Abstract
Mis-regulation (e.g. overproduction) of the human Ndc80/Hec1 outer kinetochore protein has been associated with aneuploidy and tumourigenesis, but the genetic basis and underlying mechanisms of this phenomenon remain poorly understood. Recent studies have identified the ubiquitous Ndc80 internal loop as a protein-protein interaction platform. Binding partners include the Ska complex, the replication licensing factor Cdt1, the Dam1 complex, TACC-TOG microtubule-associated proteins (MAPs) and kinesin motors. We review the field and propose that the overproduction of Ndc80 may unfavourably absorb these interactors through the internal loop domain and lead to a change in the equilibrium of MAPs and motors in the cells. This sequestration will disrupt microtubule dynamics and the proper segregation of chromosomes in mitosis, leading to aneuploid formation. Further investigation of Ndc80 internal loop-MAPs interactions will bring new insights into their roles in kinetochore-microtubule attachment and tumourigenesis.
Keywords: cancer, ch-TOG, Kinesin-8, loop, Ndc80/Hec1, overexpression, TACC
Introduction
Mitosis plays a central part in accurate segregation of genetic materials (i.e. chromosomes) into the two daughter cells (Fig. 1). Chromosome missegregation results in genome instability such as aneuploidy, which is a hallmark of cancer. Mis-regulation in the form of up- or down-regulation of the genes/proteins involved in microtubule (MT) dynamics, mitotic checkpoint or kinetochore-MT attachment are known to cause chromosome instability (CIN) [1,2]. Examples include the colonic and hepatic tumour overexpressed gene (ch-TOG) [3] and the genes encoding the transforming acidic coiled-coil proteins (TACCs) [4], spindle assembly checkpoint (SAC) proteins Mad2 [5] and Bub1 [6], the Ska kinetochore complex [7], kinesin-8 motors [8,9], and the outer kinetochore protein Ndc80/Hec1 (Highly Expressed in Cancer) [10–12], which are all upregulated in several cancers. Although overexpression of these genes may not have direct causative effects on the proliferative nature of cancerous cells, these studies suggest a correlation between increased expression of mitotic genes and cancer formation. Whilst these results are consistent with the idea that overproduction of these proteins is linked to tumourigenesis, there is also a view opposing this, supported by examples such as the absent or reduced expression of TACCs in ovarian and thyroid cancer tissues [13]. In addition, Mad2 and BubR1 confer tumour-suppressor activities [14,15]. Overall, previous and current studies strongly suggest a functional relationship between the stoichiometry of kinetochore proteins, SAC components and MT-associated proteins (MAPs) and tumourigenesis.
Figure 1.

Chromosome segregation during mitosis. Left: The diagram summarises chromosome movements in different stages of mitosis. In prometaphase, spindle microtubules emanate from the centrosome/SPB to bind to chromosomes at the kinetochore region. Attached chromosomes are aligned in the metaphase plate during metaphase. Sister chromatids are segregated apart towards the opposite poles during anaphase. Right: Ndc80 attaches to the spindle microtubule through its N-terminal tail. In addition, the internal loop region of Ndc80 binds to different proteins to regulate proper spindle-microtubule attachment.
Hypothesis
Despite several previous studies on the collaborative relationships between mitotic gene expression and cancer, the mechanistic details of tumourigenesis induced by coordinated transcriptional up-regulation remain poorly understood. Here, we propose a new hypothesis for this phenomenon: elevated levels of a mitotic protein will sequester away (‘absorb’) its binding partners, thereby disrupting the protein equilibrium during mitosis and consequently leading to abnormal mitotic progression and chromosome missegregation. To compensate for the loss of this protein equilibrium, expression of genes encoding the kinetochore, MAPs and SAC components may subsequently be altered (i.e. up- or down-regulated). This scenario at least in part accounts for how mis-regulation of a cluster of mitotic genes is observed in various cancer cells.
Many mitotic proteins are genetically and functionally related to each other
A myriad of structural and regulatory factors are involved in mitosis to ensure faithful chromosome segregation into two daughter cells. Mitotic proteins often genetically, functionally and physically interact with each other. Here, we provide two examples – Ndc80 and the TACC proteins, and show how mis-regulation (mutation or deletion) of these proteins results in mitotic defects leading to aneuploid formation. We will focus on the recent advances made in the fission yeast Schizosaccharomyces pombe and human cell lines that support our proposed hypothesis (see Table1 for summary of protein nomenclatures and functions).
table 1.
List of proteins and their functions discussed in this review/hypothesis
| Human | Fission yeast | Budding yeast | Functions |
|---|---|---|---|
| Ndc80/Hec1 | Ndc80 | Ndc80 | A component of the Ndc80 outer kinetochore complex |
| ch-TOG | Alp14 Dis1 | Stu2 | Microtubule (MT) polymerase |
| TACC1, 2 | — | — | MT-associated proteins (MAPs), localise to the centrosome |
| TACC3 | Alp7 | Slk19* | MAP, localises to the centrosome/SPB, spindle MT and the kinetochore |
| Cdt1 | Cdt1 | Cdt1/TAH11 | DNA replication licensing factor |
| Ska complex (Ska1,2,3) | — | — | Binds to the MT and localises to the kinetochore |
| — | Dam1 | Dam1 | A component of the Dam1 complex that binds to the MT and localises to the kinetochore (Assumed to be the yeast homolog of the Ska complex) |
| Kif18A | Klp5 Klp6 | Kip3 | Kinesin 8, regulates MT dynamics (Kip3 is a MT depolymerase) |
The Ndc80/Hec1 outer kinetochore protein
Ndc80 binds to Nuf2, Spc24 and Spc25, forming a dumbbell-shaped complex of ∼57 nm in length with two globular heads connected through a long coiled-coil region (Fig. 2A) [16–19]. This complex localises to the outer kinetochore and directly binds to MTs through the N-terminal tail and calponin-homology (CH) domain of Ndc80 [20–23] (Fig. 1). A number of biochemical studies have identified several phosphorylation sites within this N-terminal region of Ndc80. Aurora B kinase, which localises to the inner centromere region, has been shown to phosphorylate Ndc80, both in vitro [18,24,25] and in vivo [25,26]. This phosphorylation results in deterioration of the Ndc80-MT interaction, thereby interfering with the spatiotemporal control of kinetochore-MT attachment. Non-stabilised kinetochore-MT attachment (e.g. incorrect attachment) leads to SAC activation and mitotic delay. Once mal-attachment is corrected, tension is exerted to the kinetochore and anaphase initiates, leading to chromosome segregation [27–29]. It is worth noting that a recent study using the budding yeast Saccharomyces cerevisiae, on the other hand, has challenged this model for which Aurora B localisation to the inner centromere is a prerequisite [30]. An alternative model proposes that tension sensing and consequent chromosome biorientation are successfully established via Aurora B that localises to the mitotic spindle, not the inner centromere, as long as Aurora B is activated [30]. In addition to Aurora B, phosphorylation of the budding yeast Ndc80 by Mps1 has been implicated in SAC signalling [31] and human Ndc80 phosphorylation (at serine 165 in the CH domain) by Nek2 kinase plays a critical role in faithful chromosome segregation [32].
