Abstract
Cells are constantly changing their state of equilibrium in response to internal and external stimuli. These changes in cell identity are driven by highly coordinated modulation of gene expression. This coordinated regulation is achieved in large part due to changes in the structure and composition of the chromatin, driven by epigenetic modulators. Recent discoveries in cellular and genomic reprogramming have highlighted the importance of chromatin modifications to reach and uphold the fidelity of target cell states. In this review, we focus on the latest work addressing the mechanisms surrounding the epigenetic regulation of various types of reprogramming, including somatic cell nuclear transfer (SCNT), cell fusion and transcription factor- and microRNA-induced pluripotency. The studies covered herein showcase the interplay between these epigenetic pathways, and highlight the importance of furthering our understanding of these connections to form a clearer picture of the mechanisms underlying stable cell fate transitions.
INTRODUCTION
Cells have a specific molecular and physiological identity that dictates their function. However, many cell types are highly plastic and can transition efficiently from one type to another. This process requires loss of the molecular characteristics of the original cell, and acquisition of an entirely new but heritable molecular signature, in the context of an unchanging genomic sequence. This process, known as epigenetic reprogramming, often involves changes in transcription and chromatin structure as a result of changing covalent modifications on chromatin. Epigenetic reprogramming is highly temporally and spatially regulated, and a plethora of players cooperate to carefully orchestrate this process. There has recently been a large push towards understanding how to manipulate epigenetic changes to help convert one cell type into another in vitro.
Historically, the study of embryonic development, including fertilization of an oocyte and specification of primordial germ cells, has informed our view of epigenetic reprogramming. Recently, with the discovery of somatic cell reprogramming, studies have expanded to analyzing epigenetic reprogramming of diverse cell types in vitro. In vitro studies have made understanding of the molecular mechanisms of epigenetic reprogramming more attainable. This review focuses on recent progress made in understanding the dynamic epigenetic changes that are required to accurately and efficiently reprogram the epigenome of one cell type into another. We compare different methods of reprogramming cells from one type to another, and identify key epigenetic players that regulate these transitions. There are certain broad changes during reprogramming that have been identified in recent years, including genomic demethylation (both histone and DNA), histone acetylation and loss of heterochromatin (Fig. 1 and summarized in two excellent recent reviews [1,2]). The exact mechanisms by which these changes are achieved and the detailed interplay between the players responsible however remain relatively unclear. And while the pathways utilized during different forms of experimental reprogramming are not necessarily the same, there are emerging patterns common to most if not all cell state transitions.
Fig. 1.
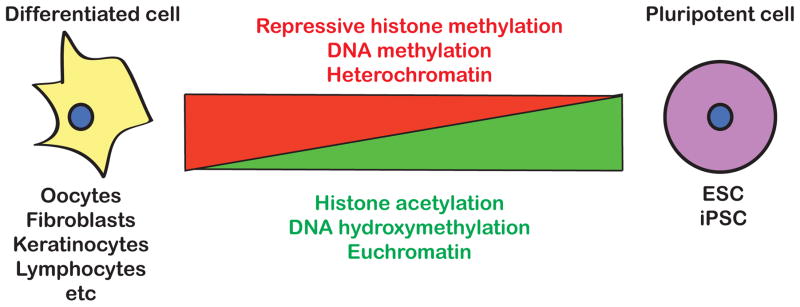
Key epigenetic changes during the transition between differentiated and pluripotent cells.
SOMATIC CELL NUCLEAR TRANSFER
Somatic cell nuclear transfer (SCNT), involving the transfer of a somatic nucleus into an enucleated oocyte to produce cloned animals, is the earliest example of experimentally induced programming [3]. Interestingly, cloned animals have reduced survival relative to naturally fertilized embryos, and it is widely hypothesized that this lethality is due to improper epigenetic reprogramming in both the embryonic and extra-embryonic lineages [4]. Aberrant DNA methylation has been observed in swine, bovine and murine cloned embryos compared to their fertilized counterparts [5–7]. Repeat regions are especially susceptible, both in cow and mouse embryos. In both cases, satellite repeats (Satellite I repeat in cows, and LINEs and LTRs in mice) remain hypermethylated in SCNT embryos relative to wild-type counterparts. The transfer of globally hypomethylated somatic nuclei cells improves the efficiency of reprogramming of those nuclei by SCNT [8]. Additionally, fertilization-specific demethylation at specific promoters fails to occur during SCNT in mouse, suggesting the absence of signals directing specific demethylation events [5,6]. Together, these data support the notion that defects in DNA demethylation in part underlie the reduced survival of cloned embryos.
