Summary
The Rpd3S histone deacetylase complex utilizes two subunits, Eaf3 and Rco1, to recognize nucleosomes methylated at H3K36 (H3K36me) with high affinity and strong specificity. However, the chromobarrel domain of Eaf3 (CHD) that is responsible for H3K36me recognition only binds weakly and with little specificity to histone peptides. Here, using Deuterium Exchange Mass Spectrometry (DXMS), we detected conformational changes of Rpd3S upon its contact with chromatin. Interestingly, we found that the Sin3-Interacting Domain of Rco1 (SID) allosterically stimulates preferential binding of Eaf3 to H3K36-methylated peptides. This activation is tightly regulated by an auto-inhibitory mechanism to ensure optimal multi-valent engagement of Rpd3S with nucleosomes. Lastly, we identified mutations at the interface between SID and Eaf3 that do not disrupt complex integrity but severely compromise Rpd3S functions in vitro and in vivo, suggesting that the nucleosome-induced conformational changes are essential for chromatin recognition.
Introduction
Histone post-translational modifications (PTM) are important cellular signals that can be read by a large repertoire of PTM recognition modules (Jenuwein & Allis, 2001) to direct many DNA template-dependent activities. These PTM reader domains have been well-documented for their ability to distinguish differently modified residues or unmodified residues (Kouzarides, 2007; Yun et al, 2011). A recent study showed that the PWWP domain of a tumor-suppressor protein ZMYND11 preferentially binds to histone H3.3 that is methylated at lysine 36 but not to methylated H3.1 (Wen et al, 2014), suggesting that the reader domain can even select for modified histone variants. The prevalent notion is that each chromatin regulator is equipped with a different combination of the PTM reading modules (Ruthenburg et al, 2007), which allows complexes that engage with nucleosomes in a multi-valent fashion to achieve robust binding and high specificity (Yun et al, 2011). The nucleosomal surface targets for these readers can be on one histone; for instance, Trim24 utilizes a tandem PHD-Bromo domain to recognize H3K4me0 and H3K23Ac on the same histone tail (Tsai et al, 2010). The targets can also be within one nucleosome, such as in the case of the PRC2 complex, which binds to a nucleosome through multiple contacts, including H3K27me, the H3 tail and H4 tails (Margueron et al, 2009; Murzina et al, 2008). Finally, nucleosomal targets can be spread over multi-nucleosomes, as has been shown for the SIR complex (Martino et al, 2009) and L3MBTL1 (Trojer et al, 2007). Another feature of chromatin structure that has emerged as a key recognition site for the chromatin complex is the linker DNA and the space between adjacent nucleosomes. Among three examples reported so far: the PRC2 histone methyltransferase complex prefers dense nucleosome arrays (Yuan et al, 2012); the Rpd3S histone deacetylase complex favors di-nucleosome units that are spaced about 30-40 bp apart (Lee et al, 2013); and the SWR1remodeler targets the longer linker and nucleosomal free regions (Ranjan et al, 2013). However, how combinations of these rather static interactions are coordinated to achieve synergetic binding remains largely unknown.
The Set2-Rpd3S pathway is one of the well-characterized chromatin signaling systems. It is responsible for maintaining stable chromatin structure in the wake of elongating RNA polymerase II to suppress cryptic transcriptional initiation, recombination, etc. (Carrozza et al, 2005; Joshi & Struhl, 2005; Keogh et al, 2005; Li et al, 2007a). The Rpd3S histone deacetylase complex utilizes the combined actions of two subunits, Eaf3 and Rco1, to recognize H3K36me nucleosomes in a robust and specific manner (Li et al, 2007b). The estimated dissociation constant of Rpd3S binding to H3K36 methylated di-nucleosomes is about 100 pM (Huh et al, 2012), making it one of the strongest chromatin binders. However, the isolated chromobarrel domain of Eaf3 (CHD), which is responsible for H3K36me recognition, binds to histone peptides poorly with little specificity (Kumar et al, 2012; Xu et al, 2008). CHD contains an optimal methyl-lysine binding pocket that is formed by four well-positioned aromatic residues (Xu et al, 2008) that seem to undergo significant rearrangements when the domain is bound to H3K36-methylated peptides (Sun et al, 2008; Xu et al, 2008). However, the rest of the domain doesn’t make extensive contact with histone peptides, which makes the binding less sequence-specific (Kumar et al, 2012; Xu et al, 2008). Another essential chromatin-interacting subunit, Rco1, contains the PHD domain and binds to histone peptides with very low affinity (Li et al, 2007b; Shi et al, 2007). Given this drastically different affinity between complex-nucleosome binding and domain-peptide interactions, we speculate that the chromatin reader within a complex may undergo nucleosome-contact-dependent conformational changes that can alter the reader domain’s ability to recognize histone PTM.
Results
Rpd3S undergoes conformational changes upon contact with nucleosomes
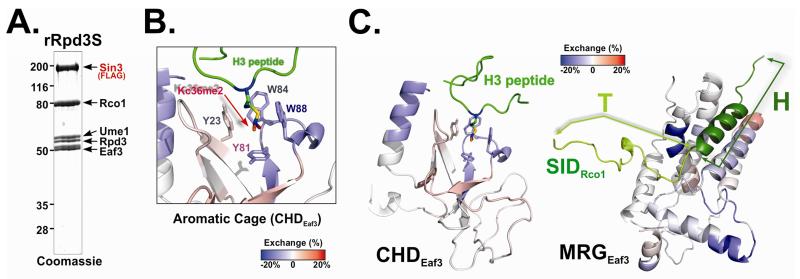
To monitor the dynamic structural changes of Rpd3S upon nucleosome contact, we employed deuterium exchange mass spectrometry (DXMS). This technique measures the hydrogen/deuterium exchange rates at each residue, which correlate with the solvent accessibility at the region (Engen, 2009). This assay can provide dynamic conformational and high-order structural information about macro-molecule complexes (Engen, 2009). We thus decided to measure the deuterium exchange profiles of Rpd3S in the absence and presence of nucleosomal substrates. Recombinant Rpd3S (rRpd3S; reconstituted in a baculovirus over expression system and purified to homogeneity as shown in Figure 1A) (Govind et al, 2010) was used because a large quantity of high-quality complex is needed for this assay. To stabilize the Rpd3S-nucleosome interaction and monitor the K36me-specific binding, we chose to use mono-nucleosomes containing methyl-lysine analogs (H3K36me3) (Huh et al, 2012). The samples of Rpd3S alone and Rpd3S mixed with nucleosomes were processed in parallel. To visualize the conformational difference between free Rpd3S and the nucleosome-bound form, the deuterium levels of free rRpd3S was subtracted from those of rRpd3S-nucleosomes (Figure 1B-C and S1B-D). Given that CHD binds to H3K36me, we expect to detect some protection at the aromatic residues of the H3K36me binding pocket. Indeed, we observed that three key residues became less solvent-exposed upon Rpd3S nucleosome binding (Figure 1B), which supported the previous observations that the pocket residues undergo conformational rearrangement upon binding to K36me (Sun et al, 2008; Xu et al, 2008). These results essentially validated our approach to detect dynamic changes of Rpd3S upon binding to nucleosomes. Other regions of CHD also undergo dramatic conformation changes, with some areas being more protected (blue) and others being more exposed (red) (Figure 1C). Furthermore, due to the highly conserved nature of the C-terminal MRG domain of Eaf3 (MRG), we were able to generate a SWISS-Model of MRG using the structure of human MRG15 bound to the Sin3-interacting domain of Pf1, the human homolog of Rco1 (Xie et al, 2012) as a template (Figure 1C). When the DXMS data were threaded on this structural model, marked conformational changes were also observed, particularly at the site of MRG that is supposed to bind to the SID domain of Rco1(Figure 1C and S1C). Therefore, we conclude that conformational changes of Eaf3, the critical subunit for nucleosome binding, can be detected upon Rpd3S chromatin substrates.