Figure 2.
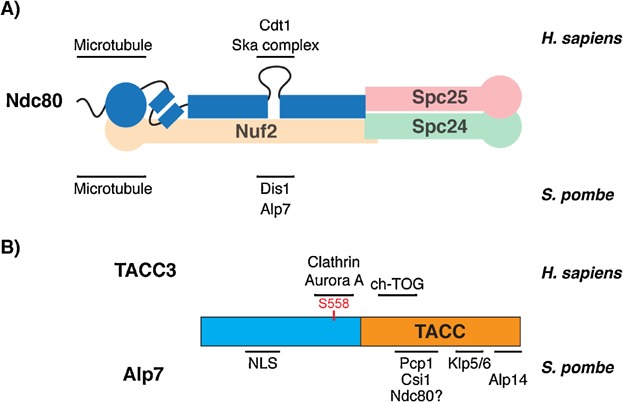
Binding partners of Ndc80 and TACC proteins. The diagram summarises binding partners of (A) Ndc80 and (B) TACC proteins in H. sapiens and S. pombe. A: Ndc80 forms a heterotetramer complex with Nuf2, Spc24 and Spc25. The N-terminal tail and Calponin-homology (CH) domain of Ndc80 bind directly to microtubules. In H. sapiens, the Ndc80 internal loop binds to Cdt1 and Ska1, whereas the loop binds to Dis1 and Alp7 in S. pombe. Note that the component of the Ska complex that interacts with the loop remains to be established. B: In H. sapiens, S558 (depicted in red) of TACC3 is phosphorylated by Aurora A. This phosphorylation is important for clathrin binding activity of TACC3. The TACC domain of TACC3 binds to ch-TOG. In S. pombe, the N-terminal of Alp7/TACC contains a nuclear localisation signal (NLS). The TACC domain of Alp7 binds to Pcp1 and Csi1 for SPB-targeting and Alp14 for microtubule localisation. The TACC domain is also deemed to bind to Ndc80, using the similar region responsible for Pcp1 and Csi1 binding.
Intriguingly, ultrastructural studies of the reconstituted budding yeast Ndc80 complex have revealed the presence of a kink (or loop) in the middle of the internal coiled-coil region within the Ndc80 protein [33–35]. More recently, the kink/loop has also been identified in human Ndc80 [36,37]. This loop was originally proposed to provide flexibility to the complex, allowing an angular bending of the two flanking rods, by which it potentially serves as a tension-sensing mechanism at the kinetochore. Although this notion remains promising [38–40], several recent studies in different organisms have identified the unexpected roles of the Ndc80 loop as a protein-protein interaction motif. To date, the Ndc80 loop has been found to associate with the Dam1 MT-binding kinetochore complex in S. cerevisiae [41], Dis1/TOG and Alp7/TACC-Alp14/TOG MAPs in S. pombe [42,43], and the Cdt1 licensing factor and the Ska kinetochore complex in human cell lines [36,44]. Extensive reviews on the roles of the Ndc80 loop can be found in [45,46].
In S. pombe, localisation of Dis1 to the kinetochore through the Ndc80 loop stabilises the mitotic spindle at the kinetochores [42]. This binding in turn allows loading of the Alp7-Alp14 complex to the Ndc80 loop to ensure faultless chromosome segregation in mitosis [43]. Our recent study has shown that the binding between the Ndc80 loop and Alp7-Alp14 is crucial for subsequent recruitment of the kinesin 8-protein phosphatase I complex (Klp5-Klp6-PP1) to the kinetochore [47]. This recruitment plays critical roles in ensuring timely mitotic progression and anaphase chromosome movement. In human cells, the Ndc80 internal loop recruits the Cdt1 licensing protein to facilitate stable kinetochore-MT attachment [44]. Furthermore, recruitment of the Ska complex to the Ndc80 internal loop plays important roles in the establishment of end-on kinetochore-MT binding and promotion of mitotic exit [36,48]. Taken together, these studies imply that by binding to various mitotic factors (Fig. 2A), the Ndc80 internal loop provides an indirect MT-binding domain in the Ndc80 complex and plays multiple roles in chromosome segregation, including the establishment of bipolar kinetochore-MT attachment and proper mitotic progression.
The TACC and TOG proteins
TACC family proteins, characterised by the presence of the TACC domain in their C-termini, are conserved from yeast to humans. Three isoforms (TACC1-3) encoded by different genes have been identified in humans [4,49], whilst only one has been found in other organisms (Alp7/Mia1 in S. pombe; TAC-1 in C. elegans, Maskin/TACC3 in X. laevis and D-TACC in D. melanogaster) [50–53]. Several mitotic proteins, including ch-TOG, Aurora A kinase and clathrin, have been identified as TACC interactors [52,54–61]. In S. pombe, the Alp7/TACC-Alp14/TOG complex localises to the spindle pole body (SPB, the functional equivalent of the animal centrosome) to regulate bipolar spindle assembly and dynamics during mitosis [52,62]. Recent studies have identified two SPB components, the pericentrin-like Pcp1 and Csi1, as recruiters of the Alp7-Alp14 complex to the SPB [63,64]. In addition, the Alp7-Alp14 complex also plays critical roles at the kinetochore through interaction with the Ndc80 internal loop (see above) [43]. In human cell lines, formation of the TACC3-ch-TOG complex plays important roles in spindle pole organisation and the regulation of centrosomal MTs [65]. In addition, binding of TACC3 to clathrin allows localisation of this complex onto the kinetochore MTs, thereby stabilising the kinetochore fibres by inter-MT bridging [54,56]. It is also reported that this complex is critical for centrosome integrity and bipolar spindle formation [59,61,66]. Collectively, these studies indicate that TACC proteins may well act as a hub to interact with multiple proteins to play distinct roles in different stages during mitosis (Fig. 2B).
