Abstract
PURPOSE
Improvements are needed for the early detection of breast cancer, as current imaging methods lack sensitivity to detect small tumors and assess their disease phenotype.
PROCEDURES
To address this issue the dual reporter adenoviral vector (Ad5/3-Id1-SEAP-Id1-mCherry) was produced with a cancer specific Id1 promoter driving expression of a blood-based screening reporter (secreted embryonic alkaline phosphatase, SEAP) and a fluorescent imaging reporter (mCherry). This diagnostic system was assessed for its screening potential on breast cancer cell lines of various aggressive phenotypes. Reporter expression was measured and correlated with promoter level expression using western blot. Adenovirus receptor expression was normalized against reporter expression with luciferase infectivity assays. Ad5/3-Id1-SEAP-Id1-mCherry infected MDA-MB-231 cells combined with uninfected cells were implanted into the mammary fat pad of athymic nude mice to recapitulate low dose tumor delivery. Id1 driven SEAP expression and mCherry imaging were monitored to validate diagnostic sensitivity and efficacy.
RESULTS
Infected breast cancer cell lines displayed SEAP levels in the media that were 10-fold above background by 2 days after infection. Ad5/3-Id1-SEAP-Id1-mCherry infected cells (MOI=10) implanted in athymic nude mice demonstrated a 14-fold increase in serum SEAP levels over baseline when as little as 2.5% of the tumor contained infected cells. This robust response was also found for the mCherry reporter which was clearly visible in tumor xenografts on day 2 post implantation.
CONCLUSIONS
This diagnostic system that combines screening with imaging for early detection and monitoring of breast cancer can be easily extended to other reporters/modalities and cancer-targeting methods. Combining screening with imaging in a genetic, cancer-specific mechanism allows sensitive multi-modal detection and localization of breast cancer.
Introduction
In 2009 the American Cancer Society estimated 192,370 new cases of invasive breast cancer (plus 62,280 cases of in situ cancer) and 40,170 deaths, making breast cancer the most common noncutaneous cancer in U.S. women. Early diagnosis along with opportune treatment leads to a decrease in mortality of breast cancer patients with a five-year relative survival of 96.8% when detected early as opposed to only a 22.5% five-year survival when diagnosed at late stage [1]. Traditional modalities of breast cancer detection include screening by breast self-examination, screening by clinical breast examination, and mammography. A comprehensive study in 2002 evaluating the effectiveness of self-breast examination demonstrated no difference in breast cancer mortality after 10 years compared to the control group [2]. For clinical breast examinations, a separate study demonstrated an increased rate of false negatives were common with 17% to 43% of cancer diseased patients being diagnosed as negative [3]. Success in mammography is subject to patient heterogeneity with common screening aberrations resulting from differences in breast tissue density and body mass index [4, 5]. Each of these features leads to a decrease in sensitivity and specificity. These conditions also adversely affect detection efficiency with an estimated 33% of breast cancer detected with mammogram representing over-diagnosis [6]. Another mammography study also predicted that 6% to 46% of women with invasive cancer will have false-negative mammograms [4]. Particular groups at high risk are young women with dense breast tissue or populations with mucinous, lobular, or aggressive cancers [7, 8]. While full-field digital mammography is growing in use and improving the overall sensitivity of detection, patient factors will continue to be a barrier for accurate and unbiased screening of breast cancer. Other modalities such as MRI, PET, and ultrasound can be used to detect breast cancer with greater sensitivity; however they are not routinely employed due to their use of radioactive contrast agents and high cost. MRI affords greater sensitivity to the more common mammography however it is less specific as contrast-enhanced foci are common in normal mammary tissue leading to routine false positives [9, 10]. The use of ultrasound in characterization of breast cancer has been limited to examination of palpable masses and there is no supporting evidence for the successful incorporation of ultrasound as a routine early screening tool for breast cancer [11].
Considering the complications associated with current strategies of cancer detection, improved methods for breast cancer screening are desperately needed for the detection of first time breast cancer, and also for cancer recurrence in women that have already been treated. A recent study in 2008 found that 1 in 5 breast cancer survivors suffer recurrence within ten years of a five-year adjuvant therapy regimen [12]. The advantages for improved screening methods also benefit high risk populations such as patients who are BRCA+, have an increased family history of incidence, or those with cancer syndromes. As genetic profiling of healthy patients becomes more prevalent, the elucidation of high-risk individuals will support a more aggressive screening strategy for the early detection of cancer. Improvements in the field of early detection should include systems that are unbiased to patient differences while relying on the genotypic characteristics of cancer and not simply the size of the mass. Considering these objectives, the current report shows progress with early diagnosis and monitoring of breast cancer using a dual reporter adenovirus that includes a secreted human embryonic alkaline phosphatase (SEAP) reporter for blood-based screening and a mCherry reporter for localized imaging, both under the control of a phenotype-specific Id1 promoter. Since dual reporters linked traditionally by internal ribosome entry sites can result in inefficient expression of the second reporter, an Id1 promoter upstream of each reporter was introduced thereby allowing both reporters to be under equal and effective promoter control. This dual reporter system enables two methods of detection that can be used in collaboration or independently. The blood based SEAP reporter allows non-invasive detection in blood with high sensitivity while the imaging reporter allows for localized visualization which can be utilized for obtaining biopsies, operative tumor resection, or monitoring treatment response. Importantly, both reporters are under control of Id, specific for aggressive breast cancer phenotypes.
