Abstract
We have achieved protein imaging mass spectrometry capabilities at sub-cellular spatial resolution and at high acquisition speed by integrating a transmission geometry ion source with time of flight mass spectrometry. The transmission geometry principle allowed us to achieve a 1 μm laser spot diameter on target. A minimal raster step size of the instrument was 2.5 μm. Use of 2,5-dihydroxyacetophenone robotically sprayed on top of a tissue sample as a matrix together with additional sample preparation steps resulted in single pixel mass spectra from mouse cerebellum tissue sections having more than 20 peaks in a range 3–22 kDa. Mass spectrometry images were acquired in a standard step raster microprobe mode at 5 pixels/s and in a continuous raster mode at 40 pixels/s.
Keywords: MALDI, transmission geometry, high spatial resolution, MS imaging, protein imaging
Introduction
Imaging mass spectrometry is a mature technology that produces ion maps or images from the direct desorption of molecules from cells in tissues [1]. Spatial resolutions of up to about 20 μm can be achieved routinely by standard commercial instruments that ablate cellular material using lasers that irradiate the topside of the tissue. For laser spot sizes on target of 5 μm or less, we have developed a new ion source for the imaging mass spectrometry (IMS) termed transmission geometry that irradiates the tissue from the backside.
A transmission geometry MALDI source has multiple advantages in imaging applications. It allows high spatial resolution with laser beam sizes on target approaching the wavelength of the laser. In the ion source, the ion optics is spatially separated from the laser optics, which opens possibilities to optimize both independently to achieve high sensitivity. The transmission geometry LDI source for non-imaging MS applications was initially introduced by Fenner and Daly [2], improved by Hillenkamp group [3] and implemented in LAMMA 500 instrument (Leybold-Heraeus GmbH, Ke1n, Germany) [4]. A sub-micron laser spot size was achieved in this scheme at 266 nm wavelength. Although playing important role in establishing the transmission geometry LDI sources, this setup was not readily applicable for imaging applications. There were also several reports of using the laser focusing optics inside the vacuum chamber by employing reflective optics in a Cassegrain scheme, providing a longer working distance, for example LIMA instrument (Cambridge Mass Spectrometry, Cambridge, UK) [5]. The reflective objectives however introduce additional diffraction limits and aberrations and the minimum laser spot sizes are larger, for example 1–3 μm at 266 nm wavelength in LIMA-2A [6] and were useful only for profiling applications. For imaging MS applications, a transmission geometry vacuum MALDI source was built in Caprioli laboratory [7,8] on a modified AB4700 MALDI instrument (Applied Biosystems/Thermo Fisher Scientific, Waltham, MA, USA). The laser focusing microscope objective in this source is placed in vacuum, although a residual part of the optical scheme is mounted outside of the sample chamber [7]. Its advantages for submicron spatial resolution MALDI IMS at 349 nm laser wavelength have been demonstrated for various tissue types and single cells [7]. In the current work, we have integrated a transmission geometry source for IMS based on the similar principles into a new generation TOF instrument that has advanced delayed ion acceleration, high laser repetition rates, and continuous laser raster sampling [9,10].
In conjunction with the instrument modifications, we have significantly improved protein sample preparation by using 2,5-dihydroxyacetophenone (2,5-DHA) as a matrix for protein imaging for the first time. This matrix provides a boost in sensitivity for proteins and also stable under normal instrument vacuum conditions.
Materials and Methods
Instrument modification
We constructed a new transmission geometry MALDI source and incorporated it into a linear MALDI TOF MS SimulTOF Combo 200 instrument (SimulTOF Systems/Virgin Instruments, Sudbury, MA, USA). The main advantages of the Simultof MALDI mass spectrometer are speed and sensitivity. Speed is achieved by employing a 5 kHz pulse repetition rate laser, model Explorer 349 from Spectra-Physics/Newport (Irvine, CA, USA). Complementing the laser with a custom made sample stage capable of velocities up to 20 mm/s, and a high-speed digitizer with on-board signal processing (model Acqiris U1082A-001, Keysight Technologies, Inc., Santa Rosa, CA, USA), highly efficient Continuous Laser Raster Sampling (CLRS) is possible. The system is capable of acquisition rates up to 100 spectra/s with on the fly peak detection and automatic internal calibration, ideal for imaging MALDI MS applications. A sensitivity boost is possible with state of the art gridless ion optics allowing ions generated at the source to be detected efficiently at the detector. A high voltage pulsed set of parallel plates are employed to reject highly abundant low mass ions from reaching the detector while simultaneously discriminating neutrals generated, a result of the ionization process.
