Abstract
Bacteria and archaea insert spacer sequences acquired from foreign DNAs into CRISPR loci to generate immunological memory. The Escherichia coli Cas1–Cas2 complex mediates spacer acquisition in vivo, but the molecular mechanism of this process is unknown. Here we show that the purified Cas1–Cas2 complex integrates oligonucleotide DNA substrates into acceptor DNA to yield products similar to those generated by retroviral integrases and transposases. Cas1 is the catalytic subunit, whereas Cas2 substantially increases integration activity. Protospacer DNA with free 3'-OH ends and supercoiled target DNA are required, and integration occurs preferentially at the ends of CRISPR repeats and at sequences adjacent to cruciform structures abutting A-T rich regions, similar to the CRISPR leader sequence. Our results demonstrate the Cas1–Cas2 complex to be the minimal machinery that catalyzes spacer DNA acquisition and explain the significance of CRISPR repeats in providing sequence and structural specificity for Cas1–Cas2-mediated adaptive immunity.
Prokaryotic adaptive immunity relies on clustered regularly interspaced short palindromic repeats (CRISPRs) together with CRISPR associated (Cas) proteins to detect and destroy foreign nucleic acids1,2. CRISPR loci contain an A-T-rich leader sequence followed by repetitive sequence elements flanking ~30 base pair (bp) spacer segments that are transcribed to produce precursor CRISPR RNAs (pre-crRNAs)3-5. Spacers are frequently virus- or plasmid-derived, although “self”-derived spacers from the host chromosome are present in some CRISPR loci6. After pre-crRNA processing and assembly with Cas proteins, the resulting surveillance complexes target and cleave foreign nucleic acids bearing sequences complementary to the crRNA spacer sequence7-12. How spacer DNA sequences, termed protospacers, are acquired into the host CRISPR locus remains unknown.
Overexpression of Cas1 and Cas2 nucleases, the only Cas proteins found in all CRISPR–Cas systems, leads to the site-specific acquisition of 33 bp protospacers at the leader end of the CRISPR locus in E. coli13-15. Furthermore, Cas1 and Cas2 function as a complex in vivo16, suggesting that the Cas1–Cas2 complex might possess DNA recombination activity. We reconstituted CRISPR spacer acquisition using purified Cas1 and Cas2 proteins, protospacers and acceptor plasmid DNA, revealing an elegant mechanism in which both the sequence and structural elements of the CRISPR repeats specify spacer integration sites.
Protospacer DNA integration by Cas1–Cas2
To test whether the Cas1–Cas2 complex is sufficient to catalyze DNA recombination in vitro, assays were conducted using purified Cas1–Cas2 complex, 33 bp protospacer DNA and an acceptor “target” plasmid consisting of the pUC19 backbone with an inserted CRISPR locus (pCRISPR) (Fig 1a). Co-incubation of these reagents converted the supercoiled plasmid into three main products: relaxed and linear plasmid species, and a fast-migrating species we term Band X (Fig. 1b, c and Extended Data Fig. 1a). Product formation required Cas1, Cas2 and the protospacer DNA (Extended Data Fig. 1b-d), and was consistent with previous divalent metal ion-dependent and sequence-nonspecific in vitro activity requirements of Cas117-19 and Cas220-22. Product DNA migration was not affected by treatment with EDTA, EDTA and phenol-chloroform extraction or Proteinase K in the presence of EDTA and detergent (Extended Data Fig. 1e), indicating that product DNAs are unlikely to be bound to Cas1 and/or Cas2. Consistent with product DNA resulting from covalent integration of protospacer DNA into the plasmid, the relaxed and linear forms of pCRISPR became radiolabeled in reactions containing 32P-labeled protospacer DNA (Fig. 1d and Extended Data Fig. 2). Although Cas1 alone catalyzed a low level of protospacer integration in the presence of Mn2+, the reaction was enhanced significantly by the presence of Cas2 (Extended Data Fig. 2b).
Figure 1. The Cas1–Cas2 complex integrates protospacers in vitro.
a, Schematic of the in vitro integration assay (PDB code 4P6I for Cas1–Cas2). b, The presence of Cas1, Cas2 and a protospacer results in the conversion of the supercoiled pCRISPR into relaxed, linear and Band X products. c, Neither the Cas1 H208A active site mutant nor and the complex formation-defective Cas2 β6–β7 deletion mutant support the reaction. The Cas2 E9Q active site mutant (lane 5 from the marker) is as active as the wild-type. d, Salt- and metal-dependence of radiolabeled protospacer integration into pCRISPR. e, Same as c except using radiolabeled protospacers. The data presented in b-e are representative of at least three replicates.
Bacteria expressing Cas1 active-site mutants, but not active-site mutants of Cas2, are incapable of acquiring new spacers in vivo, demonstrating the catalytic role of Cas1 during spacer acquisition13,14,16. Consistent with these data, Cas1 active site mutants H208A and D221A were defective for protospacer integration in vitro, whereas the Cas2 E9Q active-site mutant supported integration (Fig. 1c, e and Extended Data Fig. 3). The Cas2 C-terminal β6–β7 deletion mutant, which is defective for complex formation with Cas1 and spacer acquisition in vivo, failed to support Cas1-mediated integrase activity (Fig. 1c, e). We conclude that our in vitro assay recapitulates the in vivo functions of Cas1 and Cas2 during spacer acquisition.
