Abstract
The t haplotype in house mice is a well-known selfish genetic element with detrimental, nonadditive fitness consequences to its carriers: recessive lethal mutations cause t/t homozygotes to perish in utero. Given the severe genetic incompatibility imposed by the t haplotype, we predict females to avoid fertilization by t haplotype incompatible males. Indeed, some of the strongest evidence for compatibility mate choice is related to the t haplotype in house mice. However, all previous evidence for compatibility mate choice in this system is based on olfactory preference. It is so far unknown how general these preferences are and whether they are relevant in an actual mating context. Here, we assess female compatibility mate choice related to t haplotypes in a setting that – for the first time – allowed females to directly interact and mate with males. This approach enabled us to analyse female behaviour during the testing period, and the resulting paternity success and fitness consequences of a given choice. We show that genetic incompatibilities arising from the t haplotype had severe indirect fitness consequences and t females avoided fertilization by t incompatible males. The results are inconclusive whether this avoidance of t fertilization by t females was caused by pre- or post-copulatory processes.
Keywords: genetic compatibility, mate choice, polyandry, selfish genetic elements, sexual selection, t Haplotype
Introduction
Female mate choice is recognized as a powerful evolutionary process, potentially explaining the origin of extravagant male ornaments that remain puzzling under the concept of natural selection. However, the question of why females choose mates at all, especially in the absence of direct fitness benefits (indirect selection), remains a much debated topic in sexual selection research. Indirect selection on female mate preference occurs if the preference trait is genetically correlated to a trait under direct selection. A variety of mechanisms have been proposed about how such a correlation can arise, ranging from Fisher's runaway hypothesis (Fisher, 1930), where a male display trait becomes genetically correlated with the female preference for that trait, to indicator or ’good genes’ models where the male display trait is used as a signal of male genetic quality (Andersson, 1994). Selection should also favour female preference for partners that produce offspring with the most adaptive gene combinations. This concept of mate choice for ’compatible genes’ – originally advanced by Trivers (1972) – has only recently received attention in empirical studies (Mays & Hill, 2004). Under this paradigm, the best mate for a given female does not only depend on the male's genotype (such as in ’good genes’ models), but also on her own genotype (Zeh & Zeh, 1996,1997,2003; Tregenza & Wedell, 2000). In contrast to ’good genes’ hypotheses that assume additive gene action (such that an optimal choice for a female is independent of her own genotype), mate choice for compatibility is based on nonadditive genetic effects such as dominance or epistasis (Kotiaho et al., 2008). On a between-species-level, mate choice for compatibility, that is preference for conspecific partners, is well documented and is important in the process of sympatric speciation (Butlin & Ritchie, 1989). On the within-population level, however, the importance of genetic compatibility remains unclear. Nonadditive effects are likely to be complex if many genes are involved. Selection for genetic compatibility is therefore likely to be constrained to specific genetic systems with potentially large fitness effects (Puurtinen et al., 2005).
Selfish genetic elements and genetic incompatibility
A promising class of genetic systems to drive the evolution of mate choice for genetic compatibility are selfish genetic elements [SGEs henceforth, Tregenza & Wedell (2000); Zeh & Zeh (1996)]. SGEs are broadly defined as stretches of DNA that promote their own transmission at the expense of rival genes (Burt & Trivers, 2006). Most known SGEs are associated with substantial fitness costs to their carriers (Burt & Trivers, 2006). Despite these detrimental effects on carrier fitness, SGEs often stably persist in populations as a result of their systematic transmission advantage. The stable presence of a SGE may select for female mating strategies to avoid substantial, SGE-related fitness losses. Females could avoid carrier males at a pre- or post-copulatory stage. There is only limited evidence for discrimination against SGE-carriers prior to mating. In stalk-eyed flies, females prefer to mate with males with long eye-stalks. In populations that harbour a sex-ratio distorter, it has been shown that long male eye-stalks indicate a genetic suppressor of drive (Cotton et al., 2014). A preference for long eye-stalks will hence ensure that females produce both sons and daughters (Wilkinson et al., 1998). Because SGEs often target male spermatogenesis to achieve transmission advantage (termed drive), male SGE-carriers are often compromised in their sperm competitiveness (Price & Wedell, 2008). Females may capitalize on this link and avoid SGE-fertilization by mating with multiple males (polyandry), thereby enhancing the importance of sperm competition (Haig & Bergstrom, 1995). Empirical evidence for the importance of such post-copulatory SGE-avoidance is numerous, especially in insect species [see Wedell (2013) for a recent review]. For example, in sex-ratio drive systems of Drosophila simulans and Drosophila pseudoobscura, distorters were shown to considerably reduce competitive ability of gametes (Atlan et al., 2004; Price et al., 2008a). In the latter case, female flies were even found to evolve higher remating rates in the presence of the distorter (Price et al., 2008b).
In the previous examples, all females are susceptible to the negative fitness effects of the SGE and hence, all females should avoid carriers. They can therefore not be considered cases of choice for compatibility. However, SGE-related fitness costs are often nonadditive, hence causing incompatibilites between maternal and paternal genotypes (Zeh & Zeh, 1996). As a consequence, the fitness consequences of a given female mating decision is not only a function of her partner's genotype, but also of her own. In Drosophila paulistorum flies, both males and females mate assortatively based on the intracellular bacterium Wolbachia (Miller et al., 2010), helping them to avoid Wolbachia-induced embryo mortality and male sterility. Such Wolbachia-related assortative mating has also been reported in the spider mites Tetranychus urticae, where uninfected females avoided incompatible infected males in mate choice tests (Vala et al., 2004).
