Abstract
This paper describes a methodology for redesigning the clinical processes to manage diagnosis, follow-up, and response to treatment episodes of breast cancer. This methodology includes three fundamental elements: (1) identification of similar and contrasting cases that may be of clinical relevance based upon a target study, (2) codification of reports with standard medical terminologies, and (3) linking and indexing the structured reports obtained with different techniques in a common system. The combination of these elements should lead to improvements in the clinical management of breast cancer patients. The motivation for this work is the adaptation of the clinical processes for breast cancer created by the Valencian Community health authorities to the new techniques available for data processing. To achieve this adaptation, it was necessary to design nine Digital Imaging and Communications in Medicine (DICOM) structured report templates: six diagnosis templates and three summary templates that combine reports from clinical episodes. A prototype system is also described that links the lesion to the reports. Preliminary tests of the prototype have shown that the interoperability among the report templates allows correlating parameters from different reports. Further work is in progress to improve the methodology in order that it can be applied to clinical practice.
Keywords: Structured reporting, Breast cancer, BI-RADS, Clinical oncology, Information storage and retrieval
Introduction
The influence of digital medical images in the improvement of clinical diagnostic process has caused a major revolution in medicine. However, despite the widespread use of computer-based systems like picture archiving and communication system (PACS) [1], radiology information system (RIS) [2], or hospital information system (HIS) [3], these systems do not totally fit the requirements for collaboration among different administrative domains (e.g., sharing information about relevant cases between two hospitals). However, research projects based on grid computing [4], peer to peer [5], and cloud computing [6] propose collaborative frameworks among unaffiliated centers [7–9] addressing this issue.
The exploitation of such distributed information has led to new research lines, which are mainly focused on the guidance, organization, and extraction of knowledge to assist radiologists and researchers to access and use existing medical information repositories. This constitutes a major advantage when compared to other approaches relying on less information on which to base their decisions and will be translated in the improvement of the clinical management of patients. In this process, the quality of input data is a key factor to achieve the best results.
Medical informatics has long ago focused on the identification and definition of standard terminologies to provide physicians with an unambiguous and precise schema to create reports that can be processed by computers. They have proven to be useful to facilitate data mining procedures on the coded reports and images [10]. Natural text searching presents many ambiguities and false positives [11, 12], which reduce their applicability in patient health care. In this sense, the use of ontologies can improve the way in which data is organized [13], reducing the ambiguities of plain text descriptions [14–17]. Structuring radiology reports constitutes a step beyond coding and defining terminologies, as they are bound to the clinical pathway and objectives of the study.
However, the definition of general-purpose structured reports is often ineffective due to the particularities of different image modalities, clinical protocols, and medical disciplines [18]. Notwithstanding their difficulties, structured reports offer a homogeneous way to define the reports, enhancing the capability of computer-enabled tools to extract knowledge and to search and compare reports.
To cope with this limitation, the present work defines six templates for structured reports for six diagnostic modalities present in clinical pipelines of breast cancer (clinical examination, mammography, ultrasound, magnetic resonance imaging (MRI), pre- and postsurgical biopsy), ending up with one single, comprehensive report.
The structured reports provide the basis on which a knowledge database was built. The objective of this database is to facilitate the search for relevant reports and, in the future, to support training and clinical decision. Currently, a prototype implementation exists that automatically identifies retrospective cases together with the associated diagnosis and other relevant information. This is currently done by comparing a case provided by the radiologist with the reports stored in the database, which is possible through the use of the fields of the report that are annotated with standard terminologies. The system returns additional evidence that the radiologist can use for the assessment of the diagnostic.
The use of such a tool can assist in identifying cases with similar (or dissimilar) clinical features. Clinical information retrieval has been revealed as a powerful tool to improve diagnosis [19]. However, searching for similarities on medical databases is often not an effective task. This is especially the case when annotations are provided in natural language, since very different annotations could lead to similar diagnosis.
Extracting all relevant concepts from a narrative report is especially challenging, since text data has a very high variability that limits its clinical usefulness. For example, searching free text for negative statements usually causes many false negatives. Also, synonyms can be used by different radiologists to annotate the same finding. For example, one radiologist may prefer the term nodule, while others may use mass or lesion. Searching for reports annotated in this manner will reduce the chance to find all relevant information when not all synonyms are considered, which is impractical in many cases. Another challenge is to deal with semantic aspects of the information. For example, a report can be annotated as mass in the upper outer quadrant, but a user can search for cancer in the right breast. The software that implements the search must then identify whether this annotation belongs to the right breast or not.
Basing searches on standard annotations is of great value, since it produces more clinically relevant results than free text searches [20]. For example, many relationships can be expressed in terms of standard terminologies and ontology [21].
On the other hand, dealing with structured information retrieval is a complex task, which often requires a significant effort to establish a direct correspondence between the user needs and the content of the document [22]. Therefore, structured reports have been often criticized as impractical for complex cases [23]. Usability evaluates whether users perform tasks in an effective manner through a tool in a specific environment [24]. In this work, usability was used to measure the success of the clinical processes redesign.
This paper presents an overview of the work done at the clinical and information technology sides to define the clinical processes, data items, and applicable terminologies, reporting schemes and using cases to systematize breast cancer diagnosis and follow-up through structured reports. It focuses especially on the definition of report templates and the redesign of clinical processes, covering also the implementation of a prototype system on top of the TRENCADIS (Towards a gRid Environment for proCessing and shAring DIcom objectS) framework. Additional references are given for specific technical details and usability tests, already published in other journals.
The rest of this paper is structured as follows: “Background” section introduces the concepts in which the redesign of the clinical process of breast cancer is based. “Clinical Process Redesign Overview” section presents an overview of the methodology used in the redesign of the clinical process and the actors that are involved in it. The new clinical process is described in “Enhanced Clinical Process” section, while “Lessons Learned from Redesigning a Clinical Process” section presents a review of our experience, developing this new process. Conclusions and future lines of work are presented in “Conclusion” section.
Background
Terminologies and Ontologies
SNOMED CT (Systematized Nomenclature of Medicine Clinical Terms) aims at providing a comprehensive terminology for encoding electronic health records [25]. SNOMED CT also provides an ontological layer and a mechanism to build customized vocabularies. In addition to SNOMED CT, there are several projects trying to develop modularized SNOMED CT ontologies in order to increase computability (e.g., scalability for querying data) and usability (e.g., understandability or context awareness) [26]. These subsets of SNOMED CT ontologies should facilitate the development of more usable applications.