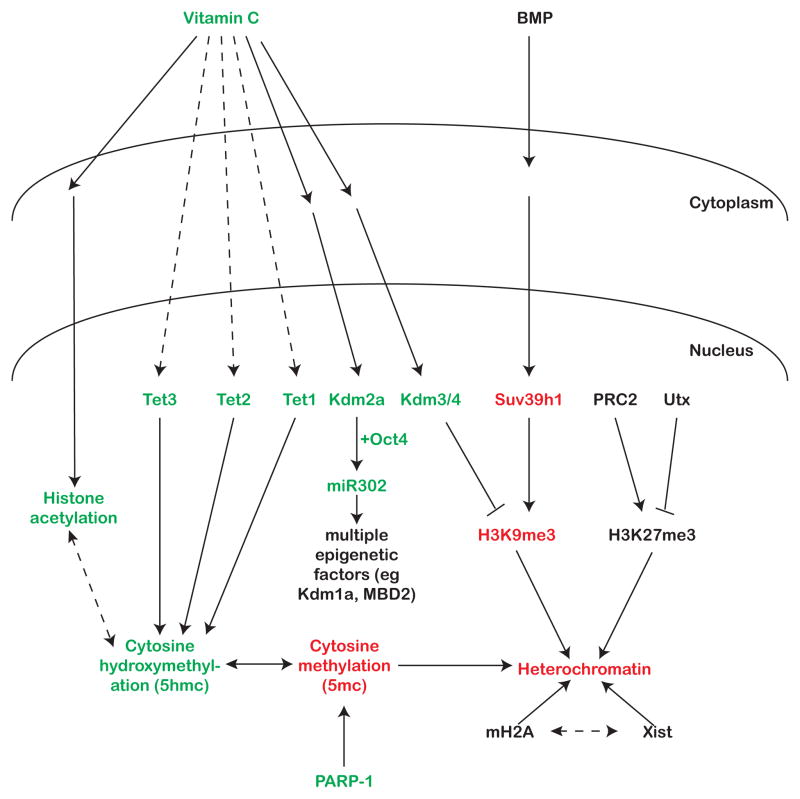
In recent years, genome-wide methylation has been shown to be more dynamic than previously thought, providing insights on how regulation of methylation contributes to epigenetic plasticity. During DNA demethylation, 5-methyl cytosines (5mc) can be converted to 5-hydroxymethyl cytosines (5hmc) by the Tet family of proteins [9]. Tet3, which is expressed in the oocyte, localizes to the somatic pseudo-pronucleus upon SCNT and Tet3 knockout oocytes fail to de-repress somatic Oct4 following SCNT [10]. 5hmc may be more than just a demethylation intermediate as it has been shown to have specific binding partners [11,12]. Furthermore, changing DNA methylation has to be accompanied by modulation of other epigenetic modifications in order to achieve a reprogrammed epigenome, and it is likely that these events are co-regulated (Fig. 2).
Fig. 2.
Pathways influencing the epigenetics of reprogramming. Green text -known enhancers of reprogramming. Red text - known inhibitors of reprogramming. Black text - unknown, unclear, or many different effects on reprogramming. Solid lines represent known connections and dashed lines represent speculative connections between factors.
One way to achieve coordinated regulation of different epigenetic factors is to have common signaling molecules. It has been postulated that Vitamin C, a known co-factor for histone demethylases, might also work through the Tet family of DNA demethylating enzymes [13,14]. Vitamin C can enhance blastocyst formation following SCNT and is associated with enhanced histone acetylation and increased expression of pluripotency markers such as Sox2 and Klf4 [15] (Fig. 2). Indeed, histone deacetylase (HDAC) inhibitors improve SCNT efficiency [16,17]. Vitamin C is unlikely to directly influence histone acetylation as it has not been shown to be a cofactor of acetyltransferases. Rather it may influence acetylation indirectly by acting through demethylase, tying together several epigenetic pathways (Fig. 2)
Tet-dependent hydroxymethylation of the somatic genome during reprogramming may result in recruitment of additional epigenetic modifiers. It is known that embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) have higher levels of hydroxymethylation than somatic cells, further supporting a functional role for that modification and suggesting that the presence of 5hmc might help drive the unique epigenetic environment required to achieve pluripotency [18]. So the defects in the Tet3 mutant could be either due to persistence of 5mc or absence of 5hmc (or perhaps a combination of both). Efficiency of SCNT also seems to be associated with the histone modification status of the donor genome. Using nuclei from fibroblasts pre-treated with ooplasm greatly improves SCNT efficiency, specifically reducing histone H3K9 methylation and increasing acetylation [19]. Taken together, these papers showcase the importance of multiple pathways cooperating to coordinate reprogramming of various epigenetic modifications during SCNT (Fig 2.).