The t haplotype
Some of the strongest evidence for compatibility mate choice is related to the t haplotype system in house mice. The t haplotype in house mice is a classical example of a SGE. It consists of a whole set of genes occupying about one-third of mouse chromosome 17 and is protected from recombination by a large inversion system (Silver, 1993). The t haplotype is thought to have existed for more than 3 million years (Burt & Trivers, 2006) and occurs in populations of all four house mouse subspecies (Silver, 1993). It has the properties that make it a promising candidate for compatibility mate choice. First, the t haplotype is associated with substantial nonadditive fitness costs. Most t haplotypes carry recessive embryonic lethal mutations, causing t/t homozygotes to perish in utero (Klein et al., 1984). +/t heterozygotes, on the other hand, are fully viable. Second, several genes within the complex ensure that this genetic entity is passed in a non-Mendelian manner from one generation to the next (drive). As a result, heterozygote +/t males typically transmit their t gametes to 90% of their offspring (Lyon, 2003; Lindholm et al., 2013). +/t females show normal Mendelian segregation. The resulting genetic incompatibilities are severe. A +/t female that mates with a +/t male is expected to lose up to half (depending on levels of drive) of her offspring from t lethal effects. Indeed, Lindholm et al. (2013) recently reported a litter size reduction of 40% at birth in controlled laboratory crosses. Hence, selective pressures on +/t heterozygote females to avoid t fertilization are substantial. Lenington and colleagues showed a consistent odour preference of +/t females towards +/+ males in a series of experiments [see Lenington (1991) for a review]. Results for +/+ females were not as clear: in some studies, they exhibited the same preference for +/+ males (Lenington, 1983; Lenington & Egid, 1985), whereas in others, preferences were not found (Coopersmith & Lenington, 1992; Williams & Lenington, 1993). It remains unclear whether these olfactory preferences reflect actual mating preferences and whether they are generalizable to different t haplotype variants [16 t haplotype variants have been described so far, Klein et al. (1984)]. The role of polyandry and sperm competition in the t haplotype system is largely unknown. High multiple paternity rates in wild populations suggest that sperm competition plays an important role in house mice (Dean et al., 2006; Firman & Simmons, 2008b; Manser et al., 2011). However, there is only very limited data on sperm competitiveness of t-carrying males. In the three previous studies looking at fertilization success of +/t males when competing with +/+ males, paternity shares of +/t males ranged between 0.17 and 0.36, but were based on very limited sample sizes (Olds-Clarke & Peitz, 1986; Ardlie & Silver, 1996; Carroll et al., 2004).
In the present study, we aimed to experimentally test female mate choice in relation to the t haplotype using an elaborate choice device. The device allowed females to freely associate and mate with either or both males of different genetic background (+/t and +/+, termed t and w for wild type hereafter) without interference of direct male–male competition. Paternity analyses of the resulting offspring as well as behavioural data during the experiment allowed us to address the following three questions. (i) What are the fitness consequences of female mating decisions? (ii) Do t females avoid fertilization of incompatible t males? (iii) Are potential fertilization biases caused by pre- or post-copulatory processes?
Materials and methods
Animals
The animals used for mating preference tests were all descendants from a wild house mouse (Mus musculus domesticus) population outside Zurich, Switzerland [see König & Lindholm (2012) for details]. They were kept under standard conditions (22–25∘C, 40–50% humidity, 14:10 h light:dark cycle starting at 7:30) with ad libitum food (laboratory diet for mice and rats, no. 3804 and 3336, Provimi Kliba SA, Kaiseraugst, Switzerland), water, nesting material and standard animal bedding (Lignocel Hygienic Animal Bedding, JRS). Altogether, 65 females and 45 males were used in the experiments. All males and females were typed at the Hba-ps4 locus – a marker containing a 16-bp t haplotype-specific insertion (Hammer et al., 1989) – to determine genotype at the t locus. The level of segregation distortion for the laboratory population has been estimated previously and is 90% (Lindholm et al., 2013). No females, but all males had mating experience prior to the experiment.
Choice device
An elaborate choice device developed in our group (Rüsch, 2002) was used to assess female mating strategy (see Fig. 1). The device consisted of three Macrolon II cages (A–C) connected with tunnels separated by doors. Only the female was allowed free access to all three cages (A–C). Males on the other hand were confined to their own, outer cages (A and C). To achieve this, all individuals used in the experiment were tagged individually with transponders (glastag micro read only, article no. 860-0220; IQ Automation GmbH, Eching, Germany). Transponders were recognized by specific readers (1–4, easy key Standalone module, article no. A402-0031; IQ Automation GmbH) over antennae that were wound manually around the tunnels at positions (i)–(iv). Each of these readers was further connected to specific doors (readers 1 and 4 to doors I and IV, respectively, and readers 2 and 3 to doors I + II and III + IV, respectively). Readers were specifically programmed to open/close the associated doors after the recognition of transponder numbers of choice. Free female movement (a) and male confinement to their own cage (b) were guaranteed by the following settings.
Figure 1.
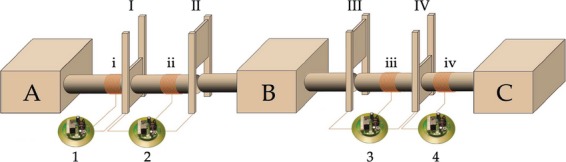
Scheme of choice device with the three cages (A–C), four doors (I–IV), four antennae (i–iv) and their corresponding readers (1–4). Males were confined to cages A and C, respectively. The female had access to all three cages.
In its initial state, inner doors (II and III) were open and outer doors (I and IV) were closed and the female was placed in her home cage (B). Inner antennae (ii and iii) only reacted to female transponders. When the female passed by an inner antenna, for example (ii), her transponder was read by the antenna device (2) and the door status was switched (the inner door II was closed and the outer door I was opened). The female could hence freely access a male cage (A in this example). The same process applied for the opposite direction and allowed the female to return to the middle cage (B).
Outer doors were only open when a female was visiting, entering or leaving. To prevent male escape, outer antenna (i and iv) were programmed specifically on male transponders, causing an immediate closing of the outer door as soon as the male was read by its antenna. The door remained closed as long as the male was within the reach of this antenna.
Experimental procedure
Females of both genetic backgrounds (t and w) were given the choice between a t male and a w male. Weight difference between males never exceeded 2 g, and no combination of males was used twice. Most males were used several times with different females. None of the individuals used in the same test were siblings. As we had five choice devices at our disposal, up to five tests could be run at the same time. All cages (A–C) were equipped with standard animal bedding, nesting material and ad libitum food and water. Choice tests were divided into two phases.
Priming
To habituate females to the device and to initiate oestrous, females were first put into the device without males in the side cages for 1 day. The side cages had previously housed the test males for 1 day and thus contained soiled bedding. Allocation of genotypes to positions A and C was randomized. Males were removed just before the start of the trial. In the presence of male urine, females initiate an oestrous cycle (Bronson, 1979). Wild house mice quickly learned to open doors by moving through the fields of the antennae.
Choice test
After the priming phase, males were put back into their cages and sides were swapped systematically. Male tests lasted 5–12 days so as to encompass at least one full oestrous cycle. An oestrous cycle typically lasts four days, with oestrous occurring during one night (Bronson, 1979). We did not use invasive assessments to determine oestrous (vaginal smears) to minimize disturbance and repeated handling of females.
At the end of the preference test, cage B was removed and used as the respective female's home cage, again to minimize handling of females. Females were daily checked for birth of a litter and offspring were counted when present. Tissue sampling of the pups for paternity analysis was done by an ear punch at the age of 13 days or when pups were found dead. This experiment was approved by the Veterinary Office Zürich, Switzerland (permit 97/2009).
