Abstract
Recent studies suggest a role for the microbiota in autism spectrum disorders (ASD), potentially arising from their role in modulating the immune system and gastrointestinal (GI) function or from gut–brain interactions dependent or independent from the immune system. GI problems such as chronic constipation and/or diarrhea are common in children with ASD, and significantly worsen their behavior and their quality of life. Here we first summarize previously published data supporting that GI dysfunction is common in individuals with ASD and the role of the microbiota in ASD. Second, by comparing with other publically available microbiome datasets, we provide some evidence that the shifted microbiota can be a result of westernization and that this shift could also be framing an altered immune system. Third, we explore the possibility that gut–brain interactions could also be a direct result of microbially produced metabolites.
Keywords: autism, GI problems, gut microbiome, gut microflora, Prevotella, next-generation sequencing, meta-analysis, gut–brain interaction
There is a general consensus that gastrointestinal (GI) problems are common in individuals with autism spectrum disorders (ASD) (1), although the exact percentage varies from study to study and depends on the age of the study population. Population-based studies, which do not directly select or bias their samples, are the best way to determine incidence. In a study of 137 children with ASD, 24% had a history of at least one GI symptom, with diarrhea being the most prevalent – occurring in 17% of individuals (2). Similarly, a study of 172 children with ASD found 22.7% were positive for GI distress, primarily with diarrhea and/or constipation (3). A study of 160 children with ASD found 59% had GI dysfunction with diarrhea or unformed stools, constipation, bloating, and/or gastroesophageal reflux (GERD) (4). A study of 150 children [50 children with ASD, 50 controls, and 50 children with other developmental disabilities (DD)] found that 70% of children with ASD presented GI symptoms, compared to 28% of typically developing children and 42% of children with DD (5). A study by Adams et al. (6) where 51 children with ASD were compared to 40 typical healthy controls aged 3–15 found that 63% of children with autism had moderate or severe chronic diarrhea and/or constipation, versus 2% of the control children. A study of 960 children with ASD, DD, and typical development found that children with ASD and DD had at least one reported GI symptom, and that children with ASD that had common occurrences of abdominal pain, gases, diarrhea, and constipation; scored worse on four behavioral measurements assessed by the Aberrant Behavior Checklist (ABC) (7). A study that compared 121 children with ASD to 242 matched controls found significantly higher constipation (33.9% vs. 17.6%) and food selectivity (24.5% vs. 16.1%) but not diarrhea, abdominal bloating, or reflux in the ASD cohort (8). Based on this finding, the authors suggested that GI issues with ASD may be primarily neurobehavioral rather than of a primary organic GI etiology (8), although this conclusion is not consistent with the other reported studies that did find increased incidence of diverse GI problems. A recently published meta-analysis of GI symptoms in ASD by McElhanon et al. (9) that gathered data from published peer-reviewed journals concluded that GI symptoms are more common in children with ASD than control children, although they identified high methodological variability between the studies. In summary, these studies demonstrate that GI symptoms are common in children with ASD but that the degree and nature of these differences have varied across studies.
GI complications in children with ASD may contribute to the severity of the disorder. One study (10) found a strong correlation between GI symptoms and autism severity in a group of 58 children with ASD, consistent with the CHildhood Autism Risks from Genetics and Environment (CHARGE) study (7). Abdominal pain, constipation, and/or diarrhea are unpleasant and likely to produce frustration, decreased ability to concentrate on tasks, behavior problems, and possibly aggression and self-abuse, especially in children unable to communicate their discomfort. These problems also result in a decreased ability to learn toilet training, leading to increased frustration for the child and their parents/caregivers. However, given many recent studies that have linked the gut with the brain (11–13), there is also the intriguing possibility that correlations between ASD and GI symptoms may not alone be driven by discomfort, but rather by differences in function of the microbiota, such as the metabolites that they produce, that may affect neuronal processes.
The cause of these GI problems in ASD is unclear, but it appears to partly relate to abnormal gut microbiota and possibly to the excessive use of oral antibiotics which can alter gut flora. Several studies have reported significantly higher oral antibiotic use in children with autism versus typical children (6, 14–17). Oral antibiotics were primarily used for treating otitis media (ear infections), which may suggest an impaired immune system. Commonly used oral antibiotics alter the normal gut microbiota (18), which play an important role in metabolizing plant polysaccharides, promoting GI motility, maintaining water balance, producing some vitamins, and competing against pathogenic bacteria. Loss of normal gut flora can result in the overgrowth of pathogenic flora, which can in turn cause constipation and other problems. However, it is also worth noting that in one small study of children with autism, treatment with the minimally absorbed glycopeptide antibiotic vancomycin resulted in short-term improvement in ASD symptoms, supporting a direct role for the antibiotic-sensitive gut bacteria in ASD (19).
