Abstract
Background
The Wnt signaling pathway is involved in the control of cell proliferation and differentiation during skeletal muscle development. Secreted frizzled-related proteins (SFRPs), such as SFRP1, function as inhibitors of Wnt signaling. MicroRNA-1/206(miRNA-1/206) is specifically expressed in skeletal muscle and play a critical role in myogenesis. The miRNA-mRNA profiles and bioinformatics study suggested that the SFRP1 gene was potentially regulated by miRNA-1/206 during porcine skeletal muscle development.
Methods
To understand the function of SFRP1 and miRNA-1/206 in swine myogenesis, we first predicted the targets of miRNA-1/206 with the TargetScan and PicTar programs, and analyzed the molecular characterization of the porcine SFRP1 gene. We performed a temporal-spatial expression analysis of SFRP1 mRNA and miRNA-206 in Tongcheng pigs (a Chinese indigenous breed) by quantitative real-time polymerase chain reaction, and conducted the co-expression analyses of SFRP1 and miRNA-1/206. Subsequently, the interaction between SFRP1 and miRNA-1/206 was validated via dual luciferase and Western blot assays.
Results
The bioinformatics analysis predicted SFRP1 to be a target of miRNA-1/206. The expression level of the SFRP1 was highly varied across numerous pig tissues and it was down-regulated during porcine skeletal muscle development. The expression level of the SFRP1 was significantly higher in the embryonic skeletal compared with postnatal skeletal muscle, whereas miR-206 showed the inverse pattern of expression. A significant negative correlation was observed between the expression of miR-1/206 and SFRP1 during porcine skeletal muscle development (p <0.05). Dual luciferase assay and Western-blot results demonstrated that SFRP1 was a target of miR-1/206 in porcine iliac endothelial cells.
Conclusions
Our results indicate that the SFRP1 gene is regulated by miR-1/206 and potentially affects skeletal muscle development. These findings increase understanding of the biological functions and the regulation of the SFRP1 gene in mammals.
Keywords: SFRP1, miRNA-206, miRNA-1, Skeletal muscle, Development, Pig
Background
The Wnt signaling pathway plays an essential role during embryonic and postnatal muscle development [1,2] because it regulates the expression of myogenic regulatory factors, which are essential for myogenic lineage progression and the formation of functional multinucleated myotubes [3,4]. The Wnt signaling pathway also simultaneously promotes myogenic and inhibits adipogenic differentiation within primary adult myoblasts [5]. During adult skeletal muscle regeneration, the Wnt signaling pathway is involved in satellite cell proliferation and differentiation as well as in self-renewal [6]. Secreted frizzled-related protein 1 (SFRP1) is a member of the SFRP family that inhibits Wnt signaling [7]. The SFRPs inhibit Wnt receptor binding to down-regulate pathway signaling during development [8]. The SFRP gene family has five members (SFRP1-5) in the mouse and human genomes [9], which play important roles in developmental and oncogenic processes [10,11]. The addition of recombinant SFRP1 and SFRP2 to C2C12 or primary satellite cells may inhibit myotube formation; therefore, SFRP1 and SFRP2 act to prevent myoblasts from entering the terminal differentiation process [12]. Additionally, SFRP1 also controls vascular cell proliferation in vitro and in vivo [13].
MicroRNAs are a class of small, single-stranded, noncoding RNA (~21-24 nt in length) that occur in the genomes of plants and animals. They function post-transcriptionally by interacting directly with 3′-UTRs of mRNAs to repress their expression by translational inhibition, mRNA degradation, or both [14,15]. miRNAs are involved in multiple biological processes, including development [16], cancer [17,18], cell differentiation [19], apoptosis [20], and metabolism [21]. Moreover, miRNAs play a modulatory role in the development and growth of skeletal muscles [22]. Three miRNAs, miRNA-1, −133 and −206, are specifically expressed in muscle and are considered to be myomiRs [23,24]. miRNA-1 and miRNA-133 are expressed in both cardiac and skeletal muscles [25] and miRNA-206 is only expressed in skeletal muscle [26]. miRNA-1 and miRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing the paired box 7 (Pax7) gene [27]. These two miRNAs also promote myogenesis by targeting the histone deacetylase 4 (HDAC4) and the largest subunit of DNA polymerase α (Pola1), whereas miRNA-133 may inhibit myoblast differentiation and increase proliferation by repressing the serum response factor (SRF) [28,29]. Our recent study documented that miRNA-1 and miRNA-206 were abundantly and specifically expressed in porcine skeletal muscle [30].
In our previous study, we conducted mRNA and miRNA transcriptome profiling on prenatal skeletal muscle of Tongcheng pigs at 33, 65 and 90 days post-coitus (dpc) using the microarray analysis. Integrated analysis of miRNA and mRNA suggested that SFRP1 and miRNA-1/206 exhibited opposite expression patterns and potentially interacted during prenatal skeletal muscle development. To further explore the biological functions and regulatory mechanisms of SFRP1 gene and miRNA-1/206 in porcine muscle development, we analyzed the temporal and spatial expression patterns of miRNA-206 and SFRP1 in prenatal and postnatal skeletal muscle at 20 developmental stages. Subsequently, the interaction between SFRP1 and miRNA-1/206 was validated using dual luciferase and Western-blot assays.