ICD-10 (International Statistical Classification of Diseases and Related Health Problems, 10th revision) provides codes to classify diseases and a variety of related terminologies, such as symptoms or findings [27]. The ICD-10 Clinical Modification (CM) identifies diseases, disorders, symptoms, human response patterns, and medical signs, while the Procedure Coding System (PCS) identifies specific health interventions taken by medical professionals.
However, those general-purpose terminologies were found to be ineffective in radiology reports, especially in oncology where terms are highly specialized. Therefore, specific terminologies in oncology have arisen, such as BI-RADS (Breast Imaging-Reporting and Data System) [28], ICD-O-3 (ICD for Oncology, 3rd version) [29], or TNM (tumor, node, metastasis classification of malignant tumors) [30, 31]. BI-RADS is a radiology terminology that provides a standardized nomenclature or lexicon for describing breast imaging findings and assessments. BI-RADS is commonly used in mammography, ultrasound, and MRI reports. It also provides a categorization of breast lesions, depending on their suspected malignancy (BI-RADS code). Each category (between 0 and 6) leads to a specific therapeutic procedure. ICD-O-3 is used principally to code the topography and the morphology of neoplasms. Pathological TNM (pTNM) is a scoring system based on biopsied tissue, while clinical TNM (cTNM) uses other clinical information to describe the extent and spread of cancer.
The increasing variety of terminologies and standards has led to a non-coherent nomenclature that is inconsistent between terminologies and often confusing to the radiologists who use them. RadLex provides a unified language of radiology terms for standardized indexing and retrieval of radiology information resources [21]. To this end, RadLex unifies and supplements radiology terms in other terminologies, such as SNOMED CT, ICD-10, or BI-RADS.
Digital Imaging and Communications in Medicine Structured Reporting
Digital Imaging and Communications in Medicine (DICOM) is a standard for managing digital images that is widely used in clinical practice. DICOM structured reporting (SR) extends DICOM to encode structured information, allowing applications to share documental information [32], besides medical images. Furthermore, DICOM SR has proven to be particularly valuable in improving the expressiveness, precision, and comparability of documentation about diagnostic images [33].
DICOM SR supports different standard medical terminologies and ontologies that can be combined with custom codification schemes to code the structured reports. This feature facilitates indexing and searching on large databases.
Customization is required in many cases to include additional information in the reports. For example, BI-RADS divides breast density into four standardized categories. BI-RADS 3 or 4 for breast density indicates that the breasts are extremely dense, which lowers the sensitivity of mammography and can lead to a larger inter- and intra-observer variability [34]. In those cases, the reports must include customized terminologies for expressing fine-grain details, which might help to reduce ambiguous conclusions [35]. A customization is presented in this paper that complements BI-RADS with additional annotations, which helps in reducing this ambiguity in mammography reports.
DICOM SR Templates
DICOM SR also provides a means of establishing patterns of applications that are called DICOM SR templates [36]. These patterns describe and constrain the information that can be represented through a DICOM SR document. For example, the Supplement 50 to the DICOM standard defines the computer-aided detection templates for mammography. This template uses BI-RADS.
One limitation on applying DICOM SR in clinical practice is that only a few templates are available, covering a particular application or medical procedure, and therefore, the information is often reported as free text. However, even with this limitation, DICOM SR represents an important step towards achieving clinical data integration within the imaging department and with the rest of the health-care team, enhancing the quality and efficiency of diagnostic services [33].
There are efforts ongoing to provide an XML schema that will be flexible enough to represent DICOM SR. XML is the standard format for data exchange between applications and for format-independent data storage. Health Level Seven (HL7) is an example of medical standard that specifies the structure and semantics of clinical documents encoded in XML [37]. The Supplement 135 to DICOM provides the guidelines to transform DICOM SR diagnostic imaging reports to and from HL7.
TRENCADIS Framework
TRENCADIS is a grid software framework and development toolkit that aims at sharing and processing distributed DICOM objects in virtual storages that are built from ontology-based searches, performed in a secure context [38, 39].
The group of experts who are collaborating in the development of TRENCADIS includes physicians and radiologists from several hospitals and medical schools. They provide an excellent level of expertise and knowledge, which enables TRENCADIS developers to build highly polished data models and to focus their efforts in developing end user services and applications. TRENCADIS was first deployed in the project CVIMO, which demonstrated the usefulness of this approach for securely sharing medical images in a multi-institutional environment [38]. More recently, TRENCADIS was extended to include breast cancer [40]. Also, an international collaboration was started in the context of the IBERGRID initiative towards the implementation of a transnational federated database in breast cancer [41].
Structured Document Retrieval
The goal of a structured document retrieval system is to retrieve relevant parts of a document, instead of complete documents, as conventional methods do. This approach is particularly advantageous when the documents cover a wide variety of topics, since it can reduce the effort required to locate relevant information [42]. This is of great interest for computer-aided diagnosis, since it provides a means of comparing report parts with respect to known patterns, drawing radiologist’ attention to possible targets in the interpretation of a study.
However, despite the potential of this approach to improve the quality and the efficiency of diagnosis, it has not been widely adopted in clinical practice. Structured document retrieval has been most commonly criticized in terms of its lack of compatibility with the information systems (e.g., PACS) used in the hospitals [43]. To cope with this limitation, several grid-based DICOM storages have been proposed as an alternative to PACS for sharing findings and other results among radiologists and physicians. TRENCADIS is an example of such a framework that provides developers with a means for creating virtual storages outside the PACS environment. TRENCADIS DICOM storages support advanced features to index, search, and share DICOM objects.
Clinical Process Redesign Overview
The redesign of the clinical process to manage diagnosis, follow-up, and response to treatment episodes of breast cancer is mainly driven by three processes: (1) the analysis of the existing clinical processes, (2) the results of a prototype for the creation and retrieval of structured reports, and (3) a usability study performed by 11 radiologists working on selected cases.
Figure 1 shows the framework that captures the aspects of information processing and decision support that were used to redesign and evaluate the clinical processes as well as the different experts who participated in this activity.
Fig. 1.