Figure 1. Rpd3S undergoes conformation changes upon Rpd3S contact with nucleosomes.
(A) Coomassie staining of the recombinant Rpd3S complex used in DXMS experiments. (B-C) Changes in the deuteration levels of Eaf3 upon Rpd3S binding to nucleosomes suggest conformational changes of Rpd3S; (B) Deuterium exchange results were mapped to 3D structure of the chromo domain of Eaf3 (PDB-2K3X). Blue color indicates slower deuterium exchange rates upon Rpd3S contact with nucleosomes, while red areas represent increased exchanges; Four aromatic residues that form the methyl-lysine binding pocket were labeled; (C) A zoom-out view of the deuterium exchange results of CHD (PDB: 2K3X) (Left) and MRG/SID (a molecular model based on PDB-2LKM using SWISS-MODEL)(the right panel). SID is represented in cartoon and green. The helix region of SID (dark green) is defined as the “H” region and the turn region (light green) is referred to as “T”. See also Figure S1.
Eaf3 can be allosterically activated to recognize H3K36me
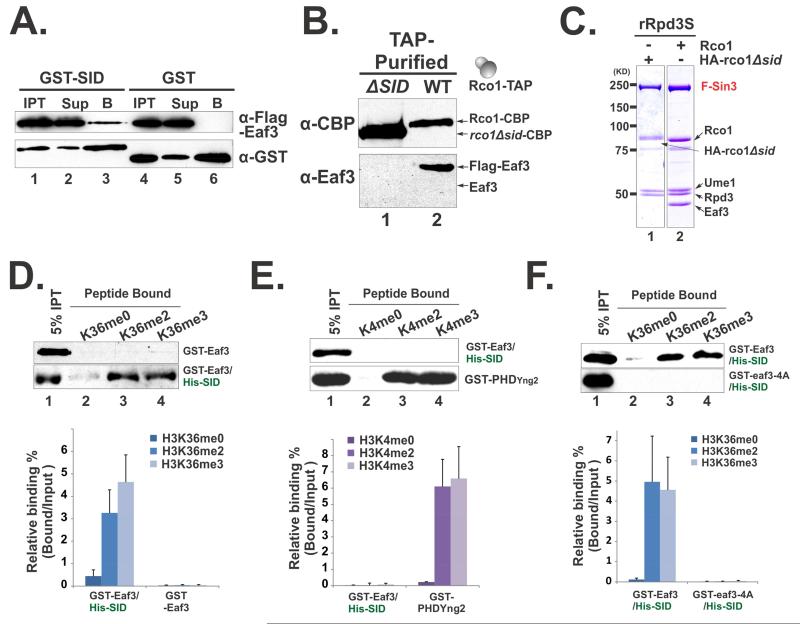
To find out the causes of the conformational changes of Eaf3 and their functional consequences, we focused on the Rco1 subunit of Rpd3S, which makes direct contact with Eaf3 and is also important for chromatin binding (Carrozza et al, 2005; Li et al, 2007b). We first sought to establish whether SID and Eaf3 interact similarly to their human counterparts (Xie et al, 2012). Using a GST pull-down assay, we showed that GST-SID can efficiently interact with FLAG-tagged Eaf3 protein (Figure 2A), suggesting that SID directly binds to Eaf3. Since Rco1 is required for incorporation of Eaf3 into Rpd3S (Carrozza et al, 2005), we asked if SID is responsible for this essential function. We first deleted SID at endogenous locus of Rco1 in yeast cells, and purified native Rpd3S mutant complex though TAP-purification. As shown in Figure 2B (Lane1), SID deletion led to Eaf3 dissociated from the complex. We then prepared recombinant Rpd3S with SID deletion from insect cells. Similarly, we found that Eaf3 is also released from the complex, while all other subunits remain bound (Figure 2C). Based on the consistent results from two independent systems, we thus conclude that SID is indispensable for tethering Eaf3 to Rpd3S.
Figure 2. Eaf3 can be allosterically activated to recognize H3K36me.
(A-C) SID is required for incorporation of Eaf3 in Rpd3S; (A) GST-SID interacts with Flag-Eaf3 in vitro as shown by GST pull-down experiments. “IPT”-Input, “Sup”-Supernatant, “B”-Bound to beads; (B) Western blots of native Rpd3S that were TAP-purified from yeast strains YCR353(ΔSID) and YBL583(WT); (C) Coomassie staining of recombinant Rpd3S complexes purified from insect cells. (D-F) SID stimulates Eaf3 to recognize H3K36me preferentially. Histone peptide pull-down assays were performed using indicated proteins or protein complexes. The upper panels in each figures show representative western blot results, while the lower panels display quantification of the western results based on at least four independent experiments. Data are represented as mean ± SEM; (D) SID increases the binding of Eaf3 to H3K36 methylated peptides; (E) SID/Eaf3 heterodimer does not bind to H3K4 methylated peptides. The PHD domain of Yng2 was used as a positive control; (F) The elevated binding of SID/Eaf3 to H3K36 methylated peptides relies on the aromatic cage of Eaf3 CHD. pBL1290 was used to purify the cage mutant of SID/Eaf3, in which all four aromatic residues were mutated to alanine (GST-eaf3-4A). See also Figure S2.
Given that SID is one of the main contacts between Eaf3 and Rco1, we hypothesized that SID may contribute to the observed Eaf3 conformational changes and potentially alter its histone binding properties. To test this, we prepared a SID/Eaf3 heterodimer using a polycistronic expression system (Figure S2A). To rule out the possibility that the SID/Eaf3 heterodimer may be contaminated with free GST-Eaf3, we showed that GST-Eaf3 was not retained on the NTA-Ni resin in the absence of HIS-SID (Figure S2B, Lane 1). We then performed peptide pull-down experiments using the histone peptides that were either unmodified or methylated at H3K36. GST-Eaf3 binds to all three peptides very weakly (Figure 2D). However, SID association dramatically increased the affinity of Eaf3 to histone peptides, particularly H3K36 di-methylated and tri-methylated peptides (Figure 2D). This result suggests that a chromatin binding domain can be allosterically activated to recognize modified histone peptides. Importantly, SID/Eaf3 does not bind to H3K4 methylated peptides (Figure 2E), suggesting that this activation is site-specific and physiologically relevant. Moreover, we showed that mutating one of the four aromatic residues within CHD abolished the binding of SID/Eaf3 to histone peptides (Figure 2F and S2C). This result indicated that elevated PTM recognition of CHD relied on previously identified methyl-lysine binding pocket.
The DNA- and histone-binding abilities of Eaf3 are self-contained
SID-mediated Eaf3 activation can explain the conformational changes of Rpd3S upon contacting nucleosomes. However, within the Rpd3S complex, SID should bind to MRG at all times in order to maintain Eaf3 association (Figure 2B and C). This implies that the SID/MRG contact alone should not automatically activate Eaf3 in the complex context. To explore the detailed mechanism underlying the dynamic changes of Rpd3S upon nucleosome contact, we decided to further investigate the properties of the PTM-reading subunit Eaf3.