The XMAP215/ch-TOG family proteins are conserved from yeast to humans, and are characterised by the presence of TOG domains in the N-terminal region. The number of TOG domains present varies amongst different organisms: two in Stu2 (S. cerevisiae), Dis1 and Alp14 (S. pombe), three in Zyg9 (C. elegans), and five in Msps (D. melanogaster), XMAP215 (X. laevis) and ch-TOG (H. sapiens) [67,68]. Several lines of evidence have shown that XMAP215/ch-TOG proteins possess MT polymerase activities [68–72]. Recent structural studies have solved the structure of TOG domains, giving further insights into how these domains recognise and bind to free α/β tubulin dimers [73–77]. These studies report that both TOG1 and TOG2 from Stu2 [73,74] or TOG1-4 from Msps [77] bind preferentially to curved α/β tubulin dimers, thereby selectively recognising the growing MT ends and facilitating polymerisation of the MTs. Intriguingly, S. pombe cells contain two XMAP215/ch-TOG members – Dis1 and Alp14 [78–81], which have both been shown to localise to the kinetochore through the Ndc80 internal loop domain (see above) [42,43]. Cumulatively, not only do the XMAP215/TOG family proteins regulate MT dynamics through the TOG domains, but they also play crucial roles in the regulation of kinetochore-MT attachment and mitotic progression.
Does overproduction of Ndc80 in fission yeast recapitulate analogous defects seen in human cancerous cells?
Based on these studies, we propose that overproduced mitotic proteins will result in dominant negative sequestration of their interactors, and subsequently interfere with the normal functions of these proteins [82]. This spatial segregation may then elevate gene expression of the interactors, causing a coordinated up-regulation of a cluster of genes. We used Ndc80 as an example to test our hypothesis, by overproducing different Ndc80 constructs in S. pombe cells. Previous studies have shown that approximately 20–25 Ndc80 complexes bind to each kinetochore MT [83,84]. Fission yeast contains three chromosomes, in which each sister kinetochore attaches to two or three MTs [85,86], and the total copy number of the Ndc80 complex per cell is 500 to 1,600 molecules [87,88]. We envision that overproduced Ndc80 will generate an excess Ndc80 pool that cannot be incorporated into the complex, yet this ‘free’ Ndc80 population will nonetheless, bind to its binding partners (Fig. 3).
Figure 3.

Overproduction of Ndc80 in S. pombe cells. The outer kinetochore Ndc80 complex is made up from Ndc80, Nuf2, Spc24 and Spc25. Under ‘overexpressed’ condition, two populations of Ndc80 will be formed. One propotion will be incorporated into the Ndc80 complex in place of endogenously produced Ndc80 and localise to the outer kinetochore, whereas the excess ‘free’ Ndc80 may be dispersed or form large polymers/aggregates in the cytoplasm.
As previously mentioned, the internal loop region of Ndc80 has been shown to be an important protein-protein interacting platform [45,46], so we retained the presence of the internal loop in each of our constructs. To this end, we created four different Ndc80 constructs, namely Ndc80-FL (full-length), Ndc80-ΔNCH, Ndc80-ΔNCH(F420S) and Ndc80-ΔNCH(L405P). Ndc80-FL binds to MTs through its N-terminal tail and CH domain (Fig. 4A) [20–23]. This MT-binding region is truncated in the Ndc80-ΔNCH constructs. We also introduced either the mutation F420S (defective in Alp7-binding) or L405P (defective in Dis1-binding) in the Ndc80-ΔNCH constructs to create Ndc80-ΔNCH(F420S) and Ndc80-ΔNCH(L405P), respectively (Fig. 4A). These constructs were then cloned into the pREP1 (or pREP41-GFP) vector under control of the inducible nmt1 (or nmt41) promoter [89]. In the presence of thiamine, gene expression from the nmt promoter is repressed, whilst removal of thiamine from the medium allows high-level expression of the ndc80 constructs.
Figure 4.

Excess ‘free’ Ndc80 absorbs Dis1 in the cytoplasm. A: A schematic showing the ability of different Ndc80 constructs to bind to microtubule (MT), Dis1 and Alp7-Alp14 complex. B: Localisation of overproduced GFP-Ndc80. Overproduced Ndc80-FL colocalise with the kinetochore (marked by Nuf2-mCherry). In sharp contrast, overproduced Ndc80-ΔNCH, Ndc80-ΔNCH(F420S) or Ndc80-ΔNCH(L405P) does not localise to the kinetochore, instead forming large polymers/aggregates in the cell. Scale bar, 5 µm. C: Localisation of Dis1 in cells overexpressing different Ndc80 constructs. Dis1 localises to the cytoplasmic MTs (marked by CFP-Atb2) during interphase (in vector, Ndc80-FL). Localisation of Dis1 to the cytoplasmic MTs is disrupted in cells overproducing Ndc80-ΔNCH or Ndc80-ΔNCH(F420S). This disruption is rescued by introducing L405P (defective in Dis1 binding) into the internal loop of Ndc80-ΔNCH. Scale bar, 5 µm. D: A schematic showing absorption of MAPs by overproduced Ndc80. Under normal conditions, Ndc80 localises to the kinetochore whereas MAPs localise to the MTs. When Ndc80 is overproduced, MAPs are sequestered by overproduced Ndc80, thereby disrupting the normal functions of MAPs (depicted by dimmer MTs).