Distinctive gut microbes have been associated with ASD
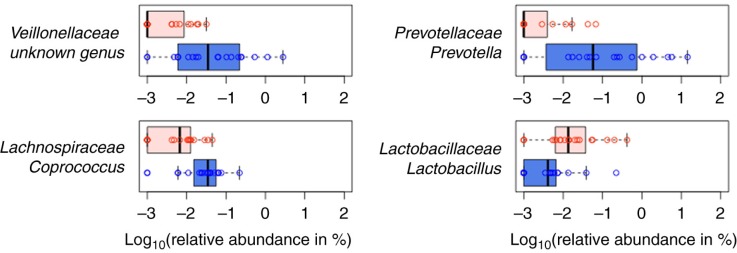
Several small pilot studies of GI bacterial composition in ASD have been conducted, and although the majority agree that gut microbiota composition is distinctive in ASD, these studies have yielded conflicting results as to the nature and/or extent of GI bacterial community differences in children with ASD compared to controls (20–24). In work recently described by Kang et al. (21), high-throughput sequencing of the 16S rDNA gene was used to compare bacterial composition in fecal samples from 19 children with ASD who had varying GI symptoms and 20 neurotypical controls with minimal GI symptoms. The presence of autistic symptoms rather than the severity of GI symptoms was associated with lower abundance of the bacterial genera Prevotella, Coprococcus, and unclassified Veillonellaceae (p<0.05 after correction for multiple hypothesis testing) (Fig. 1) and an overall less diverse gut microbiome (p<0.05). Prevotella, Coprococcus, and Veillonellaceae are intriguingly versatile carbohydrate-degrading and/or fermenting bacteria, suggesting that the observed compositional differences can result in differences in the spectrum of metabolites produced from a given diet.
Fig. 1.
Four most differentially abundant genera when children with ASD and neurotypical children are compared (blue box: neurotypical children; red box: children with ASD). (Figure from Kang et al. (21) under an open access license of PLoS journals called the Creative Commons Attribution (CC-BY) license.)
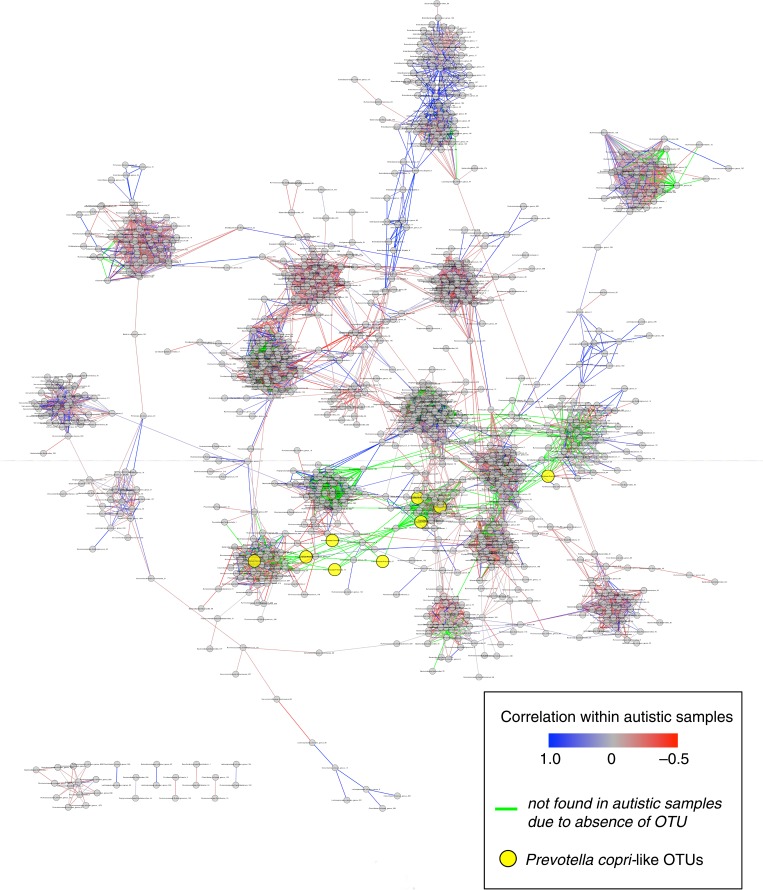
Gut microbes interact heavily through competitive and cooperative interactions, the latter of which allows for the metabolism of complex components of our diets. It is thus of interest to not only look at how ASD associates with change in the relative abundance of particular types of bacteria, but also to understand how ASD affects the complex interaction network that occurs in health. Kang et al. (21) performed pairwise Pearson correlation tests across the samples from only the neurotypical individuals to identify possible microbial interactions that occur in neurotypical individuals. For instance, with this analysis, Prevotella copri-like operational taxonomic units (OTUs) (yellow circles) were connected to multiple different OTUs mapping to other divergent types of bacteria in the network, showing that their abundance in the gut is often associated with the abundance of other types of bacteria (Fig. 2). The network also reflects the degree to which the observed connections are also present or absent in the ASD cohort samples (Fig. 2). Although some OTU pairs had highly correlated relative abundances within both the neurotypical and independently in the ASD cohort (blue lines in Fig. 2), many of the relationships that were observable in neurotypical individuals were not detectable in the ASD cohort, either because the OTUs were both present in the ASD cohort but their relative abundances were not positively correlated (grey and red lines in Fig. 2), or because at least one OTU in the pair was absent from the ASD cohort altogether (green lines in Fig. 2). Thus, the differences in microbiota composition that occurred with ASD may possibly have led to the loss of important microbial interactions that result in a decreased overall diversity and reduced function.