Id1 is a helix-loop-helix protein, a member of the inhibitor of differentiation family of transcription factors. They form inactive heterodimers with basic helix-loop-helix (bHLH) family of transcription factors that control cellular processes such as cell-fate determination, proliferation, cell-cycle regulation, angiogenesis, invasion, and migration [13, 14]. The basic DNA binding domain is absent in Id1 proteins. When bound to bHLH monomers they form inactive heterodimers leading to a dominant negative regulation of gene expression. Aberrant Id1 protein expression has been found in most cancers however it is not expressed in normal tissue [15, 16]. Considering the effects of irregular Id1 expression, it is not surprising to discover that expression is directly associated with an aggressive cancer phenotype and has been implicated in metastasis [17, 18]. An up-regulation in Id1 activity has also been discovered in the endothelial cells that compose the tumor vasculature, however Id1 expression is absent in normal vasculature [19]. While Id1 expression varies between cell lines and tissue type, expression in the tumor vasculature has been confirmed in all cancers, regardless of stage and grade [20, 21]. Id1 expression is also serum dependent in vitro, with a correlative response between Id1 activity and serum concentration [22-24]. The Id1 promoter was utilized in this breast cancer screening system due to the specific nature of Id1 expression in cancer, the correlation between expression and cancer phenotype, and the robust expression in the tumor vasculature.
Secreted human embryonic alkaline phosphatase (SEAP) is a truncated form of human embryonic alkaline phosphatase, which is present in the body only during early embryonic stages. The truncated C-terminus lacks the 24 amino acid membrane-anchoring domain normally present in the placental form allowing SEAP to be secreted from the cell [25]. SEAP has the unusual properties of being extremely heat stable and resistant to the phosphatase inhibitor L-homoarginine. Therefore, endogenous alkaline phosphatase activity can be eliminated by pretreatment of samples at 65°C and incubation with this inhibitor [26]. SEAP detection is extremely sensitive with sensitivity in the picogram/ml range. The human origin of SEAP allows use of this reporter in a blood-based medium without generating an immune response. These characteristics of extreme sensitivity and absent immunogenicity make SEAP a primary candidate for a blood-based reporter and incorporation into our early screening system.
The imaging reporter, mCherry, has an excitation peak of 587 nm and emission peak of 610 nm, and is a mutated variant of the widely used mRFP1. mCherry matures more quickly and completely than mRFP1, yielding higher extinction coefficient and brightness, yet bleaches 10-times more slowly [27]. The longer wavelength of mCherry decreases the interference from tissue auto-fluorescence and allows for greater tissue penetration. A recent publication summarized the advantages of mCherry in imaging applications [28].
Here a diagnostic adenovirus (Ad5/3-Id1-SEAP-Id1-mCherry) was evaluated in multiple cancer cell lines in vitro; in vivo tests also verified the approach. The breast cancer cell lines included a range of phenotypes from those with less invasive behavior to others that are highly invasive. This dual-reporter, cancer-specific construct was designed to shuttle the reporter genes to the cancer cells more efficiently due to an Ad “serotype chimera“ modification. Due to the limited availability and low surface expression of the Ad serotype 5 (Ad5) natural receptor (CAR) in cancers [29], a hybrid Ad5/3 fiber was constructed to ablate CAR-tropism and alter it to improve the vector infectivity. The serotype 3 Ad (Ad3)-specific tropism conferred to the Ad serotype 5 fiber by its knob domain replacement with that of the Ad3 fiber. The Ad3 receptor, hypothesized to be CD46 [30], is widely expressed in cancer and allows improved infectivity with Ad vector that was produced [31].
Materials and Methods
Generation of Ad5/3-Id1-SEAP-Id1-mCherry
The mCherry sequence was procured with an MTA from Dr. Roger Tsien (University of California, San Diego). PCR was done to isolate the mCherry sequence introducing HindIII into the 5’ end and XbaI downstream of the mCherry 3’ ORF. The mCherry sequence was then cloned into the MCS of a pGL3 cloning vector after complete digestion with HindIII /XbaI to create pGL3-mCherry. The SEAP sequence was sequestered by modifying pSEAP2-Basic (purchased from Clontech, Mountain View, CA) introducing HindIII into the 5’ end and XbaI downstream of the SEAP 3’ ORF. The SEAP sequence was then cloned into the MCS of a pGL3-basic cloning vector after complete digestion with HindIII/ XbaI in both plasmids to create pGL3-SEAP. The Id1 cDNA sequence was provided by Dr. Pierre-Yves Desprez [32]. In order to reduce the size of the plasmid, the short length Id1 sequence (272bp) was used in the construction of the virus [33]. PCR was performed to isolate the Id1 sequence introducing BglII in the 5’ end and HindIII into the 3’ end. The resulting segment was then BglII/ HindIII digested along with pGL3-mCherry and pGL3-SEAP cloning vectors followed by an overnight ligation. This generated pGL3-Id1-mCherry and pGL3-Id1-SEAP single reporter vectors. The Id1-SEAP cassette was removed from the pGL3-Id1-SEAP cloning vector along with the addition of 5’KpnI upstream of Id1 and 3’XhoI downstream of SEAP. The sequence was then digested with KpnI/XhoI and cloned into a pShuttle MCS generating pId1-SEAP. The Id1-mCherry cassette was excised and isolated with PCR introducing 5’ Id1 SalI and 3’ mCherry BglII restriction sites. Following SalI/BglII digestion, the Id1-mCherry sequence was cloned downstream of the Id1-SEAP sequence in the pId1-SEAP shuttle cloning vector. The following steps produced pId1-SEAP-Id1-mCherry which was confirmed by sequencing. Following Pme1 digestion of the dual-reporter shuttle and its homologous recombination in E.Coli (BJ5183) using standard methods [34] with an E1/E3-deleted, replication-defective Ad5/3 backbone (AdEASY-1 derivative), containing a chimera fiber with the knob domain from the Ad3 fiber [35], a bicistronic Ad5/3-Id1-SEAP-Id1-mCherry was generated. Following molecular validation of the vector structure by PCR, restriction analysis and partial sequencing, the recombinant genomes were digested with PacI and transfected in HEK293 helper cells using lipofectamine 2000 (Invitrogen, Carlsbad, CA). Following fluorescent plaque purification the virus was amplified and purified by double cesium chloride gradient centrifugation. A standard agarose-overlay plaque assay was conducted with HEK-293 cells to determine the infectious titer of the viral preparation (3.2×109 plaque forming units per mL).