Overproduced Ndc80 sequesters its binding partner in the cells
We first examined the localisation patterns of the individual Ndc80 constructs. Ectopically produced Ndc80-FL colocalises with the kinetochore, whilst the Ndc80-ΔNCH constructs fail to localise to the kinetochore (Fig. 4B). This observation is consistent with a recent study showing the importance of the CH domain in the hetero-dimerisation of Ndc80 and Nuf2 [38]; without the function of this domain, Ndc80 cannot be incorporated into the Ndc80 complex. Intriguingly, we observed the formation of large polymers/aggregates in cells overproducing the Ndc80-ΔNCH constructs (Fig. 4B). This localisation pattern is highly unusual, suggesting a potential self-polymerisation activity of the constructs lacking the NCH domain. We did not however observe large polymers in cells overexpressing Ndc80-FL. We consider two possibilities to explain this observation. Firstly, Ndc80-ΔNCH fails to localise to the kinetochore, thus all the Ndc80-ΔNCH overproduced is freely available to bind to other MAPs and disrupt their functions. In contrast, Ndc80-FL forms a complex with endogenous Nuf2 and localises to the kinetochore. This localisation may have a dilution effect on the overproduced Ndc80-FL and reduce the population of excess ‘free’ Ndc80 present in the cells. Secondly, we envision that the absence of the NCH domain in Ndc80-ΔNCH may lead to non-physiological large polymers/aggregates in the cells by an unknown mechanism. These possibilities are not exclusive, and further experiments are required to test this notion.
Having observed the distinct localisation patterns of each individual Ndc80 construct, we then examined the localisation of Dis1, a known interacting partner of Ndc80 through the internal loop region [42]. As expected, Dis1 localises to the cytoplasmic MTs during interphase in cells expressing empty vectors or Ndc80-FL (Fig. 4C). Intriguingly, Dis1 forms large polymers/aggregates in cells expressing Ndc80-ΔNCH (Fig. 4C). Although colocalisation between Ndc80-ΔNCH and Dis1 is not formally proven, this localisation pattern is similar, if not identical, to that of Ndc80-ΔNCH. Further experiments on colocalisation or binding between Ndc80-ΔNCH and Dis1 should clarify this phenomenon. We noted that MT intensity is reduced in cells overexpressing Ndc80-ΔNCH, suggesting a disrupted MT structure possibly ascribable to malfunctioning of aggregated Dis1. We also observed similar localisation in cells expressing Ndc80-ΔNCH(F420S), consistent with the notion that the F420S mutation does not disrupt the ability to bind to Dis1 in the cells, but instead is defective in binding to the Alp7/TACC-Alp14/TOG complex [43]. In fact, localisation of Alp7 is not noticeably altered under the same condition, the reason for which is currently not explored further (unpublished). Interestingly, the defective phenotypes were rescued by introducing into the construct the L405P mutation with impaired Dis1 binding activity (Fig. 4C) [42]. In line with this observation, Dis1 localises on the MTs in the L405P mutant, in a similar manner to that seen in cells containing empty vectors or overproducing Ndc80-FL. Overall, these experiments strongly suggest that overproduced Ndc80 absorbs its binding partners (e.g. Dis1), thereby disrupting their functions (Fig. 4D). It would be of interest to test whether overproduction of other regions within Ndc80, including the N-terminal tail/CH domain and the C-terminal coiled coil domain could also sequestrate their individual interactors.
Can in vivo overexpression be used as a tool to identify interacting partners?
We have shown that overproducing Ndc80-ΔNCH results in large aggregates in the cells, and that these aggregates abduct its interacting partner Dis1 (Fig. 4). This ability to form large aggregates suggests that the coiled-coil and loop domains of Ndc80 have the potential to self-polymerise, and that this polymerisation may play important roles in the functions of Ndc80. Furthermore, overexpression of the TACC domain in HeLa cells produces large polymers that appear to associate with MTs and tubulin in the cytoplasm [90]. It would be intriguing to test whether other TACC interactors such as ch-TOG and clathrin are also sequestered under these conditions. As the formation of large aggregates is easily observable and recognisable in the cells, we propose that this in vivo protein overproduction system could provide an excellent tool to identify the interacting partners of a protein (e.g. coimmunoprecipitation followed by mass spectrometry analysis). It is of note that overproduction of a protein may lead to unspecific binding of other functionally non-related proteins. Therefore, further in vivo validations are required upon the identification of interacting partners using this method.
The Ndc80 internal loop has been shown to bind several proteins in different organisms, but the Ndc80 loop-interacting partners known to date do not share homology amongst these organisms. For example, it is unknown whether TACC-TOG binds to the Ndc80 internal loop in vertebrates, though it is firmly established that TACC-TOG localisation to the Ndc80 internal loop in fission yeast plays important roles in mitosis [42,43] and meiosis [91]. We could now use this in vivo overproduction system to identify interacting partners of Ndc80 in different model organisms. We envision that this system could also be applied to identify interactors of other mitotic proteins.
A new hypothesis: How does Ndc80 overproduction cause cancer?
Overexpression of Ndc80/Hec1 has long been implicated in tumourigenesis [10–12]. Interestingly, the expression of Ndc80/Hec1-associated genes (i.e. Nuf2, Spc24, Spc25 and Nek2) is coordinately up-regulated in Ndc80/Hec1-overexpressed cancer cells [92]. Recent analyses of gene expression profiles in cancer patients have suggested that altered expression of individual kinetochore genes is unlikely to cause cancer on its own [93]. Instead, it is proposed that these altered expression patterns in cancer cells arise as a consequence of an altered cell division programme. These studies indicate that the overexpression of a cluster of genes, rather than a single gene, affects the potential and severity of tumourigenesis. Nonetheless, it is possible that this coordinated expression of mitotic genes does promote the process of tumourigenesis, or contributes to the functional compensation for up-regulation of some mitotic proteins such as Ndc80. In line with this notion, overexpressing Ndc80 in mice displays increased levels of Mad2, leading to checkpoint hyperactivation and aneuploidy [12]. Furthermore, breast cancer patients with both Ndc80 and Nek2 overexpression display shorter survival compared to patients with only either Ndc80 or Nek2 overexpression [94]. In fission yeast, we observed prolonged mitotic arrest in cells overproducing Ndc80-ΔNCH or Ndc80-ΔNCH(F420S), suggesting that the SAC is continuously active in these cells. These data support the notion that overexpression of a single gene may induce similar overexpression of its genetic or functional partners, thereby leading to tumourigenesis.
In brief, we hypothesise that overproduced Ndc80/Hec1 may sequestrate its binding partners through the Ndc80 internal loop domain. This absorption will result in altered mitotic progression and defective chromosome segregation, leading to aneuploidy. On the other hand, in response to these defects, the transcription programme of Ndc80 and its interacting partners may also be altered as a result of different cell cycle profiles and/or cellular compensatory mechanism. This alteration may further promote growth of aneuploid cells (summarised in Fig. 5).