Integration and disintegration products
We tested whether the reaction products of Cas1–Cas2-mediated DNA integration resemble those formed by the strand transfer activity of retroviral integrases and cut-and-paste transposases23-26. These enzymes generate two main products in vitro corresponding to half-site and full-site integration events (Fig. 2a). We observed similar gel mobility of the slowly migrating DNA product generated by Cas1–Cas2 and Nb.BbvCI nickase-digested pCRISPR, consistent with the slow-migrating relaxed DNA species corresponding to half-site products and/or products resulting from full-site integration of one protospacer molecule (Extended Data Fig. 1a). Digestion with EcoRI, which cuts pCRISPR once, converted the reaction products to linear DNAs (Fig. 2b, compare lane 4 to lane 2, and Fig. 2c). We therefore conclude that both the relaxed and Band X DNA products comprise unit-sized pCRISPR circles.
Figure 2. Half-site, full-site integration and pCRISPR topoisomer products.
a, Schematic of half-site and full-site integration products. b, Linearization of the integration products (lane 4). Lane 3 is the un-treated reaction products. c, Linearization of integration products from radiolabeled protospacer reactions. d, The time course reveals the initial formation of relaxed products, followed by Band X. The inset reveals the products detected using 32P-labeled protospacers. e,f, Analysis of gel-purified relaxed and Band X on agarose gels pre-stained with ethidium bromide (e) or post-stained after electrophoresis (f). g, Schematic of the disintegration reaction. h, Native polyacrylamide gel analysis of the disintegration reaction. The data presented in b-f, h are representative of at least three replicates.
We observed that Band X did not become radiolabeled in reactions conducted with 32P-labeled protospacer DNA. A time course analysis revealed relaxed DNA product formation within the first minute, followed by accumulation of Band X between 10 and 30 min (Fig. 2d). To determine the properties of Band X, the purified product was analyzed in two different types of agarose gels – one pre-stained with ethidium bromide, similar to the gels presented thus far, and the other stained with ethidium bromide after electrophoresis (post-stained) (Extended Data Fig. 4a). Although Band X migrated as a single species in the pre-stained gel, a ladder of species that migrated faster than the relaxed products was observed in the post-stained gel (Fig. 2e, f). These intermediates are reminiscent of plasmid topoisomers27,28. The same pre- and post-stained agarose gel analysis was performed on the entire integration reaction, generating similar results to those observed with purified Band X (Extended Data Fig. 4b, c). PCR analysis of various segments of pCRISPR using gel-purified Band X as the template yielded amplification products indistinguishable from those generated using unreacted supercoiled pCRISPR or relaxed integration products, supporting the conclusion that Band X corresponds to pCRISPR topoisomers (Extended Data Fig. 4d).
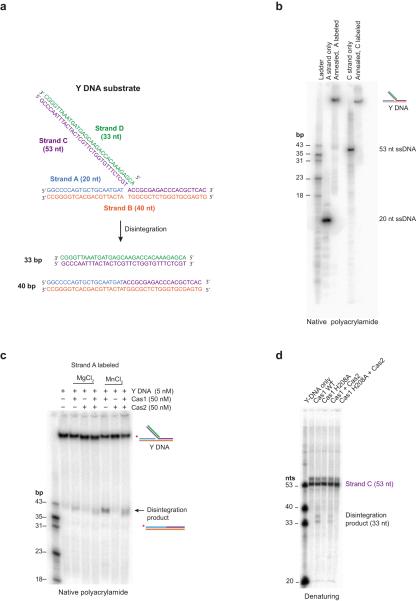
We wondered whether Band X arose from protospacer excision from half-site integration products to regenerate pCRISPR in different supercoiled states, analogous to the in vitro disintegration activities of retroviral integrases and transposases (Fig. 2g)29,30. To test this hypothesis, a synthetic Y-structured DNA intermediate that mimics the half-site integration product (Extended Data Fig. 5a,b) was radiolabeled such that the liberated 33 bp protospacer DNA could be detected following disintegration activity. Using this substrate, we observed that Cas1 catalyzed disintegration activity either by itself or in the presence of Cas2 (Fig. 2h). Disintegration activity was confirmed by radiolabeling the 20-nt target DNA strand and monitoring the formation of the joined 40 bp target DNA product (Extended Data Fig. 5c, d). Thus, Cas1–Cas2 integration and disintegration activities are similar to those of retroviral integrases and transposases.
Integration requires 3'-OH protospacer ends
We next investigated the DNA protospacer and target DNA requirements for integration. Single-stranded protospacer DNA failed to support the reaction (Fig. 3a, b). The Cas1–Cas2 complex accommodated various protospacer lengths in vitro despite the strict 33 bp requirement for spacer acquisition in vivo (Extended Data Fig. 6a), suggesting that protospacer length is pre-determined before integration in vivo by an unknown mechanism. The Cas1–Cas2 complex integrated DNA substrates with blunt-ends or with 3'-overhangs up to 5 nt in length (Extended Data Fig. 6b). In contrast to retroviral integrases31, substrates with 5'-overhangs were nonviable (Extended Data Fig. 6b).
Figure 3. Integration requires 3'-OH protospacer ends and supercoiled target DNA.
a,b, Integration assays using single-stranded DNAs and either –OH or –PO4 at the 3' or 5' ends of (a) unlabeled or (b) radiolabeled protospacers. S1 corresponds to one strand of the protospacer and S2 corresponds to the complementary strand. c, Comparison of protospacer integration into different DNA targets. d,e, Restriction enzyme digestion of pCRISPR, either in a pUC19 (d) or pACYC backbone (e), after the integration assay detects integration into the CRISPR fragment (green arrows). The data presented in a-e are representative of at least three replicates.