A high mass bipolar detector (Photonis USA, Inc., Sturbridge, MA, USA) employs 3 gain stages. The 1st stage is a microchannel plate (MCP) operating from ground with a bias up to 1000 V. The 2nd stage is a fast scintillator operating up to 20 kV. The 3rd stage is a fast photomultiplier tube (PMT) operating down to negative 1000 V. Incoming ions strike the MCP generating an electron shower. The energetic electrons strike the scintillator, and because the light output from the scintillator is proportional to the energy lost to electronic collisions, high isolation voltages translate into intense flashes of light at the output. The PMT detects these flashes of light, amplifies and produces a high current output pulse at ground potential regardless of the voltage setting at the front of the detector [11]. Operating the MCP at low voltage and taking advantage of the high output current of the PMT high gain and dynamic range, saturation effects of highly abundant low mass ions are minimized. High light output for single-ion pulses far exceeds system and digitizer electronic noise relative to thermionic emission of electrons from the PMT photocathode. The net result is a sensitive high gain high dynamic range detector, ideal for protein tissue imaging.
The modified delayed ion extraction does not facilitate the detection of proteins independently. The geometry of ion source in conjunction with the downstream ion optics complemented with the high mass bi-polar detector facilitates the detection of proteins. The geometry of the ion source is optimized for space and velocity focus at the plane of the detector while the aperture sizes and the length of the 2nd stage of the ion source is optimized for the spatial focusing conditions at the detector. Einzel lenses are employed downstream of the ion source to further optimize spatial focusing conditions of the ion beam. The specific matrix used has an influence on the limitations on resolving power of high mass proteins and analytes, compared to space and velocity focusing. Different matrices impact ionization behavior, formation of adducts, stability, or fragmentation of analytes. Another consideration in MALDI IMS applications could be the influence or interference of the tissue itself.
The original translation stage in the instrument provided a 2.5 μm minimal step size in the XY plane and was open frame type, typically used in inverted optical microscopes. The opening in the center of the stage was made large enough to incorporate the microscope objective underneath the target in order to raster an entire standard microscope slide (75 mm × 25 mm × 1 mm). The commercial frontside geometry source provided a 50 μm laser spot diameter on the target for use in comparative studies. The instrument has 2 modes of MS imaging: a step raster mode and a continuous raster mode. The step raster mode, called a “drill mode” in the instrument control software operates in a conventional MS microprobe mode, i.e. the XY stage is moved to a specific coordinate on the sample, stops, the sample is ablated by the laser firing a number of shots, and the mass spectrum is acquired. In contrast, in the continuous raster mode, the XY stage moves without stopping for each pixel, the laser fires continuously, and the MS spectra are collected continuously. A pixel is therefore defined by the laser repetition rate and the speed of movement of the sample stage. A comparison of these two modes of MS imaging for the instrument has been reported [9,12]. In the current work, both modes were used.