Fig. 2.
Schematic of relationships among pairs at the 97% OTU level. Each grey or yellow circle represents an individual OTU, and paired OTUs are connected by a line if there is a high positive correlation (Pearson's R>0.8) among neurotypical samples. When paired OTUs were also present from autistic samples, lines connecting each OTU pair are highlighted either in blue or red depending on their correlation among autistic samples (either positive or negative correlation, respectively). If at least one OTU in the pair was absent from autistic samples, the connecting line is colored in green to show the changes in microbial interactions that resulted from the absence of the OTUs in children with ASD. Prevotella copri-like OTUs are highlighted in yellow to demonstrate how often their abundance is associated with the other in the network. (Figure from Kang et al. (17) under an open access license of PLoS journals called the Creative Commons Attribution (CC-BY) license.)
A decreased abundance in Prevotella in children with ASD was reported when compared with their siblings in one other study (23), but its prevalence was not significantly different in other studies of gut microbiota in ASD (25). Moreover, differences not found in Kang et al. (21), such as increases in Clostridium and Sutterella with ASD, were reported in other studies (20, 26, 27). The study that detected differences in Sutterella was unique in the use of mucosal-biopsies (20) rather than fecal samples, which results in the observation of bacteria at different prevalence (28).
Observed microbiota differences with ASD have also been inconsistent with regard to overall microbiota (alpha) diversity. Kang et al. (21) observed significantly decreased alpha diversity in children with ASD, which is interesting in light of the fact that low microbiome diversity has been associated with increased vulnerability to various chronic disorders, including a lower resistance to pathogenic bacteria (29). However, Finegold et al. (30) reported higher microbial diversity in feces from children with ASD. The differences in currently reported findings could be attributed to: the heterology of the disorder, small sampling sizes, different sample types (e.g. fecal versus mucosal), different methods of characterizing the microbiota (e.g. culture-based versus culture-independent), and to different bioinformatics analysis methods. These differences may also reflect different degrees to which the studies considered potentially confounding factors such as diet, oral antibiotic use, prebiotics, probiotics, GI problems, and autistic symptoms. Differences also may reflect whether siblings or unrelated individuals are used as the control population. There is one published study that reported to find no significant differences between the gut microbiome of individuals with ASD and controls that used only neurotypical siblings as a control population (22). Interestingly, one study that compared the fecal microbiome of children with ASD, neurotypical siblings of children with ASD, and unrelated neurotypical controls, found the sibling controls to have a microbiome more similar to children with ASD than to control subjects without a neurotypical sibling (23). This may indicate either a scenario whereby a shared environment produces the development of a microbiome that leads to autism in only the genetically susceptible sibling or may also be related to the shared genetics of siblings. Differences between cohorts may also reflect the natural human microbial diversity both within and between individuals. Inconsistent results with regards to the nature of microbiota differences observed across different studies of a disorder are not unique to the study of ASD. For instance, there is strong consensus from both human and mouse studies of obesity, another complex disease with varied etiology, that microbial composition is altered, but little consensus on the nature of these changes (31). Overall, the study of microbiota associations with complex diseases is difficult and requires large and carefully characterized subject cohorts.