Modulation of histone and DNA marks goes hand-in-hand with changes in chromatin structure, which impacts the expression of underlying genes, and therefore, the state of the cell. Epigenetic reprogramming to pluripotency requires the dramatic loss of heterochromatin, which includes the re-activation of the inactive X chromosome that is characteristic of somatic cells. Intriguingly, oocytes are relatively ineffective at re-activating the X-chromosome following SCNT (as measured by aberrant expression of Xist RNA, the non-coding RNA responsible for coating and inactivating one X), suggesting that other areas of heterochromatin may also be poorly resolved [20]. The knockdown of the histone variant macroH2A facilitates X-reactivation through loss of heterochromatin, and improves pluripotency gene expression, in the context of SCNT in Xenopus oocytes [21]. However, the role of canonical X-inactivation factors does not appear to be restricted to reactivation of the X chromosome during epigenetic reprogramming. SCNT-derived embryos have ectopic expression of Xist associated with global mis-regulation of many genes until the morula stage in SCNT-derived embryos. RNAi-mediated knockdown of Xist improves the survival of mice cloned by SCNT[20]. This result suggests that preventing Xist expression during pre-implantation development helps SCNT-derived embryos obtain an epigenetic state beneficial for survival, and provides a yet unknown and uncharacterized role for Xist that extends beyond X-inactivation. Further investigation into the connection between chromatin modifications and general chromatin structure and architecture will shed light on how global changes in chromatin are coupled with highly specific changes in gene expression (Fig. 2).
REPROGRAMMING BY CELL FUSION
Oocytes are not the only cell type capable of reprogramming other cells. As early as half a century ago, scientists discovered that fusion of two somatic cell types resulted in epigenetic reprogramming, highlighting the epigenetic plasticity of terminally differentiated cells [22]. About fifteen years ago, Surani and colleagues discovered that fusion of embryonic germ cells (EGCs – pluripotent cells derived from primordial germ cells) with thymic lymphocytes leads to the demethylation of imprinted loci as would normally occur during PGC reprogramming [23]. This experiment showed that these pluripotent cells possess the ability to override a somatic cell’s inherent program providing an exciting platform to identify the factors that contribute to reprogramming. For example, the ability of ESCs to reprogram fibroblasts is dependent on the cell cycle stage of the ESCs in question. Specifically, cells in the S/G2 phases are more likely to successfully reprogram somatic cells than cells in G1, suggesting that the ongoing DNA synthesis is required to erase the memory of the fibroblast genome [24].
In fact, ESCs reprogram somatic cells in a bi-phasic manner, the second of which appears to require DNA synthesis. The somatic nucleus first undergoes a ‘trans-reprogramming’ that occurs with rapid kinetics, and has been hypothesized to require the rapid action of trans-activating transcription factors present in the ESCs. Following this, the somatic genome experiences ‘cis-reprogramming’, which requires a round of replication and has much slower kinetics, suggesting that some parts of the somatic chromatin are resistant to trans-reprogramming until they have been ‘rebooted’ by DNA replication [25]. However, it has recently been shown that pluripotency in heterokaryons can be achieved rapidly without cell division or DNA replication showing that cis-reprogramming is not always required [26].
Key trans-factors for reprogramming are now beginning to be identified, including a number of epigenetic factors. For example, two members of the Tet family of DNA demethylases, Tet1 and Tet2, have critical yet distinct roles in reprogramming during cell fusion. While Tet2 has a global demethylation role during cell fusion, and depletion of Tet2 in EGCs greatly reduces the efficiency with which they reprogram human B cells, Tet1’s role appears to be focused on the demethylation of imprinted control regions [27]. This difference highlights once again how coordinated and regulated the process of reprogramming is, to the extent that two highly related proteins perform different non-redundant functions, both of which contribute to the ultimate success of reprogramming (Fig. 2).
