near real time. This manuscript reports the results of this evaluation. Our objectives were to formally assess the performance of several automated algorithms to identify metastatic status from radiology reports, select the algorithm with the optimal measures of accuracy, and to identify methods to improve this automated process.
Methods
Study overview
The Clinical Trial Eligibility Database Integrated System (CTED) was utilized for identifying patients with metastases in radiology reports.24-26 Search algorithms (queries) within CTED were developed and applied to radiology reports completed at our Institution. The performance of each algorithm was compared to a reference standard which consisted of a physician’s review and validation of the metastatic status from radiology reports.
Brief overview of the CTED system
The CTED Integrated system is an Investigator-developed set of software tools to aid in the clinical trial recruitment process at the Massey Cancer Center, Virginia Commonwealth University (VCU). It consists of three components: the CTED Tracking System, the CTED Automated Matching Tool (CTED-AMT) and the MD/Clinical Research Staff (CRS) Alert Notification System. Detailed information on the CTED system has been reported in prior publications.24-26 The CTED-AMT is the component used in this study to automate patient identification. This tool searches the VCU patients’ data collected from multiple electronic sources and maintained longitudinally in a data warehouse. It allows for automatic selection of potentially eligible study patients based on protocol inclusion/ exclusion criteria. The electronic sources include data from the scheduling system, billing data, and all clinical notes including surgical pathology reports, radiology reports and clinic visits or hospital admissions. The system searches discrete data including demographics, billing diagnoses (cancer and other diagnoses related to comorbidity), prior treatment for cancer (including specific and generic categories of treatment), and laboratory test results indicating current and prior measures of disease status (e.g. metastatic disease, recurrence, disease progression, tumor markers). It also searches unstructured text documents based on the National Cancer Institute Enterprise Vocabulary Services (NCI EVS) meta-thesaurus27 or user-defined search terms and text strings. A list of patients who meet a set of protocol-specific eligibility criteria is created each time a query is executed. This list is matched with the patient scheduling system for automated notification of physicians and research staff regarding visits for potentially eligible patients.
Study sample
The study sample consists of all patients who had one or more radiology reports completed during 2 consecutive weeks (10/31/11 to 11/14/11) at VCU Health System. A total of 5,523 patients with 10,492 radiology reports were included in the analysis. We chose not to limit the algorithms only to patients with cancer diagnosis because some categories of patients with metastases may not be included (for example, patients who present with advanced metastatic disease at the first encounter or patients who were not receiving cancer care at our institution but had only radiology tests/consults). In order to automatically subset patients based on cancer diagnosis, the billing ICD-9 diagnosis code or EHR problem list must be used. However, the billing data usually lag in time and the problems list are not routinely updated. Thus, we chose to include all patients because for oncology clinical trials patients must be identified at the time metastases are initially detected. All diagnostic radiology reports were included in the evaluation (plain radiographs, fluoroscopic studies, ultrasound exams, computed tomography (CT) scans, magnetic resonance imaging (MRI), positron emission tomography (PET), scintigraphic tests, and angiographic studies). Although some of the radiologic diagnostic tests such as fluoroscopic exam have limited sensitivity to detect metastases, we included all types of reports to assure that we are capturing any evidence of metastasis and to test how our algorithms perform against the entire range of radiographic diagnostic tests.
Algorithms for identifying metastatic disease in unstructured radiology reports using CTED
We used eight different search algorithms (queries) to identify patients with metastatic disease from radiology reports. These queries represented two types of approach, term/string matching and document indexing. Each approach was then combined with ICD-9 cancer diagnosis and/or the “Ignore phrase” feature in CTED-ATM (see below), Table 2.
Table2.