ASD-associated fecal microbiota evaluated in a broader context
The prevalence of ASD in developed/Western populations has risen dramatically over the past decade and a half (32), leading some to suggest that cultural/ethnic, environmental, and/or socioeconomic factors in the developed world are to blame. This theory has been hard to test because of poor data on ASD prevalence trends in the developing world and controversy over the degree to which the observed rise in Western populations is due to differences in diagnostic criteria (33). However, in one intriguing study, pervasive developmental disorder (PDD) prevalence rates in children who were currently living in Israel was much lower in immigrant Ethiopian children compared to native Israeli children of Ethiopian heritage (34), giving some evidence that there is an environmental risk factor for PDD that is unique to industrialized countries. This notion and the fact that the bacterial genus that was most strongly depleted in the analysis of the fecal microbiome of children with ASD by Kang et al. 21, Prevotella, is highly enriched in the fecal microbiota in populations in Africa including agrarian societies in Malawi (35) and Burkina Faso (36), and the Hadza hunter–gatherers in Tanzania (37) intrigued us and inspired us to perform comparative analyses. Since Prevotella is only one genus in the very diverse gut microbiota and has a tendency to co-occur with a complex collection of other bacteria species (38, 39), we wanted to determine whether Prevotella depletion in children with ASD is an indicator that the gut microbiome of children with ASD who live in the United States differs even more from individuals in the developing world than does the gut microbiome of neurotypical children in the US, providing evidence of the gut microbiota as an environmental factor that may correlate with increased rates of ASD in industrialized countries.
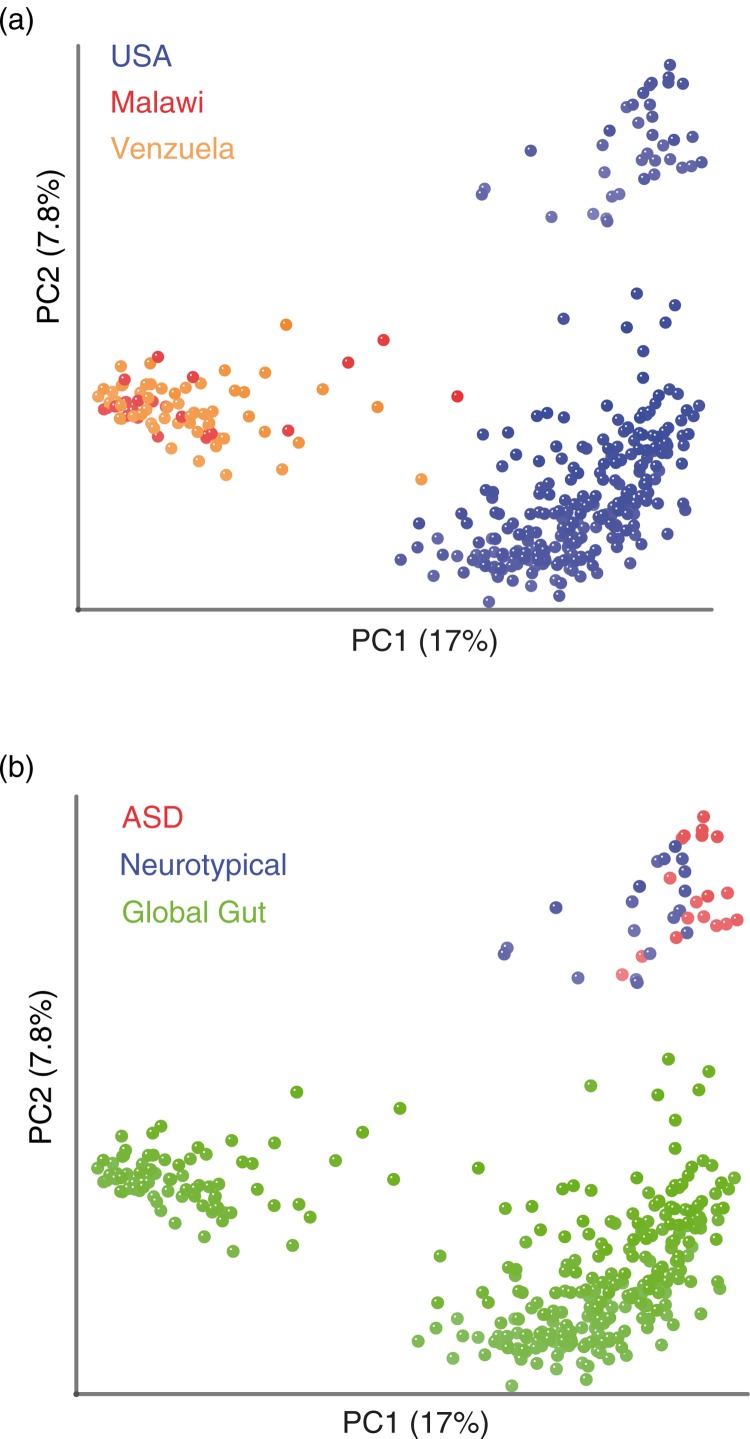
To directly relate the gut microbiota of children with ASD to those in agrarian cultures, we combined 16S rRNA genomic data from the fecal samples sequenced in Kang et al. (21) from children with ASD and neurotypical controls, with data from another published study that used 16S rRNA gene sequencing to survey the fecal microbiota of individuals from Malawi, the Amazonas State of Venezuela, and the US (35) (Fig. 3). This latter paper found the fecal microbiota of individuals in agrarian cultures in Malawi and Venezuela to have a similar Prevotella-rich composition that was highly divergent from individuals living in the US.