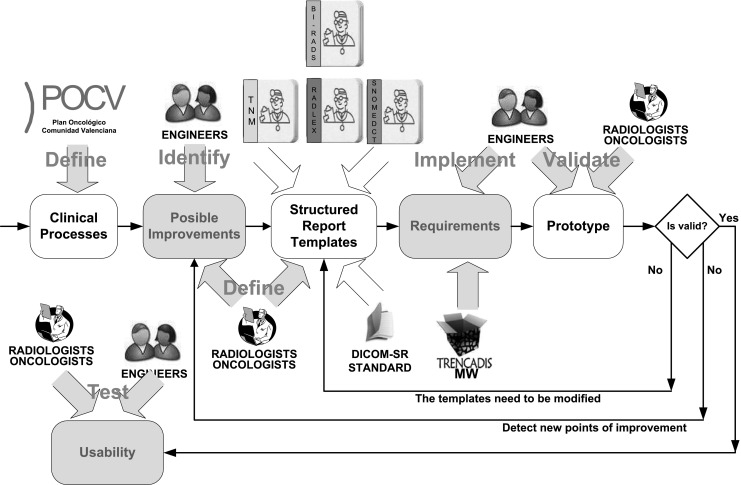
Actors and processes involved in the redesign of the clinical pipelines
In summary, medical experts from diagnostic imaging and oncology identified the issues that can be improved in the existing clinical processes through the introduction of structured reports and terminologies. They also defined the DICOM SR templates and the terminologies and ontology to be used. At this point, a group of software analysts gathers the requirements and implemented the first prototype with TRENCADIS.
Approaching clinical process analysis and redesign around this prototype allowed the medical experts to interact with intermediate versions of the system and to be more involved in the development [44]. Also, the software developers could experiment with alternative system designs.
Once the implementation of the first prototype was completed, a validation study was performed by the team of medical experts. The objective of this study was to test the completeness and correction of the protocols, templates, and terminologies deployed with the prototype. Potential improvements were fed back in the system, which iteratively improved the prototype.
The final prototype integrated a clinical information retrieval tool to search for cases based on fields selected from the structured reports.
A combination of system testing and customer acceptance testing was applied to the final prototype in order to reduce inter-operator variability in the assignment of the codes to the report. In particular, these tests were applied to breast composition, tumor stage, and prognosis factors.
Finally, a test was performed to evaluate the usability of the system.
These steps are further described in the following sections.
Original Clinical Processes
The clinical processes for diagnosis, follow-up, and response to treatment in breast cancer currently in practice in the University Hospital Dr. Peset are shown in Figs. 2 and 3. These processes are based upon the oncological guide for breast cancer from the Valencian Community health authorities [45], which defines the health-care guidelines for public hospitals in the Spanish autonomous community known as Valencian Community (the fourth largest region with 10.6 % of the total population of Spain—over 4.5 million people).
Fig. 2.
Free text-based clinical process for the management of breast cancer
Fig. 3.
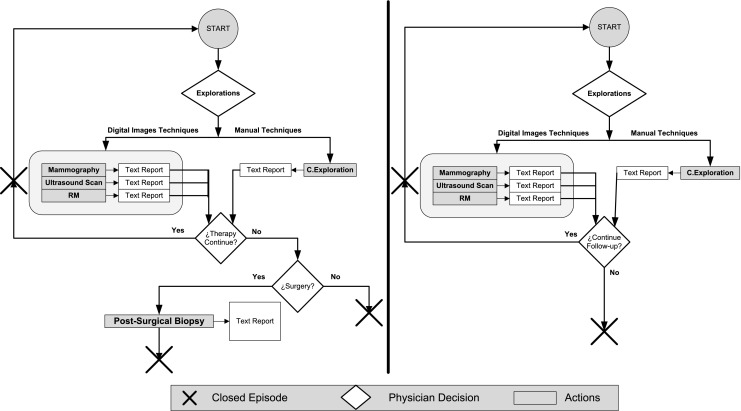
Clinical process for the follow-up (left) and response to treatment (right) in breast cancer
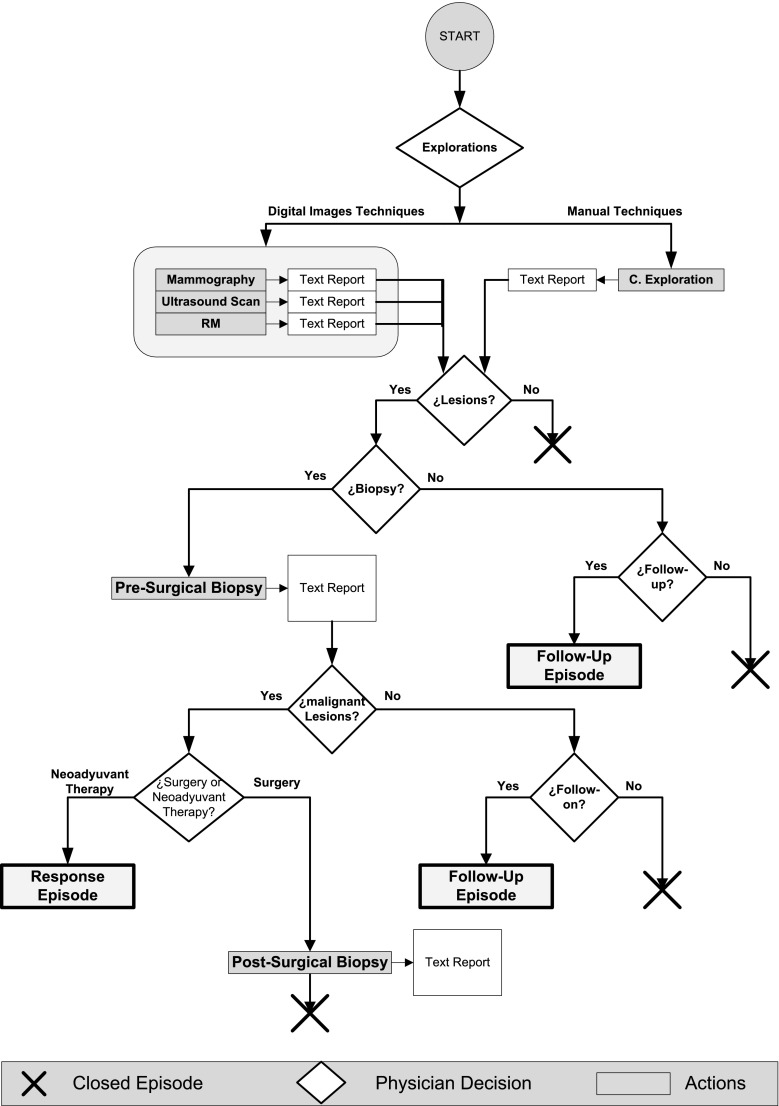
Figure 2 shows a diagram of the complete clinical process. In this paper, we focus on the diagnosis of breast cancer through different image modalities (mammography, ultrasound, and MRI) and clinical examination. The results of these explorations are originally documented in free text format. Further confirmation of the diagnosis includes invasive studies, such as presurgical biopsy, and noninvasive studies, such as mammography, ultrasound, and MRI. A treatment plan is formulated by the physician when the malignancy of a tumor is surgically confirmed in a biopsy. In contrast, follow-up studies evaluate other unconfirmed lesions (including those that are discarded in biopsy). Figure 3 (left) shows the data and clinical path for a follow-up episode.