Characteristics of different approaches to detect presence of metastasis in free text radiology reports a
| N patients identified |
N true positive |
Sensitivity, 95 % CI |
Specificity, 95 % CI |
Positive predictive value, 95 % CI |
Negative predictive value, 95 % CI |
Likelihood ratio positive, 95 % CI |
Likelihood ratio negative, 95 % CI |
Accuracy, 95% CI |
|
|---|---|---|---|---|---|---|---|---|---|
| 1. Term search of radiology reports (RR) b | 330 | 199 | 94.8 [90.8, 97.4] |
97.5 [97.1, 97.9] |
60.3 [54.8, 65.6] |
99.8 [99.6, 99.9] |
38.4 [32.4, 45.6] |
0.05 [0.03, 0.1] |
97.4 [97.0, 97.8] |
| 2. Term search of RR AND billing ICD-9 diagnosis for any cancer |
284 | 179 | 85.2 [79.7, 89.7] |
98.0 [97.6, 98.4] |
63.0 [57.1, 68.7] |
99.4 [99.2, 99.6] |
43.1 [35.4, 52.6] |
0.15 [0.11, 0.21] |
97.5 [97.1, 97.9] |
| 3. Term search AND use of “Ignore phrases” feature in CTED |
268 | 198 | 94.2 [90.2, 97.0] |
98.7 [98.3, 99.0] |
73.9 [68.2, 79.0] |
99.7 [99.6, 99.9] |
71.6 [56.6, 90.5] |
0.06 [0.03, 0.10] |
98.5 [98.2, 98.8] |
| 4.Term search AND use of “Ignore phrases” feature in CTED AND billing ICD-9 code for any malignancy |
239 | 179 | 94.8 [90.8, 97.4] |
97.5 [97.1, 97.9] |
60.3 [54.8, 65.6] |
99.8 [99.6, 99.9] |
38.4 [32.4, 45.6] |
0.05 [0.03, 0.1] |
97.4 [97.0, 97.8] |
| 5. Metastatic algorithm (programmed in CTED) |
267 | 198 | 94.3 [90.2, 97.0] |
98.7 [98.4, 99.0] |
74.2 [68.5, 79.3] |
99.8 [99.6, 99.9] |
72.6 [57.3, 92.0] |
0.06 [0.03, 0.1] |
98.5 [98.2, 98.8] |
| 6. Metastatic algorithm AND billing ICD-9 diagnosis for any cancer |
232 | 178 | 84.8 [79.2, 89.3] |
99.0 [98.7, 99.2] |
76.7 [70.7, 82.0] |
99.4 [99.1, 99.6] |
83.4 [63.6, 109.4] |
0.15 [0.11, 0.21] |
98.4 [98.1, 98.8] |
| 7. Metastatic algorithm AND “Ignore phrases” feature in CTED |
220 | 197 | 93.8 [89.6, 96.7] |
99.6 [99.4, 99.7] |
89.5 [84.7, 93.3] |
99.8 [99.6, 99.9] |
216.7 [143.9, 326.3] |
0.06 [0.04, 0.11] |
99.3 [99.1, 99.5] |
| 8. Metastatic algorithm AND “Ignore phrases” feature AND billing ICD-9 diagnosis for any cancer |
187 | 171 | 81.4 [75.5, 86.4] |
99.7 [99.5, 99.8] |
91.4 [86.5, 95.0] |
99.3 [99.0, 99.5] |
270.4 [165.1, 442.9] |
0.19 [0.14, 0.25] |
99.0 [98.7, 99.2] |
Presence of metastases was determined based on manual review of radiology reports by physicians
Terms used: metastatic, metastasis, metastases, carcinomatosis
The details for each query are provided below.
Term search only: This method used the “Term search” feature in CTED-AMT. The Term Search feature allows for an unlimited number of terms or text strings to be entered and to establish relational operands (AND, OR, NOT) between each of the terms or strings specified. In addition to user-defined terms, some or all of NCI EVS metathesaurus27 related terms may be included with or in lieu of the specified term. Further, the system allows selection of the report type to search as well as date ranges. For the “term search only” algorithm, we performed the search in the radiology reports within the study period using the terms: “metastatic”, “metastasis”, “metastases” and “carcinomatosis”. These 4 terms were found to capture the majority of the radiology reports with metastatic disease findings during the pre-testing and development of the CTED metastatic algorithm (approach # 5).
Term search and a cancer diagnosis based on International Classification of Diseases, Ninth Revision (ICD-9) diagnosis codes in billing: Limiting the search to those patients that had prior diagnosis for cancer may decrease the number of false positive cases. Thus, we added searching for any ICD-9 cancer diagnosis codes (ICD codes 140-239) in the billing claims data to algorithm 1.
Term search and use of the “Ignore Phrases” feature in CTED-AMT: This feature allows the user to enter an unlimited number of phrases or sentences in conjunction with the primary term or text string. Any terms, phrases or sentences specified as “Ignore Phrases” are ignored by the system in searching and selecting cases with the specified term or phrase in the narrative reports. The “Ignore Phrases” were identified during the manual review of radiology reports. An example of an “ignore phrase” is “evaluate for metastasis”. In this context, the patient may or may not have metastatic disease. Without further positive corroboration in the same report, it will not be categorized as positive for metastases (i.e. it will be ignored). The ignore phrases were added to algorithm 1.
Term search and any billing ICD-9 cancer diagnosis, and CTED-ATM “Ignore Phrases” feature (Algorithm 1, 2 and 3 combined).
Metastatic algorithm programmed in CTED: This is a hard-coded option algorithm that is a component of the CTED system. It automatically screens incoming radiology reports and indexes each report as positive or negative based on a sequence of positive and negative relations using metastatic terms.
CTED metastatic algorithm and a billing ICD-9 cancer diagnosis code: As for strategy 2 above, a billing code for cancer was added to the algorithm to reduce false positives.