Fig. 3.
The fecal microbiota of individuals with ASD in the U.S. shows a greater divergence from individuals in agrarian cultures compared to neurotypical controls. Unweighted UniFrac PCoA plot comparing data from children with ASD and neurotypical control in the U.S. (21) with a global survey of fecal microbial community composition conducted in individuals from Malawi, the Amazonas State of Venezuela, and the U.S. (35) (global gut). (a) Points colored by country of residence. (b) Same plot, but with points colored by study/ASD status.
We first assigned sequences from both studies to 97% identity (ID) OTUs (clusters in which sequences have ≥97% ID over their aligned 16S rRNA genes). This is a method that is typically used by microbial ecologists to use 16S rRNA gene sequences to approximate counts of different species in each sample, where each 97% ID OTU is a unique species-like unit (40). Specifically, we assigned our sequences to their closest relative in a database of previously characterized 16S rRNA 97% ID OTUs (February 4, 2011, Greengenes database) (41) using UCLUSTref (42) as described in Lozupone et al. (43). Unassigned sequences, which typically were only <5% of each sample, were dropped from the analysis. The degree of difference in the collection of OTUs found in each pair of samples was then estimated with the unweighted UniFrac distance metric. UniFrac is similar to algorithms that estimate distance between communities based on degree to which species are shared versus unique, such as the Sorenson index, except that it also considers phylogenetic relationships between species when performing the calculations (44). The relationship between the microbial diversity in the different samples was visualized using Principal Coordinate Analysis (PCoA) (Fig. 3). Only samples from individuals in Yatsunenko et al. who were between the ages of 4 and 60 were included in the analysis. We excluded the samples for children younger than 4 to be consistent with the population assessed in Kang et al. (21), in which study participants were 4–16 years old. Since the microbiota is highly variable in early life but stabilizes to an adult-like configuration in children aged 4 and older (35), we did not exclude the adults in the cohort of Yatsunenko et al. from the analysis.
As expected based on the analysis conducted by Yatsunenko et al. (35), the individuals from the US separated from individuals from Malawi and Venezuela along PC1 (Fig. 3a). The second PC axis separated US individuals evaluated in the two different studies from each other (Fig. 3b). This separation is not surprising because of methodological differences between the studies, such as the use of different techniques for extracting DNA out of the fecal samples and using pyrosequencing 454 versus Illumina technology for the sequencing (43). Interestingly, along PC1, the samples from children with ASD from Kang et al. (21) showed a significantly greater deviation from individuals from agrarian cultures than did the neurotypical children evaluated in Kang et al. (21) (Fig. 3b).
The implication of our finding of a ‘hyper-Westerization’ of the fecal microbiota of children with ASD is hard to ascertain. Since Prevotella-prevalence has been linked with dietary patterns within the US population, with Prevotella being high in individuals who consumed diets poor in animal protein and saturated fats and rich in carbohydrates (as assessed over the prior year with a food frequency questionnaire) (45), we cannot rule out that this observation is due to a distinct aspect of the diets of the children with ASD. However, dietary factors, such as adherence to a gluten and casein free diet, could not explain the microbiota differences between children with ASD and neurotypical controls (21). Another fascinating idea is that this ‘hyper-Westernization’ could also be linked with differences in the immune system. This notion is supported by the observation that individuals with untreated HIV infection have uniformly high Prevotella and in a similar meta-analysis with the dataset of Yatsunenko et al. (35), showed the opposite pattern of children with ASD, in having a fecal microbiota with a striking resemblance to that of individuals in the developing world (46). Evaluation of dietary questionnaires indicated that Prevotella-richness in HIV patients occurred independently of diet (47). The greater deviation of children with ASD from individuals with HIV that have a suppressed adaptive immune system may indicate that this deviation is driven by individuals with ASD having a hyper-active adaptive immune system. This is consistent with the observation that individuals with ASD have high adaptive immune cytokine responses in stimulations of their peripheral blood mononuclear cells (48, 49) and high amounts of the Th1 cytokine INF-gamma in the brain (50).