While reprogramming to pluripotency is often associated with an increase in active chromatin marks, ESCs and EGCs lacking functional polycomb complex 2 (PRC2, responsible for depositing the repressive mark H3K27me3) are unable to reprogram B-cells upon fusion [28]. This suggests that the active repression of lineage-specific genes is also important. However, knockout of PRC2 components (and therefore loss of H3K27me3) does not cause the loss of pluripotency in ESCs [29,30]. PRC is therefore critical for the transition of a somatic genome to a pluripotent one, but not for its maintenance, suggesting that resulting de-repression of lineage-specific genes is not sufficient to exit pluripotency.
TRANSCRIPTION FACTOR- AND MIRNA-INDUCED PLURIPOTENCY
Cell fusion experiments highlight the importance of ESC trans-factors for reprogramming the somatic genome. This concept led Yamanaka and colleagues to identify key pluripotency-related transcription factors that when overexpressed can reprogram somatic cells into induced pluripotent stem cells (iPSCs)[31]. This exciting discovery not only led to an explosion in the field of reprogramming, but also considerably advanced our understanding of stem cell biology. In the last few years, many studies have focused on elucidating the mechanisms by which exogenously introduced transcription factors can reprogram somatic cells to a pluripotent state. A recent review by Papp and Plath summarizes in great depth the relationship between chromatin state and reprogramming to pluripotency, as well as the role of various chromatin-modifying factors on the efficiency of reprogramming [1]. Here, we will focus on the interplay between transcription factors, microRNAs and epigenetic modifiers during this cell state transition.
There appear to be multiple stages in preparing the chromatin for pluripotency, each of which requires different players. Recent work has shown the broad requirement for a number of epigenetic modifiers for preparing cells for pluripotency [32], including identifying individual roles for specific proteins. For example, the histone H3K27 demethylase Utx is not necessary for maintaining pluripotency, but is required for somatic cells to transition to iPSCs [33]. Similarly, the interplay between the methyltransferases and demethylases of histone H3K9 establish a pre-pluripotency state that is responsive to BMP signaling [34]. The down-regulation of BMP signaling causes a switch of the H3K9 methylation status of pluripotency genes, resulting in their expression. The authors suggest that demethylation of repressive histone marks can establish a ‘pluripotency primed’ state during reprogramming. This state is not fully pluripotent, but in response to vitamin C (which is likely regulating the H3K9 demtheylases), the cells overcome the barrier separating the “primed” and fully pluripotent states (Fig. 2). It is conceivable that before iPSCs can acquire the appropriate transcriptome, their epigenome must be prepared in advance to initiate and sustain the expression of key genes [35].
Establishing the correct DNA methylation state of specific loci is vital to set up this pluripotency-permissive epigenetic state. Tet2 is required to increase 5-hydroxymethyl (5hmc) levels at critical loci. 5-methylcytosine (5mc) on the other hand is also regulated independently by additional factors such as the chromatin-binding protein PARP-1 [36]. This uncoupling of 5hmc and 5mc lends support to the growing theory that 5hmc is its own epigenetic modification rather than just a demethylation intermediate. It suggests that the factors recruited by 5hmc during reprogramming might be required in establishing an epigenetic state that allows complete reprogramming to pluripotency. Furthermore, recent work has shown that Oct4 can be replaced by Tet1 during reprogramming of mouse fibroblasts. Both methylation and hydroxymethylation are increased during the intermediate stages of reprogramming [37]. Interestingly, while Tet1 can replace Oct4, physical interaction with Nanog appears to be important for Tet1 function during reprogramming. In a recent study, Nanog is proposed to recruit Tet1 to pluripotency loci for hydroxymethylation and activation of gene expression [38]. These papers suggest that tight regulation of the balance between methylation and hydroxymethylation is critical for successful transition of the epigenetic landscape towards pluripotency.