CTED metastatic algorithm and “Ignore phrases” feature in CTED: The same ignore phrases used in approach 3 were included here to reduce false positives.
CTED metastatic algorithm and any billing ICD-9 cancer diagnosis, and CTED-ATM “Ignore Phrases” feature (Algorithm 5, 6, and 7 combined).
Review of Electronic Medical Records (Validation)
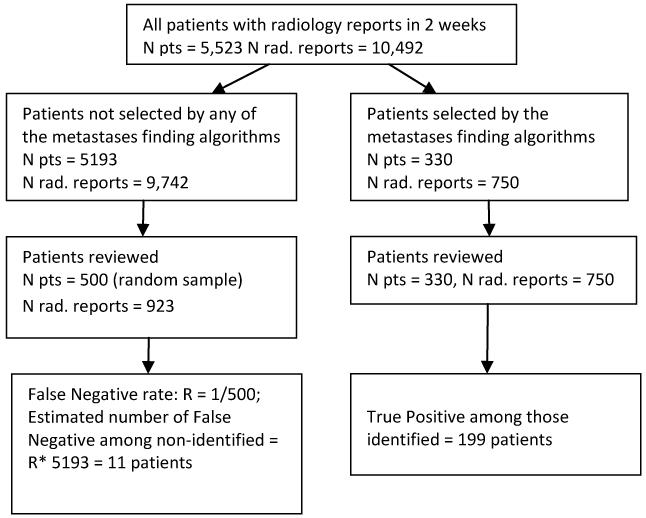
Figure 1 outlines the selection of patients for review. The consensus opinion of 3 physicians, one of whom is an oncologist, served as a gold standard for final determination of a radiology report as being positive or negative for metastatic disease. One physician reviewed all radiology reports of patients identified by any of the queries as positive in order to verify metastatic disease and classify cases as True Positive (TP) or False Positive (FP). These 750 reports represented 7.1% of all radiology reports received during the two week study period, and they are for 330 unique patients. Twenty percent of all patients reviewed by the first reviewer were reviewed by a second physician to determine inter-rater agreement. Disagreements between the two physicians were adjudicated by a medical oncologist and a final decision was reached. Any radiology report that was suggestive but not conclusive as to the presence of metastatic disease (e.g. consistent with, cannot be ruled out) was considered a positive report because in practice all potentially eligible patients identified by CTED will be followed up by the research nurse or the Principal Investigator.
Figure 1.
Selection of patients for manual review
Estimating the number of false negative cases (FN)
The FN cases are those cases that were not identified by any of the algorithms although their radiology reports indicated that they had metastatic conditions. We estimated the number of FN cases among patients that were not selected by any of the algorithms (Npatients = 5193) as follows: A simple random sample of 500 patients (approximately 10% out of 5193) was selected and all of their radiology reports completed in the 2 week study period were reviewed (Nreports = 923). The rate of FN cases was estimated as number of FN found in the random sample divided by 500. The total number of false negatives in the complete sample (FN) was estimated by multiplying this rate by the 5193 sample size. Further, for each query, the number of estimated FNs was adjusted to reflect positive cases that were identified by other queries but not the query in question. The patients and TP identified by the second through eighth queries were subsets of query 1.
Analyses
We categorized patients into four categories using the following definitions for each query: True positive (TPquery) - patients identified by a query as positive for metastases that were confirmed by the manual review to have metastases; False positive (FPquery) – patients identified by a query as positive for metastases but not confirmed by the manual review; False Negative (FNquery) – estimated number of patients with a radiology report positive for metastatic disease that were not identified by a query; True Negative (TNquery) - estimated as the number of patients not selected by a search algorithm minus the estimated number of FN for that algorithm. The exact numbers of TP and FP were determined by the manual review of all patients selected by the different approaches. The FNs and TNs were estimated for each query separately using the methods shown in Table 1.
Table 1.
Methods used for the estimation of False Negative and True Negative cases in each query
| Gold standard (Manual Review) | Total | ||
|---|---|---|---|
| Positive | Negative | ||
| Algorithm Positive | a = TPquery | b = FPquery | TPquery+FPquery |
| Algorithm Negative | c = FNquery = 210 − TPquery | d = TNquery = 5523−( TPquery + FPquery + FNquery) | 5523 − ( TPquery + FPquery ) |
| 199 + 11 = 210 | 5523 − 210 = 5313 | 5523 | |
The performance of each algorithm was assessed by calculating sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) along with 95% confidence interval using physician expert classification as a reference standard. Since PPV is affected by the prevalence of the condition of interest, specifically when the prevalence is low, we calculated the likelihood ratio positive (LR+ = sensitivity/false positive error rate) and likelihood ratio negative (LR− = false negative error rate/specificity) for each metastatic finding algorithm. The likelihood ratio provides a more accurate estimate of the usefulness of a test in low prevalent conditions such as in this study where the prevalence of metastatic disease in radiology reports was <3%.28,29 The overall accuracy of each approach was calculated as (TP +TN)/ (sample size).