The clinical path for response to treatment episodes in patients with treated tumors is described in Fig. 3 (right).
TRENCADIS Structured Report Management Prototype
Radiology reports are often stored along with provenance information (author, creation time, etc.). Such a description does not describe the content of the report and inherently fails to reveal any discernible similarity between the different reports stored with the repository.
The prototype for managing structured reports described in this paper uses a series of conventional feature extraction routines to classify the images into different groups, which are provided by the DICOM SR templates. For example, the DICOM content items are examined to extract breast composition, tumor stage, and prognosis factors. These fields are then used to build automatic indexes that reference the reports and their associated images.
Depending on the context of a study, a radiologist may need to study more cases to confirm a conclusion. DICOM SR indexing facilities introduced by the prototype also allow radiologists to use additional content items to build more indexes, facilitating the classification and comparison of reports.
The prototype followed the reference implementation described in the study of Blanquer et al. [39] to implement the redesigned clinical process. It deploys two TRENCADIS sites: one at a hospital and the other at a university.
A user interface was also developed which allows the creation of new reports in the system and also permits radiologists to update the reports with new findings and studies. Guidance is one of the fundamental premises of this interface, with the purpose of assisting radiologists to reach a minimum standard of training and comply with the best practices for the use of the system.
Assessing the Impact: a Usability Study
A usability study was conducted to validate the effectiveness, efficiency, and usability of the prototype and the methodology. This study was conducted by 11 radiologists from different profiles and backgrounds. The methodology used for evaluation of the usability was previously described by Maestre et al. [46], and the metrics used in this study are described in Table 1.
Table 1.
Usability metrics
| Metric | Description |
|---|---|
| Effectiveness | Degree of success achieved in the execution of a task, measuring the deviation of the user result from the reference result and differentiating between tasks that have reached any of their objectives and tasks that have failed |
| Relative user efficiency | Deviation with respect to the efficiency of the expert user that performs the same task, with the same results, under the same conditions |
| User-perceived usability | User satisfaction and perception of the degree of success achieved in operating the tool, measured in the Computer System Usability Questionnaire (CSUQ) [47] |
Enhanced Clinical Process
Structured Report Templates Specific to Breast Cancer Diagnosis
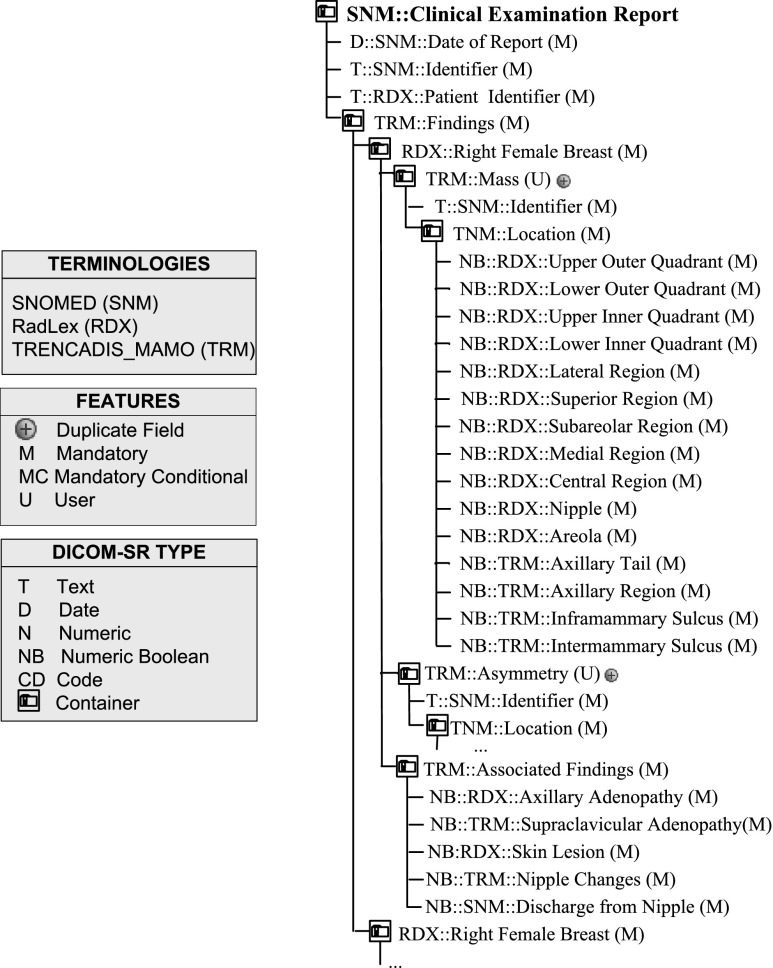
Figures 4, 5, and 6 show the new DICOM SR templates developed to support radiologist in the diagnosis of breast cancer, which are applicable to six different types of explorations: Fig. 4 shows clinical examination; Fig. 5 shows mammography, ultrasound, and MRI, and Fig. 6 shows biopsy and surgical biopsy. These templates include a standardized terminology that is common to all the reports produced, regardless of the type of exploration used. This terminology is a subset of the RadLex terminology (including BI-RADS to describe lesions), combined with a subset of terms selected from SNOMED CT and TNM to encode clinical reports. Also, a small set of customized terminologies is used to report internal parameters of the hospital, which are necessary to integrate the structured reports in the HIS system.
Fig. 4.
SR templates for clinical examination
Fig. 5.
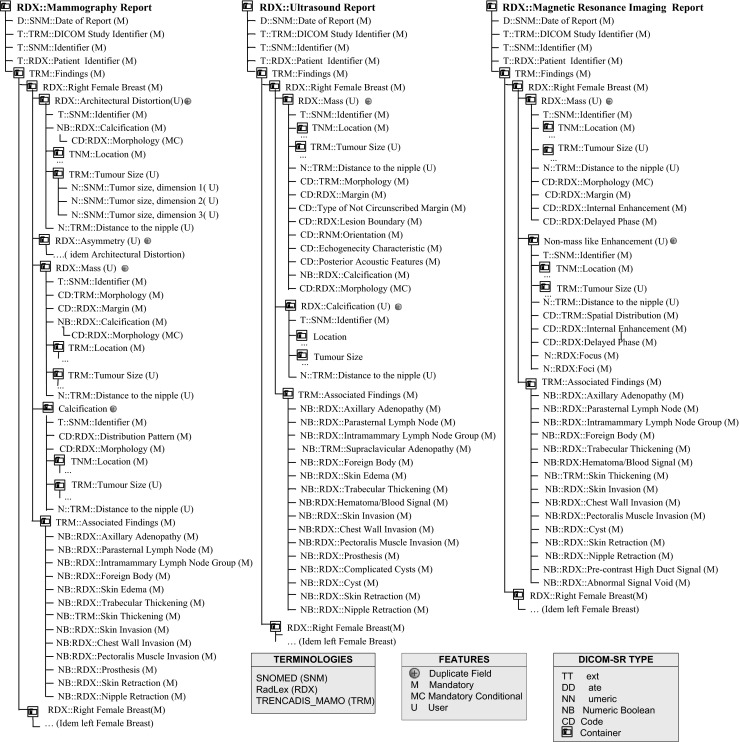
SR templates for mammography, ultrasound, and MRI
Fig. 6.