Epigenetic modifiers are sensitive to environmental stimuli, raising the question of how external factors can affect somatic cell reprogramming. Recent work reveals that reprogramming is greatly improved by addition of vitamin C, as is the case with SCNT [13,14,34,39,40]. This improved efficiency could be through the action of histone demethylases, as suggested by the requirement of H3K9 and H3K36 demethylation for iPSC formation [34,41]. Intriguingly, the H3K36 demethylase Kdm2a is vitamin C-dependent and promotes expression of cell cycle genes by demethylating and repressing Ink4a/Arf (Fig. 2). This highlights the importance of repressing of the somatic program in addition to activating the pluripotency program to achieve iPSC status. Indeed, during de-differentiation, a vast majority of cells are refractory to reprogramming, suggesting inadequate silencing of areas of chromatin regulating the somatic transcriptional program [42,43].
Kdm2a also cooperates with Oct4 to activate the miR302 family of microRNAs, which might further promote the transition to pluripotency. The miR302 family is part of a broader family of miRNAs, the ESCC miRNAs, known to regulate the unique cell cycle properties of ESCs [41,44]. Recent work has shown that miR302 and other ESCC family members, are capable of reprogramming mouse and human somatic cells to iPSCs, either alone or in concert with other factors [45–48]. In fact, among the targets of miR302 in human ESCs are a number of chromatin regulators such as MECP2, MBD2, LSD1 and SMARCC2, further highlighting the link between different methods of epigenetic modulation (for example, miRNAs and methylation) [48,49].
Covalent modification of chromatin is only one step in establishing the appropriate epigenetic environment. Modified chromatin recruits a multitude of other factors that regulate the structure of chromatin as well as the transcription of underlying genes. One example of a newly discovered chromatin-binding factor that affects reprogramming is the telomere-associated Zscan4 [50]. Although it is known that pluripotent cells maintain longer telomeres, and that Zscan4 has been shown to extend telomere length, it only enhances somatic cell reprogramming in the first 3 days of the process, suggesting roles that potentially extend beyond the maintenance of telomere length, as is the case in ESCs [50,51]. Indeed, Zscan4 has been shown to increase genomic stability during iPSC formation, and although it is known that telomeres rejuvenate during reprogramming, Zscan4 may also be acting via additional mechanisms [52]. For example, greatly altering the epigenetic state of a cell may cause undesirable transcriptional instability that can be buffered by factors such as Zscan4 [53].
FUTURE PERSPECTIVES
The highly complex regulatory mechanisms that define a cell state are carefully orchestrated. In attempting to replicate a specific cell state it is critical to mimic these mechanisms as closely as possible. Central to achieving that goal is a more thorough understanding of the epigenetic state of fully reprogrammed cells. For example, in order to achieve stable and high-fidelity iPSCs, it is essential to be able to reproduce the epigenetic environment of a pluripotent cell as accurately as possible, thus arming the iPSCs with the potential to produce normal downstream lineages.
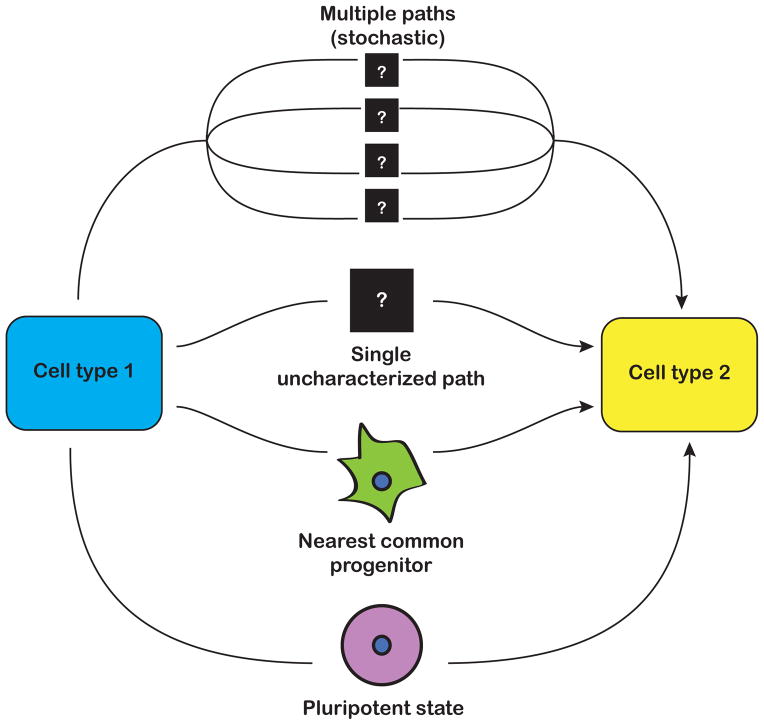
While making ESC-like cells from somatic cells could transform the landscape of regenerative medicine, an alternative strategy is to directly convert one somatic cell type to another, termed transdifferentiation. Epigenetic reprogramming almost certainly plays an equally important role in transdifferentation as it does in dedifferentiation to iPSCs, although very little is known in this context. One recent example is the transdifferentiation of pre-B cells to macrophages, during which key genes are hydroxymethylated. The transition fails in the absence of Tet2, suggesting that demethylation is critical for transdifferentiation, as is the case with reprogramming to pluripotency [54]. It will be important to determine whether there are intermediate or transitory epigenetic states during transdifferentiation, and how fully reprogrammed the final products are (Fig. 3). For example, are there certain molecular benchmarks that need to be reached, as is the case with reprogramming to pluripotency? What are the roles of stochasticity and hierarchy? How much memory of the originating fate remains following transdifferentiation? Similar to the production of iPSCs, answering these questions will allow for optimization of current methods of transdifferentiation.