Sample sizes
In total, we tested 31 t and 34 w females (see Table1 for an overview). Some females were tested again if previous tests did not result in offspring. Overall, we conducted 83 mate choice tests on 65 females.
Table 1.
Overview of choice test sample sizes
| t females | w females | total | |
|---|---|---|---|
| Females tested | 31 | 34 | 65 |
| Test repeats | 14 | 4 | 18 |
| Total number of tests | 45 | 38 | 83 |
| Females producing offspring | 19 | 15 | 34 |
| Behavioural data available | 16 | 20 | 36 |
| Offspring and behavioural data | 9 | 9 | 18 |
Paternity analysis
All offspring produced in the experiment, their mothers and the two candidate fathers of the relevant trials were genotyped at five neutral microsatellite loci in a single multiplex reaction (average He = 0.75, average number of alleles per locus was 6.2). If variation at these markers was insufficient to unambiguously assign paternity in any trial, an additional six neutral microsatellite loci were amplified in a second multiplex reaction. Paternity analyses were performed using maximum likelihood as implemented in Cervus 3.0 (Kalinowski et al., 2007). Paternity assignments were made at a confidence level of 95%.
Statistical analysis
Litter sizes
Litter sizes were analysed as a function of female genotype (t and w), their genetic fathers (three categories: t male, w male, and multiple paternity) and their interaction in a generalized linear model using an exponential link function and a Poisson error distribution.
Fertilization bias
The results of the paternity analyses were used to estimate fertilization bias, that is the proportion of offspring sired by the t male given female genotype i,Fi ∈ [0,1]. Any systematic deviation from no preference (Fi = 0.5) can be the result of pre- or post-copulatory mate choice. Hence, we were interested in two pieces of information: (i) The mean prediction 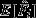 indicates whether there is a systematic deviation from no choice expectations (H0: Fi = 0.5). This information will not help to distinguish between pre- and post-copulatory choice processes. For this, we need to estimate (ii) variance
indicates whether there is a systematic deviation from no choice expectations (H0: Fi = 0.5). This information will not help to distinguish between pre- and post-copulatory choice processes. For this, we need to estimate (ii) variance
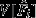 (see below for more details). However, a binomial generalized linear model (GLM) – the standard approach for this type of data – will not estimate the variance (because binomial errors directly follow from binomial parameters p and n). Therefore, we ran three custom null models using computer simulations. Each null model assumed a specific fertilization probability (see below), for which we derived the expected probability distribution of Fi (see Fig. 3b). From these expected distributions, we drew 105 values based on our sample sizes (nt = 19, nw = 15), giving us a population of realized mean E[Fi] and variance V[Fi] expectations. These expectations were compared to observed mean
(see below for more details). However, a binomial generalized linear model (GLM) – the standard approach for this type of data – will not estimate the variance (because binomial errors directly follow from binomial parameters p and n). Therefore, we ran three custom null models using computer simulations. Each null model assumed a specific fertilization probability (see below), for which we derived the expected probability distribution of Fi (see Fig. 3b). From these expected distributions, we drew 105 values based on our sample sizes (nt = 19, nw = 15), giving us a population of realized mean E[Fi] and variance V[Fi] expectations. These expectations were compared to observed mean 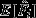 and variance
and variance 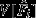 . Here is a short description of the three null models.
. Here is a short description of the three null models.
Null model 1: Precopulatory process only; no choice. In this null model, no polyandry occurs and females have no preference for either male genotype. All offspring will hence be sired completely by one of the two males, Fi for an individual female represents a Bernoulli trial with probability P = 0.5, i.e. Fi∼Bernoulli(P = 0.5).
Null model 2: Postcopulatory process only; no choice. Here, all females mate with both males at equal frequency, but t males do not suffer from sperm competition disadvantages. Fi hence follows a binomial distribution with Fi∼Binom(pi,n), where n denotes litter size and pi the fertilization probability of a t male in a female of genotype i. In w females, both males have equal chances of fertilization, that is pw = 0.5. In t females, however, Ft will be reduced as a proportion of sired zygotes will die during embryogenesis due to t/t recessive lethal effects. This proportion is a function of segregation distortion τ. For the mice used in this study, τ = 0.9 (Lindholm et al., 2013). We hence have pt = (1−τ/2)/(2−τ/2), as τ/2 t sired embryos perish in utero. Observed average litter size was
 and followed a Poisson distribution (mean-to-variance ratio
and followed a Poisson distribution (mean-to-variance ratio 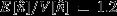 ). Thus, litter sizes n were drawn from a Poisson distribution with λ = 6.11.
). Thus, litter sizes n were drawn from a Poisson distribution with λ = 6.11.Null model 3: Postcopulatory processes only; t sperm disadvantage; no choice. In the third null model, females again mate invariably with both males, so again Fi∼Binom(pi,n). This time, however, t males do worse in sperm competition. If sperm number is an important determinant of a male's sperm competitive ability, t males are expected to be poor sperm competitors as a substantial fraction 1−1/(2τ) of their gametes are rendered dysfunctional by the distorter. Based on the difference in functional sperm number only, we expect pw = 1/(1+2τ) and pt = (2 − τ)/(τ/2 + 3τ) (Manser et al., 2011).
Figure 3.
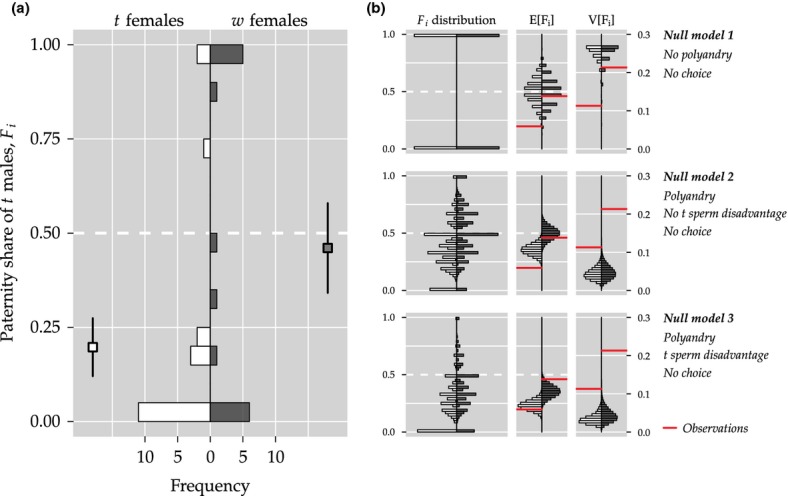
Observed (a) and expected (b) distributions of fertilization biases Fi for t (white) and w (dark-grey) females. Dashed horizontal lines represent no fertilization bias (Fi = 0.5). Squares in (a) depict mean observed values 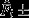 standard error of the mean (SEM). (b) Left panels show Fi distributions based on the three different null models. Centre and right panels show the resulting mean E[Fi] and variance V[Fi] expectations for 19 t and 15 w females (based on 105 draws). Observed values are shown as red horizontal lines.
standard error of the mean (SEM). (b) Left panels show Fi distributions based on the three different null models. Centre and right panels show the resulting mean E[Fi] and variance V[Fi] expectations for 19 t and 15 w females (based on 105 draws). Observed values are shown as red horizontal lines.