SR templates for biopsy and surgical biopsy reports
Along with the terminology, the DICOM SR templates of clinical examination, mammography, ultrasound, and MRI share a common tree structure, as depicted in Fig. 4. This tree has two main branches that describe left and right breasts. Each breast branch has one branch per finding type, and each finding type branch may have none or several lesion descriptions.
Similarly, the DICOM SR templates of biopsy and surgical biopsy share some common elements. Reports generated with these templates document the study of one single lesion. In particular, the biopsy template reports the study of material obtained in a presurgical biopsy and the surgical biopsy reports the study of material removed in a surgical intervention. The field procedure identifies the type of surgical procedure performed in a surgical biopsy, for example, a mastectomy.
One of the main features of the DICOM SR templates is the use of BI-RADS codes to quantify the malignancy grade of breast tumors and to record all features associated, which provide a quantitative key indicator for the definition of the actions to be taken for the treatment of the patient. Depending on the BI-RADS code, some actions still need an evaluation of the physician and the consensus with the patient, but in other cases, therapeutic actions are totally determined, which could enable the systematization of the process.
Redesigned Clinical Process
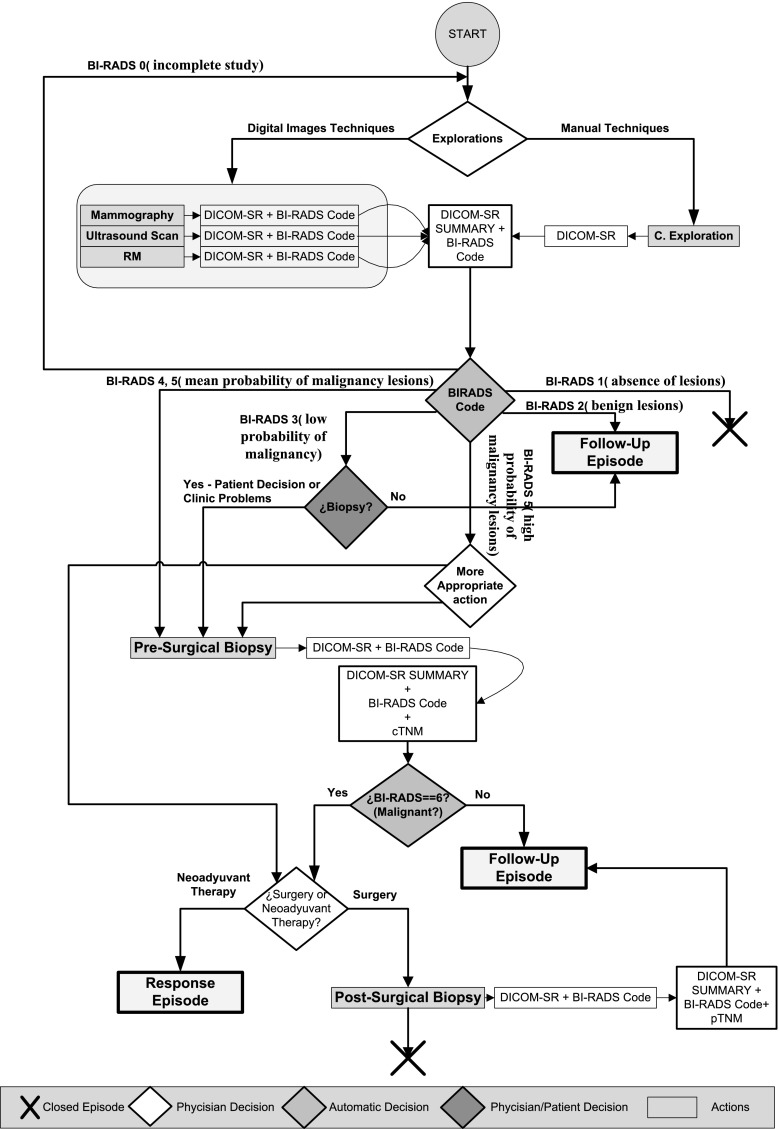
Figure 7 shows the new clinical process for the management of breast cancer, redesigned from the original process to include the new DICOM SR templates. Figure 8 shows the extensions made to the original formulation of the follow-up and response to treatment procedures.
Fig. 7.
SR-based clinical process for the management of breast cancer
Fig. 8.
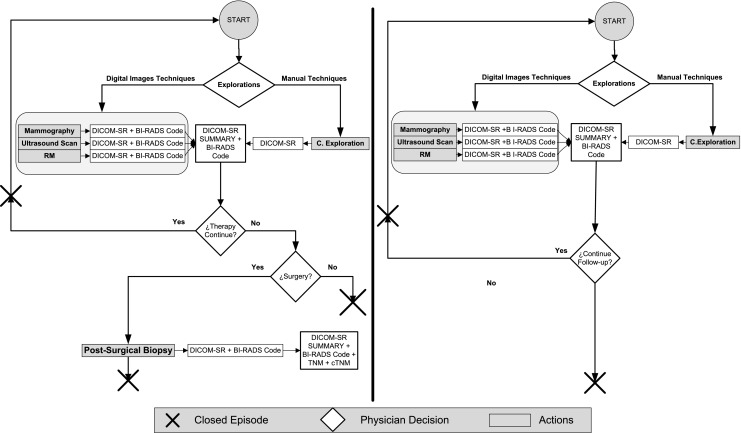
SR-based clinical process for the follow-up (left) and response to treatment (right) in breast cancer
In the case of diagnosis, the reports generated from the different explorations (mammography, ultrasound, MRI, and clinical examination) are automatically combined in a summary report. This template merges the findings of all explorations in one unique document. In particular, a summary of each lesion is included in the summary report as well as a reference to the original report where the complete information of the lesion is found. A final BI-RADS classification is computed as the BI-RADS of the lesion with the higher likelihood of malignancy. The BI-RADS computed in this manner defines the conclusion of the summary report and the recommended action.