In vitro experiments
All in vitro experiments were conducted in triplicate using 6 well plates and cell numbers were determined using a hemocytometer. Cell viability was measured by trypan dye exclusion. Cells were plated 24 hrs prior to adenovirus infection. Multiplicity of infection (MOI) was based on plaque forming units determined by a standard plaque assay. All media was removed after SEAP analysis at every time point and replaced with fresh media to account for the cumulative effects of SEAP production in culture. Background SEAP levels represent uninfected MDA-MD-231 and MCF7 (3.5×105 cells/well in triplicate) measurements at parallel time points. A total of 4 in vitro experiments were conducted. Experiment 1: MDA-MB-231 and MCF7 cell lines were plated (3.5×105 cells/well) in triplicate and infected with Ad5/3-Id1-SEAP-Id1-mCherry at an MOI of 50. SEAP levels were measured on days 1, 2, 8, and 13 post infection. Experiment 2: MDA-MB-231 and MCF7 cell lines were plated (3.5×105 cells/well) in triplicate and infected with Ad5/3-Id1-SEAP-Id1-mCherry (MOI=50) under high serum (10%) conditions. SEAP levels were measured on days 1 and 3 after infection. The 10% media was then replaced with 2% serum media to simulate serum starvation (ss) conditions. Experiment 3: In order to qualify SEAP reporter expression as a singular function of Id1 promoter activity, an Ad3 receptor normalization assay was performed using an Ad5/3-CMV-Luc adenovirus. This adenovirus was generated using the same Ad5/3 adenoviral backbone used to construct the Ad5/3-Id1-SEAP-Id1-mCherry adenovirus. MCF10a, Dy36T2, MCF7, MDA-MB-468, LCC6, and MDA-MB-231 cell lines were plated (3.5×105 cells/well) in triplicate and infected with Ad5/3-CMV-Luc (MOI=300). 24hrs post infection all wells were assayed for luciferase activity. An in vitro SEAP assay was then performed on the Ad5/3-Id1-SEAP-Id1-mCherry infected (MOI=50) mammary cell panel and SEAP levels were measured. These values were then divided by the luciferase counts, determined for each Ad5/3-CMV-Luc –infected cell line, which represented a relative surface expression of the Ad3 receptor of each cell type. Experiment 4: SEAP assays were performed with Ad5/3-Id1-SEAP-Id1-mCherry infected (MOI=10) MDA-MB-231 and MCF7 cell lines using either 1.0×106 or 3.5×106 cells for each cell type. SEAP amounts were measured at several time points from day 0 to day 13 post infection.
Cell lines and culture methods
LCC6 (a subclone of MDA-MB-435), MDA-MB-468, MCF7, and Dy36T2 (a subclone of MDA-MB-361) breast cancer cell lines were kindly provided by Dr. Donald Buchsbaum (UAB). MDA-MB-231 and MCF10a were purchased from the American Tissue Type Collection (Manassas, VA). MDA-MB-231 and Dy36T2 cell lines were maintained in DMEM, 10% FBS, and 1% L-glutamine. LCC6 cells were grown in DMEM, 15% FBS, 1% L-glutamine, and 10ug/mL insulin. MDA-MB-468 cells were maintained in DMEM, 10% FBS, 1% NEAA, 1% MEM vit, and 1% NaPr. MCF7 cells were maintained in eMEM, 10% FBS, 1% L-glutamine, 1mM NaPr, 0.1mM NEAA, 20 μg/mL insulin, and 1.5g/L Na Bi. MCF10a cells were grown in DMEM/F12, 5% horse serum, 20 μg/mL EGF, 0.5 μg/mL Hydrocortizone, 0.1 μg/mL CholeraToxin, and 10 μg/mL insulin. All cells were maintained to 70%-90% confluency before passaging. Cell lines were cultured at 37°C and 5% CO2. The serum-starvation conditions were created by supplementing 2% FBS for 10% FBS in all culture media.
Western blot
Protein lysates from both serum-supplemented (10% FBS) and serum starved (2% FBS) breast cancer cells were collected with RIPA-modified buffer (Sigma-Aldrich) with 1% SDS and phosphatase inhibitors (1mM sodium orthovanadate, 25mM b-glycerophosphate, 100mM sodium fluoride) and protease inhibitors (10 mg/ml leupeptin, 10 mg/ml aprotinin, 1mM PMSF). Protein lysates (20μg, determined by Lowry assay) were separated with 4%-20% Tris-glycine electrophoresis gels (Invitrogen, Carlsbad, CA) followed by transfer to PVDF membranes (Millipore Immobilon, Billerica, MA). Membranes were blocked with 5% BSA and probed with rabbit monoclonal anti-mouse Id1, clone 195-14 (CalBioreagents, San Mateo, CA) followed by an HRP-conjugated goat anti-rabbit Ig (SouthernBiotech, Birmingham, AL). All membranes were washed 3 times with TBST buffer for 20min per wash. Id1 protein was visualized using chemiluminescent substrate (SuperSignal West Pico Chemiluminescent Substrate, ThermoScientific, Rockford, IL).
Bioluminescent analysis
Breast cancer cells were seeded (3.5×105cells/well) in 6-well plates in triplicate 24hr prior to adenoviral infection. Cells were infected (MOI=300) with Ad5/3 CMV-Luc 24hr post infection. Substrate luciferin was added (15μg/well) and plates were imaged for 60 seconds at a binning of 8 with an IVIS-100 CCD imaging system (Caliper Life Sciences, Mountain View, CA). ROI analysis was performed with the instrument software (Living Image 3.2) to quantify total luciferase counts per well.