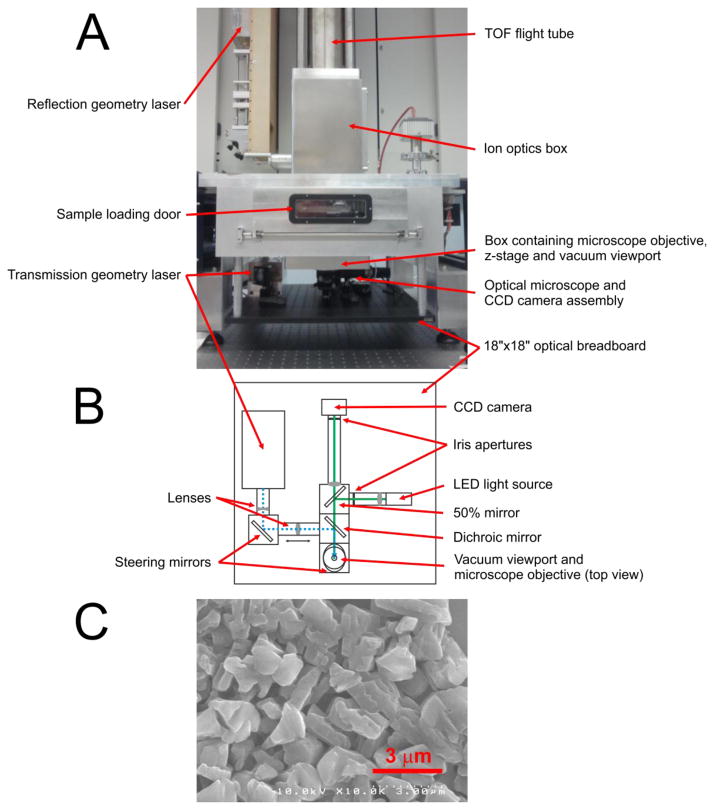
A transmission geometry ion source was installed based on our modified earlier design [7]. An inverted optical microscope was built into the source for monitoring the ablation process and the laser spot size on target. A photograph of the SimulTOF 200 Combo instrument with the transmission geometry assembly installed underneath the sample chamber is shown in Fig. 1. Panel A of the Figure shows a front view of the instrument and panel B shows a scheme of the optical path and elements of the transmission geometry assembly. The setup was built on an 18″×18″ optical breadboard using opto-mechanical elements from Thorlabs Inc. (Newton, NJ, USA). Laser pulses at 349 nm wavelength were generated by the third harmonic of a Nd:YLF separate laser, model Explorer 349 from Spectra-Physics/Newport (Irvine, CA, USA), the same model as the original instrument laser. Both lasers were controlled through the instrument software. The laser pulses had a duration < 5 ns at up to 5 kHz repetition rate. Typically used pulse energy was 3–7 μJ. The laser beam then passed a variable beam expander, was reflected by a dichroic mirror, and was positioned on the target by a mirror, as observed through a fused silica viewport. Inside the source, the laser beam entered a microscope objective, model Mitutoyo LCD Plan Apo NUV 100X (Mitutoyo Corp., Takatsu-ku, Kawasaki-shi, Kanagawa, Japan), having a numeric aperture 0.5 and a working distance 11.03 mm. The objective was mounted on a Z-stage, which had a linear motor actuator to focus the microscope objective on the target and to move the objective away during the sample load/unload sequence to avoid damage to the objective. Software control of the actuator was integrated into the instrument controller software. In order to monitor the laser focusing and ablation process, a custom built optical inverted microscope was used. The dichroic mirror allowed a view path collinear with the laser beam. A light illuminator (LED illuminator model MCWHL5 from Thorlabs Inc.) was also collinear and was coupled to the view path using a 50% transmission mirror. The LED light illuminated the target surface through the focusing objective. The light reflected from the target was focused by the same objective and an additional focusing lens to form a sharp optical image on a CMOS camera chip surface. The CMOS color camera (model Sentech STC-MCA5MUSB3 with USB 3.0 interface, (Sensor Technology Co., Ltd., Atsugi-City, Kanagawa, Japan) allowed video monitoring of the target during MS imaging with full HD resolution at 30 fps or with VGA resolution at 123 fps. To avoid interference with the MS image acquisition process, the CMOS camera live video was viewed using a separate computer workstation.
Figure 1.
SimulTOF 200 Combo instrument with transmission geometry assembly installed below the sample chamber and an SEM micrograph of 2,5-DHA matrix layer: A – front view photo of the instrument, B – top view scheme of the optical path and elements of the transmission geometry assembly, C – SEM micrograph of 2,5-DHA matrix layer.
The inverted optical microscope, built into the source especially was handful during the optics alignment, when the laser spot characteristics were studied by examination of ablation patterns and craters. Such examinations were conducted in a metal film, blue Sharpie marker layer, highly sensitive DHB matrix layer prepared by sublimation [13], or in a real tissue sample. The ablated patterns were analyzed by the built-in microscope in real time or by laboratory optical microscope. Under optimal focusing conditions, the observed craters had round shape with diameters, depending on the layer properties and the laser pulse energy, due to Gaussian beam profile. The typical alignment procedure on the real sample was as follows: the laser was optimally focused by observing the spot sizes in real time, and then the laser pulse energy was varied while the signal to noise ratio (SNR) for MS peaks was monitored. Once the signal to noise ratio was near 10, the crater sizes were evaluated by the built-in microscope and the laser focusing spot adjustment was attempted again. This sequence was repeated several times until an optimal set of focusing position and laser pulse energy was reached. Under such criteria, for gold films and Sharpie marker layers, the minimum spot size was at sub-micron level. For protein analysis in tissues, however the optimal laser energy was higher and the spot size was near 1 μm.