In all cases that go through a presurgical biopsy, the results of the biopsy are documented using the presurgical DICOM SR template. Also in these cases, the summary report is automatically compiled from the available reports, but it also includes the SNOMED CT terms from the presurgical reports. A cTNM stratification index is automatically computed (except for the M term, which requires information about the existence of additional sites of cancer) and added to the summary report. Biopsy discriminates between malignant and benign lesions (BI-RADS 6). When a lesion is classified as benign, it is then necessary to open a follow-up episode. Otherwise, in the case of a malignant lesion, the physician has to assess the adequacy of a neoadjuvant treatment followed by a response episode or a surgical treatment followed by a postsurgical biopsy. Similar to presurgical biopsy, the results of the postsurgical biopsy are documented using the postsurgical DICOM SR template. Also in this case, the summary SR is automatically computed and includes the SNOMED CT terms and the computed pTNM.
In the case of the follow-up and response episodes, the clinical process was modified to include the DICOM SR templates previously described for diagnosis. In addition, two new summary DICOM SR templates were created for follow-up and response, respectively. The system computes automatically the cTNM scores from the exploration reports and writes them to the summaries.
A significant feature of the designed DICOM SR templates is the possibility of linking the exploration findings with the different reports that are produced in the complete clinical process. To this end, the same identifier assigned to a lesion in an exploration study is maintained in the subsequent studies. This feature is supported in the prototype with a set of tools that provide users with the means to select and visualize previous lesions. For example, one of these tools allows users to load all the lesions found in a breast quadrant in previous explorations. Users can browse the lesions and the associated information. Also, a lesion can be selected and a new report can be added to that lesion in particular.
Usability
The usability study was performed simultaneously by 11 radiologists. More details of this study are presented in the study of Maestre et al. [46]. Eight of the 11 radiologists who participated in the study reported to reach 60–99 % of completeness of the study goals. Several metrics were used in this study to measure usability, such as effectiveness and efficiency. In general, no case exceeded a deviation from the measured value greater than 27 %. All radiologists reached an effectiveness rate greater than 85 %, and the relative user efficiency was greater than 52 % (see Table 1).
The average usability perceived by all participants in this study was 5.3073, which exceeds the minimum threshold level of 4.3726 Computer System Usability Questionnaire (CSUQ) units (mean item score), proposed by Lewis et al. [48].
Additionally, the Shapiro-Wilk test [49] of normality (significance at 0.05) demonstrated that the data for CSUQ did followed a normal distribution (Shapiro-Wilk statistic = 0.879, df = 11, sig = 0.098).
Lessons Learned from Redesigning a Clinical Process
During the clinical process redesign, nine new DICOM SR templates were created specific to breast cancer diagnosis: one template for clinical exploration, three templates for imaging-based studies, one for each imaging modality (mammography, ultrasound, and MRI), one template for presurgical biopsy, and one for postsurgical biopsy. Finally, three summary templates were created, one for each episode type (diagnosis, follow-up, and response to treatment).
Although the structure of the summary templates is not shown in the paper, their content is redundant with respect to the templates that originate them. Instead, a description of the generation process is included in the previous section.
In the case of diagnosis, the main contributions with respect to the existing clinical process are the use of DICOM SR templates with standardized terminology to document the diagnosis and definition of decision rules, depending on the classification of the tumors. These features were positively evaluated by the team of radiologists who collaborated in the development and validation of the system. In particular, they were found useful to assist radiologists in their decisions.
DICOM SR templates for diagnosis are identical to those for follow-up and response to treatment episodes, and therefore, they are completely interoperable. This improves the existing report templates, allowing radiologists to correlate parameters between different reports, even from different exploration techniques or different patients. For example, it allows the reports generated on diagnosis to be searched and compared to the reports generated on follow-up episodes.
The redesigned clinical process automates several clinical decisions, whenever is possible. This was also evaluated positively, since radiologists found it useful to improve their efficiency.
One goal of the work was to demonstrate an effective methodology for accomplishing breast cancer diagnosis. This included adding new information to the reports and searching for relevant information on remote DICOM storages.
Virtual storages built from ontologies were proven useful in improving the relevance of search results. This would not be the case if the structured reports do not reflect the objectives of the clinical studies that they represent. In other words, in our experience, the careful selection of terminologies and ontologies that fit the requirements of the radiologists is a key to correlate the results acquired with different techniques, possibly in different hospitals. This should help in creating new models that improve the accuracy of computer-assisted breast cancer diagnosis [50].
Prototyping the clinical process allowed radiologists and developers to gain appreciation of the system through active engagement with iterative prototypes, while usability studies provided developers with information about the effectiveness, efficiency, and user-perceived usability of the system.
The final prototype also provides radiologists with the means to search structured report databases for similar and contrasting cases (or even possibly conflicting interpretations) that may be of clinical relevance based upon a target study. The user interface allows end users to select a field of interest from the list of fields available in the structured reports and to enter the query in an input text box. Different fields can be combined in a single query by using logical operators like AND, OR, and NOT. The user interface sends the query to the service back end, which executes the query and returns the reports that match the search parameters.
Tests provided valuable information to medical experts and developers about the suitability of potential changes to improve the system. Relying on clinical aspects, such as inter-operator variability, has proven to shorten development cycle time and to enhance the quality of the final prototype. For example, the time it takes to complete an iteration cycle (to develop a new version of the prototype) is shorter when our development team receives instructions that include the opinion of our medical experts (e.g., the query interface must filter out the lesions before presenting them to the user to avoid that the server response would include any unwanted items) than when our development team only receives technical instructions (e.g., the server response includes invalid items). Changes to the prototype were applied when they were approved by the medical experts in oncology and radiology who participated in the development of the system.
On the other hand, the results obtained in the usability study have corroborated our initial hypotheses that the redesign of the clinical process may lead to a significant improvement in accuracy and reproducibility of automatic identification of similar and contrasting cases of breast cancer. In particular, the high level of effectiveness (over 85 % in all cases, regardless of the professional experience of the participants) achieved by the radiologists who participated in the study in the three different test scenarios has confirmed the validity of the DICOM SR templates and the redesigned clinical process. Similarly, the consistency of the results (normally distributed with reasonable variability) obtained from the 11 participants demonstrated a low inter-operator variability.