SEAP analysis
Blood-based reporter analysis was conducted using a Great EscAPe™ Fluorescence Detection Kit (Clontech). All steps were performed according to the established protocol (New Fluorescent Great EscAPE SEAP Assay (January 1997) CLONTECHniques XII (1):18-19).
Fluorescence imaging
Representative in vitro fluorescent images were acquired on day 3 post infection with Ad5/3-Id1-SEAP-Id1-mCherry. Breast cancer cell images (200x) were rendered using an mCherry 587nm excitation/600nm long pass filter on an inverted microscope with a halogen light source and Nuance multi-spectral camera (Cri, Woburn, WA). A liquid-crystal tunable wavelength filter in the camera was set for collection of emission images from 600 to 720 nm in 5 nm increments. Composite images (unmixed composites) were generated for each image cube by unmixing the spectral signature of the mCherry reporter proteins from those of background auto-fluorescence using a spectral library that was compiled from numerous spectral profiles collected from uninfected control cells under the same conditions. For in vivo fluorescent images, mice were anesthetized with isoflurane and tumors were imaged on day 2 post implantation with a Leica stereomicroscope (Model MZ-FLIII, Vashaw Scientific Inc., Norcross, GA). Filter, camera, and image processing from in vitro fluorescence methods were used along with the stereomicroscope. After final imaging of intact animals, mice were sacrificed and tumors were excised and imaged.
In Vivo Evaluation (Experiment 5)
Athymic female nude mice were obtained from Frederick Cancer Research (Hartford CT). Human breast cancer cells MDA-MB-231 were plated using standard tissue culture techniques and 24 hrs later 5×106 cells were infected with Ad5/3-Id1-SEAP-Id1-mCherry (MOI=10), while 4.0×107 cells remained uninfected and were maintained in a separate culture. All cells were harvested 48hr post infection, washed, and counted. Female nude mice (n=9) were implanted with 5.0×104 infected cells (2.5% of total cells) along with 1.95×106 uninfected cells in the mammary fat pad of each mouse. A second group of 9 mice were implanted with 3.5×105 infected cells (17.5% of total cells) and 1.65×106 uninfected cells in the mammary fat pad. Appropriate numbers of infected and uninfected cells were mixed in the syringe prior to implantation. A third group of 5 athymic mice were implanted in the mammary fat pad with 2.0×106 uninfected cells to serve as an uninfected, tumor bearing control group. Blood plasma SEAP levels and tumor fluorescence in the control group represent background levels for statistical comparison. All groups were then fluorescently imaged and blood plasma collected on days 2,5,10, and 13 post-implantation. Blood was obtained from anesthetized mice by retro-orbital collection with heparin treated capillary tubes. Collections were then spun and blood plasma was obtained for SEAP analysis. After day 14, mice were sacrificed and tumors collected for fluorescence imaging. Quantitative region of interest analysis for mCherry fluorescence in tumors was performed with Image J software using the original TIFF images. Matching ROIs were compared for mean counts/pixel. The uninfected tumor group was compared to the 2.5% infected tumor group.
Statistical analysis
Statistical comparisons were performed using Minitab 15 statistical software (State College, PA). In Exp. 1, MDA-MB-231 and MCF7 SEAP data were compared at each day collected. For Exp. 2, SEAP levels from the MDA-MB-231 cell line on day 3 are compared to SEAP levels on day 8. Day 3 and day 8 MCF7 SEAP levels are also compared to determine statistical significance. In Exp. 4, statistical analysis compares SEAP values from background MDA-MD-231 levels versus day 3 levels in addition to background versus day 13 values. MCF7 background SEAP levels are also compared to day 3 MCF7 values. Statistical analysis of Exp. 5 data compare background plasma SEAP levels to both 17.5% infected tumor group and 2.5% infected tumor group at day 2 post implantation. Additional analyses with ImageJ compared mean counts/pixel for fluorescence for tumors with 2.5% infected cells versus control (no infected cells).
Results
SEAP levels in the media correlate with endogenous Id1 expression in MDA-MB-231 (high level Id1) and MCF7 (low level Id1) cells, and are serum-dependent
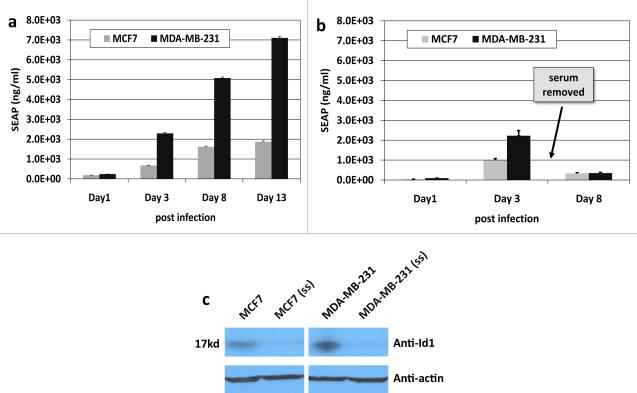
As shown in Figure 1a, the SEAP enzyme levels (ng/ml) in the MDA-MB-231 cells were significantly (p<0.05) higher by at least 3-fold as compared to the MCF7 cell line at each measured time point (Methods, Exp.1). This difference in reporter expression correlated with the relative Id1 protein levels, revealed by an Id1 protein immunoblot (fig.1c). The SEAP levels increased significantly (p<0.05) over time for both cell lines. Since media in each well was completely replaced after sampling for SEAP analyses, the SEAP levels represent robust expression from the cells during the time represented.
Fig 1.
Studies with MCF7 and MDA-MB-231 for. (a) media SEAP over time, (b) media SEAP level as influenced by serum starvation (ss), (c) Id1 protein by Western blot analyses.. Data are means +/− SD.