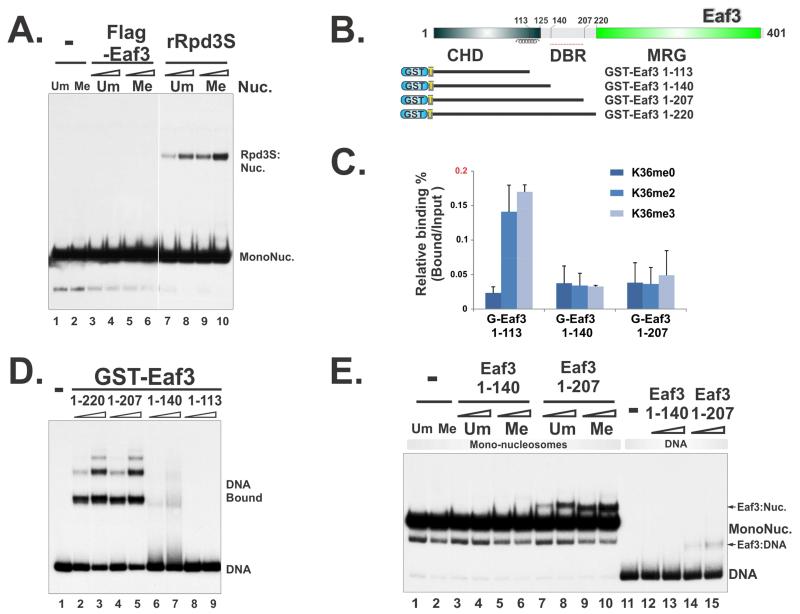
Full-length Eaf3 was purified using two independent systems. Surprisingly, neither baculovirus-expressed Eaf3 (Figure 3A) nor bacterially produced GST-Eaf3 (Figure S4A) could bind to nucleosomes or DNA. Considering the earlier data showing that Eaf3 does not bind to histone peptides (Figure 2D), the full-length Eaf3 appeared to be in a self-contained state, which allows very little affinity toward any part of the nucleosomes. To reconcile the seeming conflicts between our results and previous publications showing that weak interactions were detected using various Eaf3 CHD constructs (Carrozza et al, 2005; Sun et al, 2008; Xu et al, 2008), we systematically mapped the peptide-binding regions of Eaf3. Unlike full-length Eaf3, the modest binding of H3K36me peptides from Eaf3 (1-113) (amino acids 1-113) was also detected (Figure 3B-C), despite at a much lower lever than that of SID/Eaf3 (the relative binding of Eaf3 (1-113) to K36me3 peptide is less than 0.2%, whereas the relative binding of SID/Eaf3 to same peptides is ~5%). Comparing to Eaf3 (1-113), Eaf3 (1-124) has slightly reduced affinity to K36me peptides as reported previously (Sun et al, 2008). We therefore tested Eaf3 (1-140) and Eaf3 (1-207) and found no detectable binding (Figure 3C), suggesting that weak K36me binding of Eaf3 (1-113) was also auto-inhibited by a small extension at the C-terminus of CHD.
Figure 3. DNA and histone binding abilities of Eaf3 are self-contained.
(A) Full-length Eaf3 protein, purified from an insect-cell system, does not bind to nucleosomes and DNA. (B) Constructs for mapping Eaf3 DNA-binding regions and the histone H3K36me-binding subunit. (C) The binding of Eaf3 to histone H3K36me is auto-inhibited; Histone peptide binding assays were quantified based on three repeats. Data are represented as mean ± SEM. (D) The region of 140-207 of Eaf3 is a potential DNA-Binding Region. EMSA assay using 32P labeled 196-1X probe and GST-fused Eaf3 truncations. (E) The Eaf3 truncations that include CHD and DBR can weakly bind to nucleosomes as measured by EMSA using mono-nucleosome substrates. * indicates partially disintegrated nucleosomes, likely to be hexasomes. See also Figure S3 and S4.
We showed previously that the binding of Rpd3S to nucleosomes requires linker DNA (Li et al., 2007). However, neither the complex as a whole nor the PHD domain binds to DNA (Li et al., 2007). Since the region between CHD and MRG of Eaf3 is predicted to be a potential DNA-binding region (Figure S3), we tested the binding of the truncated constructs described above to DNA in gel-shift assays. Indeed, we observed robust DNA binding for Eaf3 (1-207) and Eaf3 (1-220), but not Eaf3 (1-140) (Figure 3D). The 140-207 segment of Eaf3 was thus defined as a potential DNA-Binding Region (DBR). Once again, similar to its affinity to histones, the DNA binding capacity of Eaf3 appears to be also blocked in the presence of the MRG domain as in the full-length Eaf3.
We next asked if any of those truncations were able to bind to nucleosomes. To avoid introducing artificial multivalent-binding potentials for nucleosomes, which can be caused by GST-dimerization (Figure S4), GST-tags on those Eaf3 truncations were removed by TEV protease digestion. His-tagged TEV proteases were subsequently depleted through Ni-NTA resins. Interestingly, monomeric Eaf3 (1-207) only weakly binds to DNA (Figure 3E, lanes 14-15), compared to GST-Eaf3 (1-207) (Figure 3E, lanes 4-5). However, the combination of this weak affinity to DNA and the low-affinity peptide binding of CHD (Figure 3C) gave rise to a weak binding of Eaf3 (1-207) to nucleosomes (Figure 3E, lanes 7-10). Collectively, these results suggest that Eaf3 possesses the potential capacity of binding to DNA and histone peptides; however, all affinities were secured in a self-contained state as an individual full-length protein.
The minimal chromatin recognition module of Rpd3S is controlled by an auto-inhibitory mechanism
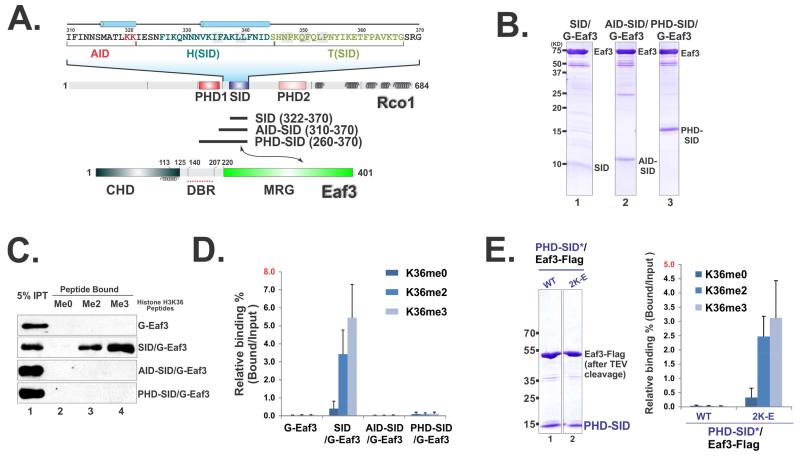
We have shown that SID alone can activate Eaf3’s histone peptide binding capacity (Figure 2D). Given that PHD and SID are closely linked in Rco1 (Figure 4A), we asked whether PHD-SID in complex with Eaf3 (PHD-SID/Eaf3) can recapitulate the multi-valent binding nature of Rpd3S. To this end, we prepared three heterodimers (as illustrated in Figure 4A) through tandem-purification to ensure the uniform stoichiometry of each component in these subcomplexes (Figure 4B). Surprisingly, PHD-SID/Eaf3 heterodimers can no longer bind to H3K36me peptides (Figure 4C-D). Indeed, the 12 amino acids between PHD and SID were sufficient to suppress SID-mediated Eaf3 activation (Figure 4C-D) without disrupting SID-MRG association (Figure 4B). This region was therefore referred to as the auto-inhibition domain (AID). To further dissect the function of AID, we identified two evolutionarily conserved lysines that could be crucial for its auto-inhibitory ability based on our molecular model at this region (Figure S5). When those residues of AID in PHD-SID/Eaf3 were mutated, we observed significantly increased H3K36me-dependent histone peptide binding (Figure 4E), which further confirmed the autoinhibitory function of this region.