Due to the limited number of animals available, some males were used repeatedly across tests (on average, males were used 1.9 ± 1.1 times). Note that we had no repeated measures of Fi for females, as females were only tested again if the previous trial did not result in offspring. Our custom model approach did not allow us to account for the repeated use of males in our experiment. To account for this nonindependence, we additionally analysed fertilization biases Fi as a function of female genotype (t or w) in a generalized linear mixed effects model (GLMM) with a logit link function and a binomial error distribution, using t and w male identity separately as random effect variables.
Behavioural preference
Every time a transpondered animal was read by an antenna in the choice device system, a data string containing information about transponder number, time and origin was sent to a computer and logged with a specific program (Advanced Serial Data Logger, AGG Software, Kolchugino, Russia). The positional information was used to assess female behavioural preference. An entry into one of the male cages was scored if a female log entry of the inner antenna was followed by a record of the outer antenna. An exit was scored in case of the opposite order. With this information in hand, the duration of female visits in the t and w males cage was determined. The analysis of the visit duration was only started after the female had visited both male cages at least once. Behavioural preference of a female of genotype i was defined as
where Tt and Tw is the total time a female spent in the t and w male cage, respectively. Observed behavioural preferences 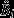 were logit-transformed (Warton & Hui, 2011) and analysed as a function of female genotype using a linear mixed effects model (with H0: Bi = 0.5). To account for the repeated use of males, t and w male identity were used as seperate random effect variables. There was no repeated measure of Bi for females.
were logit-transformed (Warton & Hui, 2011) and analysed as a function of female genotype using a linear mixed effects model (with H0: Bi = 0.5). To account for the repeated use of males, t and w male identity were used as seperate random effect variables. There was no repeated measure of Bi for females.
Influence of behavioural preference on fertilization biases
In order to find out if behavioural preference Bi were predictive for t paternity shares Fi, we analysed Fi as a function of female genotype i and behavioural preference Bi and their interaction. In contrast to the analysis above, we used an arcsine-square-root transformation of the response variable here (as Fi-values of 0 and 1 transform to undefined values −∞ and ∞, respectively, when logit-transformed). The full model was reduced to the minimal adequate model using likelihood-ratio tests.
Data processing, statistical analysis and computer simulations were carried out in R 3.0.0 for Mac (R Core Team, 2014) lme4 package (Bates et al., 2014).
Results
Litter sizes
34 of 83 mate choice tests (41%) resulted in offspring. Average litter size was 6.11 ± 2.21 (mean ± SD). In t females, litter sizes were significantly lower when mating with a t male than a w male (95% CI for difference between t and w sired litters: [1.24,5.94], z = 2.17, P = 0.030; see Fig. 2). Multiply sired litters on the other hand were not reduced in litter sizes (95% CI for difference between w and multiply sired litters: [−2.04,2.83], z = 0.63, P = 0.528). In w females, litter sizes were also smaller when t sired, but this was not statistically significant (95% CI w vs. t sired: [−0.76,4.11], z = 1.52, P = 0.129; 95% CI w vs. multiply sired: [−4.43,2.55], z = −0.10, P = 0.922).
Figure 2.
Litter sizes ± standard errors of the mean (SEM) as a function of mother genotype (two panels) and the paternity of the litter, that is the genotype of the genetic father of the litter (either exclusively w, exclusively t, or both). The surface of the circles is proportional to the number of observations.
Fertilization biases
Figure 3a shows distributions of observed paternity biases Fi for t and w females. 3 shows the expected distributions of Fi based on the three null model simulations as well as distributions of mean E(Fi) and variance V(Fi) predictions for the given sample sizes (nt = 19, nw = 15) resulting from 105 simulation runs. Table2 summarizes the comparisons between the observed data and expectations of the null model simulations. As mentioned in the Methods, mean predictions may help us understand possible deviations from no choice expectations, whereas variance predictions may help to distinguish between pre- and postcopulatory processes.
Table 2.
Observed mean and variance of t paternity shares ( ,
, 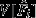 ) in comparison to the three null model expectations.
) in comparison to the three null model expectations.
|
t females |
w females |
|||
|---|---|---|---|---|
| E[Ft] | V[Ft] | E[Fw] | V[Fw] | |
| Observations | 0.20 | 0.11 | 0.46 | 0.21 |
| Null model 1 | Precopulatory processes only; no choice | |||
| Ft∼Bernoulli (P = 0.5) | Fw∼Bernoulli (P = 0.5) | |||
| 0.50 | 0.25 | 0.50 | 0.25 | |
| P–value | 0.020 | < 0.001 | 0.607 | 0.119 |
| Null model 2 | Postcopulatory processes only; no choice | |||
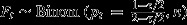 |
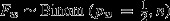 |
|||
| 0.36 | 0.044 | 0.50 | 0.049 | |
| P–value | 0.001 | < 0.001 | 0.489 | < 0.001 |
| Null model 3 | Postcopulatory processes only; t sperm disadvantage; no choice | |||
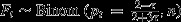 |
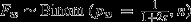 |
|||
| 0.23 | 0.035 | 0.36 | 0.045 | |
| P–value | 0.393 | < 0.001 | 0.059 | < 0.001 |
Mean predictions: The observed t females’ paternity bias
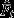 deviates significantly from ’no choice’ expectations, either solely based on precopulatory (null model 1) or postcopulatory processes (null model 2).
deviates significantly from ’no choice’ expectations, either solely based on precopulatory (null model 1) or postcopulatory processes (null model 2). 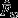 , on the other hand, do not deviate from ’no choice’ expectations (null models 1 and 2). This leaves null model 3 as the only one fully compatible with mean
, on the other hand, do not deviate from ’no choice’ expectations (null models 1 and 2). This leaves null model 3 as the only one fully compatible with mean 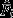 observations.
observations.Variance predictions: Predicted variance patterns strongly differ between the different null models: null model 1, which assumes precopulatory processes only, predicts substantially higher variances in Fi than the null models 2 and 3, which are based on postcopulatory processes. A comparison to the observed data may therefore help us to distinguish between the two processes. In fact, none of the null models is in line with the observed variance patterns, the observed variance lies between the low variance predictions of null model 2 and 3 and the high variance predictions of null model 1.