Retroviral integration and transposition reactions proceed via nucleophilic attack of DNA 3'-OH groups at target DNA phosphodiester bonds31,32. We found that phosphorylation of both 3'-ends of the protospacer ablated integration, whereas phosphorylation of only one 3' end strongly limited integration (Fig. 3a, b). By analogy to known integrase enzyme mechanisms, DNA integration could proceed by Cas1-catalyzed direct nucleophilic attack of the substrate 3'-OH on the target DNA, or by formation of a Cas1–DNA intermediate, as occurs in the serine and tyrosine families of recombinases33. Four tyrosine residues in the vicinity of the Cas1 active site17-19 could be involved in forming such a covalent intermediate (Extended Data Fig. 7a,b). Purified Cas1 mutant proteins in which each tyrosine was individually changed to alanine each supported protospacer integration in vitro at levels comparable to wild type Cas1–Cas2 (Extended Data Fig. 7c). Thus, the integration reaction likely proceeds via direct nucleophilic attack of protospacer 3'-OH ends onto the target DNA phosphodiester bonds, a mechanism previously hypothesized to occur in vivo34.
Supercoiled DNA and CRISPR locus requirements
Cas1 and Cas2 overexpression leads to site-selective spacer acquisition proximal to the leader end of the CRISPR locus, a result consistent with observations in native populations of CRISPR-containing bacteria13-15,35. To determine what drives such site-specific integration, we first tested various forms of the pCRISPR plasmid to determine target DNA requirements. Integration requires target DNA supercoiling, as neither relaxed nor linear pCRISPR, nor the isolated 1 kb CRISPR locus, supported integration (Fig. 3c and Extended Data Fig. 6c,d).
As a control, we tested supercoiled pUC19 DNA, the parental plasmid of pCRISPR that lacks a CRISPR locus, and were surprised to observe integration products upon incubation with Cas1 and Cas2 in the presence of protospacer DNA (Fig. 3c and Extended Data Fig. 6e). This finding raised two possibilities: 1) in vitro spacer integration is non-specific with respect to target DNA sequence or 2) structures and/or sequence(s) favoring integration are present in the pUC19 plasmid. To determine if integration preferentially occurred at the CRISPR locus of pCRISPR, products of radiolabeled reactions were double-digested to separate the CRISPR locus (960 bp) from the pUC19 plasmid backbone (~2.27 kb). Suggestive of CRISPR-specific integration, the 32P-radiolabel migrated solely with the CRISPR locus fragment (Fig. 3d). The same result was observed when the experiment was conducted using a target plasmid containing the CRISPR locus and a different backbone sequence (pACYC) (Fig. 3e).
CRISPR repeats provide specificity
To determine the exact sites of protospacer integration in these reactions, we performed high-throughput sequencing of reaction products that resulted from using either pCRISPR or the parental pUC19 vector as the target of integration (Extended Data Fig. 8a). Of the 7,866 protospacer-pCRISPR junctions retrieved, ~71% mapped to the CRISPR locus (Fig. 4a and Extended Data Fig. 8b). Protospacer insertion occurred at the borders of each repeat, with the most preferred site at the first repeat adjacent to the leader (Fig. 4b). The minus strand of each repeat (the bottom strand in Fig. 4a,b that runs 5'-to-3' towards the leader sequence) is also highly preferred, highlighting the role of CRISPR repeats in providing sequence specificity for the Cas1–Cas2 complex (Fig. 4b). Sequence alignment of the integration sites revealed strong preference for sequences resembling the CRISPR repeat on both strands of pCRISPR, further supporting the selection of CRISPR repeat borders by the Cas1–Cas2 complex (Extended Data Fig. 8d-f).
Figure 4. Protospacers are specifically integrated into the CRISPR locus.
a, Integration sites along pCRISPR. b, Magnified view of the integration sites along the ~1 kb CRISPR locus. The cyan peaks represent positions where the 3' T of the protospacer DNA was integrated whereas the black peaks represent the C 3'-OH integration events. The protospacer sequence is depicted above the plot. c, Integration sites along pUC19. d, Comparison of C 3'-OH or T 3'-OH selection in the total reads from pCRISPR and pUC19 targets (n=7,866 reads for pCRISPR and n=5,524 reads for pUC19, Chi-square test, *p<0.0001). e, Schematic of DNA cruciform formation of the repeat sequences. The orange arrows depict the cleavage sites.
The most frequent integration site in the pUC19 control plasmid mapped to the amp resistance gene adjacent to the A-T rich promoter sequence (~8.8% of 5,524 total retrieved junctions, Fig. 4c and Extended Data Fig. 8c). An inverted repeat sequence with a propensity to form a DNA cruciform36 occurs 9 nt adjacent to this integration site (plus strand sequence: 5'-TTCAATATTATTGAA-3'), suggesting that potential DNA cruciform formation adjacent to A-T rich sequences is important for protospacer integration. Sequence analysis of pUC19 target sites revealed the propensity for a G nucleotide to occur at the −2 and +1 positions of the protospacer insertion site, similar to the preferred pCRISPR sites (Extended Data Fig. 8g,h). These observations imply that in addition to sequence, pCRISPR repeat selectivity stems from the unique structural features of these sites, such as their ability to form cruciforms (Fig. 4a, b, e).