Figure 4. SID-induced Eaf3 activation is controlled by an auto-inhibition mechanism.
(A). An illustration of domain structures in Eaf3 and Rco1. (B) Coomassie staining of tandem purified Eaf3-Rco1 heterodimers. (C) Histone peptide pull-down assay show that AID suppresses the SID-mediated activation of Eaf3. (D) Quantification of (C) based on three repeats. Data are represented as mean ± SEM. (E) Coomassie staining of tandem purified AID mutated PHD-SID/Eaf3 heterodimers (left). Histone peptide binding assay (right). See also Figure S4 and S5.
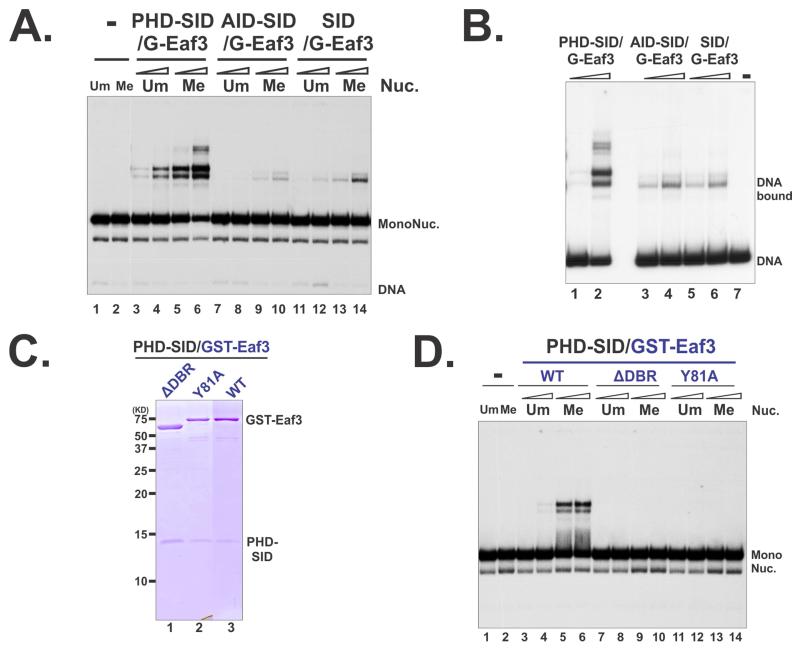
We next tested if above Rco1/Eaf3 heterodimers can bind to nucleosomes. PHD-SID/Eaf3, despite its low affinity to peptides, could bind to nucleosomes in a H3K36me-dependent fashion (Figure 5A and S6A). On the other hand, SID/Eaf3, a strong K36me-peptide binder, only displayed very weak interaction with nucleosomes (Figure 5A), underscoring that high affinity for peptides alone was not sufficient for nucleosome engagement. We then asked if the association of PHD-SID could possibly alter the Eaf3 DNA binding capacity. The results from gel-shift assays suggested that the presence of PHD in PHD-SID/Eaf3 allowed the heterodimer to bind to DNA (Figure 5B, lanes 1-2); whereas the binding of AID-SID and SID to Eaf3 did not release the DNA binding capacity of Eaf3 (Figure 5B, lanes 3-6). Consistently, PHD-SID/Eaf3 only interacted with nucleosomes containing linker DNA (Figure S4C). Therefore, one of the contributions of PHD within PHD-SID/Eaf3 towards overall binding is to allow Eaf3 contacting DNA. We further tested this possibility by examining the roles of the DBR of Eaf3 in chromatin recognition. Deletion of DBR abolished the binding of PHD-SID/Eaf3 to mono-nucleosomes (Figure 5C-D) and di-nucleosomes (Figure S6B), confirming that the DNA ability of this heterodimer is critical for its engagement with chromatin. Moreover, we showed that the Y81A mutation at the aromatic cage of CHD was also detrimental to nucleosome binding of this heterodimer (Figure 5C-D and S6B), which was consistent with the results obtained from the intact Rpd3S (Huh et al, 2012). Taken together, these results suggest that PHD-SID/Eaf3 constitutes the minimal recognition module of Rpd3S for H3K36-methylated nucleosomes. It should be noted that PHD-SID/Eaf3 nucleosome binding is still weaker than rRpd3S (Figure S4A and S4D), suggesting that other parts of Rpd3S may also make contact with nucleosomes.
Figure 5. PHD-SID/Eaf3 is the minimal nucleosome-binding module of Rpd3S.
(A) The binding Rco1/Eaf3 heterodimers were tested in EMSA using mono-nucleosome substrates. (B) EMSA using DNA alone. (C-D) The DBR and the aromatic pocket of CHD are required for the binding of PHD-SID/Eaf3 heterodimers to nucleosomes; (C) Coomassie staining of tandem purified wild type and mutant PHD-SID/Eaf3 heterodimers; Plasmids pBL1291 and pBL1296 were used to purify PHD-SID/eaf3-Y81A and PHD-SID/ eaf3ΔDBR (116-206) respectively; (D) EMSA using mono-nucleosome substrates. See also Figure S4 and S6.
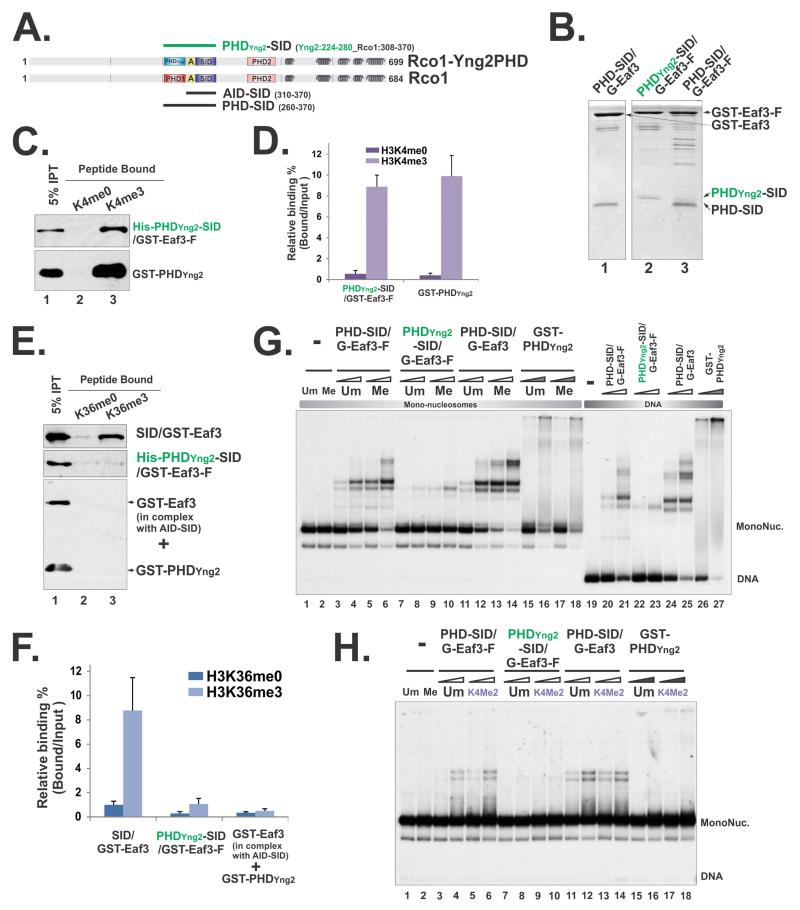
To evaluate the specific roles of PHD in this minimal chromatin recognition module, we thought to disrupt the functions of PHD without changing its structural integrity. Since the contacting residues of PHD with nucleosomes are not known, we developed an alternative approach. We have shown previously that replacement of PHDRco1 with its closest homolog PHDYng2 abrogates the functions of the Rpd3S complex both in vitro and in vivo (Li et al, 2007b). Therefore, we created a hybrid heterodimer (PHDYng2-SID/Eaf3) based on the same domain-swapping construct used in the Rpd3S mutant (Li et al, 2007b) (Figure 6A), and tested its binding to histone peptides and nucleosomes. We first examined if PHDYng2 within the hybrid heterodimer could still bind to H3K4me. As shown in Figure 6C and D, the hybrid heterodimer specifically interacted with H3K4 methylated peptides similarly to GST-PHDYng2 domain alone, suggesting the structural integrity of PHDYng2 was well maintained in this heterodimer. However, the hybrid PHDYng2-SID/Eaf3 did not bind to H3K36me peptides (Figure 6E-F), indicating that the AID domain could still effectively repress SID-mediated Eaf3 activation. We next sought to test if PHDYng2-SID/Eaf3 could bind to nucleosomes using gel-shift assays. Despite the strong interaction between H3K4me and PHDYng2-SID/Eaf3, this heterodimer did not efficiently bind to DNA nor nucleosomes that are methylated at H3K36 (Figure 6G) or H3K4 (Figure 6H). This result suggested that replacing PHD with a stronger histone PTM reader (PHDYng2) did not lead to more efficient nucleosome engagement; rather it disrupted the coordinated actions between Rco1 and Eaf3 that are essential for engaging to nucleosomal substrates.