Fig. 3.
Unanswered questions in the field of transdifferentiation. Transitioning from one cell type to another could involve going through a pluripotent state, the nearest common progenitor state, or one or more unknown states.
Acknowledgments
The authors would like to thank R. Parchem, A. Shenoy, M. LaRussa, J. Freimer, A. Chen and J. Vincent for critical reading of the manuscript. RB is funded by California Institute of Regenerative Medicine (RN2-00906-1), NIH (R01 NS057221, R01 GM101180, U54 HD055764), DOD (PC110465), and the Leona M. and Harry B. Helmsley Charitable Trust. RK is supported by an A.P. Giannini post-doctoral fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Papp B, Plath K. Epigenetics of Reprogramming to Induced Pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Narbonne P, Miyamoto K, Gurdon JB. Reprogramming and development in nuclear transfer embryos and in interspecific systems. Curr Opin Genet Dev. 2012;22:450–458. doi: 10.1016/j.gde.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin J, Shi L, Zhang M, Yang H, Qin Y, Zhang J, Gong D, Zhang X, Li D, Li J. Defects in trophoblast cell lineage account for the impaired in vivo development of cloned embryos generated by somatic nuclear transfer. Cell Stem Cell. 2011;8:371–375. doi: 10.1016/j.stem.2011.02.007. [DOI] [PubMed] [Google Scholar]
- **5.Chan MM, Smith ZD, Egli D, Regev A, Meissner A. Mouse ooplasm confers context-specific reprogramming capacity. Nat Genet. 2012;44:978–980. doi: 10.1038/ng.2382. This study compares DNA methylation of the genome following fertilization and SCNT, and identifies certain loci that are demethylated after fertilization but resistant to demethylation during SCNT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Couldrey C, Wells DN. DNA Methylation at a Bovine Alpha Satellite I Repeat CpG Site during Development following Fertilization and Somatic Cell Nuclear Transfer. PLoS One. 2013;8:e55153. doi: 10.1371/journal.pone.0055153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen CJ, Cheng WT, Wu SC, Chen HL, Tsai TC, Yang SH, Chen CM. Differential differences in methylation status of putative imprinted genes among cloned swine genomes. PLoS One. 2012;7:e32812. doi: 10.1371/journal.pone.0032812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blelloch R, Wang Z, Meissner A, Pollard S, Smith A, Jaenisch R. Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem Cells. 2006;24:2007–2013. doi: 10.1634/stemcells.2006-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **10.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 477:606–610. doi: 10.1038/nature10443. This paper shows that Tet3 is required for demethylation of the paternal genome after fertilization. Additionally, Tet3-deficient oocytes have greatly reduced efficiency of SCNT, highlighting the importance of genomic demthylation during this process. [DOI] [PubMed] [Google Scholar]
- 11.Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung TL, Brena RM, Kolle G, Grimmond SM, Berman BP, Laird PW, Pera MF, Wolvetang EJ. Vitamin C promotes widespread yet specific DNA demethylation of the epigenome in human embryonic stem cells. Stem Cells. 2010;28:1848–1855. doi: 10.1002/stem.493. [DOI] [PubMed] [Google Scholar]
- 14.Esteban MA, Pei D. Vitamin C improves the quality of somatic cell reprogramming. Nat Genet. 2011;44:366–367. doi: 10.1038/ng.2222. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y, Tang X, Xie W, Zhou Y, Li D, Zhu J, Yuan T, Lai L, Pang D, Ouyang H. Vitamin C enhances in vitro and in vivo development of porcine somatic cell nuclear transfer embryos. Biochem Biophys Res Commun. 2011;411:397–401. doi: 10.1016/j.bbrc.2011.06.160. [DOI] [PubMed] [Google Scholar]
- 16.Bui HT, Seo HJ, Park MR, Park JY, Thuan NV, Wakayama T, Kim JH. Histone deacetylase inhibition improves activation of ribosomal RNA genes and embryonic nucleolar reprogramming in cloned mouse embryos. Biol Reprod. 2011;85:1048–1056. doi: 10.1095/biolreprod.110.089474. [DOI] [PubMed] [Google Scholar]