In E. coli, newly acquired spacers harbor a 5' G as the first nucleotide flanking the leader-proximal end of the repeats, which originates from the last nucleotide of the AAG protospacer-adjacent motif (PAM) from foreign DNA13-15,37-39. Such positional specificity is critical for crRNA-guided interference, as a mutation in this position of the corresponding crRNA disrupts PAM binding and subsequent target destruction40-42. We found that ~73% of all integration events into pCRISPR utilized the 3' C end instead of the 3' T end of protospacer DNA during integration (see Fig. 4b for protospacer sequence), and there was a strong preference for this nucleotide to attack the minus strand of the repeat sequence (Fig. 4b,d,e). A similar nucleotide bias was observed in the pUC19 target plasmid sequence data (Fig. 4d). This preference positions the G at the 5' end of the protospacer substrate as the first nucleotide of the newly integrated spacer in the CRISPR locus (Fig. 5). When we used protospacer DNAs lacking a 3' C or bearing 3' C on both ends, the preference for integration into the minus strand of the CRISPR locus was significantly decreased (Extended Data Fig. 9). Thus, the Cas1–Cas2 complex plays a critical role in correctly orienting the C 3'-OH end of protospacer DNA substrates for incorporation within the CRISPR locus.
Figure 5. Model of protospacer integration during CRISPR–Cas adaptive immunity.
The first nucleophilic attack occurs on the minus strand of the first repeat, distal to the leader, by the C 3'-OH end of the protospacer. After half-site intermediate formation, the second integration event occurs on the opposite strand at the leader-repeat border. The resulting single-stranded DNA gaps are repaired by yet uncharacterized mechanisms and the protospacer is fully integrated with the G as the first nucleotide at its 5' end. The asterisk denotes the duplication of the first repeat, as previously observed in vivo13-15.
Mechanism of protospacer integration
The results presented here explain the mechanistic basis for foreign DNA acquisition during CRISPR–Cas adaptive immunity (Fig. 5). The Cas1–Cas2 complex catalyzes integration of protospacers at the leader-end of the CRISPR locus and also selects the terminal C 3'-OH as the attacking nucleophile, resulting in the 5' G on the opposite strand of the protospacer becoming the first nucleotide of the newly integrated spacer. This orientation bias, previously observed in vivo39, is a key step during immunity for productive downstream foreign DNA targeting by the Cascade complex and Cas3 effector nuclease (Extended Data Fig. 10). Interestingly, the presence of the complete AAG PAM in the protospacer is not required for in vitro integration, suggesting that a highly specific selection or processing step occurs in vivo to exclude the AA nucleotides from the mature protospacer prior to integration.
We propose a two-step integration mechanism in which the C 3'-OH first attacks the minus strand of the CRISPR repeat to produce a half-site intermediate (Fig. 5). The 3'-OH on the opposite strand of the integrating DNA then attacks the target DNA 28 bp away on the opposite side of the repeat on the plus strand, leading to full integration of the protospacer (Fig. 5). Our in vitro system predominantly traps the first step of this two-step integration mechanism, suggesting that the second nucleophilic attack is greatly accelerated in vivo in the presence of cellular factors. This model is consistent with spacer integration intermediates that are observed in vivo, in which protospacers are integrated such that staggered cleavage at each end of the repeat generates single-stranded gaps that ensure repeat duplication34. The in vivo conditions could also promote the high specificity of integration to occur solely downstream of the first repeat of the CRISPR locus in E. coli, instead of at every repeat, as observed in our in vitro assay.
CRISPR spacer integration shares mechanistic similarities with retroviral integration and DNA transposition, where the integrase/transposase enzyme uses donor DNA 3'-OH ends to make a staggered cut at the DNA target site, which concurrently joins the donor DNA to target DNA 5'-phosphates31,32. Completion of the integration reaction requires a DNA polymerase to fill in sequence gaps and a DNA ligase to seal the phosphodiester backbone43. Similar polymerase and ligase functions are required to complete CRISPR spacer acquisition in vivo, although the specific enzymes involved have not yet been identified. Despite these similarities, we note that the Cas1 active site does not harbor the RNase H fold that defines the retroviral integrase enzyme superfamily44. This structural difference could explain the unexpected production of different topoisomers of pCRISPR (Band X) in vitro, although the physiological significance of Band X production remains unclear.
Our results highlight the fundamental role of repeat sequences at multiple stages of CRISPR–Cas adaptive immunity. In addition to creating structures within nascent CRISPR transcripts that ensure correct RNA processing during crRNA maturation45, the repeats operate at the DNA level to recruit the Cas1–Cas2 complex for sequence- and structure-specific protospacer integration. We envision that this recruitment involves transient DNA cruciform formation within the CRISPR inverted repeats that occurs as a function of target DNA supercoiling46. The observation that a preferred non-CRISPR site of Cas1–Cas2-mediated DNA integration is proximal to an inverted repeat adjacent to an A-T-rich sequence suggests the fascinating possibility that CRISPR loci arise in naïve genomes through integration events that become self-propagating through creation of repetitive sequences with properties that ensure continual recognition and activity by the Cas1–Cas2 integration machinery.