Conclusion
Having redesigned the clinical process to manage breast cancer in the Valencian Community, we have developed and validated a prototype of clinical information retrieval system, which implements the new process. This system aims at becoming a practically useful tool for medical experts who treat, teach, or research in breast cancer areas. To this end, it was designed with usability in mind, with particular attention to aspects of the current data and clinical path that can be improved to facilitate diagnosis. Although the preliminary studies presented in this paper have probed the capability of the system to identify similar and contrasting (dissimilar) cases of breast cancer from a large dataset of medical reports of this disease, further studies must be undertaken before the system receives the approval of the public health authorities and can be used in routine practice. This is a common requirement in health applications.
Although focused in diagnosis, the clinical process presented in this paper has potential for radiation therapy planning and surgical planning. Furthermore, the presented methodology is not specific to any particular disease and can be applied to improve other clinical processes.
Acknowledgments
We thank the subject matter experts for sharing their insights through this study. We are especially appreciative of the efforts of the Radiology Unit and Medical Oncology Unit teams at the University Hospital Dr. Peset. This work was partially supported by the Vicerectorat d’Investigació de la Universitat Politècnica de València (UPVLC) to develop the project “Mejora del proceso diagnóstico del cáncer de mama” with reference UPV-FE-2013-8.
References
- 1.Ratib O: Imaging informatics: From image management to image navigation. Yearb Med Inform 2009; 167–172 [PubMed]
- 2.Oakley J. Digital Imaging: A Primer for Radiographers, Radiologists and Health Care Professionals. Cambridge University Press, 2003.
- 3.Prokosch HU, Dudeck J: Hospital information systems: Design and development characteristics, impact and future architecture. Elsevier health sciences, 1995
- 4.Foster I, Kesselman C, Tuecke S. The anatomy of the grid: Enabling scalable virtual organizations. Int J High Perform Comput Appl. 2001;15(3):200–222. doi: 10.1177/109434200101500302. [DOI] [Google Scholar]
- 5.Oram A: Peer-to-Peer: Harnessing the power of disruptive technologies. O’Reilly Media, 2001
- 6.National Institute of Standards and Technology. The NIST Definition of Cloud Computing. 2011. http://csrc.nist.gov/publications/nistpubs/800-145/SP800-145.pdf (accessed 29 Jan 2013)
- 7.Oster S, Langella S, Hastings S, Ervin D, Madduri R, Phillips J, Kurc T, Siebenlist F, Covitz P, Shanbhag K, Foster I, Saltz J. caGrid 1.0: An enterprise grid infrastructure for biomedical research. J Am Med Inform Assoc. 2008;15:138–149. doi: 10.1197/jamia.M2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Natter MD, Quan J, Ortiz DM, et al. An i2b2-based, generalizable, open source, self-scaling chronic disease registry. J Am Med Inform Assoc. 2013;20:172–179. doi: 10.1136/amiajnl-2012-001042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohno-Machado L, Bafna V, Boxwala AA, et al. iDASH: Integrating data for analysis, anonymization, and sharing. J Am Med Inform Assoc. 2012;19:196–201. doi: 10.1136/amiajnl-2011-000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Channin DS, Mongkolwat P, Kleper V, Rubin DL. Computing human image annotation. Conf Proc IEEE Eng Med Biol Soc. 2009;1:7065–7068. doi: 10.1109/IEMBS.2009.5333365. [DOI] [PubMed] [Google Scholar]
- 11.Sittig DF, Wright A, Osheroff JA, et al. Grand challenges in clinical decision support. J Biomed Inform. 2008;41(2):387–392. doi: 10.1016/j.jbi.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagholikar KB, Sundararajan V, Deshpande AW. Modeling paradigms for medical diagnostic decision support: a survey and future directions. J Med Syst. 2012;36(5):3029–3049. doi: 10.1007/s10916-011-9780-4. [DOI] [PubMed] [Google Scholar]
- 13.Rubin DL. Creating and curating a terminology for radiology: Ontology modeling and analysis. J Digit Imaging. 2008;21(4):355–362. doi: 10.1007/s10278-007-9073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahn CE, Jr, Langlotz CP, Burnside ES, Carrino JA, Channin DS, Hovsepian DM, et al. Toward best practices in radiology reporting. Radiology. 2009;252(3):852–856. doi: 10.1148/radiol.2523081992. [DOI] [PubMed] [Google Scholar]
- 15.Taira PK, Soderlang SG, JAbovits RM. Automatic structuring of radiology free-text reports. Radiographics. 2001;21(1):237–245. doi: 10.1148/radiographics.21.1.g01ja18237. [DOI] [PubMed] [Google Scholar]
- 16.Fujii H, Yamagishi H, Ando Y, Tsukamoto N, Kawaguchi O, Kasamatsu T, et al. Structuring of free-text diagnostic report. Stud. Health Technol. Inform. 2007;129:669–673. [PubMed] [Google Scholar]
- 17.Murff HJ, FitzHenry F, Matheny ME, Gentry N, Kotter KL, Crimin K, Dittus RS, Rosen AK, Elkin PL, Brown SH, Speroff T. Automated identification of postoperative complications within an electronic medical record using natural language processing. JAMA. 2011;306(8):848–855. doi: 10.1001/jama.2011.1204. [DOI] [PubMed] [Google Scholar]
- 18.Clunie DA: DICOM structured reporting. PixelMed Publishing, 2000
- 19.D’Avolio LW, Nguyen TM, Farwell WR, Chen Y, Fitzmeyer F, Harris OM, Fiore LD. Evaluation of a generalizable approach to clinical information retrieval using the automated retrieval console (ARC) J Am Med Inform Assoc. 2012;17:375–382. doi: 10.1136/jamia.2009.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Napel SA, Beaulieu CF, Redriguez C, Cui J, Xu J, Grupta A, et al. Automated retrieval of CT images of liver lesions on the basis of image similarity: Method and preliminary results. Radiology. 2010;256(1):243–252. doi: 10.1148/radiol.10091694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langlotz CP. RadLex: A new method for indexing online educational materials. Radiographics. 2006;26(6):1595–1597. doi: 10.1148/rg.266065168. [DOI] [PubMed] [Google Scholar]
- 22.Crestania F, Vegas J, de la Fuente P. A graphical user interface for the retrieval of hierarchically structured documents. Inf Process Manag. 2004;40(2):269–289. doi: 10.1016/S0306-4573(02)00120-6. [DOI] [Google Scholar]