Figure 6. Perturbation of PHD domain within the minimal nucleosome binding module compromised its nucleosome engagement.
(A) A schematic illustration of the PHDYng2-SID/Eaf3 construct. “A” framed in the yellow box represents the AID domain. Amino acids that were included in the hybrid protein were indicated by the residue numbers behind each constructs. (B) Coomassie staining of tandem purified hybrid Rco1-Eaf3 heterodimers. (C) Histone peptide pull-down using unmethylated and methylated H3K4 peptides. (D) Quantification of (C) based on three repeats. Data are represented as mean ± SEM. (E) Histone peptide pull-down using unmethylated and methylated H3K36 peptides. The low level binding of PHDYng2-SID/Eaf3 was likely due to PHDYng2-somehow slightly compromises AID function, because when AID-SID/Eaf3 heterodimers and GST-PHDYng2 were mixed together, no binding was detected. (F) Quantification of (E) based on three repeats. (G) EMSA using mono-nucleosome substrates that unmethylated or tri-methylated at H3K36 and DNA. Two concentrations of heterodimers were 15pM, 30pM respectively and indicated as open triangles. 3.2μM and 6.4μM of GST-PHDYng2 were used and labeled as filled triangles. (H) EMSA using mono-nucleosome substrates that unmethylated or di-methylated at H3K4. 1.6μM and 3.2μM of GST-PHDYng2 were used.
Induced conformational changes are essential for Rpd3S function in vivo
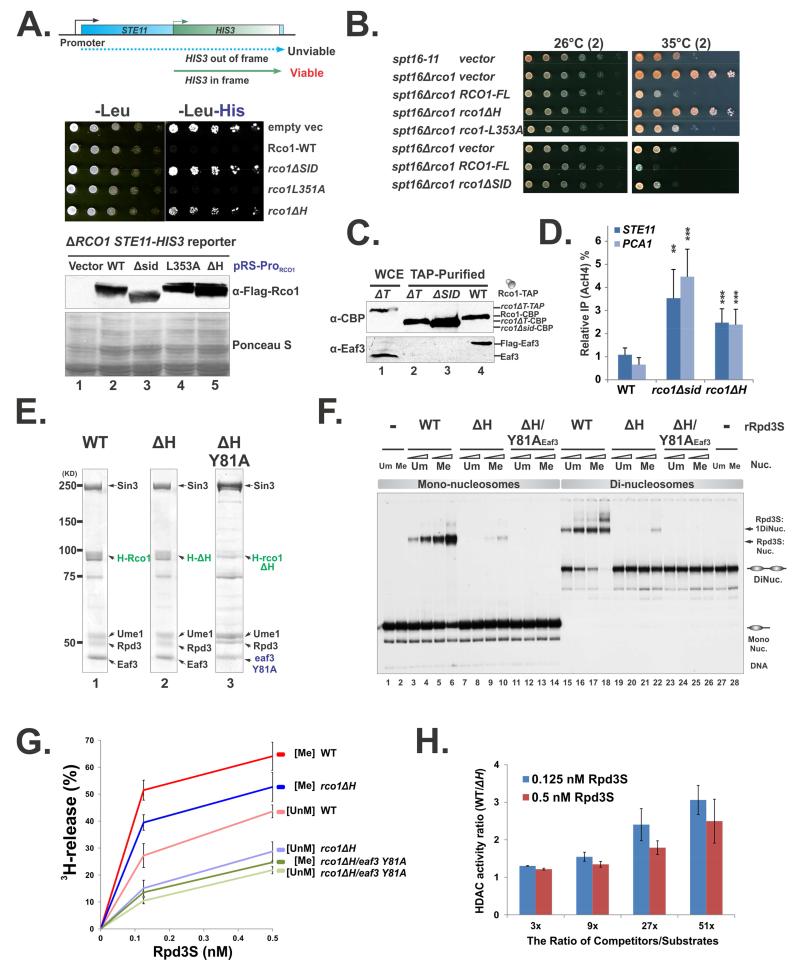
We next investigated the physiological importance of the SID-induced Eaf3 activation. We rationalized to identify a mutation at the SID/MRG binding interface that abolishes nucleosome-induced conformational changes without disrupting the complex so that we can test whether those mutations affect Rpd3S functions both in vitro and in vivo. To this end, we designed two independent genetic systems to test all selected Rco1 mutants. The first system was adapted from a previous strategy that utilized FLO8-HIS3 reporter genes to detect cryptic transcription phenotype (Cheung et al, 2008). Since STE11 is more sensitive to defects in the Set2-Rpd3S pathway (Carrozza et al, 2005), we generated a genome-integrated STE11-HIS3 reporter yeast strain to test our mutants (Figure 7A). In this system, the functional His3 protein can only be produced when the HIS3 transcript initiates at the cryptic promoter of STE11 (Figure 7A). Therefore, the reporter strain can only grow on histidine-depleted plates when RCO1 is deleted. Introduction of a plasmid that carries the wild-type RCO1 gene that is driven by its own promoter suppressed the growth of the reporter strain (Figure 7A row 2). When mutant Rco1 plasmids were transformed into the reporter strain, different phenotypes were observed even when all proteins were expressed at comparable levels (Figure 7A, the lower panel). As expected, deletion of the entire SID caused the loss of Eaf3 from Rpd3S and exhibited the cryptic transcription phenotype (Figure 7A). However, even with removal of the helical part of SID (defined as the “H” region, Figure 1C), the ΔH mutant, clear Rpd3S pathway defects were also detected (Figure 7A). L353 is a critical interface residue in the Rco1 mammalian counterpart, and the mutation of this residue decreases the MRG/SID interaction (Xie et al, 2012). However, incorporation of the L353A mutation to Rco1 did not lead to a detectable phenotype (Figure 7A). The second complementary system we used took advantage of the fact that deletion of RCO1 can partially rescue defects caused by the FACT mutation (spt16-11) (Figure 7B) (Biswas et al, 2008). Consistent with the trend of the mutant phenotype described above (Figure 7A), this genetic system also revealed that ΔH severely compromised the function of Rpd3S in vivo (Figure 7B). A similar mutation- ΔT (deletion of the “T” region of SID, Figure 1C) also displayed a strong phenotype. However, TAP-purification showed that this mutation caused Eaf3 to dissociate from Rpd3S (Figure 7C, Lane2). Therefore, the ΔT mutant did not meet the criteria that we established above, and this mutation was not further investigated. Furthermore, we performed chromatin immunoprecipitation experiments to confirm that ΔH disrupts the HDAC activity of Rpd3S in vivo. As shown in Figure 7D, elevated levels of histone acetylation (AcH4) were observed at the coding regions of two model genes PCA1 and STE11, which were shown to be the Set2-Rpd3S regulated genes (Li et al, 2007c). Collectively, these three lines of evidence suggest that ΔH disrupts Rpd3S function in vivo.