Methods
Cas1, Cas2 and DNA preparation
The cas1 and cas2 genes from E. coli K12 (MG1655) were cloned into expression vectors and the proteins were separately purified as previously described16. The proteins were stored in 100 mM KCl, 20 mM HEPES-NaOH, 5% glycerol and 1 mM TCEP at −80 °C prior to use. Single-stranded DNAs were synthesized (Integrated DNA Technologies). Double-stranded DNA protospacers were annealed in 20 mM HEPES-NaOH, pH 7.5, 25 mM KCl, 10 mM MgCl2 or MnCl2, 1 mM DTT, 10% DMSO by heating at 95 °C for 3 min and slow cooling to room temperature. The sequence of the 33 bp protospacer used in this study was shown to be the most acquired in vivo in E. coli K12 after M13 bacteriophage infection14: Strand 1 (5'-GCCCAATTTACTACTCGTTCTGGTGTTTCTCGT-3') and Strand 2 (5'- ACGAGAAACACCAGAACGAGTAGTAAATTGGGC-3'). The pCRISPR target plasmid was constructed by PCR amplifying the E. coli BL21-AI genomic CRISPR locus and cloning the fragment into pUC19 using the following primers: Forward/EcoRI: 5'-ACGTCGAATTCTACCTTTTTAATCAATGG-3' and Reverse/AflIII: 5'-ACGTCACATGTGGTTATATGGTGGTTTATCC-3'. The pACYC CRISPR plasmid was constructed by cloning the CRISPR fragment into a pACYCDuet-1 vector using the EcoNI and AvrII restriction sites.
In vitro integration assays
The integration reactions were performed in 20 mM HEPES-NaOH, pH 7.5, 25 mM KCl, 10 mM MgCl2 or MnCl2, 1 mM DTT and 10% DMSO. There was little difference when DMSO was omitted from the reaction (Fig. 1d), in contrast to its in vitro integration enhancement with HIV-1 integrase47. All of the reactions were conducted with MgCl2 unless otherwise noted. For reactions with the Cas1–Cas2 complex, separately purified Cas1 and Cas2 were pre-incubated for 20–30 min at 4 °C to allow complex formation. The protospacer DNAs were incubated with the protein(s) for 10–15 min at 4 °C, followed by the addition of the target pCRISPR or pUC19 plasmid DNA. The reactions were conducted at 37 °C for 1 h and quenched with DNA loading buffer containing a final concentration of 50 mM EDTA. The products were analyzed on 1.5% agarose gels pre-stained with ethidium bromide. All of the reactions, except those shown in Fig. 1 and Extended Data Fig. 1a, c–e, were conducted with 75 nM protein, 200 nM protospacers and 7.5 nM pCRISPR to clearly visualize Band X from pCRISPR. Reactions in Fig. 1 and Extended Data Fig. 1a,c,e were performed with 50 nM protospacers. Each integration and disintegration assay was performed a minimum of three times.
Radiolabeled protospacer integration assays
Pre-annealed double-stranded protospacer DNA substrates were 5'-radiolabeled using [γ-32P]-ATP (PerkinElmer) and T4 polynucleotide kinase (New England Biolabs). Protospacers with 3'-PO4 ends were 5'-radiolabeled using T4 polynucleotide kinase with 3' phosphatase minus activity (New England Biolabs). The reactions were carried out in the same buffer as above. Unless otherwise noted, 200 nM of Cas1–Cas2 was first incubated with 20 nM protospacers at 4 °C for 10–15 min, followed by the addition of 200 ng (~5 nM) of pCRISPR. The reactions were conducted at 37 °C for 1 h and quenched with 25 mM EDTA and 0.4% SDS. The DNAs were deproteinized with 30 μg of Proteinase K for 1 h at 37 °C and ethanol precipitated. The reactions were analyzed on 1.5% agarose gels. After electrophoresis, the gels were dried onto positively charged nylon transfer membrane (GE Healthcare) and imaged using Phosphor Screens (GE Healthcare). The restriction enzyme digest experiments were performed by first conducting the integration reaction, followed by addition of the respective enzymes (New England Biolabs), which were allowed to digest for an additional 1 h at 37 °C.
Disintegration assays
The four single stranded DNA substrates were annealed to form the Y DNA in a stepwise manner: 95 °C for 3 min, 65 °C for 20 min, 50 °C for 20 min, and gradual cooling to room temperature. The annealing reactions were analyzed on a 15% native polyacrylamide gel to confirm the formation of the Y DNA (Extended Data Fig. 5b). The disintegration assay was performed in the integration reaction buffer with 50 nM protein and 5 nM Y DNA at 37 °C for 1 h. For native polyacrylamide gel analysis, the reaction was quenched with DNA loading buffer with 50 mM EDTA and analyzed on 15% polyacrylamide gels. For denaturing polyacrylamide gel analysis, the reactions were quenched with formamide buffer and heated at 95 °C prior to loading on 15% 8M urea-polyacrylamide gels. The sequences of the four strands are as follows: A (5'-GGCCCCAGTGCTGCAATGAT-3'); B (5'- GTGAGCGTGGGTCTCGCGGTATCATTGCAGCACTGGGGCC-3'); C (5'- GCCCAATTTACTACTCGTTCTGGTGTTTCTCGTACCGCGAGACCCACGCTCAC-3'); and D (5'- ACGAGAAACACCAGAACGAGTAGTAAATTGGGC-3').