ASD possible gut–brain connection might be driven by microbial metabolites
Differences reported on microbial diversity and composition (20, 21, 23, 24) can also be attributed to the fact that several microorganisms can perform the same function. This metabolic function can be assessed by measuring metabolites produced. Several published metabolomics analyses of urine and fecal metabolites have revealed differential abundance of bacteria-produced metabolites that have the potential to directly affect neural processes (51–54). Published urinary metabolites that positively correlate with ASD symptoms include 1) 3-(3-hydroxyphenyl)-3-hydroxypropionic acid (HPHPA), a probable metabolite of a tyrosine analog that depletes catecholamines and causes symptoms of autism, such as stereotypical behavior, hyperactivity, and hyper-reactivity in experimental animals (51), 2) p-cresol, which can compete with neurotransmitters for enzymes and co-factors essential for sulfonation reactions in the liver (52, 53), and 3) urocanate, which is regulated by the enzyme urocanase (55). Deficiency of urocanase raises levels of urocanate in the urine and leads to urocanic aciduria that is associated with neurological disorders (54). Evidence of a role for bacterial metabolites in eliciting neurobehavioral symptoms of ASD also comes from a recent study of a maternal immune activation (MIA) model of ASD in mice, in which the MIA mice had significant increases in 8% of 322 serum metabolites, with one particular metabolite, 4-ethylphenylsulfate (4EPS), displaying a striking 46-fold increase. 4EPS is produced by gut bacteria, and treatment with a particular gut bacterial strain (Bacteroides fragilis) in early life changed the microbiome structure of MIA offspring mice, restored levels of 4EPS to normal, and greatly improved ASD symptoms. Furthermore, injection of 4EPS into naïve mice resulted in anxiety-like behavior, providing clear evidence that at least one metabolite produced by gut bacteria can substantially affect behavior in mice (56). Although these studies provide evidence that particular microbially produced metabolites may directly affect ASD symptoms, an important deficiency in our current understanding is which microbes may be responsible for the production of which metabolites, how these microbes interact with each other and with the host, and how feasible is it to modify ASD symptoms by managing these microbial interactions. Studies that explore links between microbiota composition, functional potential (gene content), gene expression (transcriptomics), metabolomics, presence and severity of ASD disease symptoms, or neurotransmitter signaling in the subject have promise for yielding a mechanistic understanding of a link between gut microbiota and ASD that will pave the way for therapeutics that target the microbiota in the treatment of ASD.
Prospective
A high rate of GI problems in children with ASD, correlations between symptom severity and GI symptoms within children with ASD, distinctive profiles of gut microbes and their metabolites in children with ASD, and a growing appreciation of a link between the gut and the brain for many neurological disorders, all point towards the potential for a role for gut microbiota in the presentation and severity of ASD symptoms. Furthermore, our observation of a ‘hyper-Westernization’ of the gut microbiota of children with ASD could indicate that gut microbiota differences that are driven by unique aspects of the Western lifestyle compared to the developing world lead to the association of unique gut microbiota composition with ASD. The complexity of the symptoms and the etiology of ASD coupled with the complexity of the microbiota and its functions has presented challenges in establishing the nature of an association between gut microbiota and ASD, pinning down whether a link even exists and for which individuals with ASD, and in producing a mechanistic understanding of the nature of this association. Further work in this field that apply cutting edge ‘omics’ technologies coupled with confirmatory experiments in mouse models, as has been done more extensively with other complex diseases such as obesity, have the promise to address these pressing questions for the millions of people affected by ASD worldwide.
This paper is part of the Supplement: The Microbiome in Autism Spectrum Disorder. More papers from this supplement can be found at http://www.microbecolhealthdis.net
Conflict of interest and funding
The authors have not received any funding or benefits from industry. The authors thank the Bhare autism and Emch foundations for funding some of this work.