The conclusions are unaffected when we account for the repeated use of males across tests. The mixed effects model (GLMM) confirmed that Ft was significantly different from no choice expectations (z = −2.49, n = 34, P = 0.013). Fw, on the other hand, did not deviate from no choice expectations (z = −1.04, n = 34, P = 0.300). The data were not overdispersed (dispersion parameter: 0.89).
Behavioural preference
The software recording of female behaviour was available for 71 choice tests, of which data from 36 tests had to be discarded due to technical problems in the recording of the data (Table 1). For the remaining 35 tests, a total 6 414 744 log entries accumulated. Females visited either male cage on average 84.4 ± 44.8 (mean ± SD) times per day. Average visit duration was 9.52 ± 9.48 min. They spent 37.27 ± 17.14% of their time in a male's cage (see also Figure S1). Figure 4a shows behavioural preferences for the t male 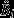 for t and w females. According to the linear mixed effects model using a logit-transformation, the 95% confidence bands of
for t and w females. According to the linear mixed effects model using a logit-transformation, the 95% confidence bands of  did not fall outside the no choice predictions (Bi = 0.5) both in t females (95% CI: [0.10, 0.64]) and w females (95% CI: [0.16, 0.75]).
did not fall outside the no choice predictions (Bi = 0.5) both in t females (95% CI: [0.10, 0.64]) and w females (95% CI: [0.16, 0.75]).
Figure 4.
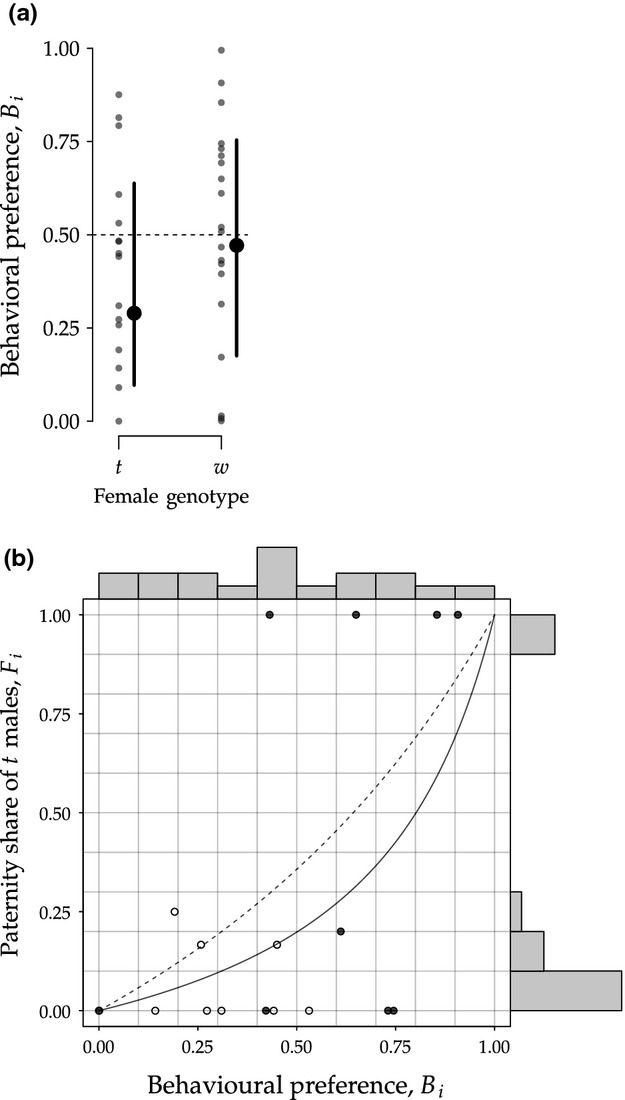
(a) Behavioural preference for t males Bi as a function of maternal genotype including mean and 95% confidence interval estimates. The dashed horizontal line depicts the null hypothesis, that is no choice (Bi = 0.5). (b) Paternity share of t males Fi as a function of behavioural preference Bi. The colour of the dots represent female genotype (white: t, dark-grey: w). The histograms at the figure margins depict the distribution of the data in both x- and y-direction, illustrating that nearly uniformly distributed behavioural preference Bi translate into clear fertilization biases Fi. Dotted and solid lines show expected paternity shares for t and w females, respectively, based on null model 3. It is assumed that a female's behavioural preference Bi is proportional to the number of matings as well as the number of competing sperm of a given male. For example, if Bi = 0.5, both males contribute equally to the competing sperm pool. However – according to null model 3 – a proportion 1−1/(2τ) of a t male's sperm is dysfunctional.
Influence of behavioural preference on fertilization biases
For 9 t and 9 w females, estimates for both behavioural preference Bi and genetic preference Fi were available. According to the model selection procedure using likelihood-ratio tests, behavioural preference Bi was significantly associated with t paternity shares Fi (see Fig. 4b, t15 = 2.46, P = 0.026). Female genotype, on the other hand, was removed by model selection.
Discussion
This study showed that (i) genetic incompatibilities arising from the t haplotype had severe indirect fitness consequences and (ii) t females avoided fertilization by t-locus incompatible males. This is the first experimental evidence to show that t females avoid t incompatible males in an actual mating context. The results are inconclusive whether this avoidance of t fertilization by t females was caused by pre- or postcopulatory processes.
The cost of genetic incompatibility
We found a severe litter size reduction of about 60% if t females mated with genetically incompatible t males rather than a compatible w male (Fig. 2). However, this estimate is based on a very low sample size (as a consequence of free partner choice, only two females had completely t-sired offspring). In a previous experimental study on the same mouse population, litter sizes were reduced by 40% (Lindholm et al., 2013). In that study, examination of uterine scars confirmed that this reduction is a result of prenatal mortality. The difference in litter size reduction to the present study is likely a consequence of sampling error, that is the low sample size in the present study. w females also suffered from a slight reduction in litter size when mating with t males, but this reduction was not statistically significant (P = 0.13). Based on a larger sample size, Lindholm et al. (2013) found a similar, nonsignificant trend towards reduced t male fertility. This trend would suggest a fertility difference between t and w males, which has been reported previously (Lenington et al., 1994; Ardlie & Silver, 1998; Carroll et al., 2004). In contrast to Carroll et al. (2004), we found no indication of reduced t female fertility, both in this study and Lindholm et al. (2013). Furthermore, we show that females can avoid litter losses arising from genetic incompatibility by mating with both males (polyandry): litter sizes of multiply sired litters did not differ from litters exclusively sired by w males. This result suggests that t males are poor sperm competitors (see also below). Overall, litter size results nicely outline the two possible female strategies to avoid fertilization by incompatible t males, that is maximize litter size: females can either avoid t males prior to mating or mate multiply and rely on sperm competition to reduce t fertilization.