Serum starvation (ss) was utilized to confirm that SEAP was specific for the Id1 promoter (Methods, Exp. 2). SEAP levels obtained after serum starvation were significantly (p<0.05) lower for both cell lines as compared to cells grown under 10% serum conditions (fig. 1b, day 8). This decrease is in stark contrast to the higher SEAP values observed with 10% serum on day 8 post infection (fig. 1a). This difference in reporter expression between 10% serum and ss conditions was found to correlate with Id1 protein levels, determined by western blot for Id1 in ss MDA-MB-231 and ss MCF7 cell lines (fig.1c). A similar effect was observed in the mCherry reporter expression (data not shown) as fluorescence dramatically decreased in both cell lines during serum starvation. These results were consistent with expression of both reporters being driven by Id1 promoter activity.
Expression of the Id1-driven reporters from Ad5/3-Id1-SEAP-Id1-mCherry shows correlation with both endogenous Id1 levels and invasiveness of the infected cell’s phenotype
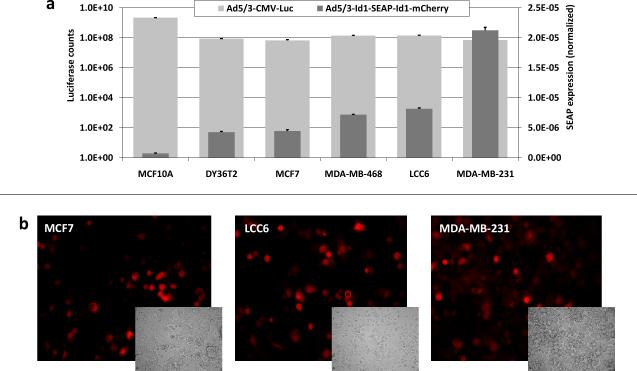
A difference between cell lines in their susceptibility to infection with tropism-modified Ad5/3-luc vector, caused by possible differences in their Ad3 receptor levels, could account for the observed differences in expression of the Id1-driven reporters. To rule out the infectivity factor a quantitative luciferase (luc) assay (Methods, Exp. 3), determining the relative infectivity of the tropism-modified Ad5/3-CMV-Luc vector for each cell type, was performed, allowing subsequent normalization of each reporter expression by the Ad3 receptor levels. All breast cancer cell lines in the panel displayed comparable susceptibilities to Ad5/3 CMV-Luc infection owing to similar levels of the Ad3 receptor expression (fig. 2a). An in vitro SEAP assay was performed on the panel of breast cancer cells following infection with the Ad5/3-Id1-SEAP-Id1-mCherry and the SEAP values were divided by luciferase counts for each cell line to normalize for infectivity. The normalized SEAP levels were consistent with cancer phenotypes of the cells, increasing from left to right (fig. 2a). For example, the lowest SEAP level was with MCF10A cells, an epithelial cell line that is not tumorigenic. mCherry fluorescence showed the same trend as SEAP for all the tested cell lines (fig. 2b). The intensity of fluorescence was also found to increase over time (data not shown).
Fig. 2.
Comparison of breast cancer cell lines for (a) Id1 expression (via SEAP) and infectivity (via luciferase) and (b) fluorescence imaging of representative cell lines for Id1-specific mCherry. Data are means +/− SD.
The Id1-driven expression of mCherry and SEAP provides a sufficient sensitivity to detect low number of Ad5/3-Id1-SEAP-Id1-mCherry–infected cells in vitro
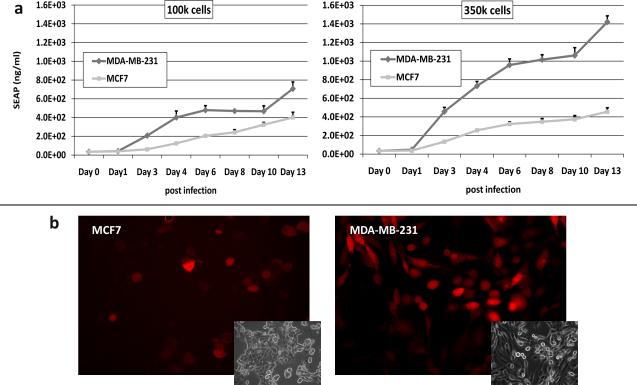
In order to demonstrate the ability of Id1 promoter to drive reporter expression and to assess the reporter detection sensitivity, SEAP and fluorescence assays were performed using a relatively low number of infected cells and low MOI (Methods, Exp. 4). Low cell numbers simulated the conditions of a small tumor mass, whereas low MOI represented conditions of low level virus delivery to the tumor. MDA-MB-231 cells demonstrated the greatest levels of SEAP reporter expression at all time points (fig 3a.). The level of SEAP expression by 1.0 × 105 MDA-MB-231 cells showed a statistically significant difference (p<0.05) over the background at day 3 and peaking at 5-fold level above the baseline on day 13 (fig 3a left). The larger population of infected MDA-MB-231 cells (3.5×105) produced a 10-fold increase in the SEAP reporter level on day 13 (fig 3a. right). These levels were found to be lower in the less invasive, low Id1-expressing MCF7 cell line, although, the reporter expression was still significantly (p<0.05) detectable at all time points beyond day 1 post infection, demonstrating the sensitivity of the diagnostic system. The fluorescent reporter expression under these conditions was also detectable at day 3 post infection (fig 3b.). The mCherry reporter expression in these cell lines correlated with SEAP reporter expression leading to MDA-MB-231 cells displaying the strongest overall fluorescence.
Fig 3.
Time course sensitivity study of MCF7 and MDA-MB-231 cells (100K and 350K) following infection with Ad5/3-Id1-SEAP-Id1-mCherry for (a) media SEAP, and (b)fluorescence (Representative fluorescent and inset bright field images are included). Images (20x) were collected using a 587nm excitation/600nm long pass filter and Nuance multi-spectral camera at 100ms exposure. Data are means +/− SD.