References
- 1.Buie T, Fuchs GJ, Furuta GT, Kooros K, Levy J, Lewis JD, et al. Recommendations for evaluation and treatment of common gastrointestinal problems in children with ASDs. Pediatrics. 2010;125:S19–29. doi: 10.1542/peds.2009-1878D. [DOI] [PubMed] [Google Scholar]
- 2.Molloy CA, Manning-Courtney P. Prevalence of chronic gastrointestinal symptoms in children with autism and autistic spectrum disorders. Autism. 2003;7:165–71. doi: 10.1177/1362361303007002004. [DOI] [PubMed] [Google Scholar]
- 3.Nikolov RN, Bearss KE, Lettinga J, Erickson C, Rodowski M, Aman MG, et al. Gastrointestinal symptoms in a sample of children with pervasive developmental disorders. J Autism Dev Disord. 2009;39:405–13. doi: 10.1007/s10803-008-0637-8. [DOI] [PubMed] [Google Scholar]
- 4.Ming X, Brimacombe M, Chaaban J, Zimmerman-Bier B, Wagner GC. Autism spectrum disorders: concurrent clinical disorders. J Child Neurol. 2008;23:6–13. doi: 10.1177/0883073807307102. [DOI] [PubMed] [Google Scholar]
- 5.Valicenti-McDermott M, McVicar K, Rapin I, Wershil BK, Cohen H, Shinnar S. Frequency of gastrointestinal symptoms in children with autistic spectrum disorders and association with family history of autoimmune disease. J Dev Behav Pediatr. 2006;27:S128–36. doi: 10.1097/00004703-200604002-00011. [DOI] [PubMed] [Google Scholar]
- 6.Adams JB, Holloway CE, George F, Quig D. Analyses of toxic metals and essential minerals in the hair of Arizona children with autism and associated conditions, and their mothers. Biol Trace Elem Res. 2006;110:193–209. doi: 10.1385/BTER:110:3:193. [DOI] [PubMed] [Google Scholar]
- 7.Chaidez V, Hansen RL, Hertz-Picciotto I. Gastrointestinal problems in children with autism, developmental delays or typical development. J Autism Dev Disord. 2014;44:1117–27. doi: 10.1007/s10803-013-1973-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ibrahim SH, Voigt RG, Katusic SK, Weaver AL, Barbaresi WJ. Incidence of gastrointestinal symptoms in children with autism: a population-based study. Pediatrics. 2009;124:680–6. doi: 10.1542/peds.2008-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McElhanon BO, McCracken C, Karpen S, Sharp WG. Gastrointestinal symptoms in autism spectrum disorder: a meta-analysis. Pediatrics. 2014;133:872–83. doi: 10.1542/peds.2013-3995. [DOI] [PubMed] [Google Scholar]
- 10.Adams JB, Johansen LJ, Powell LD, Quig D, Rubin RA. Gastrointestinal flora and gastrointestinal status in children with autism-comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011;11:22. doi: 10.1186/1471-230X-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert JA, Krajmalnik-Brown R, Porazinska DL, Weiss SJ, Knight R. Toward effective probiotics for autism and other neurodevelopmental disorders. Cell. 2013;155:1446–8. doi: 10.1016/j.cell.2013.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al. Microbiota-generated metabolites promote metabolic benefits via gut–brain neural circuits. Cell. 2014;156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 13.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–12. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 14.Adams JB, Romdalvik J, Levine KE, Hu LW. Mercury in first-cut baby hair of children with autism versus typically-developing children. Toxicol Environ Chem. 2008;90:739–53. [Google Scholar]
- 15.Adams JB, Romdalvik J, Ramanujam VMS, Legator MS. Mercury, lead, and zinc in baby teeth of children with autism versus controls. J Toxicol Environ Health A. 2007;70:1046–51. doi: 10.1080/15287390601172080. [DOI] [PubMed] [Google Scholar]
- 16.Konstantareas MM, Homatidis S. Ear infections in autistic and normal children. J Autism Dev Disord. 1987;17:585–94. doi: 10.1007/BF01486973. [DOI] [PubMed] [Google Scholar]
- 17.Niehus R, Lord C. Early medical history of children with autism spectrum disorders. J Dev Behav Pediatr. 2006;27:S120–7. doi: 10.1097/00004703-200604002-00010. [DOI] [PubMed] [Google Scholar]
- 18.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108:4554–61. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandler RH, Finegold SM, Bolte ER, Buchanan CP, Maxwell AP, Vaisanen ML, et al. Short-term benefit from oral vancomycin treatment of regressive-onset autism. J Child Neurol. 2000;15:429–35. doi: 10.1177/088307380001500701. [DOI] [PubMed] [Google Scholar]
- 20.Williams BL, Hornig M, Parekh T, Lipkin WI. Application of novel PCR-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. MBio. 2012;3:e00261–11. doi: 10.1128/mBio.00261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang DW, Park JG, Ilhan ZE, Wallstrom G, Labaer J, Adams JB, et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068322. e68322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gondalia SV, Palombo EA, Knowles SR, Cox SB, Meyer D, Austin DW. Molecular characterisation of gastrointestinal microbiota of children with autism (with and without gastrointestinal dysfunction) and their neurotypical siblings. Autism Res. 2012;5:419–27. doi: 10.1002/aur.1253. [DOI] [PubMed] [Google Scholar]