Materials
Ethanol, acetonitrile (ACN), acetic acid (AcOH), chloroform, trifluoroacetic acid (TFA) and 2,5-DHA were purchased from Fisher Scientific (Suwanee, GA, USA). 2,5-DHA was recrystallized twice with 90% ACN. Xylene was purchased from Acros (Morris Plains, NJ, USA). Conductive indium tin oxide (ITO) coated microscope glass slides were purchased from Delta Technologies (Stillwater, MN, USA).
Carnoy’s fluid was prepared as a 6:3:1 solution of 100% ethanol: chloroform: acetic acid. Fresh frozen mouse brain was purchased from Pel-Freez Biologicals (Rogers, AZ, USA).
Sample preparation
Tissue preparation, rinsing steps and matrix spraying protocols were optimized to maximize ionization efficiency and to reduce matrix grain size, delocalization, and laser induced forward transfer (LIFT) [14] effects. We found that 2,5-DHA matrix material was superior in sensitivity to other common matrix materials, for example sinapinic acid [15,16]. In contrast to the work where 2,6-dihydroxyacetophenone (2,6-DHA) was used for lipids imaging [17], we used the 2,5 isomer that provided significantly higher sensitivity for the imaging of proteins. This compound is significantly more stable under the instrument vacuum than 2,6-DHA.
Further optimization was performed for the 2,5-DHA deposition protocol. During the optimization, the matrix grains were inspected using an optical microscope. For the optimized coatings used for MS imaging, approximately 95% of the grains were 1–3 μm size, and 5% were up to 5 μm in size. A Scanning Electron Microscopy (SEM) micrograph of the matrix layer before the ablation in shown in Fig. 1, panel C. Delocalization was tested by ion imaging of the tissue edge.
For protein imaging, fresh frozen tissues were sectioned at 5–12 μm thickness using a Leica CM3050 cryostat (Leica Microsystems GmbH, Wetzlar, Germany). The sections were thaw mounted on ITO-coated microscope slides, dried under ambient conditions for 10 minutes for the next step, or stored in a slide mailer box (from Electron microscope science (Hatfield, PA, USA), catalog # 71548-01) at −80°C. For analysis, the slides were placed into a vacuum desiccator at room temperature for 30 minutes to allow them to reach room temperature while preventing water condensation on the sections that may cause delocalization of analytes. As a pretreatment, the sections were sequentially rinsed in a series of petri dishes (100 × 15 mm, Fisher Scientific (Suwanee, GA, USA)) containing 10 mL of each solution: 70% ethanol for 30 s, 100% ethanol for 30 s, Carnoy’s fluid for 2 min, 100% ethanol for 30 s, 40% ethanol for 30 s, 100% ethanol for 30 s. For matrix application, robotic spraying was performed using a TM-Sprayer (HTX Technologies (Carrboro, NC, USA)). Slides with pretreated sections were placed in the TM-Sprayer chamber. The 2,5-DHA solution was prepared using 150 mg 2,5-DHA, 6 mL ACN, 3 mL H2O, 200 μL TFA, and 200 μL AcOH. Robotic spraying was performed using 8 passes using a nozzle velocity of 1300 mm/min, flow rate of 0.085 mL/min, a track spacing of 2.5 mm, 90 degree rotation of the nozzle motion direction at passes 2, 4, and 6, and a 1.2 mm track offset at passes 3, 4, 7, and 8. This protocol resulted in a coating layer of 0.4 mg/cm2 across the slide surface, determined by weighing the sample before and after spraying. We check the layer thickness using a Zeta-20 optical profiler (Zeta Instruments, San Jose, CA, USA).