- 23.Weiss DL, Langlotz CP. Structured reporting: Patient care enhancement or productivity nightmare? Radiology. 2008;249(3):739–747. doi: 10.1148/radiol.2493080988. [DOI] [PubMed] [Google Scholar]
- 24.Yen PY, Bakken S. Review of health information technology usability study methodologies. J Am Med Inform Assoc. 2012;19(3):413–422. doi: 10.1136/amiajnl-2010-000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patrick R, Julien G, Christian L, Antoine G. Automatic medical encoding with SNOMED categories. BMC Med Inform Decis Mak. 2008;8(Suppl 1):S1–S6. doi: 10.1186/1472-6947-8-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez-Garcia P, Boeker M, Illarramendi A, Schulz S. Usability-driven pruning of large ontologies: The case of SNOMED CT. J Am Med Inform Assoc. 2012;19:e102–e109. doi: 10.1136/amiajnl-2011-000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.World Health Organization. International Statistical Classification of Diseases and Related Health Problems 10th Revision. http://apps.who.int/classifications/apps/icd/icd10online/ (accessed 29 Jan 2013)
- 28.American College of Radiology (ACR) Breast Imaging Reporting and Data System Atlas (BI-RADS® Atlas) [DOI] [PubMed]
- 29.World Health Organization. International Classification of Diseases for Oncology, 3rd Edition (ICD-O-3). http://www.who.int/classifications/icd/adaptations/oncology/en/index.html (accessed 29 Jan 2013)
- 30.Greene FL. TNM: Our language of cancer. CA Cancer J Clin. 2004;54(3):129–130. doi: 10.3322/canjclin.54.3.129. [DOI] [PubMed] [Google Scholar]
- 31.American Joint Committee of Cancer (AJCC). AJCC Cancer Staging Manual. Seventh Edition. Springer, 2010
- 32.Hussein R, Engelmann U, Schroeter A, Meinzer HP. DICOM structured reporting: Part 1. Overview and characteristics. Radiographics. 2004;24(3):891–896. doi: 10.1148/rg.243035710. [DOI] [PubMed] [Google Scholar]
- 33.Sluis D, Lee KP, Mankovich N. DICOM SR - integrating structured data into clinical information systems. Medicamundi. 2002;46(2):31–36. [Google Scholar]
- 34.Percha B, Nassif H, Lipson J, Burnside E, Rubin D. Automatic classification of mammography reports by BI-RADS breast tissue composition class. J Am Med Inform Assoc. 2012;19(5):913–916. doi: 10.1136/amiajnl-2011-000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciatto S, Houssami N, Apruzzese A, Bassetti E, Brancato B, Carozzi F, Catarzi S, Lamberini MP, Marcelli G, Pellizzoni R, Pesce B, Risso G, Russo F, Scorsolini A. Reader variability in reporting breast imaging according to BI-RADS assessment categories (the Florence experience) Breast. 2006;15(1):44–51. doi: 10.1016/j.breast.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 36.National Electrical Manufacturers Association (NEMA). Digital Imaging and Communications in Medicine (DICOM). Part 16: Content Mapping Resource. http://medical.nema.org/dicom/2004/04_16PU.PDF (accessed 29 Jan 2013)
- 37.Dolin RH, Alschuler L, Boyer S, Beebe C, Behlen FM, Biron PV, Shvo AS. HL7 clinical document architecture, release 2. J Am Med Inform Assoc. 2006;13:30–39. doi: 10.1197/jamia.M1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blanquer I, Hernández V, Meseguer JE, Segrelles D. Content-based organisation of virtual repositories of DICOM objects. Future Gener Comput Syst. 2009;25(6):627–637. doi: 10.1016/j.future.2008.12.004. [DOI] [Google Scholar]
- 39.Blanquer I, Hernández V, Segrelles D, Torres E. Enhancing privacy and authorization control scalability in the grid through ontologies. IEEE Trans Inf Technol Biomed. 2009;12(1):16–24. doi: 10.1109/TITB.2008.2003369. [DOI] [PubMed] [Google Scholar]
- 40.Salavert J, Maestre C, Segrelles D, Blanquer I, Hernández V, Medina R, Martí L: Grid prototype to support cancer of breast diagnostics in clinic practice. Proc of the 4th. Iberian Grid Infrastructure Conf. Netbiblo, 2010
- 41.Segrelles D, Franco JM, Medina R, Blanquer I, Salavert J, Hernandez V, Martí L, Díaz G, Ramos R, Guevara MA, González N, Loureiro J, Ramos I. Exchanging data for breast cancer diagnosis on heterogeneous grid platforms. Computing and Informatics. 2012;31(1):3–15. [Google Scholar]
- 42.Ali MS, Consens M, Lalmas M. Extended structural relevance framework: A framework for evaluating structured document retrieval. Inf Retrieval. 2012;15:558–590. doi: 10.1007/s10791-012-9192-1. [DOI] [Google Scholar]
- 43.Welter P, Riesmeier J, Fischer B, Grouls C, Kuhl C, Deserno TM. Bridging the integration gap between imaging and information systems: A uniform data concept for content-based image retrieval in computer-aided diagnosis. J Am Med Inform Assoc. 2011;18:506–510. doi: 10.1136/amiajnl-2010-000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenkins CW. Application prototyping: A case study. Perform Eval Rev. 1981;10(1):21–27. doi: 10.1145/1010627.807905. [DOI] [Google Scholar]
- 45.Generalitat Valenciana. Conselleria de Sanitat. Oncoguía de Cáncer de Mama Comunidad Valenciana. http://publicaciones.san.gva.es/publicaciones/documentos/V.2478-2006.pdf (accessed 29 Jan 2013)
- 46.Maestre C, Segrelles-Quilis JD, Torres E, Blanquer I, Medina R, Hernández V, Martí L. Assessing the usability of a science gateway for medical knowledge bases with TRENCADIS. J Grid Computing. 2012;10:665–688. doi: 10.1007/s10723-012-9243-2. [DOI] [Google Scholar]
- 47.Lewis J. IBM computer usability satisfaction questionnaires: Psychometric evaluation and instructions for use. Int J Hum-Comput Interact. 1995;7(1):57–78. doi: 10.1080/10447319509526110. [DOI] [Google Scholar]
- 48.Lewis JR. Psychometric evaluation of the PSSUQ using data from five years of usability studies. Int J Hum-Comput Interact. 2002;14(3–4):463–488. doi: 10.1080/10447318.2002.9669130. [DOI] [Google Scholar]
- 49.Shapiro SS, Wilk MB. An analysis of variance test for normality (complete samples) Biometrika. 1965;52(3–4):591–611. doi: 10.1093/biomet/52.3-4.591. [DOI] [Google Scholar]
- 50.Chhatwal J, Alagoz O, Lindstrom MJ, Kahn CE, Jr, Shaffer KA, Burnside ES. A logistic regression model based on the national mammography database format to aid breast cancer diagnosis. AJR Am J Roentgenol. 2009;192:1117–1127. doi: 10.2214/AJR.07.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]