Expression of SEAP and mCherry can be detected in vivo following implantation of only 5×104 Ad5/3-Id1-SEAP-Id1-mCherry virus-infected cells
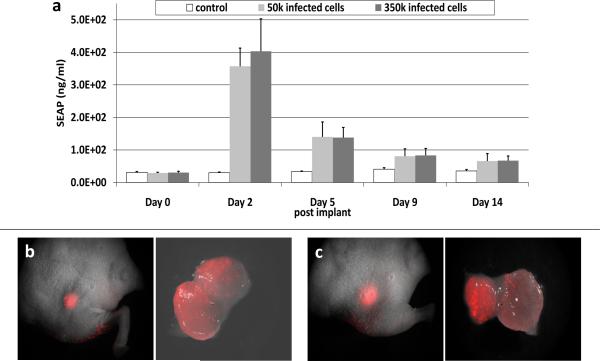
The sensitivity of the breast cancer screening system was tested in an animal model (Methods, Exp. 5). SEAP levels on day 2 post implantation were 403.5ng/ml +/− 99.4, for the mice implanted with 3.5×105 infected cancer cells (17.5% of total tumor) displaying a significant (p<0.05), 16-fold increase in SEAP levels above background (fig4.a). For the mice implanted with 5×104 infected cells (2.5% of total tumor), the average was 357.1ng/ml +/−55.7, i.e. a 14-fold increase (p<0.05) in SEAP level above background. SEAP production was found to return to near baseline by day 14 post implantation, while the tumors continued to grow for the duration of the experiment. A robust mCherry reporter expression was detected on day 2 post implantation in the tumors implanted with 3.5×105 infected cells (fig4.c) and also in the tumors with 5×104 infected cells (fig4.b). Quantitative analysis of TIFF pixel intensity with “Image J” demonstrated a significantly higher (p<0.05) fluorescence on day 2 post implantation for the tumors with 2.5% infected cells (50.6 +/− 12.9 counts/pixel) as compared with the control tumor group (12.1 +/− 4.8 counts/pixel). The fluorescence imaging confirmed the utility of a mCherry reporter for visual detection of a suspected lesion in animal model of cancer. Imaging of excised tumors from the two implantation groups on day 14 demonstrated a continuous mCherry expression, and the potential of the system in surgical resection applications (fig4.b-c).
Fig 4.
In vivo experiment with MDA-MB-231 tumor xenograft showing (a) SEAP levels (ng/ml of blood plasma, mean +/− SD), and imaging of mCherry on day 2 post implantation in tumor with (b) 5.0×104 infected cells with matching excised tumor (right) removed 14 days post implantation. (c) 3.5×105 infected cells with matching excised tumor (right) removed 14 days post implantation. Whole body images (0.63x) and excised tumor images (2.5x) were collected using a 587nm excitation/600nm long pass filter and Nuance multi-spectral camera at 4s exposure.
Discussion and Conclusions
A dual-reporter vector Ad5/3-Id1-SEAP-Id1-mCherry was constructed to allow noninvasive monitoring of breast cancer-specific expression of SEAP compatible with blood-based detection, and the mCherry optimized for fluorescence imaging in mammalian tissues. These studies showed a 3-fold higher SEAP and mCherry levels in MDA-MB-231 cells relative to MCF7. The MDA-MB-231 breast cancer cell line is classified as a more invasive and aggressive cell line displaying a triple negative (ER−, PR−, Her-2−) phenotype [25]. The Id1 expression level has been shown to be relatively higher in MDA-MB-231 cell line than in MCF7 (ER+, PR+, Her-2+) breast cancer cell line, which consistently portrays a less invasive and aggressive phenotype [25]. It has been reported that Id1 promoter activity is serum-dependent [22-24] and the results of our experiments are fully consistent with this finding.
Low Id1 expression was reported previously for the MCF10a, a non-tumorigenic human mammary epithelial cell line [25, 36]. Importantly, in the current study, the SEAP levels for the MCF10a cell line were the lowest, i.e. approximately 20-fold lower than those in the most aggressive MDA-MB-231 cell line. As suggested by the data in figure 2, both mCherry and SEAP reporter expression show relevance to invasiveness of the cancer cell phenotype, rather than to their Ad3 receptor levels. The observed increase in SEAP expression levels correlated with the increasingly aggressive behavior of the tested breast cancer cells [25, 36-38]. It was previously reported that Id1 expression directly correlates with an invasive and aggressive cellular phenotype [39]. The screening and imaging system tested here utilizes the Id1 promoter as a sensor to activate expression of two different reporters. In the future this approach could be used to alert oncologists to the presence of aggressive breast cancer cells during early screening.
Figure 3 experiments demonstrate the sensitivity of the dual reporter system under conditions of low cell numbers. The observed high sensitivity can be related to the strength of the Id1 promoter to drive reporter expression even for low endogenous Id1 expression, and the power of the SEAP assay to detect minute amounts of the enzyme. In terms of the localized imaging potential, it was demonstrated that the longer wavelength of the mCherry fluorescent reporter offered improved tissue penetration with low background.
In order to accurately estimate the minimum amount of cancer cells that can be detected in vivo, experiments were conducted with a known number of infected cancer cells representing various but minor fractions of the total tumor mass by mixing infected and uninfected cells in different proportions. This approach could also reflect the efficiency of targeted delivery of the vector to the tumors in the clinical context. Two levels were tested, namely: tumors, which contained either 2.5% or 17.5% of infected cells. With just 2.5% of the tumor containing adenovirus infected cells, SEAP levels were 14 fold above background. Owing to consistency in the background levels, it is clear that the 2-fold excess of the SEAP level over background could be considered as a threshold value for a positive signal. In the mouse model this level would represent approximately 7000 cells infected with the Ad vector. Together with the Id1 sensor of aggressive phenotype, these calculations support the potential of Ad5/3-Id1-SEAP-Id1-mCherry to provide sensitive and applicable means for the early detection of aggressive breast cancer.