Mate choice for genetic compatibility
The litter size results highlight the severe selective pressure on t females to avoid fertilization by genetically incompatible t males. Indeed, we found that t females successfully avoided t male paternity when given free choice between a t and a w male. Only 20% of all t female offspring were sired by the t male. This proportion was significantly different from no choice expectations of scenario 1 (50%) and scenario 2 (36%). There is ample experimental evidence for olfactory preference for w males (Lenington et al., 1992). Our study, for the first time, provides experimental evidence that t females avoid t male fertilization in an actual mating context.
In w females, on the other hand, t paternity shares did not deviate from the random 50% expectations. This seems surprising, given that discriminating against t males would help w females to avoid producing sons with impaired sperm competitive ability and/or low attractiveness to females. It is, however, difficult to assess whether t avoidance is beneficial without knowing the precise underlying cost/benefit structure of the behaviour. Let bi be the benefits of avoiding t males, where i ∈ [t, w] denotes female genotype. Because of genetic incompatibility, we have bt > bw > 0. Let us further assume that avoiding t males is associated with a genotype-independent cost c (cost of preference or multiple mating). Even though t avoidance is beneficial for females of both genotypes (bi > 0), the evolutionary relevant question is whether the overall pay-off of t avoidance, that is bi − c, is positive. Now, the asymmetry in fitness consequences for t and w females (bt ≠ bw) may well be adaptively meaningful. Overall, pay-offs depend on the relative magnitude of costs c. If c < bw < bt, t male avoidance is beneficial for both female genotypes (likewise, if c > bt > bw, it is detrimental to both). There is, however, also the possibility that bt > c > bw, in which case t avoidance is adaptive for t females (as bt − c > 0), but not for w females (as bw − c < 0). More precise theoretical models are clearly necessary to investigate whether such a scenario can be evolutionarily stable and whether there are genetic mechanisms that could maintain such systematic preference differences between t and w females.
Pre- or postcopulatory processes?
The observed fertilization bias in t females reported here can principally be the result of both pre- and/or postcopulatory processes. We did not systematically observe matings during preference tests. Instead, we used two indirect approaches to investigate whether the observed fertilization bias in t females are the result of pre- or postcopulatory mechanisms.
First, we compared observed fertilization distributions against specific, customized null models. Despite the fact that not all multiple matings will result in multiple paternity, we expected a systematic variance difference between a scenario where all females mate with one male only (null model 1) and a scenario where all females mate multiply (null models 2 and 3). Observed variance estimates fell between these extreme scenarios, suggesting that both pre- and postcopulatory processes are important here. Overall, the multiple paternity rate in the experiments was 29%. This value is consistent with previous estimates on house mice (Dean et al., 2006; Firman & Simmons, 2008a; Manser et al., 2011) and confirms that female mice are actively polyandrous. It is unknown whether t and w females differ systematically in levels of polyandry. The low sample sizes did not allow for a systematic test for such a difference here. In terms of mean predictions, scenario 3 (assuming full polyandry and t sperm disadvantage) was most compatible with our data. This indicates, as suggested previously (Olds-Clarke & Peitz, 1986; Ardlie & Silver, 1996), that t males are poor sperm competitors. However, we could not quantify the fraction of males that mated with a female, but did not successfully sire offspring. The expected mean distribution of null model 3 (in Fig. 3b) shows that the fraction of such unsuccessful t males can be quite large. Another class of a postcopulatory mechanism by which females could bias fertilization towards w males is cryptic female choice (Eberhard, 1996). Our experimental design did not allow disentanglement of intramale sperm competition from cryptic female choice and our results are compatible with one and/or the other. Controlled sperm competition experiments are clearly needed for reliable estimates of t sperm disadvantage as well as to identify the precise mechanisms that determine fertilization success.
Second, we analysed female behaviour during the preference tests to investigate the importance of precopulatory preference. The variation in visiting preference both within (over the course of an experiment, see Figure S1) and between females was substantial. Overall, we did not find any indication that t females avoided t males prior to mating. In fact, most females actively visited both males over the course of an experiment (see Figure S1). The lack of clear precopulatory preferences is surprising given the series of studies repeatedly reporting t female olfactory preferences for w males (Lenington et al., 1992). It remains unclear whether females of our population are able to distinguish between t and w conspecifics. t haplotypes carry several MHC loci that could principally serve as indicators of male t status. Lindholm et al. (2013) found that t haplotypes were – as a consequence of recombination suppression – associated with a single, unique MHC allele. However, the role of MHC in mate choice remains controversial, both related to t haplotypes (Lenington, 1991) and in general (Cheetham et al., 2007). In a very similar experiment, Rolland et al. (2003) gave females the choice between dominant and subordinate males. The resulting behaviour was analogous to the one observed here. Females actively visited and mated with both males. The authors argued that the preference for the preferred male (in this case the dominant male) was only apparent in a narrow time window during oestrous. Additionally, they found that females accepted more intromissions from preferred males and mated last with the preferred males. We cannot exclude such subtle differences in female behaviour towards t versus w males here. Based on a limited sample size, we did find a weak correlation between female behavioural preference throughout the preference tests and the resulting fertilization bias. However, the fact that female visiting patterns are, to some degree, predictive of paternity outcomes, is by no means an indicator of precopulatory choice as long as there are no clear behavioural preferences. In any case, the fact that nearly uniformly distributed behavioural preferences translate into clear fertilization biases (see histograms in Fig. 4b) makes multiple mating and sperm competition the likelier candidates to drive the observed t fertilization bias. The fact that null model 3 best describes the distribution of t paternity shares (discussed above) confirms this conjecture. Having said that, the data do not allow us to categorically rule out precopulatory choice.