Inter-rater reliability was estimated with Cohen’s kappa statistics.
IBM SPSS Statistics version 20 and R version 2.15.0 (R Foundation of Statistical Computing) were used in the analysis.
Ethical considerations
The CTED system is maintained under IRB Protocol HM 11089. This project was performed as a component of our continuous quality assessment and improvement of the CTED integrated system in supporting the cancer center research enterprise.
Results
During the two week evaluation period, 5,523 patients had a total of 10,492 radiology reports. The number of patients identified as having metastatic disease according to each approach ranged from 187 to 330 (Table 2). Patients selected by the second through eighth queries represented a subset of patients identified by the first query (terms search). The total number of patients validated as having metastatic disease among these patients was 199 but the number of true positive cases identified varied by query. The review of the 500 randomly selected patients not identified by any of the algorithms found only 1 case positive for metastases. The rate of FN cases was 0.002, 95% CI [0, 0.012]. Based on this FN rate, the estimated numbers for FN cases ranged from 11 (Query 1) to 39 (Query 8). The inter-rater agreement between the 2 reviewers was excellent with a kappa value of 0.90, 95 % CI [0.80, 0.98].
Sensitivity, specificity, PPV, NPV, LR+ and LR−, and accuracy are also shown in Table 2. While specificity and NPV were very high for each of the search strategies, query # 7 (based on the CTED programmed metastatic algorithm and “Ignore phrases” feature) had the most favorable combination of sensitivity (93.8 %), PPV (89.5%) and LR+ (216.7). Adding any billing ICD-9 diagnosis for cancer improved PPVs by 2-3 % but decreased the sensitivity by almost 10 %.
Because a focus of this project was to improve the system, we performed a detailed review of all 69 false positive (FP) cases identified by the CTED programmed metastatic algorithm (query #5) to identify methods to improve the accuracy of the system. The results are provided in Table 3. The majority of the FP cases were eliminated when the “ignore phrase” feature is used and were due to negation expressions or a diagnostic test indicators in the report such as “evaluate for”, “assess”, or “rule out metastases”. Applying the “Ignore phrases” feature decreased the number of false positive cases by 46, resulting in an increase of 15% in the PPV. A slight drop in sensitivity by 0.5% was due to 1 true positive case eliminated by the “Ignore phrases” function.
Table 3.
Reasons for false positive cases (FP) of the metastatic algorithm programmed in CTED
| Type of False Positive reports |
Reason for misclassification | Number of FP subjects (N = 69) a |
Possible solutions | Number of FP subjects after applying solutions (N =20) a |
|---|---|---|---|---|
| FP1 | Clinical history states evaluation/ assessment/ rule-out metastatic disease |
36 | use “Ignore phrase” feature in Terms search |
7 |
| FP2 | Diagnostic test name: “Radioiodine metastatic thyroid cancer imaging” |
3 | use “Ignore phrase” feature in Terms search |
0 |
| FP3 | Newly discovered negative expressions for metastasis |
39 | use “Ignore phrase” feature in Terms search |
7 |
| FP4 | Metastatic calcification due to ESRD |
2 | use “Ignore phrase” feature in Terms search |
1 |
| FP5 | Convoluted/complex language used to express negation |
2 | None identified | 2 |
| FP6 | Discussion of technical limitations | 3 | None identified | 3 |
| FP7 | Recommendations for future studies | 3 | None identified | 3 |
The column sum is greater than the total because some patients’ radiology reports had more than one reason for misclassification
Discussion
As a screening tool to identify cancer patients with metastatic disease, the automated system using the CTED programmed algorithm and “ignore phrases” (approach 7) was shown to have excellent results with sensitivity of 94 %, specificity of 99 %, PPV- 90 %, NPV- 100 % and a LR+ of 216. The CTED programmed metastatic algorithm had excellent sensitivity and specificity (94 %, and 99 % respectively), but the PPV was somewhat lower (74.2 %). Because the goal is to quickly and accurately identify patients with metastatic disease, the approach using the algorithm programmed in CTED in conjunction with the “ignore phrases” was optimal as it identified the most patients with the lowest false positive rate.