Figure 7. Conformational changes are essential for Rpd3S function.
(A) Test cryptic transcription phenotype caused by Rco1 mutants in an STE11-HIS reporter strain (YCR239). The bottom panel: Western blot shows that all mutant proteins are expressed at similar levels. (B) Test Rco1 function in FACT mutants. Plasmids carrying wild type or mutant RCO1 under the control of its native promoter (parental vector pBL1114) were transformed into YBL823 (spt16-11 ΔRCO1). The resulting strains were subjected to spotting assays and grown at semi-permissive temperature. (C) Deletion of the “T” region (ΔT) results in the loss of Eaf3 from Rpd3S in vivo. TAP purified Rpd3S complexes were subjected to western blot to monitor the association of Eaf3 with Rpd3S. Noted that wild type Rco1 strain containing a Flag-Eaf3 (YBL768), therefore the Eaf3 bands as detected by a polyclonal antibody against Eaf3 migrates slower than untagged version (Lane 1). (D) ChIP assay using an antibody against AcH4 show that disruption of SID/Eaf3 interaction interface results in increased acetylation levels at coding regions of the STE11 and PCA1 gens. IP efficiency of each gene was normalized to AcH4 IP efficiency at the Y region (a gene desert on Chromosome 6 which serves as an internal control). Data are represented as mean ± SEM, N>3, *(P<0.05) **(P<0.01), ***(P<0.001) based on 2-tailed Student T-Test. (E-H) Deletion of the “H” region (ΔH) does not disrupt complex integrity but compromises Rpd3S functions in vitro; (E) Coomassie staining of Rpd3S ΔH mutants; (F) EMSA assays using mono- and di-nucleosomes; (G) Nucleosome-based HDAC assay for indicated Rpd3S complexes. The deacetylation activity, which is indicated by the amount of free 3H release, was plotted against the concentration of Rpd3S; (H) Defects caused by ΔH are more severe under stringent competition. Increasing amount of competitors (DNA and HeLa oligonucleosomes) were added into each HDAC reactions as shown in (G), The ratio of the HDAC activity of wild type Rpd3S over the ΔH mutant was shown as a function of the amount of competitors.
Having mutant candidates that showed functional defects of Rpd3S in vivo, we next examined their biochemical properties. We prepared rRpd3S that contained the ΔH mutation and demonstrated that ΔH did not disrupt the integrity of the complex (Figure 7E, Lane 2). Gel-shift experiments using 32P-labeled mono-nucleosomes and di-nucleosomes (Huh et al, 2012) were then performed to test if the mutant complex binds to nucleosomes. Consistent with previous results (Huh et al, 2012; Lee et al, 2013), wild type Rpd3S can recognize K36-methylated nucleosomes (Figure 7F, lanes 3-4 vs. 5-6) and prefer di-nucleosomes (Figure 7F, lanes 15-18). Importantly, the binding of ΔH mutant complex to both mono- and di-nucleosomes was dramatically reduced (Figure 7F Lane 7-11 and Lane 19-22). We attributed the weak association of ΔH rRpd3S with nucleosomes to the low-affinity state of CHD. Previously, all four aromatic residues in CHD that form the H3K36me binding pocket have been shown to be essential for the binding of CHD to histone peptides (Xu et al, 2008). We demonstrated here that mutations at any of these residues also give rise to cryptic transcription phenotype in vivo (Figure S2C). Therefore, we introduced Y81A mutation to the ΔH rRpd3S (Figure 7E) and found that this mutation eliminated the residual binding seen above from both mono-nucleosomes and di-nucleosomes (Figure 7F, lanes 11-14 and 23-26).
Once we established that the ΔH mutation compromises the binding of Rpd3S to nucleosomes, we asked if this mutation also influences the HDAC activity of Rpd3S in a similar manner using nucleosome-based histone deacetylase assays that we developed previously (Huh et al, 2012). We showed previously that Rpd3S displays stronger HDAC activity toward methylated nucleosomes, and it also favors di-nucleosomes over mono-nucleosomes, when each single parameter was evaluated (Huh et al, 2012). Interestingly, although Rpd3S binds to unmodified di-nucleosomes with higher affinity than to methylated mono-nucleosomes, it shows stronger HDAC activity towards K36 methylated mono-nucleosomes (Huh et al, 2012), suggesting that K36me may potentially stimulate Rpd3S catalytic activity as well (Drouin et al, 2010). Here, Similar to the binding defects of these mutant complexes (ΔH and ΔH-Y81A), we found that their HDAC activities were also compromised on both methylated and unmethylated mono-nucleosomes (Figure 7G). It was noted that ΔH Rpd3S on methylated nucleosomes showed more HDAC activity than that by wide type Rpd3S on unmethylated nucleosomes (Figure 7G), but the binding of ΔH Rpd3S to methylated nucleosomes (Figure 7F, Lane 9-10) is weaker than that of wild type Rpd3S. This seeming discrepancy reminisces the phenomenon described above (Huh et al, 2012), which provides another support for a role of H3K36me in Rpd3S catalytic activation. We noticed that the defects caused by these mutations were relatively subtle in the absence of competitors (Figure 7G). However, as the competitor levels increased, which more closely resembles the physiological conditions, the defects of HDAC activity caused by those mutations became more evident (Figure 7H). In summary, Rpd3S uses multiple domains to recognize nucleosomal substrates, including CHD, PHD (Li et al, 2007b) and the DBR of Eaf3 (Figure 3). In the ΔH mutant, all these known chromatin-contacting modules remain intact in the complex, but the complex is not functional. These results strongly suggest that SID-mediated allosteric activation of CHD plays pivotal roles in regulating Rpd3S functions.
Discussion
Allosteric activation is an important regulatory mechanism to control enzyme activity. Previously it has been shown that the histone methyltransferase activity of EZH2 can be allosterically stimulated through another subunit of PRC2-EED through its contact with H3K27-methylated peptides (Margueron et al, 2009). Here, we report a molecular mechanism by which a low-affinity chromatin modification reader can be allosterically converted into a strong binder upon nucleosomes contact. This discovery has two important implications on our understanding of chromatin recognition: (1) Histone modification readers may not be the simple static units that we previously thought, and their reading properties may be dynamically regulated in different contexts. (2) Chromatin modifying complexes may also require a pivotal “nerve” system to sense the environment and direct their dynamic multi-valent interactions with nucleosomes. Simple combinations of several histone binding domains are not sufficient to efficiently engage with chromatin substrates.