The successful incorporation of this diagnostic system would impact a variety of populations including women in post treatment remission and high risk individuals who are in need of superior screening methods. The diagnostic system would allow detection at an early stage that would lead to improved treatment efficacy. Additional applications of the fluorescent component include the potential use of fluorescence imaging during surgical resection, metastasis detection, and localized tumor imaging. Advantages of using the Id1 promoter to drive the blood-based and imaging reporters include the presence of high Id1 levels in the endothelial cells of the tumor vasculature. Id1 expression in the tumor vasculature of all cancers has been documented [20]. Extravasation of adenovirus into the tumor environment is not required for successful detection and subsequent reporter expression. Systemically delivered particles need only infect the tumor endothelium to enable expression of the reporters. Id1 is also a hallmark of angiogenesis, invasion, survival, and overall cancer aggression. It was expressed at all levels of transformation and the degree of expression correlated with the degree of aggression. Hence the Id1 sensor becomes more activated with the more aggressive and invasive cancer phenotypes, leading to higher reporter signals in the dangerous tumors. This is advantageous as the more aggressive cancers require earlier detection to avoid metastasis and improve treatment success.
Another important feature of the system is the way it can be improved in the future. The SEAP could be replaced by other detection markers that may become available. Similarly, the imaging reporter cassette could be updated with reporters that can be detected by PET or MRI. Furthermore, the mCherry could be replaced with an improved optical reporter in the near infrared region. Finally, systemic delivery can be achieved with targeted Ad vectors with conditional replication for signal amplification, or via cell vehicles (stem or immune cells) with natural capability of homing to tumor sites upon their ex vivo transduction with this diagnostic system.
Acknowledgements
This work was supported the UAB Small Animal Imaging Shared Facility NIH Research Core Grant (P30CA013148) and the Department of Defense (BC050034).
References
- 1.Smith RA, Cokkinides V, Brawley OW. Cancer screening in the United States, 2009: a review of current American Cancer Society guidelines and issues in cancer screening. CA Cancer J Clin. 2009;59:27–41. doi: 10.3322/caac.20008. [DOI] [PubMed] [Google Scholar]
- 2.Thomas DB, Gao DL, Ray RM, et al. Randomized trial of breast self-examination in Shanghai: final results. J Natl Cancer Inst. 2002;94:1445–1457. doi: 10.1093/jnci/94.19.1445. [DOI] [PubMed] [Google Scholar]
- 3.Baines CJ, Miller AB, Bassett AA. Physical examination. Its role as a single screening modality in the Canadian National Breast Screening Study. Cancer. 1989;63:1816–1822. doi: 10.1002/1097-0142(19900501)63:9<1816::aid-cncr2820630926>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg RD, Hunt WC, Williamson MR, et al. Effects of age, breast density, ethnicity, and estrogen replacement therapy on screening mammographic sensitivity and cancer stage at diagnosis: review of 183,134 screening mammograms in Albuquerque, New Mexico. Radiology. 1998;209:511–518. doi: 10.1148/radiology.209.2.9807581. [DOI] [PubMed] [Google Scholar]
- 5.Banks E, Reeves G, Beral V, et al. Influence of personal characteristics of individual women on sensitivity and specificity of mammography in the Million Women Study: cohort study. BMJ. 2004;329:477. doi: 10.1136/bmj.329.7464.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zahl PH, Strand BH, Maehlen J. Incidence of breast cancer in Norway and Sweden during introduction of nationwide screening: prospective cohort study. BMJ. 2004;328:921–924. doi: 10.1136/bmj.38044.666157.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerlikowske K, Grady D, Barclay J, et al. Likelihood ratios for modern screening mammography. Risk of breast cancer based on age and mammographic interpretation. JAMA. 1996;276:39–43. doi: 10.1001/jama.276.1.39. [DOI] [PubMed] [Google Scholar]
- 8.Porter PL, El-Bastawissi AY, Mandelson MT, et al. Breast tumor characteristics as predictors of mammographic detection: comparison of interval- and screen-detected cancers. J Natl Cancer Inst. 1999;91:2020–2028. doi: 10.1093/jnci/91.23.2020. [DOI] [PubMed] [Google Scholar]
- 9.Lord SJ, Lei W, Craft P, et al. A systematic review of the effectiveness of magnetic resonance imaging (MRI) as an addition to mammography and ultrasound in screening young women at high risk of breast cancer. Eur J Cancer. 2007;43:1905–1917. doi: 10.1016/j.ejca.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Lehman CD, Gatsonis C, Kuhl CK, et al. MRI evaluation of the contralateral breast in women with recently diagnosed breast cancer. N Engl J Med. 2007;356:1295–1303. doi: 10.1056/NEJMoa065447. [DOI] [PubMed] [Google Scholar]
- 11.Teh W, Wilson AR. The role of ultrasound in breast cancer screening. A consensus statement by the European Group for Breast Cancer Screening. Eur J Cancer. 1998;34:449–450. doi: 10.1016/s0959-8049(97)10066-1. [DOI] [PubMed] [Google Scholar]