The concepts utilized herein are not entirely new, but the combined processes and potential for generalizability represent a novel use of existing methods. Previous studies have tested information extraction of various conditions of interest in different types of unstructured medical records such as identification of “medical problems” from clinical notes for the purpose of enriching the EHR problem list30, determining cancer stage from pathology reports and clinical notes to improve cancer registries31,32, or smoking status in medical free text documents.33,34 Few studies have focused on radiology reports and cancer. For those studies, our results are comparable. Hripcsak et al reported sensitivity of 81% (CI, 73% to 87%) and specificity of 98% (CI, 97% to 99%) for a natural language processor in determining 6 conditions (one of which was neoplasm) in 200 admission chest radiographs.35 Sensitivity of 80.6%, specificity 91.6%, PPV 82.4% and NPV 92.0% were found by Cheng et al in a study testing the ability of NLP to detect brain tumor progression from pre-selected unstructured brain MRI reports.36 Carrell et al used unmodified caTIES (a software developed to extract findings from pathology reports) to identify cancer in radiology reports with sensitivity of 82% and specificity of 95%.37 Our CTED built-in metastatic algorithm alone or in combination with the “Ignore phrase” feature performed somewhat better and across a broader range of cancers, clinical trials and was used to effectively screen all types of radiologic diagnostic tests.
Our estimated performance characteristics reported in Table 2 reflected the system performance on a limited cross-sectional basis. In the clinical trials recruitment practice, automated screening would occur on an ongoing basis. Thus, some FNs occurring during the 2 week period would likely be identified either before or after the study interval, as cancer patients have multiple radiology tests throughout the disease course. In order to evaluate the longitudinal performance of the system as it would be used in production, we attempted to broaden our search to identify actual patients with metastatic disease (FNs) that would be identified from data sources other than radiology reports. This included searching clinical notes for metastatic terms and screening patients with ICD-9 billing diagnoses for secondary malignancy during the 2 week study interval. Using these alternate methods, 18 FN cases were identified. These FNs cases were then run against the metastatic algorithm (Query #5) with no date limits (i.e. before and after the two week study window) to mimic ongoing longitudinal use of the system. The algorithm (using only the radiology reports) correctly identified 78 %, 95% CI [52, 93] of these 18 FN cases from radiologic studies completed either before or after the two week study interval. Of the remaining 4 FN cases, 3 had limited information in the EHR with only one radiology report. The fourth case was a widespread follicular lymphoma with organ involvement. From a practical perspective, it is unlikely that three of these patients would be eligible for a trial as they did not receive their care within our health care system. The resulting system performance measures based on this adjusted FN rate are as follows: sensitivity 98%, specificity 100%, PPV 90%, NPV 100%, LHR+ 225, and LHR− 0.02.
The major challenge in identifying patients with metastatic disease is that the earliest indicator is typically information only available through unstructured text. Thus, the ability to screen these text documents to accurately identify patients in a timely manner is critical. Identification of negation phrases in unstructured clinical reports represents the most significant challenge to this process.38 The presence or absence of metastatic disease is a component of the inclusion or exclusion criteria for the majority of cancer clinical trials, thus is a key factor that if known can quickly eliminate patients and reduce the number of patients research staff must screen to identify eligible patients. Minimizing the FP rate is important when excluding patients with possible metastatic disease, whereas maximizing the TP rate is important when using metastasis as an inclusion criterion. In oncology trials that include patients with metastases the FP rate is less crucial but still important as it will result in unnecessary review of records and increase the time spent by the research staff reviewing erroneously included patients. For clinical trials which exclude metastases the issue of negation becomes more important. In these trials, the search algorithms can be used to exclude patients during the automated pre-screening. The inclusion of the “ignore phrases” with the CTED programmed algorithm (approach 7) was a very important outcome of our evaluation as it resulted in a 15 % improvement in the PPV (from 74.2 to 89.5) while maintaining a good sensitivity and increased specificity.
The decreased sensitivity that occurred when cancer diagnosis was added may be due to several reasons. Patients may not have been treated at VCU Health System and the radiology report was performed for an outside health care provider who is treating the patient. These patients may therefore not have a cancer diagnosis in the billing records. Further, some patients were diagnosed with metastatic disease during their first encounter at our health care system. Billing data typically lags by several weeks the real time radiology dictations thus may not have been incorporated into the system until well after the radiology report was available.
There were several key benefits in the methods from this study compared with systems reported in the published literature. First, we included all radiologic diagnostic tests to maximize the ability to capture information on metastases as close to the diagnosis as possible. Other studies focused on a limited set of diagnostic radiologic procedures such as chest radiographs35,39 or brain MRI 36. Our approach based on near real time capture and screening of all reports provides information to clinical research staff at the earliest point at which metastatic disease is identified whether it is by chest x-ray, CT or MRI. Typically, radiology reports are available the day of or day after the diagnostic study is completed. Thus, the tool is optimal for rapidly including or excluding potential study eligible patients for clinical trials. The ability to accurately detect metastatic disease through radiology reports is a key step in identifying patients with a progressive metastatic disease burden. Rapid identification in real time of patients with known cancer and newly metastatic disease is critical to clinical trial enrollment, as these patients are the ones most likely to need access to a clinical trial. The near real time identification is also critical as these patients are often in need of immediate treatment. Although our evaluation did not focus on newly diagnosed metastases, the CTED system provides features that can be used to successfully identify new metastases particularly in patients that received the majority of their cancer care at the VCUHS. These features include restricting the search to recent time and excluding patients who had evidence of metastases in clinical documents or billing records prior to the specified time point. This approach has been used to identify not only new metastases but other newly diagnosed medical conditions.