- 17.Iager AE, Ragina NP, Ross PJ, Beyhan Z, Cunniff K, Rodriguez RM, Cibelli JB. Trichostatin A improves histone acetylation in bovine somatic cell nuclear transfer early embryos. Cloning Stem Cells. 2008;10:371–379. doi: 10.1089/clo.2007.0002. [DOI] [PubMed] [Google Scholar]
- 18.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 19.Bui HT, Kwon DN, Kang MH, Oh MH, Park MR, Park WJ, Paik SS, Van Thuan N, Kim JH. Epigenetic reprogramming in somatic cells induced by extract from germinal vesicle stage pig oocytes. Development. 2012;139:4330–4340. doi: 10.1242/dev.086116. [DOI] [PubMed] [Google Scholar]
- 20.Matoba S, Inoue K, Kohda T, Sugimoto M, Mizutani E, Ogonuki N, Nakamura T, Abe K, Nakano T, Ishino F, et al. RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc Natl Acad Sci U S A. 2011;108:20621–20626. doi: 10.1073/pnas.1112664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **21.Pasque V, Gillich A, Garrett N, Gurdon JB. Histone variant macroH2A confers resistance to nuclear reprogramming. EMBO J. 30:2373–2387. doi: 10.1038/emboj.2011.144. This work shows that the histone variant macroH2A prevents X-reactivation during reprogramming. Cells lacking macroH2A are therefore able to reprogram with much higher success. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tada M, Tada T, Lefebvre L, Barton SC, Surani MA. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 1997;16:6510–6520. doi: 10.1093/emboj/16.21.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsubouchi T, Soza-Ried J, Brown K, Piccolo FM, Cantone I, Landeira D, Bagci H, Hochegger H, Merkenschlager M, Fisher AG. DNA synthesis is required for reprogramming mediated by stem cell fusion. Cell. 2013;152:873–883. doi: 10.1016/j.cell.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **25.Foshay KM, Looney TJ, Chari S, Mao FF, Lee JH, Zhang L, Fernandes CJ, Baker SW, Clift KL, Gaetz J, et al. Embryonic stem cells induce pluripotency in somatic cell fusion through biphasic reprogramming. Mol Cell. 2012;46:159–170. doi: 10.1016/j.molcel.2012.02.013. This paper introduces the idea of biphasic reprogramming during cell fusion, with the second, cis-reprogramming phase being slower and replication-dependent. The authors also demonstrate that cis-reprogramming is specific to ESCs and does not occur when two somatic cells are fused. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2009;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **27.Piccolo FM, Bagci H, Brown KE, Landeira D, Soza-Ried J, Feytout A, Mooijman D, Hajkova P, Leitch HG, Tada T, et al. Different Roles for Tet1 and Tet2 Proteins in Reprogramming-Mediated Erasure of Imprints Induced by EGC Fusion. Mol Cell. 2013;49:1023–1033. doi: 10.1016/j.molcel.2013.01.032. This study shows the non-redundant functions of Tet1 and Tet2 hydroxymethyltransferases in cell fusion, addressing an important issue of whether Tet proteins perform overlapping functions during reprogramming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pereira CF, Piccolo FM, Tsubouchi T, Sauer S, Ryan NK, Bruno L, Landeira D, Santos J, Banito A, Gil J, et al. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell. 2010;6:547–556. doi: 10.1016/j.stem.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 29.Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 30.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 32.Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **33.Mansour AA, Gafni O, Weinberger L, Zviran A, Ayyash M, Rais Y, Krupalnik V, Zerbib M, Amann-Zalcenstein D, Maza I, et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature. 2012;488:409–413. doi: 10.1038/nature11272. This study focuses on the commonalities between reprogramming in PGCs and establishing pluripotency, and the authors show that Utx is involved in both. [DOI] [PubMed] [Google Scholar]