The investigation on how these conformational changes were triggered also led us to identify a minimal module of Rpd3S that can effectively recapitulate the chromatin binding capacity of the entire complex. With this reduced form, we were able to discover that the allosteric activation of Eaf3 is regulated by an auto-inhibitory mechanism. We therefore proposed a “touch-then-lock” mode of chromatin recognition: The complex first “touches” nucleosomes through several weak interactions, presumably to orient the complex to a favorable position. The high-affinity histone binding is then induced to “lock” the complex onto modified nucleosomes. Thus, it is the collective contributions of several weak and inducible strong interactions that dictate the efficient nucleosome engagement. We believe that the allosteric activation of Rpd3S can be more advantageous than a simple high-affinity reader. This mechanism may be more conducive to chromatin remodelers that modify histones along moving machinery. Accumulating evidence suggests that many chromatin-modifying enzymes are associated with traveling machinery such as RNA polymerase II, replicasome etc. In particular, Rpd3S has been speculated to travel with elongating RNA polymerase II (Drouin et al, 2010; Govind et al, 2010). The action-on-the-run type of reaction demands that the enzymes not only hold the substrates tightly but also release the products rapidly so that it will not slow down elongating polymerase II. Obviously, the price of using a constant high-affinity binder is that it would be hard to dissociate the enzymes from the products. In contrast, the allosteric activation mediated binding not only can achieve equally high affinity, but also it allows for easy enzyme release. This is because disengaging the weakly interacting “touch” components should in turn lead to a loosening of the “lock” mechanism, which will then detach enzymes from the products.
We found that PHDYng2 within the hybrid-heterodimer (PHDYng2-SID/Eaf3) can bind to H3K4me peptides similarly to GST-PHDYng2. However, this hybrid heterodimer does not bind to nucleosomes that are fully methylated at H3K4 (Figure 6H). PHDYng2/K4me interaction is widely considered as one of the strongest reader domain/histone peptides binding with a Kd in μM range. Consistently, the binding of GST-PHDYng2 to H3K4 methylated nucleosome is also at a Kd around ~10μM (Figure 6G and H). However, this affinity is far below the sub-nano Molar nucleosome binding of Rpd3S as we reported previously (Huh et al, 2012; Lee et al, 2013) and the binding of other complexes such as RSC, Chd1 etc.(Li et al, 2007b). A common feature among those strong nucleosome binders is that they all have strong affinity towards linker DNA. Therefore, we propose that it is the chromatin complex/DNA interaction that is mainly responsible for stable nucleosome engagement. The reader-histone contacts may mainly provide binding specificities with minor contribution towards overall affinity.
Experimental Procedures
Deuterium exchange mass spectrometry (DXMS)
The Rpd3S used in this assay was purified using Rco1-Flag from co-infected Sf21 insect cells (Govind et al, 2010). Mono-nucleosomes were prepared using recombinant Xenopus histone octamers that contain the methyl-lysine analog (MLA) H3K36me3 (Simon et al., 2007) and the 216 bp DNA template that includes a 601 positioning sequence as descried previously (Huh et al., 2012). Reconstituted nucleosomes were purified from 491 prep cells (Bio-Rad) (Yun et al, 2012). The ratio of Rpd3S to nucleosomes was determined via titration in an EMSA assay so that there was an excess amount of H3K36me3 nucleosomes in the reaction to ensure no free Rpd3S was present. Prior to conducting the deuterium exchange experiments, we optimized the pepsin-mediated proteolysis and quenching conditions to maximize peptide sequence coverage of mass-spec as described previously (Hsu et al., 2009; Li et al., 2011) (Figure S7A). Briefly, 2 μl of the Rpd3S stock (1.5 mg/ml in 10 mM Tris-HCl, pH7.5, 5 mM βME and 150 mM NaCl, purified through Flag-Rco1) was mixed with 6 μl of the H2O buffer (8.3 mM Tris-HCl pH7.2 and 150 mM NaCl), and 12 μl of different quench solutions (0.8 M, 1.6 M or 3.2 M GuHCl in 0.8% formic acid, 16.6% glycerol) on ice. The Rpd3S samples were then subjected to proteolysis, and the resulting peptides were separated and analyzed by mass spectrometry. The best peptide coverage maps of Rpd3S were obtained using 1.6 M GuHCl quench solution.
To prepare the samples of the Rpd3S alone and the Rpd3S-H3K36me3 nucleosome complex for functional deuteration studies, 12 μl of the Rpd3S stock was incubated with 6 μl of the nucleosome buffer (10 mM Tris-HCl pH7.5 and 5 mM βME) or 6 μl of H3K36me3 nucleosome stock (3.75 mg/ml) at 30°C for 60 min. Both samples were then cooled to 0°C. Deuterated samples were prepared by mixing 2 μl of the above mixtures (Rpd3S or Rpd3S-H3K36me3) with 6 μl of D2O buffer (8.3 mM Tris, 150 mM NaCl in D2O, pDREAD 7.2) at 0°C. Deuteration was stopped at different time points by adding 12 μl of quench buffer (1.6 M GuHCl, 0.8% formic acid and 16.6% glycerol) on ice, followed by freezing at −80°C. Duplicate samples were collected at three time points: 10 sec at 0°C and 100 sec and 10,000 sec at room temperature. Because the exchange rates of backbone amide hydrogen at 0°C is 10 times slower than that at room temperature (Hastie et al., 2011), 10 sec at 0°C is equivalent to 1 sec at room temperature. The data in Figure 1 and Figure S1 are shown as the equivalent of the deuteration time at room temperature. In addition, duplicate non-deuterated control samples (incubated in 6 μl the H2O buffer) (Figure S7B, labeled as ND) and a single equilibrium-deuterated control sample (incubated in the 6 μl D2O buffer containing 0.5% formic acid at 25°C overnight) (Figure S7B, labeled as FD) were also prepared. Upon collecting all samples, they were thawed on ice and passed over AL-20-pepsin columns (Sigma, 16 μl bed volume) at a flow rate of 20 μl/min. The resulting peptides were collected on a C18 trap (Michrom MAGIC C18AQ 0.2×2) and separated by a C18 reverse phase column (Michrom MAGIC C18AQ 0.2×50) running a linear gradient of 8–48% solvent B (80% acetonitrile and 0.01% TFA) over 30 min. The column effluents were then directly injected into an OrbiTrap Elite mass spectrometer (Thermo Fisher) for analysis. The instrument was operated in positive ESI mode with a sheath gas flow of 8 units, a spray voltage of 4.5 KV, a capillary temperature of 200°C and an S-lens RF of 67%. Mass spec data was acquired in both profile and data-dependent modes. The resolution of the survey scan was set at 60,000 at m/z 400 with a target value of 1e6 ions and 3 microscans. The maximum injection time for MS/MS was varied between 25 msec and 200 msec. Dynamic exclusion was 30 sec and early expiration was disabled. The isolation window for MS/MS fragmentation was set to 2, and the five most abundant ions were selected for product ion analysis. Proteome Discoverer software (Thermo Fisher) was used to identify the sequence of the peptide ions. The centroids of the isotopic envelopes of non-deuterated, partially deuterated and equilibrium-deuterated peptides were measured using DXMS Explorer (Sierra Analytics, Inc., Modesto, CA) (Figure S7B) and then converted to corresponding deuteration levels. The data process was essentially carried out as described previously (Burns-Hamuro et al, 2005) with more streamlined computation tools for handling larger datasets.
EMSA assays
Mono-nucleosome DNA probes containing the 601 sequence flanked by 75 bp and 26 bp linkers (196-1X) and a di-nucleosome template (196-2X, named ChrT04 (Huh et al., 2012)) were end-labeled using T4-PNK (NEB) with 32P-γATP (Yun et al., 2012). Nucleosomes were reconstituted via a salt-dilution method using unmodified Xenopus core histones and the MLA H3K36me3 core histones (Yun et al., 2012). All nucleosomes were gel-purified. EMSA reactions were carried out in a 15 l system containing 10 mM HEPES pH7.8, 50 mM KCl, 4 mM MgCl2, 5 mM DTT, 0.25 mg/ml BSA, 5% glycerol and 0.1 mM PMSF. The samples were incubated at 30°C for 45 min and run on a 3.5% acrylamide (37.5:1) gel at 4°C.