Post-imaging H&E staining
For high spatial resolution image applications, both the MS analysis and tissue staining procedure should be performed on a single section. At a 2.5 μm raster step size and 1 μm laser spot diameter, there was a significant amount of un-ablated material remaining on the slide after the imaging process. After imaging, the section was rinsed with ethanol and acetone to remove the residual matrix material. The tissue sections were then subjected to a standard hematoxylin and eosin (H&E) staining procedure [18]. H&E stained sections were scanned with a Leica SCN 400 optical slide scanner (Leica Microsystems GmbH, Wetzlar, Germany) at 20× magnification.
Results and Discussion
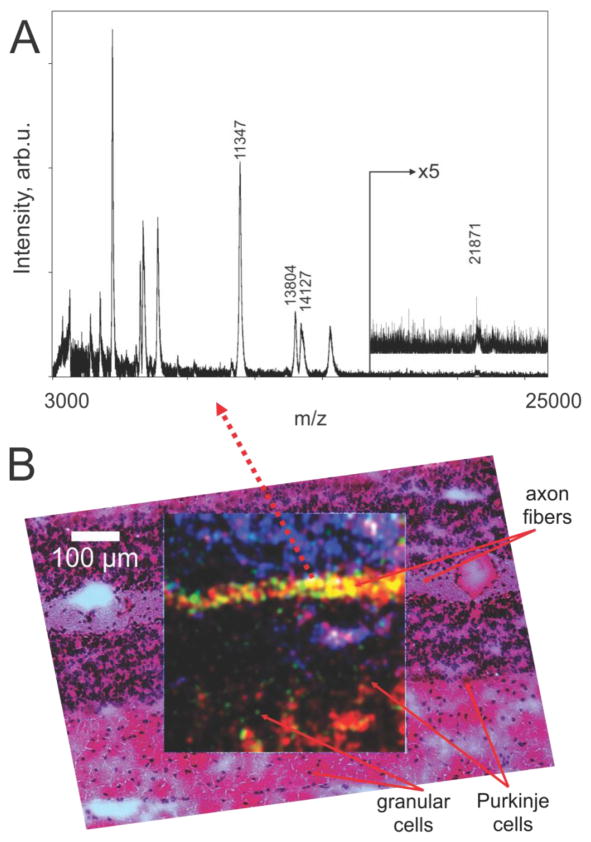
Tissue protein images at 1 μm laser spot diameter and 2.5 μm raster step sizes using transmission geometry were obtained in the step raster mode at a speed of 5 pixels/s or in the continuous raster mode at 40 pixels/s. In the step raster mode, the typical acquisition was conducted at a 2000 Hz laser repetition rate and 50 laser shots were averaged per pixel. A mass spectrum for a single pixel from mouse cerebellum tissue is shown in Fig. 2 (panel A), with the location of the pixel marked by dotted arrow line. The spectrum is unprocessed except for background subtraction. Although acquired from an extremely small area, defined by the 1 μm laser spot diameter, the mass spectrum contains 20 peaks with m/z values in a range 3–22 kDa. The resolving power of the instrument in this experiment was about 1000 FWHM.
Figure 2.
(A) - mass spectrum acquired from a single pixel in the image taken from an area of mouse cerebellum, (B) – MS ion images from the same area for 3 ions: red – m/z 14127 (myelin basic protein, isoform 8), green – m/z 13804 (histone H2B), blue – m/z 11347 (histone H4) represented as one combined RGB image. The image is overlaid with the optical microscope image of the same area that was stained with H&E following MS image acquisition. The MS imaging field is 500×500 μm scanned by a laser raster of 2.5 μm step size and laser spot diameter 1 μm on target.
In the case of a continuous raster using a stage with a minimum step size of 2.5 μm, the stage was not capable of moving with constant speed between 2.5 μm steps to accumulate a signal from 50 laser shots per pixel. Instead, the stage moved from position to position in 2.5 μm increments at a constant rate. Also in this mode the stage is not synchronized with acquisition, therefore the speed of the stage motion has to be precisely tuned to provide the same number of laser shots per each step. For 2000 Hz laser repetition rate and 50 shots per pixel, the desired speed of the stage should be tuned to 40 steps/s or averaged speed 100 μm/s. To control such speeds, the instrument control software was modified. In comparison, the images in the continuous raster mode acquired at 40 pixels/s were similar to the images obtained in step raster mode, but had higher noise and pixel crosstalk and had losses of several shots between steps. Also considering acceleration/deceleration zones at the ends of each scan line, the image raster for continuous raster mode was made wider, according to the stage parameters. In this paper we show that the image acquisition at 40 pixels/s is possible. Routine imaging at this speed will require the stage step size to decrease down to 100–200 nm.