Figure 5.

The proposed outcome of Ndc80/Hec1 overproduction. Overproducing Ndc80/Hec1 leads to the absorption of its interacting proteins by binding to the internal loop. This sequestration impedes mitotic progression and leads to chromosome mis-segregation, resulting in the formation of aneuploid progenies. Aneuploidy will, on one hand, promote tumourigenesis and yet on the other hand, trigger the cellular compensation, by which the transcription programme may alter in response to mitotic defects. Altered gene expression profiles, either up- or down-regulation of functionally related genes/proteins, will further promote or suppress growth of aneuploid cells. Cancer cells may arise from some of these populations.
Ndc80 and TACC proteins as potential drug targets
Due to their roles in controlling mitotic progression and chromosome segregation, both Ndc80/Hec1 and TACC proteins are considered potential targets for cancer drug development. The use of single vector-expressed short-hairpin RNAs (shRNAs) has shown that the depletion of Ndc80/Hec1 significantly reduces tumour size in mice [95]. Additionally, a small compound INH-1 that disrupts the Ndc80-Nek2 interaction has been developed [96] along with other INH-1 derivatives identified to show improved potency and efficacy [92,94,97]. Introducing such compounds to the cells leads to mitotic catastrophe and halts tumour progression. These studies indicate that the disruption of Ndc80 functions could provide promising therapeutic effects for cancer patients. On the other hand, a recent study has shown that depletion of TACC3 in lymphoma cells causes multi-polar spindle formation that subsequently leads to mitotic arrest and apoptosis [98]. The same group has then developed a small molecule, spindlactone (SPL) that inhibits the functions of the TACC-TOG complex [99]. Oral administration of SPL significantly reduces the tumour volume in mice [99], demonstrating the potential of SPL as a therapeutic agent for cancer chemotherapy in clinical applications.
Clinical perspectives of the hypothesis
Recent advancements in drug development against the Ndc80/Hec1 and TACC proteins seem promising for clinical use in cancer treatment. However, several key features remain unexplored. Here we propose a few questions that could be addressed in the future to provide a better understanding of a functional relationship between gene overexpression and cancer formation. For simplicity, we focus on Ndc80/Hec1 and TACC; these questions could also be applied to other mitotic genes/proteins.
Does overexpression of Ndc80 induce overexpression of its genetic and functional partners?
Recent studies have identified several proteins that interact with the Ndc80 internal loop [45,46], and we have shown that the overproduction of Ndc80 results in absorption of its interacting partner Dis1 which leads to disruption of MT structure (Fig. 4). Although we have not examined the localisation of any other Ndc80 interactors (e.g. Alp7-Alp14), our results strongly suggest that Ndc80 overproduction leads to spatial segregation of its interacting partners, thereby disrupting the functions of these proteins. To compensate for the loss of function of these proteins, the expression level of these genes may be altered (e.g. up-regulated). Intriguingly, several of the Ndc80 internal loop interactors, such as the Ska complex, TACC-TOG complex and kinesin-8, are also up-regulated in several cancers [3,4,7–9]. Despite this, whether these genes/proteins are coordinately up-regulated with Ndc80 in cancer cells remains to be examined. Further investigations into gene expression profiles of Ndc80 and its interactors will provide insight into the mechanism of Ndc80 overexpression and aneuploidy.
Does drug usage in combination give a more effective cancer treatment?
Identification of small molecules targeting Ndc80/Hec1 and TACC proteins have given promising results for translation into clinical application [92,94,96,97,99,102]. Oral administrations of these compounds have successfully been shown to reduce tumour growth in vivo, potentially through induction of apoptotic cell death [92,99]. Since recent studies have identified the functional relationships between Ndc80/Hec1 and TACC proteins (see above), we envision that administration of these drugs in combination may have a better effect in cancer treatment. Future studies using cancer cell lines or mouse models will be able to verify this proposition.
Conclusions and outlook
Genome instability is one of the major aetiologies of tumourigenesis, and is often related to defects in chromosome segregation during mitosis. Due to their roles in chromosome segregation, overexpression of mitotic genes has long been implicated in cancer formation. Although much work has focused on the defining roles of these mitotic genes in chromosome segregation, how the overexpression of these genes leads to tumourigenesis remains largely elusive. Identification of the Ndc80 internal loop as a protein-protein interaction motif has shed light on our understanding of Ndc80 overexpression and cancer formation. We propose that the expression levels of Ndc80-interacting partners may be altered to compensate for loss of functions of these proteins. Further, we propose that our in vivo overproduction system could be exploited as an efficient tool to identify the binding partners of a protein. This tool should provide an alternative way to study genetic and functional relationships between different mitotic genes/proteins. We hope that further investigations into the questions raised in this hypothesis will provide further insights into relationships between gene overexpression and tumourigenesis, leading to improved strategies for cancer treatment.
Acknowledgments
We thank Risa Mori for critical reading of the manuscript and useful suggestions. This work was supported by Cancer Research UK (T.T.).
The authors have declared no conflict of interest.