High-throughput sequencing
The integration reaction was performed with 75 nM Cas1–Cas2, 200 nM protospacer and 7.5 nM pCRISPR or pUC19 in 20 mM HEPES, pH 7.5, 25 mM KCl, 10 mM MgCl2, 10% DMSO and 1 mM DTT. The DNAs were isolated by phenol-chloroform extraction and ethanol precipitation. The excess protospacers were removed using 100K MWCO Amicon Ultra-0.5 ml centrifugal filters. The resulting integration products were digested into smaller DNA fragments using dsDNA Fragmentase (New England Biolabs) for 75 min at 37 °C and quenched at 65 °C for 15 min. Fragments were end repaired using T4 DNA Polymerase (NEB), Klenow (NEB) and T4 PNK (NEB) and A-tailed with Klenow exo (3' to 5' exo minus) (NEB). Adapters were ligated onto fragments using T4 DNA ligase (NEB) and cDNA libraries were amplified by PCR using Phusion (NEB). Libraries were sequenced on an Illumina HiSeq2500 on rapid run mode. The oligonucleotides used are: Universal adapter: 5'-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC*T-3' (*phosphorothioate bond); Indexed adapter: 5'-/5Phos/GATCGGAAGAGCACACGTCTGAACTCCAGTCAC-index-ATCTCGTATGCCGTC TTCTGCTTG-3'); PCR primers: 5'-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGA-3', 5'-CAAGCAGAAGACGGCATACGAGAT-3'.
Computational analysis
For preprocessing, 3' adapters were removed from raw Illumina reads using Cutadapt (http://code.google.com/p/cutadapt/), discarding reads shorter than 15 nt. Reads containing integrated protospacer were selected using Cutadapt, requiring the presence of at least 10 nt of protospacer sequence with no errors. After creating Bowtie48 indexes from fasta files of the pUC19 empty and pCRISPR plasmid sequences, these reads were mapped to the respective plasmids using Bowtie, allowing up to 2 mismatches and requiring unique mapping. Sequence motif analysis depicted in Extended Data Fig. 8 were generated using WebLogo, utilizing integration sites that are represented at least ten times in the sequencing data49.
Extended Data
Extended Data Figure 1. The integration reaction is dependent on the presence of protospacers, low salt and divalent metal ions.
a, In vitro integration assay alongside EcoRI- and Nb.BbvCI nickase-treated pCRISPR. b, Salt-dependence assay using Cas1 or Cas2 only and Cas1+Cas2. The titration corresponds to 0, 25, 50, 100 and 200 nM KCl, on top of the salt carried in from the reaction reagents. c, Integration assays in the presence of 10 mM EDTA, Mg2+, Mn2+ or no additive. d, Integration assays with increasing protospacer concentrations. e, A comparison of post-reaction treatments as indicated. The data presented in a-e are representative of at least three replicates.
Extended Data Figure 2. Cas1 requires Cas2 for robust protospacer integration.

a, Schematic of the integration assays using 32P-labeled protospacers (PDB code 4P6I for Cas1–Cas2). b, Integration assays in the presence of increasing protein and 10 mM MnCl2. The titration corresponds to 0, 50, 100 and 200 nM protein. c, Same as b except in the presence of 10 mM MgCl2. The data presented in b and c are representative of at least three replicates.
Extended Data Figure 3. The catalytic activity of Cas1 is required for integration.
a, Close-up view of the Cas1 active site with the conserved residues shown in stick configurations (PDB 4P6I) b, Integration assays of purified Cas1 active site mutants complexed with wild type Cas2. c, The same as b except using radiolabeled protospacers. The data presented in b and c are representative of at least three replicates.
Extended Data Figure 4. Band X corresponds to topoisomers of pCRISPR.
a, Agarose gel of purified relaxed and Band X integration products. b, Analysis of the total reaction products, after phenol chloroform extraction and ethanol precipitation, on a pre-stained agarose gel. c, Same as b except ethidium bromide staining was performed after electrophoresis. d, PCR amplification products of various segments of pCRISPR using the relaxed, Band X or pCRISPR template shown in a. The laddering effect of minor products using CRISPR locus primers likely reflects the propensity of CRISPR repeats to form DNA hairpins. The data presented in a-d are representative of at least three replicates.
Extended Data Figure 5. Cas1 catalyzes the disintegration of half-site integrated protospacers.
a, Schematic of the four strands constituting the Y DNA substrate used in the disintegration assays. b, Native polyacrylamide gel analysis of the annealing products with either Strand A or Strand C radiolabeled. c, Native polyacrylamide gel analysis of disintegration assay products using Y DNA substrates with Strand A labeled. d, Denaturing gel analysis of the disintegration assay products with Strand A labeled.
Extended Data Figure 6. Cas1–Cas2 can integrate various lengths of double-stranded DNA with blunt- or 3'-overhang ends into a supercoiled target plasmid.
a, Integration assays using the indicated lengths of protospacer DNA. b, Integration assays using varying 5' or 3' overhang lengths. c,d, A comparison of integration assays using pCRISPR or Nb.BbvCI-nicked pCRISPR target. e, Integration assay using different target plasmids with or without a CRISPR locus. The green arrows correspond to the relaxed product of each target and the cyan arrows correspond to the Band X product. The data presented in a-e are representative of at least three replicates.
Extended Data Figure 7. Cas1 tyrosine mutants support integration activity in vitro.
a, A close-up of the Cas1 active site with the tyrosine residues labeled in blue. b, Structure-based sequence alignment of Cas1 proteins, highlighting the tyrosine residues mutated to alanine in this study. c, Radiolabeled protospacer integration assay of Cas1 tyrosine mutants complexed with WT Cas2. The gel presented in b is representative of at least three replicates.