Ion images for selected m/z values are shown in Fig. 2 (panel B): red – m/z 14127 (myelin basic protein, isoform 8), green – m/z 13804 (histone H2B), blue – m/z 11347 (histone H4) overlaid to give one RGB image. Identification of the proteins was based on previous studies of similar tissues in our laboratory [19,20]. The RGB image demonstrates spatial features below the cellular level for this section. In order to demonstrate a correlation between the acquired ion images and the original tissue, the post-imaging H&E staining was performed and the RGB image was overlaid with a high spatial resolution optical microscopy image obtained from the stained section. As seen from Fig. 2 (panel B), multiple features of the mouse cerebellum, such as axon fibers, granular cells, and Purkinje cells correlate in the MS and optical images. The solid line arrows point to corresponding locations in the stained area and in the MS image.
Currently, the spatial resolution of the image is limited by the minimal step size of the original XY translation stage. Future work improving on the sub-micron spatial resolution will be achieved by employing a translation stage having a 100 – 200 nm step size.
Conclusion
We have demonstrated the IMS of tissue proteins using a 1 μm laser spot diameter and 2.5 μm raster step size as performed on a commercial instrument modified with a transmission geometry MALDI ion source. The images show spatial features significantly below cellular dimensions in mouse cerebellum tissue at a level of detail that permits visualization of axon fibers, granular cells, Purkinje cells and surrounding dendrites. Further, the MS images correlate well with images from stained sections using high resolution optical microscopy.
Acknowledgments
The authors would like to thank George Mills (SimulTOF Systems), and Boone Prentice and other members of the Vanderbilt Mass Spectrometry Research Center. This project was supported by grants from National Institutes of Health National Institute of General Medical Sciences NIH/NIGMS P41 GM103391-04 and NIH/NIGMS R01 GM058008-15.
References
- 1.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69 (23):4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 2.Wei H, Nolkrantz K, Powell DH, Woods JH, Ko MC, Kennedy RT. Electrospray sample deposition for matrix-assisted laser desorption/ionization (MALDI) and atmospheric pressure MALDI mass spectrometry with attomole detection limits. Rapid Communications in Mass Spectrometry. 2004;18 (11):1193–1200. doi: 10.1002/Rcm.1458. [DOI] [PubMed] [Google Scholar]
- 3.Zavalin A, Yang J, Hayden K, Vestal M, Caprioli RM. Tissue Protein Imaging at 2.5μm Spatial Resolution and High Speed using Transmission Geometry MALDI Source Integrated into a TOFMS Instrument. 62nd ASMS Conference on Mass Spectrometry and Allied Topics; Baltimore, MD. 2014. [Google Scholar]
- 4.Qiao H, Spicer V, Ens W. The effect of laser profile, fluence, and spot size on sensitivity in orthogonal-injection matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Communications in Mass Spectrometry. 2008;22 (18):2779–2790. doi: 10.1002/rcm.3675. [DOI] [PubMed] [Google Scholar]
- 5.Wetzel SJ, Guttman CM, Flynn KM. The influence of electrospray deposition in matrix-assisted laser desorption/ionization mass spectrometry sample preparation for synthetic polymerst. Rapid Communications in Mass Spectrometry. 2004;18 (10):1139–1146. doi: 10.1002/Rcm.1462. [DOI] [PubMed] [Google Scholar]
- 6.Lou XW, van Dongen JLJ. Direct sample fraction deposition using electrospray in narrow-bore size-exclusion chromatography/matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for polymer characterization. Journal of Mass Spectrometry. 2000;35 (11):1308–1312. doi: 10.1002/1096-9888(200011)35:11<1308::Aid-Jms64>3.0.Co;2-W. [DOI] [PubMed] [Google Scholar]
- 7.Zavalin A, Todd EM, Rawhouser PD, Yang JH, Norris JL, Caprioli RM. Direct imaging of single cells and tissue at sub-cellular spatial resolution using transmission geometry MALDI MS. Journal of Mass Spectrometry. 2012;47 (11):1473–1481. doi: 10.1002/Jms.3108. [DOI] [PubMed] [Google Scholar]
- 8.Thiery-Lavenant G, Zavalin AI, Caprioli RM. Targeted multiplex imaging mass spectrometry in transmission geometry for subcellular spatial resolution. Journal of the American Society for Mass Spectrometry. 2013;24 (4):609–614. doi: 10.1007/s13361-012-0563-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spraggins JM, Caprioli R. High-Speed MALDI-TOF Imaging Mass Spectrometry: Rapid Ion Image Acquisition and Considerations for Next Generation Instrumentation. Journal of the American Society for Mass Spectrometry. 2011;22 (6):1022–1031. doi: 10.1007/s13361-011-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.SimulTOF 200 Combo. SimulTOF Systems. http://www.simultof.com/content/simultof-200-combo.