Glossary
- ch-TOG
colonic and hepatic tumour overexpressed gene
- CIN
chromosome instability
- MAPs
microtubule associated proteins
- MTs
microtubules
- SAC
spindle assembly checkpoint
- SPB
spindle pole body
- TACC
transforming acidic coiled-coil
References
- 1.Bakhoum SF, Silkworth WT, Nardi IK, Nicholson JM. The mitotic origin of chromosomal instability. Curr Biol. 2014;24:R148–9. doi: 10.1016/j.cub.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–7. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 3.Charrasse S, Mazel M, Taviaux S, Berta P. Characterization of the cDNA and pattern of expression of a new gene over-expressed in human hepatomas and colonic tumors. Eur J Biochem. 1995;234:406–13. doi: 10.1111/j.1432-1033.1995.406_b.x. [DOI] [PubMed] [Google Scholar]
- 4.Still IH, Vince P, Cowell JK. The third member of the transforming acidic coiled coil-containing gene family, TACC3, maps in 4p16, close to translocation breakpoints in multiple myeloma, and is upregulated in various cancer cell lines. Genomics. 1999;58:165–70. doi: 10.1006/geno.1999.5829. [DOI] [PubMed] [Google Scholar]
- 5.Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ricke RM, Jeganathan KB, van Deursen JM. Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation. J Cell Biol. 2011;193:1049–64. doi: 10.1083/jcb.201012035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun W, Yao L, Jiang B, Guo L. Spindle and kinetochore-associated protein 1 is overexpressed in gastric cancer and modulates cell growth. Mol Cell Biochem. 2014;391:167–74. doi: 10.1007/s11010-014-1999-1. [DOI] [PubMed] [Google Scholar]
- 8.Zhang C, Zhu C, Chen H, Li L. Kif18A is involved in human breast carcinogenesis. Carcinogenesis. 2010;31:1676–84. doi: 10.1093/carcin/bgq134. [DOI] [PubMed] [Google Scholar]
- 9.Nagahara M, Nishida N, Iwatsuki M, Ishimaru S. Kinesin 18A expression: clinical relevance to colorectal cancer progression. Int J Cancer. 2011;129:2543–52. doi: 10.1002/ijc.25916. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Riley DJ, Chen PL, Lee WH. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol Cell Biol. 1997;17:6049–56. doi: 10.1128/mcb.17.10.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayama S, Daigo Y, Kato T, Ishikawa N. Activation of CDCA1-KNTC2, members of centromere protein complex, involved in pulmonary carcinogenesis. Cancer Res. 2006;66:10339–48. doi: 10.1158/0008-5472.CAN-06-2137. [DOI] [PubMed] [Google Scholar]
- 12.Diaz-Rodriguez E, Sotillo R, Schvartzman JM, Benezra R. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc Natl Acad Sci USA. 2008;105:16719–24. doi: 10.1073/pnas.0803504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ulisse S, Baldini E, Toller M, Delcros JG. Transforming acidic coiled-coil 3 and Aurora-A interact in human thyrocytes and their expression is deregulated in thyroid cancer tissues. Endocr Relat Cancer. 2007;14:827–37. doi: 10.1677/ERC-07-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai W, Wang Q, Liu T, Swamy M. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 2004;64:440–5. doi: 10.1158/0008-5472.can-03-3119. [DOI] [PubMed] [Google Scholar]
- 15.Michel L, Diaz-Rodriguez E, Narayan G, Hernando E. Complete loss of the tumor suppressor MAD2 causes premature cyclin B degradation and mitotic failure in human somatic cells. Proc Natl Acad Sci USA. 2004;101:4459–64. doi: 10.1073/pnas.0306069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciferri C, De Luca J, Monzani S, Ferrari KJ. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem. 2005;280:29088–95. doi: 10.1074/jbc.M504070200. [DOI] [PubMed] [Google Scholar]
- 17.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci USA. 2005;102:5363–7. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciferri C, Pasqualato S, Screpanti E, Varetti G. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–39. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–9. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 20.Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–84. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1) Curr Biol. 2008;18:1785–91. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tooley JG, Miller SA, Stukenberg PT. The Ndc80 complex employs a tripartite attachment point to couple microtubule depolymerization to chromosome movement. Mol Biol Cell. 2011;22:1217–26. doi: 10.1091/mbc.E10-07-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–97. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 24.Cheeseman IM, Anderson S, Jwa M, Green EM. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–72. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 25.DeLuca JG, Gall WE, Ciferri C, Cimini D. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–82. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 26.Zaytsev AV, Sundin LJ, DeLuca KF, Grishchuk EL. Accurate phosphoregulation of kinetochore-microtubule affinity requires unconstrained molecular interactions. J Cell Biol. 2014;206:45–59. doi: 10.1083/jcb.201312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu D, Vader G, Vromans MJ, Lampson MA. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–3. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka TU, Rachidi N, Janke C, Pereira G. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–29. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 29.Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011;21:133–40. doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell CS, Desai A. Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature. 2013;497:118–21. doi: 10.1038/nature12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemmler S, Stach M, Knapp M, Ortiz J. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 2009;28:1099–110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Riley DJ, Zheng L, Chen PL. Phosphorylation of the mitotic regulator protein Hec1 by Nek2 kinase is essential for faithful chromosome segregation. J Biol Chem. 2002;277:49408–16. doi: 10.1074/jbc.M207069200. [DOI] [PubMed] [Google Scholar]
- 33.Wang HW, Long S, Ciferri C, Westermann S. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–9. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan X, O'Quinn RP, Pierce HL, Joglekar AP. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–84. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang G, Kelstrup CD, Hu XW, Kaas Hansen MJ. The Ndc80 internal loop is required for recruitment of the Ska complex to establish end-on microtubule attachment to kinetochores. J Cell Sci. 2012;125:3243–53. doi: 10.1242/jcs.104208. [DOI] [PubMed] [Google Scholar]