Extended Data Figure 8. High-throughput sequencing of integration products reveals sequence-specific integration.
a, Schematic of the workflow for high-throughput sequencing analysis of the integration sites. b, Raw map of the total reads along pCRISPR before collapsing into single peaks of protospacer-pCRISPR junctions depicted in Fig. 4. c, Same as b, except for the pUC19 target. d, Sequence of the leader-end of the CRISPR locus in E. coli. e,f, WebLogo analysis from the −5 to +5 positions surrounding the protospacer integration sites on the (e) plus and (f) minus of pCRISPR. The arrow points to the nucleotide that is covalently joined to the protospacer. g, h, Same as e,f, except for the pUC19 target.
Extended Data Figure 9. Cas1–Cas2 correctly orients the protospacer DNA during integration.
Mapped integration sites along the CRISPR locus of pCRISPR when using protospacer DNA with nucleotide ends (a) “wild type” 3' C and 3' T, (c) 3' A and 3' T, and (e) 3' C and 3' C. The red arrow in c and e points to the nucleotide change in the protospacer DNA compared to the “wild type” sequence in a. The protospacer DNA 3' nucleotide and the CRISPR locus strand biases in a, c, e are plotted in b, d and f, respectively, as percentages of integration events within the CRISPR locus. The black and clear bars represent the (−) and (+) strands of the CRISPR locus, respectively. NS corresponds to not significant and *p<0.0001 by Chi-square test. The n values for b, d and f are 5,623, 5,685 and 12,453 reads along the CRISPR locus, respectively.
Extended Data Figure 10. Model of the CRISPR–Cas adaptive immunity pathway in E. coli.
Mature double-stranded protospacers bearing a 3' C-OH are site-specifically integrated into the leader-end of the CRISPR locus. Correct protospacer integration (left) results in the 5'G/3'C as the first nucleotide of the spacer, proximal to the leader. After transcription of the CRISPR locus and subsequent crRNA processing, foreign DNA destruction is initiated by strand-specific recognition of the 3'-TTC-5' PAM sequence in the target strand by the crRNA-guided Cascade complex. Incorrect protospacer integration (right) cannot initiate foreign DNA destruction due to the inability for the crRNA to recognize the strand with the 3'-TTC-5' PAM. Thus, foreign DNA interference during CRISPR–Cas adaptive immunity relies on the Cas1–Cas2 complex for correctly orienting the protospacer during integration.
Supplementary Material
Acknowledgements
We are grateful to M. Chung, P.J. Kranzusch and A.V. Wright for technical assistance and members of the Doudna lab and Prof. Jamie Cate for helpful discussions. This project was funded by U.S. National Science Foundation grant No. 1244557 to J.A.D. and by NIH grant AI070042 to A.E. This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303. J.K.N. is supported by a U.S. National Science Foundation Graduate Research Fellowship and a UC Berkeley Chancellor's Graduate Fellowship. A.S.Y.L. is supported as an American Cancer Society Postdoctoral Fellow (PF-14-108-01-RMC). J.A.D. is an Investigator of the Howard Hughes Medical Institute and a member of the Center for RNA Systems Biology.
Footnotes
Author Information
Sequencing data are deposited in Gene Expression Omnibus under accession number XXXXX. The authors declare competing financial interests. J.A.D. and J.K.N. have filed a related patent application. Readers are welcome to comment on the online version of the paper.
References
- 1.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. doi:10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nature reviews. Microbiology. 2014;12:479–492. doi: 10.1038/nrmicro3279. doi:10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. Journal of molecular evolution. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. doi:10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 4.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. doi:10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 5.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. doi:10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 6.Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends in genetics : TIG. 2010;26:335–340. doi: 10.1016/j.tig.2010.05.008. doi:10.1016/j.tig.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes & development. 2008;22:3489–3496. doi: 10.1101/gad.1742908. doi:10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. doi:10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. doi:10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. doi:10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. doi:10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 12.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. doi:10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic acids research. 2012;40:5569–5576. doi: 10.1093/nar/gks216. doi:10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Datsenko KA, et al. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nature communications. 2012;3:945. doi: 10.1038/ncomms1937. doi:10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 15.Swarts DC, Mosterd C, van Passel MW, Brouns SJ. CRISPR interference directs strand specific spacer acquisition. PloS one. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. doi:10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nunez JK, et al. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nature structural & molecular biology. 2014;21:528–534. doi: 10.1038/nsmb.2820. doi:10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiedenheft B, et al. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17:904–912. doi: 10.1016/j.str.2009.03.019. doi:10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 18.Babu M, et al. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Molecular microbiology. 2011;79:484–502. doi: 10.1111/j.1365-2958.2010.07465.x. doi:10.1111/j.1365-2958.2010.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim TY, Shin M, Huynh Thi Yen L, Kim JS. Crystal structure of Cas1 from Archaeoglobus fulgidus and characterization of its nucleolytic activity. Biochemical and biophysical research communications. 2013 doi: 10.1016/j.bbrc.2013.10.122. doi:10.1016/j.bbrc.2013.10.122. [DOI] [PubMed] [Google Scholar]