An additional differentiating factor of this analysis is the inclusion of cancers of any site. Previous studies have focused on one organ.35,36 While these studies have provided valuable contributions, the ability to identify metastases across all cancer sites is critical for a cancer center that typically conducts clinical trials in a broad range of cancers. Further, some studies enrolling patients with metastases may enroll patients with very different types of cancer (for example bone metastases in lung, breast and prostate cancer). The CTED system was developed to support all oncology trials, thus it was important to evaluate the system’s performance in determining metastatic status irrespective of cancer site.
Although optimized currently for cancer trials, the system is being used to identify patients with a variety of other clinical characteristics for assessing eligibility in clinical trials not only in oncology but in other clinical arenas. Examples of medical terms/text strings searches in unstructured text documents that we used in various clinical trials included specific medications use, type of sickle cell disease (Hemoglobin SS, SC, SB), eosinophilic pharyngitis, and large volume paracentesis. The latter two concepts are examples where discrete data would not be helpful since there are no specific ICD-9 or Current Procedural Terminology (CPT) codes for these conditions.
Limitations
The results from this evaluation are for a single institution. Thus, the findings may not be entirely generalizable. However, the 10,492 reports that were assessed represented dictations by 69 radiologists, who trained at a variety of institutions, thus reducing the concern that the results may be skewed by dictation patterns associated with only a limited set of radiologists.
A second limitation is the length of the study period. Although two weeks is a relatively short period, there were more than 10,000 radiology reports generated during that period, representing every radiologic diagnostic test performed at our institution. During this interval a total of 1,088 (19.7 %) of all patient records were reviewed.
Another potential limitation of the proposed approaches for identifying metastases in text documents could be misspelled words. We did not observe this to be the case in our study. This may be because the “metastatic” terms are used typically more than once in a report and frequently multiple imaging studies are performed in relatively short periods of time, thus decreasing the probability of not identifying a patient due to a typing error.
Conclusion
In summary, our evaluation process identified the optimal method for automatically screening radiology reports for rapid identification of patients with metastatic disease and identified additional methods to optimize PPV while simultaneously minimizing the FN and FP rates. The results demonstrate that these screening tools can be implemented successfully to provide critical information for identification of patients for consideration in cancer clinical trials. While we used the CTED system, similar approaches based on these results could be implemented at other institutions to enhance the efficiency of research staff in the clinical trials eligibility screening process.
Acknowledgment
Services in support of the research project were provided by the VCU Cancer Research Informatics and Services Shared Resource, supported, in part, with the funding from NIH-NCI Cancer Center Support Grant 5 P30 CA016059.”
Footnotes
Authors’ contributions
VP, LP, BD and AP contributed to the concept and study design; LP, CG, and JM were responsible for the design, development and implementation of CTED-Integrated system; VP, LP and AP reviewed radiology reports; VP and BD conducted statistical analysis; VP, LP, BD and AP interpreted the results and drafted the article; all authors approved the final manuscript.
References
- 1.Korn EL, Freidlin B, Mooney M, Abrams JS. Accrual experience of National Cancer Institute Cooperative Group phase III trials activated from 2000 to 2007. J Clin.Oncol. 2010;28:5197–5201. doi: 10.1200/JCO.2010.31.5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schroen AT, Petroni GR, Wang H, Gray R, Wang XF, Cronin W, Sargent DJ, Benedetti J, Wickerham DL, Djulbegovic B, Slingluff CL., Jr. Preliminary evaluation of factors associated with premature trial closure and feasibility of accrual benchmarks in phase III oncology trials. Clin.Trials. 2010;7:312–21. doi: 10.1177/1740774510374973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nass SJ, Moses HL, Mendelsohn J. A National Cancer Clinical Trials System for the 21st Century: Reinvigorating the NCI Cooperative Group Program. The National Accademies Press; Washington DC: 2010. [PubMed] [Google Scholar]