- 37.Petrovic A, Mosalaganti S, Keller J, Mattiuzzo M. Modular assembly of RWD domains on the Mis12 complex underlies outer kinetochore organization. Mol Cell. 2014;53:591–605. doi: 10.1016/j.molcel.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 38.Tien JF, Fong KK, Umbreit NT, Payen C. Coupling unbiased mutagenesis to high-throughput DNA sequencing uncovers functional domains in the Ndc80 kinetochore protein of Saccharomyces cerevisiae. Genetics. 2013;195:159–70. doi: 10.1534/genetics.113.152728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tien JF, Umbreit NT, Zelter A, Riffle M. Kinetochore biorientation in Saccharomyces cerevisiae requires a tightly folded conformation of the Ndc80 complex. Genetics. 2014;198:1483–93. doi: 10.1534/genetics.114.167775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aravamudhan P, Felzer-Kim I, Gurunathan K, Joglekar AP. Assembling the protein architecture of the budding yeast kinetochore-microtubule attachment using FRET. Curr Biol. 2014;24:1437–46. doi: 10.1016/j.cub.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maure JF, Komoto S, Oku Y, Mino A. The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Curr Biol. 2011;21:207–13. doi: 10.1016/j.cub.2010.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsu KS, Toda T. Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore-spindle attachment in fission yeast. Curr Biol. 2011;21:214–20. doi: 10.1016/j.cub.2010.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang NH, Takada H, Hsu KS, Toda T. The internal loop of fission yeast Ndc80 binds Alp7/TACC-Alp14/TOG and ensures proper chromosome attachment. Mol Biol Cell. 2013;24:1122–33. doi: 10.1091/mbc.E12-11-0817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varma D, Chandrasekaran S, Sundin LJR, Reidy KT. Recruitment of the human Cdt1 replication licensing protein by the loop domain of Hec1 is required for stable kinetochore–microtubule attachment. Nat Cell Biol. 2012;14:593–603. doi: 10.1038/ncb2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nilsson J. Looping in on Ndc80 - How does a protein loop at the kinetochore control chromosome segregation. BioEssays. 2012;34:1070–7. doi: 10.1002/bies.201200096. [DOI] [PubMed] [Google Scholar]
- 46.Tang NH, Toda T. Ndc80 Loop as a protein-protein interaction motif. Cell Div. 2013;8:2. doi: 10.1186/1747-1028-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang NH, Toda T. Alp7/TACC recruits kinesin-8-PP1 to the Ndc80 kinetochore protein for timely mitotic progression and chromosome movement. J Cell Sci. 2015 doi: 10.1242/jcs.160036. DOI: 10.1242/jcs.160036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sivakumar S, Daum JR, Tipton AR, Rankin S. The Spindle and kinetochore-associated (Ska) complex enhances binding of the Anaphase-Promoting Complex/Cyclosome (APC/C) to chromosomes and promotes mitotic exit. Mol Biol Cell. 2014;25:594–605. doi: 10.1091/mbc.E13-07-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Still IH, Hamilton M, Vince P, Wolfman A. Cloning of TACC1, an embryonically expressed, potentially transforming coiled coil containing gene, from the 8p11 breast cancer amplicon. Oncogene. 1999;18:4032–8. doi: 10.1038/sj.onc.1202801. [DOI] [PubMed] [Google Scholar]
- 50.Gergely F, Kidd D, Jeffers K, Wakefield JG. D-TACC: a novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J. 2000;19:241–52. doi: 10.1093/emboj/19.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Srayko M, Quintin S, Schwager A, Hyman AA. Caenorhabditis elegans TAC-1 and ZYG-9 form a complex that is essential for long astral and spindle microtubules. Curr Biol. 2003;13:1506–11. doi: 10.1016/s0960-9822(03)00597-9. [DOI] [PubMed] [Google Scholar]
- 52.Sato M, Vardy L, Angel Garcia M, Koonrugsa N. Interdependency of fission yeast Alp14/TOG and coiled coil protein Alp7 in microtubule localization and bipolar spindle formation. Mol Biol Cell. 2004;15:1609–22. doi: 10.1091/mbc.E03-11-0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Still IH, Vettaikkorumakankauv AK, DiMatteo A, Liang P. Structure-function evolution of the transforming acidic coiled coil genes revealed by analysis of phylogenetically diverse organisms. BMC Evol Biol. 2004;4:16. doi: 10.1186/1471-2148-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Booth DG, Hood FE, Prior IA, Royle SJ. A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging. EMBO J. 2011;30:906–19. doi: 10.1038/emboj.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Royle SJ. The role of clathrin in mitotic spindle organisation. J Cell Sci. 2012;125:19–28. doi: 10.1242/jcs.094607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hood FE, Williams SJ, Burgess SG, Richards MW. Coordination of adjacent domains mediates TACC3-ch-TOG-clathrin assembly and mitotic spindle binding. J Cell Biol. 2013;202:463–78. doi: 10.1083/jcb.201211127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thakur HC, Singh M, Nagel-Steger L, Prumbaum D. Role of centrosomal adaptor proteins of the TACC family in the regulation of microtubule dynamics during mitotic cell division. Biol Chem. 2013;394:1411–23. doi: 10.1515/hsz-2013-0184. [DOI] [PubMed] [Google Scholar]
- 58.Thakur HC, Singh M, Nagel-Steger L, Kremer J. The centrosomal adaptor TACC3 and the microtubule polymerase chTOG interact via defined C-terminal subdomains in an Aurora-A kinase-independent manner. J Biol Chem. 2014;289:74–88. doi: 10.1074/jbc.M113.532333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fu W, Tao W, Zheng P, Fu J. Clathrin recruits phosphorylated TACC3 to spindle poles for bipolar spindle assembly and chromosome alignment. J Cell Sci. 2010;123:3645–51. doi: 10.1242/jcs.075911. [DOI] [PubMed] [Google Scholar]
- 60.Hubner NC, Bird AW, Cox J, Splettstoesser B. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol. 2010;189:739–54. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin CH, Hu CK, Shih HM. Clathrin heavy chain mediates TACC3 targeting to mitotic spindles to ensure spindle stability. J Cell Biol. 2010;189:1097–105. doi: 10.1083/jcb.200911120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sato M, Toda T. Alp7/TACC is a crucial target in Ran-GTPase-dependent spindle formation in fission yeast. Nature. 2007;447:334–7. doi: 10.1038/nature05773. [DOI] [PubMed] [Google Scholar]
- 63.Tang NH, Okada N, Fong CS, Arai K. Targeting Alp7/TACC to the spindle pole body is essential for mitotic spindle assembly in fission yeast. FEBS Lett. 2014;588:2814–21. doi: 10.1016/j.febslet.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng F, Li T, Jin DY, Syrovatkina V. Csi1p recruits alp7p/TACC to the spindle pole bodies for bipolar spindle formation. Mol Biol Cell. 2014;25:2750–60. doi: 10.1091/mbc.E14-03-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gergely F, Draviam VM, Raff JW. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 2003;17:336–41. doi: 10.1101/gad.245603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Foraker AB, Camus SM, Evans TM, Majeed SR. Clathrin promotes centrosome integrity in early mitosis through stabilization of centrosomal ch-TOG. J Cell Biol. 2012;198:591–605. doi: 10.1083/jcb.201205116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kinoshita K, Habermann B, Hyman AA. XMA P215: a key component of the dynamic microtubule cytoskeleton. Trends Cell Biol. 2002;12:267–73. doi: 10.1016/s0962-8924(02)02295-x. [DOI] [PubMed] [Google Scholar]