- 11.Mainini V, Angel PM, Magni F, Caprioli RM. Detergent enhancement of on-tissue protein analysis by matrix-assisted laser desorption/ionization imaging mass spectrometry. Rapid Communications in Mass Spectrometry. 2011;25 (1):199–204. doi: 10.1002/Rcm.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trim P, Djidja M-C, Atkinson S, Oakes K, Cole L, Anderson DG, Hart P, Francese S, Clench M. Introduction of a 20 kHz Nd:YVO4 laser into a hybrid quadrupole time-of-flight mass spectrometer for MALDI-MS imaging. Anal Bioanal Chem. 2010;397 (8):3409–3419. doi: 10.1007/s00216-010-3874-6. [DOI] [PubMed] [Google Scholar]
- 13.Zavalin A, Yang J, Caprioli R. Laser Beam Filtration for High Spatial Resolution MALDI Imaging Mass Spectrometry. Journal of the American Society for Mass Spectrometry. 2013;24 (7):1153–1156. doi: 10.1007/s13361-013-0638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernández-Pradas JM, Colina M, Serra P, Domínguez J, Morenza JL. Laser-induced forward transfer of biomolecules. Thin Solid Films. 2004;453–454:27–30. http://dx.doi.org/10.1016/j.tsf.2003.11.154. [Google Scholar]
- 15.Dreisewerd K, Schürenberg M, Karas M, Hillenkamp F. Influence of the laser intensity and spot size on the desorption of molecules and ions in matrix-assisted laser desorption/ionization with a uniform beam profile. International Journal of Mass Spectrometry and Ion Processes. 1995;141 (2):127–148. http://dx.doi.org/10.1016/0168-1176(94)04108-J. [Google Scholar]
- 16.Guenther S, Koestler M, Schulz O, Spengler B. Laser spot size and laser power dependence of ion formation in high resolution MALDI imaging. Int J Mass Spectrom. 2010;294 (1):7–15. http://dx.doi.org/10.1016/j.ijms.2010.03.014. [Google Scholar]
- 17.Ressine A, Marko-Varga G, Laurell T. Porous silicon protein microarray technology and ultra-/superhydrophobic states for improved bioanalytical readout. In: El-Gewely MR, editor. Biotechnology Annual Review. Vol. 13. Elsevier; 2007. pp. 149–200. http://dx.doi.org/10.1016/S1387-2656(07)13007-6. [DOI] [PubMed] [Google Scholar]
- 18.Prophet EB Armed Forces Institute of Pathology, et al. Laboratory methods in histotechnology. American Registry of Pathology; Washington, D.C: 1992. [Google Scholar]
- 19.Burnum KE, Tranguch S, Mi D, Daikoku T, Dey SK, Caprioli RM. Imaging mass spectrometry reveals unique protein profiles during embryo implantation. Endocrinology. 2008;149 (7):3274–3278. doi: 10.1210/en.2008-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crecelius AC, Cornett DS, Caprioli RM, Williams B, Dawant BM, Bodenheimer B. Three-dimensional visualization of protein expression in mouse brain structures using imaging mass spectrometry. Journal of the American Society for Mass Spectrometry. 2005;16 (7):1093–1099. doi: 10.1016/j.jasms.2005.02.026. [DOI] [PubMed] [Google Scholar]