- 20.Beloglazova N, et al. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. The Journal of biological chemistry. 2008;283:20361–20371. doi: 10.1074/jbc.M803225200. doi:10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samai P, Smith P, Shuman S. Structure of a CRISPR-associated protein Cas2 from Desulfovibrio vulgaris. Acta crystallographica. Section F, Structural biology and crystallization communications. 2010;66:1552–1556. doi: 10.1107/S1744309110039801. doi:10.1107/S1744309110039801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nam KH, et al. Double-stranded endonuclease activity in Bacillus halodurans clustered regularly interspaced short palindromic repeats (CRISPR)-associated Cas2 protein. The Journal of biological chemistry. 2012;287:35943–35952. doi: 10.1074/jbc.M112.382598. doi:10.1074/jbc.M112.382598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M, Craigie R. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. The Journal of biological chemistry. 2005;280:29334–29339. doi: 10.1074/jbc.M505367200. doi:10.1074/jbc.M505367200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cherepanov P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic acids research. 2007;35:113–124. doi: 10.1093/nar/gkl885. doi:10.1093/nar/gkl885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hare S, et al. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS pathogens. 2009;5:e1000259. doi: 10.1371/journal.ppat.1000259. doi:10.1371/journal.ppat.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang JY, Jayaram M, Harshey RM. Positional information within the Mu transposase tetramer: catalytic contributions of individual monomers. Cell. 1996;85:447–455. doi: 10.1016/s0092-8674(00)81122-8. [DOI] [PubMed] [Google Scholar]
- 27.DiNardo S, Voelkel KA, Sternglanz R, Reynolds AE, Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982;31:43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- 28.Pruss GJ, Manes SH, Drlica K. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell. 1982;31:35–42. doi: 10.1016/0092-8674(82)90402-0. [DOI] [PubMed] [Google Scholar]
- 29.Chow SA, Vincent KA, Ellison V, Brown PO. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 30.Au TK, Pathania S, Harshey RM. True reversal of Mu integration. The EMBO journal. 2004;23:3408–3420. doi: 10.1038/sj.emboj.7600344. doi:10.1038/sj.emboj.7600344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 32.Mizuuchi K, Adzuma K. Inversion of the phosphate chirality at the target site of Mu DNA strand transfer: evidence for a one-step transesterification mechanism. Cell. 1991;66:129–140. doi: 10.1016/0092-8674(91)90145-o. [DOI] [PubMed] [Google Scholar]
- 33.Curcio MJ, Derbyshire KM. The outs and ins of transposition: from mu to kangaroo. Nature reviews. Molecular cell biology. 2003;4:865–877. doi: 10.1038/nrm1241. doi:10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- 34.Arslan Z, Hermanns V, Wurm R, Wagner R, Pul U. Detection and characterization of spacer integration intermediates in type I-E CRISPR-Cas system. Nucleic acids research. 2014;42:7884–7893. doi: 10.1093/nar/gku510. doi:10.1093/nar/gku510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyson GW, Banfield JF. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environmental microbiology. 2008;10:200–207. doi: 10.1111/j.1462-2920.2007.01444.x. doi:10.1111/j.1462-2920.2007.01444.x. [DOI] [PubMed] [Google Scholar]
- 36.Sheflin LG, Kowalski D. Altered DNA conformations detected by mung bean nuclease occur in promoter and terminator regions of supercoiled pBR322 DNA. Nucleic acids research. 1985;13:6137–6154. doi: 10.1093/nar/13.17.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goren MG, Yosef I, Auster O, Qimron U. Experimental definition of a clustered regularly interspaced short palindromic duplicon in Escherichia coli. Journal of molecular biology. 2012;423:14–16. doi: 10.1016/j.jmb.2012.06.037. doi:10.1016/j.jmb.2012.06.037. [DOI] [PubMed] [Google Scholar]
- 38.Savitskaya E, Semenova E, Dedkov V, Metlitskaya A, Severinov K. High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli. RNA biology. 2013;10:716–725. doi: 10.4161/rna.24325. doi:10.4161/rna.24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shmakov S, et al. Pervasive generation of oppositely oriented spacers during CRISPR adaptation. Nucleic acids research. 2014;42:5907–5916. doi: 10.1093/nar/gku226. doi:10.1093/nar/gku226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deveau H, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. Journal of bacteriology. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. doi:10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Semenova E, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. doi:10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westra ER, et al. Type I-E CRISPR-cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS genetics. 2013;9:e1003742. doi: 10.1371/journal.pgen.1003742. doi:10.1371/journal.pgen.1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Craigie R, Bushman FD. HIV DNA integration. Cold Spring Harbor perspectives in medicine. 2012;2:a006890. doi: 10.1101/cshperspect.a006890. doi:10.1101/cshperspect.a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nowotny M. Retroviral integrase superfamily: the structural perspective. EMBO reports. 2009;10:144–151. doi: 10.1038/embor.2008.256. doi:10.1038/embor.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hochstrasser ML, Doudna JA. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends in Biochemical Sciences. 2014 doi: 10.1016/j.tibs.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 46.Palecek E. Local supercoil-stabilized DNA structures. Critical reviews in biochemistry and molecular biology. 1991;26:151–226. doi: 10.3109/10409239109081126. doi:10.3109/10409239109081126. [DOI] [PubMed] [Google Scholar]
- 47.Engelman A, Craigie R. Efficient magnesium-dependent human immunodeficiency virus type 1 integrase activity. Journal of virology. 1995;69:5908–5911. doi: 10.1128/jvi.69.9.5908-5911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. doi:10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome research. 2004;14:1188–1190. doi: 10.1101/gr.849004. doi:10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.