- 68.Al-Bassam J, Chang F. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 2011;21:604–14. doi: 10.1016/j.tcb.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Widlund PO, Stear JH, Pozniakovsky A, Zanic M. XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc Natl Acad Sci USA. 2011;108:2741–6. doi: 10.1073/pnas.1016498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Al-Bassam J, Kim H, Flor-Parra I, Lal N. Fission yeast Alp14 is a dose dependent plus end tracking microtubule polymerase. Mol Biol Cell. 2012;23:2878–90. doi: 10.1091/mbc.E12-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Podolski M, Mahamdeh M, Howard J. Stu2, the budding yeast XMAP215/Dis1 homolog, promotes assembly of yeast microtubules by increasing growth rate and decreasing catastrophe frequency. J Biol Chem. 2014;289:28087–93. doi: 10.1074/jbc.M114.584300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ayaz P, Ye X, Huddleston P, Brautigam CA. A TOG:alphabeta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science. 2012;337:857–60. doi: 10.1126/science.1221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ayaz P, Munyoki S, Geyer EA, Piedra FA. A tethered delivery mechanism explains the catalytic action of a microtubule polymerase. ELife. 2014;3:e03069. doi: 10.7554/eLife.03069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Al-Bassam J, Larsen NA, Hyman AA, Harrison SC. Crystal structure of a TOG domain: conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure. 2007;15:355–62. doi: 10.1016/j.str.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 76.Slep KC, Vale RD. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol Cell. 2007;27:976–91. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fox JC, Howard AE, Currie JD, Rogers SL. The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array. Mol Biol Cell. 2014;25:2375–92. doi: 10.1091/mbc.E13-08-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nabeshima K, Kurooka H, Takeuchi M, Kinoshita K. p93dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the Cdc2 target sites. Genes Dev. 1995;9:1572–85. doi: 10.1101/gad.9.13.1572. [DOI] [PubMed] [Google Scholar]
- 79.Garcia MA, Vardy L, Koonrugsa N, Toda T. Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J. 2001;20:3389–401. doi: 10.1093/emboj/20.13.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nakaseko Y, Goshima G, Morishita J, Yanagida M. M phase-specific kinetochore proteins in fission yeast: microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr Biol. 2001;11:537–49. doi: 10.1016/s0960-9822(01)00155-5. [DOI] [PubMed] [Google Scholar]
- 81.Garcia MA, Koonrugsa N, Toda T. Spindle-kinetochore attachment requires the combined action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J. 2002;21:6015–24. doi: 10.1093/emboj/cdf611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987;329:219–22. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- 83.Coffman VC, Wu P, Parthun MR, Wu JQ. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J Cell Biol. 2011;195:563–72. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J Cell Biol. 2011;195:573–82. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol. 1993;120:141–51. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McIntosh JR, O'Toole E, Zhudenkov K, Morphew M. Conserved and divergent features of kinetochores and spindle microtubule ends from five species. J Cell Biol. 2013;200:459–74. doi: 10.1083/jcb.201209154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marguerat S, Schmidt A, Codlin S, Chen W. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–83. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carpy A, Krug K, Graf S, Koch A. Absolute proteome and phosphoproteome dynamics during the cell cycle of Schizosaccharomyces pombe (fission yeast) Mol Cell Proteomics. 2014;13:1925–36. doi: 10.1074/mcp.M113.035824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–30. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 90.Gergely F, Karlsson C, Still I, Cowell J. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc Natl Acad Sci USA. 2000;97:14352–7. doi: 10.1073/pnas.97.26.14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kakui Y, Sato M, Okada N, Toda T. Microtubules and Alp7-Alp14 (TACC-TOG) reposition chromosomes before meiotic segregation. Nat Cell Biol. 2013;15:786–96. doi: 10.1038/ncb2782. [DOI] [PubMed] [Google Scholar]
- 92.Huang LY, Chang CC, Lee YS, Chang JM. Activity of a novel Hec1-targeted anticancer compound against breast cancer cell lines in vitro and in vivo. Mol Cancer Ther. 2014;13:1419–30. doi: 10.1158/1535-7163.MCT-13-0700. [DOI] [PubMed] [Google Scholar]
- 93.Thiru P, Kern DM, McKinley KL, Monda JK. Kinetochore genes are coordinately up-regulated in human tumors as part of a FoxM1-related cell division program. Mol Biol Cell. 2014;25:1983–94. doi: 10.1091/mbc.E14-03-0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hu CM, Zhu J, Guo XE, Chen W. Novel small molecules disrupting Hec1/Nek2 interaction ablate tumor progression by triggering Nek2 degradation through a death-trap mechanism. Oncogene. 2014 doi: 10.1038/onc.2014.67. et al., in press doi: 10.1038/onc.2014.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gurzov EN, Izquierdo M. RNA interference against Hec1 inhibits tumor growth in vivo. Gene Ther. 2006;13:1–7. doi: 10.1038/sj.gt.3302595. [DOI] [PubMed] [Google Scholar]
- 96.Wu G, Qiu XL, Zhou L, Zhu J. Small molecule targeting the Hec1/Nek2 mitotic pathway suppresses tumor cell growth in culture and in animal. Cancer Res. 2008;68:8393–9. doi: 10.1158/0008-5472.CAN-08-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang LY, Lee YS, Huang JJ, Chang CC. Characterization of the biological activity of a potent small molecule Hec1 inhibitor TAI-1. J Exp Clin Cancer Res. 2014;33:6. doi: 10.1186/1756-9966-33-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yao R, Natsume Y, Saiki Y, Shioya H. Disruption of Tacc3 function leads to in vivo tumor regression. Oncogene. 2012;31:135–48. doi: 10.1038/onc.2011.235. [DOI] [PubMed] [Google Scholar]
- 99.Yao R, Kondoh Y, Natsume Y, Yamanaka H. A small compound targeting TACC3 revealed its different spatiotemporal contributions for spindle assembly in cancer cells. Oncogene. 2013;33:4242–52. doi: 10.1038/onc.2013.382. [DOI] [PubMed] [Google Scholar]
- 100.Richmond D, Rizkallah R, Liang F, Hurt MM. Slk19 clusters kinetochores and facilitates chromosome bipolar attachment. Mol Biol Cell. 2013;24:566–77. doi: 10.1091/mbc.E12-07-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zeng X, Kahana JA, Silver PA, Morphew MK. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J Cell Biol. 1999;146:415–25. doi: 10.1083/jcb.146.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ohoka N, Nagai K, Hattori T, Okuhira K. Cancer cell death induced by novel small molecules degrading the TACC3 protein via the ubiquitin-proteasome pathway. Cell Death Disease. 2014;5:e1513. doi: 10.1038/cddis.2014.471. [DOI] [PMC free article] [PubMed] [Google Scholar]