- **34.Chen J, Liu H, Liu J, Qi J, Wei B, Yang J, Liang H, Chen Y, Wu Y, Guo L, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet. 2012;45:34–42. doi: 10.1038/ng.2491. This paper highlights the importance of balanacing the H3K9 methyltransferase and demethylase in response to different external stimuli, such as BMP and Vitamin C. These findings elegantly highlight the consolidation of signaling molecules with epigenetic enzymes. [DOI] [PubMed] [Google Scholar]
- **35.Koche RP, Smith ZD, Adli M, Gu H, Ku M, Gnirke A, Bernstein BE, Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. This study probes the epigenetic events of early reprogramming, and the connection between these events and transcriptional repsonses. Their thorough analysis reveals interesting relationships between H3K4me2 and H3K27me3, as well as insights on how the enhancer lanscape transitions from a somatic to pluripotent one. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doege CA, Inoue K, Yamashita T, Rhee DB, Travis S, Fujita R, Guarnieri P, Bhagat G, Vanti WB, Shih A, et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012;488:652–655. doi: 10.1038/nature11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Y, Chen J, Li K, Wu T, Huang B, Liu W, Kou X, Zhang Y, Huang H, Jiang Y, et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell. 2013;12:453–469. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, Li W, Weng Z, Chen J, Ni S, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2009;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Stadtfeld M, Apostolou E, Ferrari F, Choi J, Walsh RM, Chen T, Ooi SS, Kim SY, Bestor TH, Shioda T, et al. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat Genet. 2012;44:398–405. S391–392. doi: 10.1038/ng.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **41.Wang T, Chen K, Zeng X, Yang J, Wu Y, Shi X, Qin B, Zeng L, Esteban MA, Pan G, et al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2012;9:575–587. doi: 10.1016/j.stem.2011.10.005. This study shows that the Vitamin C-dependent demthylase Kdm2a enhances somatic cell reprogramming in multiple ways. Kdm2a cooperates with Oct4 to activate miR302, and it also increases cell cycle-promoting genes such as Ink4/Arf. [DOI] [PubMed] [Google Scholar]
- **42.Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 151:1617–1632. doi: 10.1016/j.cell.2012.11.039. This paper dissects the elusive molecular mechanisms of different phases of somatic cell reprogramming. The authors show a biphasic pattern of chromatin modulation, the first being driven by cMyc/Klf4 and the second by Oct4/Sox2/Klf4. Cells refractory to reprogramming undergo the first but not the second phase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee MR, Prasain N, Chae HD, Kim YJ, Mantel C, Yoder MC, Broxmeyer HE. Epigenetic Regulation of Nanog by MiR-302 Cluster-MBD2 Completes Induced Pluripotent Stem Cell Reprogramming. Stem Cells. 2012;31:666–681. doi: 10.1002/stem.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin SL, Chang DC, Lin CH, Ying SY, Leu D, Wu DT. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res. 2010;39:1054–1065. doi: 10.1093/nar/gkq850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hirata T, Amano T, Nakatake Y, Amano M, Piao Y, Hoang HG, Ko MS. Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Sci Rep. 2012;2:208. doi: 10.1038/srep00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marion RM, Blasco MA. Telomere rejuvenation during nuclear reprogramming. Curr Opin Genet Dev. 2010;20:190–196. doi: 10.1016/j.gde.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 53.Jiang J, Lv W, Ye X, Wang L, Zhang M, Yang H, Okuka M, Zhou C, Zhang X, Liu L, et al. Zscan4 promotes genomic stability during reprogramming and dramatically improves the quality of iPS cells as demonstrated by tetraploid complementation. Cell Res. 2012;23:92–106. doi: 10.1038/cr.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kallin EM, Rodriguez-Ubreva J, Christensen J, Cimmino L, Aifantis I, Helin K, Ballestar E, Graf T. Tet2 facilitates the derepression of myeloid target genes during CEBPalpha-induced transdifferentiation of pre-B cells. Mol Cell. 2012;48:266–276. doi: 10.1016/j.molcel.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]