- 4.Wang-Gillam A, Williams K, Novello S, Gao F, Scagliotti GV, Govindan R. Time to activate lung cancer clinical trials and patient enrollment: a representative comparison study between two academic centers across the atlantic. J.Clin.Oncol. 2010;28:3803–07. doi: 10.1200/JCO.2010.28.1824. [DOI] [PubMed] [Google Scholar]
- 5.Schroen AT, Petroni GR, Wang H, Thielen MJ, Gray R, Benedetti J, Wang X, Sargent DJ, Wickerham DL, Cronin WM, Djulbegovic B, Slingluff CL. Achieving Sufficient Accrual to Address the Primary Endpoint in Phase III Clinical Trials from US Cooperative Oncology Groups. Clin.Cancer Res. 2011;18:256–62. doi: 10.1158/1078-0432.CCR-11-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis PM, Butow PN, Tattersall MH, Dunn SM, Houssami N. Randomized clinical trials in oncology: understanding and attitudes predict willingness to participate. J.Clin.Oncol. 2001;19:3554–61. doi: 10.1200/JCO.2001.19.15.3554. [DOI] [PubMed] [Google Scholar]
- 7.Ford JG, Howerton MW, Lai GY, Gary TL, Bolen S, Gibbons MC, Tilburt J, Baffi C, Tanpitukpongse TP, Wilson RF, Powe NR, Bass EB. Barriers to recruiting underrepresented populations to cancer clinical trials: a systematic review. Cancer. 2008;112:228–42. doi: 10.1002/cncr.23157. [DOI] [PubMed] [Google Scholar]
- 8.Howerton MW, Gibbons MC, Baffi CR, Gary TL, Lai GY, Bolen S, Tilburt J, Tanpitukpongse TP, Wilson RF, Powe NR, Bass EB, Ford JG. Provider roles in the recruitment of underrepresented populations to cancer clinical trials. Cancer. 2007;109:465–76. doi: 10.1002/cncr.22436. [DOI] [PubMed] [Google Scholar]
- 9.Lara PN, Jr., Higdon R, Lim N, Kwan K, Tanaka M, Lau DH, Wun T, Welborn J, Meyers FJ, Christensen S, O’Donnell R, Richman C, Scudder SA, Tuscano J, Gandara DR, Lam KS. Prospective evaluation of cancer clinical trial accrual patterns: identifying potential barriers to enrollment. J.Clin.Oncol. 2001;19:1728–33. doi: 10.1200/JCO.2001.19.6.1728. [DOI] [PubMed] [Google Scholar]
- 10.Townsley CA, Selby R, Siu LL. Systematic review of barriers to the recruitment of older patients with cancer onto clinical trials. J.Clin.Oncol. 2005;23:3112–24. doi: 10.1200/JCO.2005.00.141. [DOI] [PubMed] [Google Scholar]
- 11.Ulrich CM, James JL, Walker EM, Stine SH, Gore E, Prestidge B, Michalski J, Gwede CK, Chamberlain R, Bruner DW. RTOG physician and research associate attitudes, beliefs and practices regarding clinical trials: implications for improving patient recruitment. Contemp.Clin.Trials. 2010;31:221–8. doi: 10.1016/j.cct.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Beauharnais CC, Larkin ME, Zai AH, Boykin EC, Luttrell J, Wexler DJ. Efficacy and cost-effectiveness of an automated screening algorithm in an inpatient clinical trial. Clin.Trials. 2012;9:198–203. doi: 10.1177/1740774511434844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Embi PJ, Jain A, Clark J, Bizjack S, Hornung R, Harris CM. Effect of a clinical trial alert system on physician participation in trial recruitment. Arch.Intern.Med. 2005;165:2272–77. doi: 10.1001/archinte.165.19.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weng C, Batres C, Borda T, Weiskopf NG, Wilcox AB, Bigger JT, Davidson KW. A real-time screening alert improves patient recruitment efficiency; AMIA.Annu.Symp.Proc.; 2011; pp. 1489–1498. [PMC free article] [PubMed] [Google Scholar]
- 15.Rollman BL, Fischer GS, Zhu F, Belnap BH. Comparison of electronic physician prompts versus waitroom case-finding on clinical trial enrollment. J.Gen.Intern.Med. 2008;23:447–50. doi: 10.1007/s11606-007-0449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuggia M, Besana P, Glasspool D. Comparing semi-automatic systems for recruitment of patients to clinical trials. Int.J.Med.Inform. 2011;80:371–88. doi: 10.1016/j.ijmedinf.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Heinemann S, Thuring S, Wedeken S, Schafer T, Scheidt-Nave C, Ketterer M, Himmel W. A clinical trial alert tool to recruit large patient samples and assess selection bias in general practice research. BMC.Med.Res.Methodol. 2011;11:16–26. doi: 10.1186/1471-2288-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seroussi B, Bouaud J. Using OncoDoc as a computer-based eligibility screening system to improve accrual onto breast cancer clinical trials. Artif.Intell.Med. 2003;29:153–67. doi: 10.1016/s0933-3657(03)00040-x. [DOI] [PubMed] [Google Scholar]
- 19.Treweek S, Pearson E, Smith N, Neville R, Sargeant P, Boswell B, Sullivan F. Desktop software to identify patients eligible for recruitment into a clinical trial: using SARMA to recruit to the ROAD feasibility trial. Inform.Prim.Care. 2010;18:51–58. doi: 10.14236/jhi.v18i1.753. [DOI] [PubMed] [Google Scholar]
- 20.Grundmeier RW, Swietlik M, Bell LM. Research subject enrollment by primary care pediatricians using an electronic health record; AMIA.Annu.Symp.Proc.; 2007; pp. 289–93. [PMC free article] [PubMed] [Google Scholar]
- 21. [Accessed on 11/15/2012];AccelFind. http://www.cliniworks.com/index.php/products-and-solutions/clinical-research/case-finding.