- 12.Brewster AM, Hortobagyi GN, Broglio KR, et al. Residual risk of breast cancer recurrence 5 years after adjuvant therapy. J Natl Cancer Inst. 2008;100:1179–1183. doi: 10.1093/jnci/djn233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun XH, Copeland NG, Jenkins NA, et al. proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol. 1991;11:5603–5611. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pesce S, Benezra R. The loop region of the helix-loop-helix protein Id1 is critical for its dominant negative activity. Mol Cell Biol. 1993;13:7874–7880. doi: 10.1128/mcb.13.12.7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong YC, Wang X, Ling MT. Id-1 expression and cell survival. Apoptosis. 2004;9:279–289. doi: 10.1023/b:appt.0000025804.25396.79. [DOI] [PubMed] [Google Scholar]
- 16.Sikder HA, Devlin MK, Dunlap S, et al. Id proteins in cell growth and tumorigenesis. Cancer Cell. 2003;3:525–530. doi: 10.1016/s1535-6108(03)00141-7. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8:341–352. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- 18.Gupta GP, Perk J, Acharyya S, et al. ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc Natl Acad Sci U S A. 2007;104:19506–19511. doi: 10.1073/pnas.0709185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perk J, Gil-Bazo I, Chin Y, et al. Reassessment of id1 protein expression in human mammary, prostate, and bladder cancers using a monospecific rabbit monoclonal anti-id1 antibody. Cancer Res. 2006;66:10870–10877. doi: 10.1158/0008-5472.CAN-06-2643. [DOI] [PubMed] [Google Scholar]
- 20.Ruzinova MB, Schoer RA, Gerald W, et al. Effect of angiogenesis inhibition by Id loss and the contribution of bone-marrow-derived endothelial cells in spontaneous murine tumors. Cancer Cell. 2003;4:277–289. doi: 10.1016/s1535-6108(03)00240-x. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Gerald WL, Benezra R. Utilization of bone marrow-derived endothelial cell precursors in spontaneous prostate tumors varies with tumor grade. Cancer Res. 2004;64:6137–6143. doi: 10.1158/0008-5472.CAN-04-1287. [DOI] [PubMed] [Google Scholar]
- 22.Lin CQ, Singh J, Murata K, et al. A role for Id-1 in the aggressive phenotype and steroid hormone response of human breast cancer cells. Cancer Res. 2000;60:1332–1340. [PubMed] [Google Scholar]
- 23.Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 24.Fong S, Itahana Y, Sumida T, et al. Id-1 as a molecular target in therapy for breast cancer cell invasion and metastasis. Proc Natl Acad Sci U S A. 2003;100:13543–13548. doi: 10.1073/pnas.2230238100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger J, Hauber J, Hauber R, Geiger R, Cullen BR. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene. 1988;66:1–10. doi: 10.1016/0378-1119(88)90219-3. [DOI] [PubMed] [Google Scholar]
- 26.Bronstein I, Fortin JJ, Voyta JC, et al. Chemiluminescent reporter gene assays: sensitive detection of the GUS and SEAP gene products. Biotechniques. 1994;17:172–174. 176–177. [PubMed] [Google Scholar]
- 27.Shaner NC, Campbell RE, Steinbach PA, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 28.Borovjagin AV, McNally LR, Wang M. Noninvasive Monitoring of mRFP1- and mCherry-Labeled Oncolytic Adenoviruses in an Orthotopic Breast Cancer Model by Spectral Imaging. Mol Imaging. 9:59–75. [PMC free article] [PubMed] [Google Scholar]
- 29.Kanerva A, Mikheeva GV, Krasnykh V, et al. Targeting adenovirus to the serotype 3 receptor increases gene transfer efficiency to ovarian cancer cells. Clin Cancer Res. 2002;8:275–280. [PubMed] [Google Scholar]
- 30.Sirena D, Lilienfeld B, Eisenhut M, et al. The human membrane cofactor CD46 is a receptor for species B adenovirus serotype 3. J Virol. 2004;78:4454–4462. doi: 10.1128/JVI.78.9.4454-4462.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borovjagin AV, Krendelchtchikov A, Ramesh N, et al. Complex mosaicism is a novel approach to infectivity enhancement of adenovirus type 5-based vectors. Cancer Gene Ther. 2005;12:475–486. doi: 10.1038/sj.cgt.7700806. [DOI] [PubMed] [Google Scholar]
- 32.Desprez PY, Hara E, Bissell MJ, et al. Suppression of mammary epithelial cell differentiation by the helix-loop-helix protein Id-1. Mol Cell Biol. 1995;15:3398–3404. doi: 10.1128/mcb.15.6.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh J, Murata K, Itahana Y, et al. Constitutive expression of the Id-1 promoter in human metastatic breast cancer cells is linked with the loss of NF-1/Rb/HDAC-1 transcription repressor complex. Oncogene. 2002;21:1812–1822. doi: 10.1038/sj.onc.1205252. [DOI] [PubMed] [Google Scholar]
- 34.He TC, Zhou S, da Costa LT, et al. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.San Martín C, Glasgow JN, Borovjagin A, et al. Localization of the N-terminus of minor coat protein IIIa in the adenovirus capsid. J Mol Biol. 2008;383(4):923–34. doi: 10.1016/j.jmb.2008.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang YY, Brunicardi FC, Lin X. Smad3 mediates immediate early induction of Id1 by TGF-beta. Cell Res. 2009;19:140–148. doi: 10.1038/cr.2008.321. [DOI] [PubMed] [Google Scholar]
- 37.Yang X, Wei LL, Tang C, et al. Overexpression of KAI1 suppresses in vitro invasiveness and in vivo metastasis in breast cancer cells. Cancer Res. 2001;61:5284–5288. [PubMed] [Google Scholar]
- 38.Srinivasan D, Sims JT, Plattner R. Aggressive breast cancer cells are dependent on activated Abl kinases for proliferation, anchorage-independent growth and survival. Oncogene. 2008;27:1095–1105. doi: 10.1038/sj.onc.1210714. [DOI] [PubMed] [Google Scholar]
- 39.Desprez PY, Lin CQ, Thomasset N, et al. A novel pathway for mammary epithelial cell invasion induced by the helix-loop-helix protein Id-1. Mol Cell Biol. 1998;18:4577–4588. doi: 10.1128/mcb.18.8.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]