The evolutionary forces that determine t frequencies in wild populations have puzzled biologists for more than half a century (Ardlie & Silver, 1998). It is largely unknown whether the fertilization bias observed here can be an important force for t frequencies in wild populations (Burt & Trivers, 2006). Female mating decisions in wild populations may not be as unconstrained as in our experiments here. If the dominant male of her territory is a t-carrier, a female may have to mate with him to avoid infanticide (Perrigo et al., 1991). Male dominance is an unlikely factor to explain female mating patterns here, as male territories did not overlap in our experimental setup (i.e. males could not establish a dominance hierarchy). Previous studies show that preference for dominance plays an even larger role in female mate choice decisions (Coopersmith & Lenington, 1992). Nevertheless, female mating behaviour may still be important in t suppression (Manser et al., 2011). Frequencies in wild populations are typically at low, but stable levels (Ardlie & Silver, 1998). In our wild study population, of which all mice used in the present experiments originated, t frequency has decreased significantly over a period of 5.5 years. Female mating behaviour can potentially explain this decrease (Manser et al., 2011) and paternity analyses indeed revealed a weak, but significant t female mate choice bias towards t males (Lindholm et al., 2013). We hence have good indications that female mating behaviour may play an important role in suppressing t haplotypes in wild house mice. However, further analyses are necessary to specifically quantify the importance of pre- and/or postcopulatory processes in drive suppression.
In conclusion, the present study provides further evidence that genetic incompatiblities caused by SGEs, usually hidden from sight, may be an important driver of the evolution of female mating behaviour. It has been suggested that SGEs are a ubiquitous feature of life (Burt & Trivers, 2006). This study not only shows that female mating behaviour may play an important role in SGE-suppression, but also illustrates how the covert action of SGEs may help us understand important aspects of an organism's behaviour that may remain unexplained otherwise.
Acknowledgments
We thank Jari Garbely for carrying out the genetic analyses, Gabi Stichel for taking care of the laboratory stock of our wild mice. We further thank Marcel Freund, Hans-Jörg Baumann and Fabian Vögelin who were of invaluable help in setting up the mate choice device. Many thanks to Stefanie Karrer, Fabian Vögelin, Philip Wadewitz, Corinne Schnellmann, Anja Stettin and Jennifer Kappeler for their contributions and assistance in data collection. We further thank Andreas Sutter, Manuela Ferrari, Luke Holman and an anonymous reviewer for their helpful comments on previous versions of this manuscript. This study was funded by the University of Zürich, the Forschungskredit of the University of Zürich and the Swiss National Science Foundation (SNF: 310030M—138389).
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1 Visiting patterns of the 36 females for which behavioural data was available.
References
- Andersson M. Sexual Selection. Princeton: Princeton University Press; 1994. [Google Scholar]
- Ardlie K. Silver L. Low frequency of mouse t haplotypes in wild populations is not explained by modifiers of meiotic drive. Genetics. 1996;144:1787–1797. doi: 10.1093/genetics/144.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardlie K. Silver L. Low frequency of t haplotypes in natural populations of house mice (Mus musculus domes ticus. Evolution. 1998;52:1185–1196. doi: 10.1111/j.1558-5646.1998.tb01844.x. [DOI] [PubMed] [Google Scholar]
- Atlan A, Joly D, Capillon C. Montchamp-Moreau C. Sex-ratio distorter of Drosophila simulans reduces male productivity and sperm competition ability. J. Evol. Biol. 2004;17:744. doi: 10.1111/j.1420-9101.2004.00737.x. [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. Lme4: Linear mixed-effects models using Eigen and S4. 2014. R package version 1.1-7. http://CRAN.R-project.org/package=lme4. [Google Scholar]
- Bronson F. The reproductive ecology of the house mouse. Q. Rev. Biol. 1979;54:265–299. doi: 10.1086/411295. [DOI] [PubMed] [Google Scholar]
- Burt A. Trivers R. Genes in Conflict: The Biology of Selfish Genetic Elements. Cambridge: Belknap Press; 2006. [Google Scholar]
- Butlin R. Ritchie M. Genetic coupling in mate recognition systems: what is the evidence. Biol. J. Linn. Soc. 1989;37:237–246. [Google Scholar]
- Carroll L, Meagher S, Morrison L, Penn D. Potts W. Fitness effects of a selfish gene (the Mus t complex) are revealed in an ecological context. Evolution. 2004;58:1318–1328. doi: 10.1111/j.0014-3820.2004.tb01710.x. [DOI] [PubMed] [Google Scholar]
- Cheetham SA, Thom MD, Jury F, Ollier WE, Beynon RJ. Hurst JL. The genetic basis of individual-recognition signals in the mouse. Curr. Biol. 2007;17:1771–1777. doi: 10.1016/j.cub.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Coopersmith C. Lenington S. Female preferences based on male quality in house mice: interaction between male dominance rank and t-complex genotype. Ethology. 1992;90:1–16. [Google Scholar]
- Cotton A, Földvári M, Cotton S. Pomiankowski A. Male eyespan size is associated with meiotic drive in wild stalk-eyed flies (Teleopsis dalmanni. Heredity. 2014;112:363–369. doi: 10.1038/hdy.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Ardlie K. Nachman M. The frequency of multiple paternity suggests that sperm competition is common in house mice (Mus domesticus. Mol. Ecol. 2006;15:4141–4151. doi: 10.1111/j.1365-294X.2006.03068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard WG. Female Control: Sexual Selection by Cryptic Female Choice. Princeton: Princeton University Press; 1996. [Google Scholar]
- Firman R. Simmons L. The frequency of multiple paternity predicts variation in testes size among island populations of house mice. J. Evol. Biol. 2008a;21:1524–1535. doi: 10.1111/j.1420-9101.2008.01612.x. [DOI] [PubMed] [Google Scholar]
- Firman R. Simmons L. Polyandry, sperm competition, and reproductive success in mice. Behav. Ecol. 2008b;19:695–702. [Google Scholar]
- Fisher R. The Genetical Theory of Natural Selection. Oxford: Clarendon; 1930. [Google Scholar]
- Haig D. Bergstrom C. Multiple mating, sperm competition and meiotic drive. J. Evol. Biol. 1995;8:265–282. [Google Scholar]