Peptide pull-down assays
Peptide pull-down assays were performed as described previously with minor modifications (Li et al., 2003). Biotinylated histone H3K36 peptides (H3 (21-44), unmodified (me0), di-methylated (me2) and trimethylated (me3)) were custom-made by Sigma Genosys. Biotinylated histone H3K4 peptides were purchased from Millipore (H3 1-21 unmodified (12-403), di-methylated (me2) (12-460) and trimethylated (me3) (12-564)). 8 μg of each histone peptide were coupled to 0.1 mg of streptavidin-coated Dynabeads M280 in 50 μl of coupling buffer (25 mM Tris-HCl pH8.0; 1 M NaCl; 1 mM DTT; 5% glycerol; 0.03% NP-40) at 4°C for 2 hr. The beads were washed with peptide binding buffer (50 mM Tris-HCl pH7.5; 300 mM NaCl; 0.1% NP-40) then stored at 4°C. 320 ng of each peptide were used for pull-down assays with 1 μg of Eaf3 or equal molar ratio of Rco1-Eaf3 heterodimer in 25 μl peptide binding buffer. After a 2 hr incubation at 4°C on a Dyna-Mixer (Dynal), the beads were washed three times with peptide binding buffer, eluted using 10 μl 3×SDS loading buffer at room temperature for 15 min and subjected to western blotting.
Nucleosome-based HDAC assays
Recombinant Xenopus nucleosomes were reconstituted using a 248 bp DNA containing the 601 positioning sequence (601B) (Huh et al., 2012) and purified through the 491 prep cell system (Bio-Rad). The resulting nucleosomes were acetylated to saturated levels using a mixture of the histone acetyltransferase complexes (ADA2-TAP and SAGA) and 3H-acetyl-CoA (Yun et al., 2012). Then, 30-50 nmol of 3H-labeled acetylated nucleosomes was used in each HDAC reaction in the presence of HeLa oligonucleosome competitors. The final volume was adjusted to 15 l using CEB buffer (10 mM Tris-HCl pH8.0; 150 mM NaCl; 1 mM magnesium acetate; 1 mM imidazole; 2 mM EGTA pH8.0; 10 mM βME; 0.1% NP40 and 10% glycerol). After a 1 hr and 20 min incubation at 30°C, 20 l of H2O, 36 l of 1 M HCl/0.4 M acetic acid and 800 μl of ethyl acetate were added to stop the reactions. The mixtures were vigorously vortexed and centrifuged at 12,000 g at 4°C for 10 min. Then, 750 μl of supernatant was mixed with 4 ml of scintillation fluid for liquid scintillation counting.
Supplementary Material
Acknowledgements
We thank Drs. Conaway, Tan, Workman and Stillman for providing reagents and yeast strains, Dr. Xin Liu for assistance on structural modeling and Drs. Harrod for critical reading. BL is a W.A. “Tex” Moncrief, Jr. Scholar in Medical Research and is supported by grants from the National Institutes of Health (R01GM090077), the Welch Foundation (I-1713) and the American Heart Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contribution:
CR, CL, HC, SL and BL conceived the project and designed experiments. CR, CL, HC, SL and BL performed experiments. CR and BL wrote the manuscript.
References
- Biswas D, Takahata S, Stillman DJ. Different Genetic Functions for the Rpd3(L) and Rpd3(S) Complexes Suggest Competition Between NuA4 and Rpd3(S) Mol Cell Biol. 2008 doi: 10.1128/MCB.00164-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns-Hamuro LL, Hamuro Y, Kim JS, Sigala P, Fayos R, Stranz DD, Jennings PA, Taylor SS, Woods VL., Jr. Distinct interaction modes of an AKAP bound to two regulatory subunit isoforms of protein kinase A revealed by amide hydrogen/deuterium exchange. Protein Sci. 2005;14:2982–2992. doi: 10.1110/ps.051687305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008;6:e277. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin S, Laramee L, Jacques PE, Forest A, Bergeron M, Robert F. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet. 2010;6:e1001173. doi: 10.1371/journal.pgen.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engen JR. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Analytical chemistry. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JW, Wu J, Lee CH, Yun M, Gilada D, Brautigam CA, Li B. Multivalent di-nucleosome recognition enables the Rpd3S histone deacetylase complex to tolerate decreased H3K36 methylation levels. The EMBO journal. 2012;31:3564–3574. doi: 10.1038/emboj.2012.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kumar GS, Chang W, Xie T, Patel A, Zhang Y, Wang GG, David G, Radhakrishnan I. Sequence requirements for combinatorial recognition of histone H3 by the MRG15 and Pf1 subunits of the Rpd3S/Sin3S corepressor complex. Journal of molecular biology. 2012;422:519–531. doi: 10.1016/j.jmb.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Wu J, Li B. Chromatin remodelers fine-tune H3K36me-directed deacetylation of neighbor nucleosomes by Rpd3S. Molecular cell. 2013;52:255–263. doi: 10.1016/j.molcel.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007a;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007b;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 2007c;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, Ziegler M, Cubizolles F, Cockell MM, Rhodes D, Gasser SM. Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol Cell. 2009;33:323–334. doi: 10.1016/j.molcel.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, Laue ED. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A, Mizuguchi G, FitzGerald PC, Wei D, Wang F, Huang Y, Luk E, Woodcock CL, Wu C. Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell. 2013;154:1232–1245. doi: 10.1016/j.cell.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, Chan SM, Martin DG, Fingerman IM, Briggs SD, Howe L, Utz PJ, Kutateladze TG, Lugovskoy AA, Bedford MT, Gozani O. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Hong J, Zhang P, Dong X, Shen X, Lin D, Ding J. Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36. The Journal of biological chemistry. 2008;283:36504–36512. doi: 10.1074/jbc.M806564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P, Li G, Sims RJ, 3rd, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, Wang YH, Reinberg D. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, Tsai CY, Shi X, Schwarzer D, Plunkett W, Aronow B, Gozani O, Fischle W, Hung MC, Patel DJ, Barton MC. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468:927–932. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Li Y, Xi Y, Jiang S, Stratton S, Peng D, Tanaka K, Ren Y, Xia Z, Wu J, Li B, Barton MC, Li W, Li H, Shi X. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014;508:263–268. doi: 10.1038/nature13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Graveline R, Kumar GS, Zhang Y, Krishnan A, David G, Radhakrishnan I. Structural basis for molecular interactions involving MRG domains: implications in chromatin biology. Structure. 2012;20:151–160. doi: 10.1016/j.str.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Cui G, Botuyan MV, Mer G. Structural basis for the recognition of methylated histone H3K36 by the Eaf3 subunit of histone deacetylase complex Rpd3S. Structure. 2008;16:1740–1750. doi: 10.1016/j.str.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N, Li X, Xu M, Zhang Z, Niu T, Han Z, Chai J, Zhou XJ, Gao S, Zhu B. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012;337:971–975. doi: 10.1126/science.1225237. [DOI] [PubMed] [Google Scholar]
- Yun M, Ruan C, Huh JW, Li B. Reconstitution of modified chromatin templates for in vitro functional assays. Methods Mol Biol. 2012;833:237–253. doi: 10.1007/978-1-61779-477-3_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell Res. 2011;21:564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.