- 23.Finegold SM, Dowd SE, Gontcharova V, Liu C, Henley KE, Wolcott RD, et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16:444–53. doi: 10.1016/j.anaerobe.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 24.De Angelis M, Piccolo M, Vannini L, Siragusa S, De Giacomo A, Serrazzanetti DI, et al. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. 2013;8:18. doi: 10.1371/journal.pone.0076993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, Conlon MA. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl Environ Microbiol. 2011;77:6718–21. doi: 10.1128/AEM.05212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Y, Liu C, Finegold SM. Real-time PCR quantitation of clostridia in feces of autistic children. Appl Environ Microbiol. 2004;70:6459–65. doi: 10.1128/AEM.70.11.6459-6465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parracho HM, Bingham MO, Gibson GR, McCartney AL. Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. J Med Microbiol. 2005;54:987–91. doi: 10.1099/jmm.0.46101-0. [DOI] [PubMed] [Google Scholar]
- 28.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–30. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finegold SM, Molitoris D, Song YL, Liu CX, Vaisanen ML, Bolte E, et al. Gastrointestinal microflora studies in late-onset autism. Clin Infect Dis. 2002;35:S6–16. doi: 10.1086/341914. [DOI] [PubMed] [Google Scholar]
- 31.Walters WA, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014;588:4223–33. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matson JL, Kozlowski AM. The increasing prevalence of autism spectrum disorders. Res Autism Spectr Disord. 2011;5:418–25. [Google Scholar]
- 33.Elsabbagh M, Divan G, Koh YJ, Kim YS, Kauchali S, Marcin C, et al. Global prevalence of autism and other pervasive developmental disorders. Autism Res. 2012;5:160–79. doi: 10.1002/aur.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamer A, Zohar AH, Youngmann R, Diamond GW, Inbar D, Senecky Y. A prevalence estimate of pervasive developmental disorder among immigrants to Israel and Israeli natives- a file review study. Soc Psychiatry Psychiatr Epidemiol. 2004;39:141–5. doi: 10.1007/s00127-004-0696-x. [DOI] [PubMed] [Google Scholar]
- 35.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–6. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, et al. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol. 2013;9 doi: 10.1371/journal.pcbi.1002863. e1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stackebrandt E, Goebal BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–9. [Google Scholar]
- 41.DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:394–9. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 43.Lozupone C, Stombaugh J, Gonzalez A, Ackermann G, Wendel D, Vazquez-Baeza Y, et al. Meta-analyses of studies of the human microbiota. Genome Res. 2013;23:1704–14. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14:329–39. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7:983–94. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jyonouchi H, Sun S, Le H. Proinflammatory and regulatory cytokine production associated with innate and adaptive immune responses in children with autism spectrum disorders and developmental regression. J Neuroimmunol. 2001;120:170–9. doi: 10.1016/s0165-5728(01)00421-0. [DOI] [PubMed] [Google Scholar]
- 49.Molloy CA, Morrow AL, Meinzen-Derr J, Schleifer K, Dienger K, Manning-Courtney P, et al. Elevated cytokine levels in children with autism spectrum disorder. J Neuroimmunol. 2006;172:198–205. doi: 10.1016/j.jneuroim.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 50.Li X, Chauhan A, Sheikh AM, Patil S, Chauhan V, Li XM, et al. Elevated immune response in the brain of autistic patients. J Neuroimmunol. 2009;207:111–6. doi: 10.1016/j.jneuroim.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaw W. Increased urinary excretion of a 3-(3-hydroxyphenyl)-3-hydroxypropionic acid (HPHPA), an abnormal phenylalanine metabolite of Clostridia spp. in the gastrointestinal tract, in urine samples from patients with autism and schizophrenia. Nutr Neurosci. 2010;13:135–43. doi: 10.1179/147683010X12611460763968. [DOI] [PubMed] [Google Scholar]
- 52.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci USA. 2009;106:14728–33. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alberti A, Pirrone P, Elia M, Waring RH, Romano C. Sulphation deficit in “low-functioning” autistic children: a pilot study. Biol Psychiatry. 1999;46:420–4. doi: 10.1016/s0006-3223(98)00337-0. [DOI] [PubMed] [Google Scholar]
- 54.Ming X, Stein TTP, Barnes V, Rhodes N, Guo LN. Metabolic perturbance in autism spectrum disorders: a metabolomics study. J Proteome Res. 2012;11:5856–62. doi: 10.1021/pr300910n. [DOI] [PubMed] [Google Scholar]
- 55.Retey J. The urocanase story: a novel role of NAD+ as electrophile. Arch Biochem Biophys. 1994;314:1–16. doi: 10.1006/abbi.1994.1405. [DOI] [PubMed] [Google Scholar]
- 56.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–63. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]