- Hammer M, Schimenti J. Silver L. Evolution of mouse chromosome 17 and the origin of inversions associated with t haplotypes. Proc. Natl Acad. Sci. 1989;86:3261–3265. doi: 10.1073/pnas.86.9.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski ST, Taper ML. Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- Klein J, Sipos P. Figueroa F. Polymorphism of t-complex genes in European wild mice. Genet. Res. 1984;44:39–46. [Google Scholar]
- König B. Lindholm A. The complex social environment of female house mice (Mus domesticus. In: Miloš Macholán, Stuart J. E Baird, Pavel Munclinger, Jaroslav Piálek., editors; Evolution of the House Mouse. Cambridge, UK: Cambridge University Press; 2012. pp. 114–134. ed.). &. In: [Google Scholar]
- Kotiaho J, LeBas N, Puurtinen M. Tomkins J. On female choice, heterozygosity and the lek paradox. Anim. Behav. 2008;75:1–3. [Google Scholar]
- Lenington S. Social preferences for partners carrying ’good genes’ in wild house mice. Anim. Behav. 1983;31:325–333. [Google Scholar]
- Lenington S. The t complex: a story of genes, behavior, and populations. Adv. Study Behav. 1991;20:51–86. [Google Scholar]
- Lenington S. Egid K. Female discrimination of male odors correlated with male genotype at the t locus: a response to t-locus or H-2-locus variability. Behav. Genet. 1985;15:53–67. doi: 10.1007/BF01071932. [DOI] [PubMed] [Google Scholar]
- Lenington S, Coopersmith C. Williams J. Genetic basis of mating preferences in wild house mice. Integr. Comp. Biol. 1992;32:40–47. [Google Scholar]
- Lenington S, Coopersmith C. Erhart M. Female preference and variability among t haplotypes in wild house mice. Am. Nat. 1994;143:766–784. [Google Scholar]
- Lindholm AK, Musolf K, Weidt A. König B. Mate choice for genetic compatibility in the house mouse. Methods Ecol. Evol. 2013;3:1231–1247. doi: 10.1002/ece3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M. Transmission ratio distortion in mice. Annu. Rev. Genet. 2003;37:393–408. doi: 10.1146/annurev.genet.37.110801.143030. [DOI] [PubMed] [Google Scholar]
- Manser A, Lindholm AK, König B. Bagheri HC. Polyandry and the decrease of a selfish genetic element in a wild house mouse population. Evolution. 2011;65:2435–2447. doi: 10.1111/j.1558-5646.2011.01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mays HLJr. Hill GE. Choosing mates: good genes versus genes that are a good fit. Trends Ecol. Evol. 2004;19:554–559. doi: 10.1016/j.tree.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Miller WJ, Ehrman L. Schneider D. Infectious speciation revisited: impact of symbiont-depletion on female fitness and mating behavior of Drosophila paulistorum. PLoS Pathog. 2010;6:e1001214. doi: 10.1371/journal.ppat.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olds-Clarke P. Peitz B. Fertility of sperm from t/+ mice: evidence that +-bearing sperm are dysfunctional. Genet. Res. 1986;47:49–52. doi: 10.1017/s0016672300024502. [DOI] [PubMed] [Google Scholar]
- Perrigo G, Belvin L. Vom Saal F. Individual variation in the neural timing of infanticide and parental behavior in male house mice. Physiol. Behav. 1991;50:287–296. doi: 10.1016/0031-9384(91)90068-y. [DOI] [PubMed] [Google Scholar]
- Price T. Wedell N. Selfish genetic elements and sexual selection: their impact on male fertility. Genetica. 2008;134:99–111. doi: 10.1007/s10709-008-9253-y. [DOI] [PubMed] [Google Scholar]
- Price T, Bretman A, Avent T, Snook R, Hurst G. Wedell N. Sex ratio distorter reduces sperm competitive ability in an insect. Evolution. 2008a;62:1644–1652. doi: 10.1111/j.1558-5646.2008.00386.x. [DOI] [PubMed] [Google Scholar]
- Price T, Hodgson D, Lewis Z, Hurst G. Wedell N. Selfish genetic elements promote polyandry in a fly. Science. 2008b;332:1241–1243. doi: 10.1126/science.1163766. [DOI] [PubMed] [Google Scholar]
- Puurtinen M, Ketola T. Kotiaho JS. Genetic compatibility and sexual selection. Trends Ecol. Evol. 2005;20:157–158. doi: 10.1016/j.tree.2005.02.005. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. http://www.R-project.org/ [Google Scholar]
- Rolland C, MacDonald D. Berdoy M. Free female choice in house mice: leaving best for last. Adv. Study Behav. 2003;140:1371–1388. [Google Scholar]
- Rüsch E. 2002. University of Zurich Ein standartisierter für Präferenztest bei Hausmäusen (Mus domesticus). Master's thesis,
- Silver L. The peculiar journey of a selfish chromosome: mouse t haplotypes and meiotic drive. Trends Genet. 1993;9:250–254. doi: 10.1016/0168-9525(93)90090-5. [DOI] [PubMed] [Google Scholar]
- Tregenza T. Wedell N. Genetic compatibility, mate choice and patterns of parentage: invited review. Mol. Ecol. 2000;9:1013–1027. doi: 10.1046/j.1365-294x.2000.00964.x. [DOI] [PubMed] [Google Scholar]
- Trivers RL. Parental investment and sexual selection. In: Campbell B, editor. Sexual Selection and the Descent of Man. Chicago: Aldine Publishing Company; 1972. pp. 136–179. , ed.). In: [Google Scholar]
- Vala F, Egas M, Breeuwer J. Sabelis M. Wolbachia affects oviposition and mating behaviour of its spider mite host. J. Evol. Biol. 2004;17:692–700. doi: 10.1046/j.1420-9101.2003.00679.x. [DOI] [PubMed] [Google Scholar]
- Warton DI. Hui FK. The arcsine is asinine: the analysis of proportions in ecology. Ecology. 2011;92:3–10. doi: 10.1890/10-0340.1. [DOI] [PubMed] [Google Scholar]
- Wedell N. The dynamic relationship between polyandry and selfish genetic elements. Philos. Trans. R. Soc. B Biol. Sci. 2013;368:1–10. doi: 10.1098/rstb.2012.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson G, Presgraves D. Crymes L. Male eye span in stalk-eyed flies indicates genetic quality by meiotic drive suppression. Nature. 1998;391:276–279. [Google Scholar]
- Williams J. Lenington S. Factors modulating preferences of female house mice for males differing in t-complex genotype: role of t-complex genotype, genetic background, and estrous condition of females. Behav. Genet. 1993;23:51–58. doi: 10.1007/BF01067553. [DOI] [PubMed] [Google Scholar]
- Zeh J. Zeh D. The evolution of polyandry I: intragenomic conflict and genetic incompatibility. Proc. R. Soc. B Biol. Sci. 1996;263:1711–1717. [Google Scholar]
- Zeh J. Zeh D. The evolution of polyandry II: post-copulatory defences against genetic incompatibility. Proc. R. Soc. B Biol. Sci. 1997;264:69–75. [Google Scholar]
- Zeh J. Zeh D. Toward a new sexual selection paradigm: polyandry, conflict and incompatibility. Ethology. 2003;109:929–950. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Visiting patterns of the 36 females for which behavioural data was available.