- 22. [Accessed on 11/15/2012];Cerner PowerTrials. 2012 http://www.cerner.com/uploadedFiles/PowerTrials%20Marketing%20flyer%200808.pdf.
- 23.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb.Med.Inform. 2008:128–144. [PubMed] [Google Scholar]
- 24.Penberthy L, Brown R, Puma F, Dahman B. Automated matching software for clinical trials eligibility: measuring efficiency and flexibility. Contemp.Clin.Trials. 2010;31:207–17. doi: 10.1016/j.cct.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penberthy LT, Dahman BA, Petkov VI, DeShazo JP. Effort Required in Eligibility Screening for Clinical Trials. Journal of Oncology Practice. 2012 doi: 10.1200/JOP.2012.000646. epublished ahead of print 9/12/2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Penberthy L, Brown R, Wilson-Genderson M, Dahman B, Ginder G, Siminoff LA. Barriers to therapeutic clinical trials enrollment: Differences between African-American and white cancer patients identified at the time of eligibility assessment. Clin.Trials. 2012;9:788–97. doi: 10.1177/1740774512458992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Cancer Institute [Accessed on 11/15/2012];Enterprise Vocabulary Services. http://evs.nci.nih.gov.
- 28.Jekel JF, Elmore JG, Katz DL. Epidemiology, biostatistics and Preventive Medicine. W.B. Sounders Company; Philadelphia, PA: 1996. [Google Scholar]
- 29.Gallagher EJ. Clinical utility of likelihood ratios. Ann.Emerg.Med. 1998;31:391–97. doi: 10.1016/s0196-0644(98)70352-x. [DOI] [PubMed] [Google Scholar]
- 30.Meystre S, Haug PJ. Natural language processing to extract medical problems from electronic clinical documents: performance evaluation. J.Biomed.Inform. 2006;39:589–99. doi: 10.1016/j.jbi.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 31.McCowan I, Moore D, Fry MJ. Classification of cancer stage from free-text histology reports; Conf.Proc.IEEE Eng Med.Biol.Soc.; 2006; pp. 5153–56. [DOI] [PubMed] [Google Scholar]
- 32.McCowan IA, Moore DC, Nguyen AN, Bowman RV, Clarke BE, Duhig EE, Fry MJ. Collection of cancer stage data by classifying free-text medical reports. J.Am.Med.Inform.Assoc. 2007;14:736–45. doi: 10.1197/jamia.M2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savova GK, Ogren PV, Duffy PH, Buntrock JD, Chute CG. Mayo clinic NLP system for patient smoking status identification. J.Am.Med.Inform.Assoc. 2008;15:25–28. doi: 10.1197/jamia.M2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meystre SM, Deshmukh VG, Mitchell J. A clinical use case to evaluate the i2b2 Hive: predicting asthma exacerbations; AMIA.Annu.Symp.Proc.; 2009; pp. 442–46. [PMC free article] [PubMed] [Google Scholar]
- 35.Hripcsak G, Friedman C, Alderson PO, DuMouchel W, Johnson SB, Clayton PD. Unlocking clinical data from narrative reports: a study of natural language processing. Ann.Intern.Med. 1995;122:681–88. doi: 10.7326/0003-4819-122-9-199505010-00007. [DOI] [PubMed] [Google Scholar]
- 36.Cheng LT, Zheng J, Savova GK, Erickson BJ. Discerning tumor status from unstructured MRI reports--completeness of information in existing reports and utility of automated natural language processing. J.Digit.Imaging. 2010;23:119–32. doi: 10.1007/s10278-009-9215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carrell D, Miglioretti D, Smith-Bindman R. Coding free text radiology reports using the Cancer Text Information Extraction System (caTIES); AMIA.Annu.Symp.Proc.; 2007; p. 889. [PubMed] [Google Scholar]
- 38.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. Evaluation of negation phrases in narrative clinical reports; Proc.AMIA.Symp.; 2001; pp. 105–9. [PMC free article] [PubMed] [Google Scholar]
- 39.Hripcsak G, Kuperman GJ, Friedman C. Extracting findings from narrative reports: software transferability and sources of physician disagreement. Methods Inf.Med. 1998;37:1–7. [PubMed] [Google Scholar]