Methods
Bioinformatics analysis
The public TargetScan (http://www.targetscan.org/) and PicTar (http://pictar.mdc-berlin.de/cgi-bin/PicTar_vertebrate.cgi) programs were used to predict the targets and binding sites of miRNA-1/206. A DAVID functional annotation analysis (http://david.abcc.ncifcrf.gov/) was performed to investigate the potential biological function and KEGG pathways of miRNA-1/206 targets [31]. The mRNAs and protein sequences of the SFRP1 from different species were retrieved from the GenBank database. The isoelectric point and molecular weight of porcine SFRP1 were predicted using the ExPASy website (http://web.expasy.org/compute_pi/). The alignment of SFRP1sequences and generation of the phylogenetic tree were performed by MEGA5.05 [32]. The protein localization sites in cells and the porcine SFRP1 protein domains were predicted by the PSORT program using the K-NN method (http://psort.nibb.ac.jp/) and SMART software (http://smart.embl-heidelberg.de/), respectively.
Animal sample collection
The Biological Studies Animal Care and Use Committee of Hubei Province, P.R. China approved the animal procedures. In this study, all animals were sacrificed at a commercial slaughterhouse according to approved procedures. Seven tissue samples, including heart, liver, spleen, lung, kidney, small intestine and longissimus dorsi muscle, were collected from three adult Tongcheng pigs (postnatal days 240) for the spatial expression analysis. Longissimus dorsi muscle samples were collected from Tongcheng pigs for dynamic expression profile analysis and were sampled at 20 developmental stages, including embryonic days 33, 40, 45, 55, 60, 70, 75, 80, 85, 90, 95, 100, and 105 (abbreviated as E33, E40, E45, E55, E60, E70, E75, E80, E85, E90, E95, E100, and E105) and postnatal days 0, 20, 40, 60, 100, 120 and 160 (abbreviated as D0, D20, D40, D60, D100, D120 and D160). At each time point, samples from three pigs were harvested as biological replicates. All samples were stored immediately in liquid nitrogen until further use.
Isolation of RNA and reverse transcription
Total RNA was extracted according to the manufacturer’s protocol using Trizol Reagent (Invitrogen, Carlsbad, CA, USA). The total RNA concentration was determined by spectrophotometry, and sample integrity and quality were estimated by agarose gel electrophoresis and the OD260/OD280 ratio (high quality being between 1.8 and 2.0). Genomic DNA was removed using DNase I enzyme. One microgram of total RNA was reverse-transcribed into cDNA in a final volume of 20 μl using a RevertAid First Strand cDNA Synthesis Kit (MBI Fermentas, Vilnius, Lithuania) according to the manufacturer’s protocols. The cDNA was stored at −20°C.
Real-time quantitative PCR
The expression of SFRP1 mRNA and miRNA-206 was detected by real-time quantitative polymerase chain reaction (qPCR). The sequence of porcine miRNA-206 was obtained from the miRBase database (Accession ID:MI0013084) (http://www.mirbase.org/) [33]. Specific stem-looped primers were designed according to a previous study [34]. The gene-specific primers used for quantitative PCR are listed in Table 1. Each real-time PCR reaction was performed in a final volume of 20 μL containing 10 μL SYBR Premix Ex Taq (2×), 0.4 μL Rox Reference DyeII (50×)(TaKaRa, Dalian, China), 0.4 μL forward primer(10 μM), 0.4 μL reverse primer(10 μM), 2.0 μL template cDNA and 6.8 μL dH2O. PCR amplification was performed on a 7500 FAST Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) under the following cycling conditions: 30 s at 95°C, followed by 40 cycles at 95°C for 5 s, 60°C for 34 s. Porcine GAPDH and the U6 genes were amplified as reference controls for SFRP1 and miRNA-206, respectively. Each reaction was performed in triplicate, and the data were analyzed by the 2-△△Ct method using 7500 System SDS software V 1.4.0.
Table 1.
Primer information
| Gene | Primer sequence (5′-3′) | Size (bp) |
|---|---|---|
| SFRP1-CDS | F: ACCCAGGTCTTCCTCTGCTCG | 247 |
| R: TTGGAGGCTTCGGTGGCATT | ||
| SFRP1-3′UTR | F: CTCGAGTTCTTCTAGTTCCTTCCGTAGCACC | 230 |
| R: GCGGCCGCCGAGTGAATATTGATACATGGCAGG | ||
| GAPDH | F: ATGGTGAAGGTCGGAGTGAAC | 235 |
| R: CTCGCTCCTGGAAGATGGT | ||
| SFRP1 (mut-1) | F: CTCGAGTTCTTCTAGTTCCTTCCGTAGCACC | 103 |
| R: ACAACAACACACCAATGAAATAAAACGTTTTCACAGTATT | ||
| SFRP1 (mut-2) | F: AATACTGTGAAAACGTTTTATTTCATTGGTGTGTTGTTGT | 174 |
| R: GCGGCCGCCGAGTGAATATTGATACATGGCAGG | ||
| miR-206 | F: GGGTGGAATGTAAGGAA | 61 |
| R: CTCAACTGGTGTCGTGGAGTC | ||
| U6 | F:GCTTCGGCAGCACATATACTAAAAT | 89 |
| R:CGCTTCACGAATTTGCGTGTCAT | ||
| miR-206 -RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCACACAC | |
| U6-RT | CGCTTCACGAATTTGCGTGTCAT | |
Plasmid construct
A 230 bp fragment encompassing a partial SFRP1 3′-UTR containing miRNA-1/206 binding sites was cloned from a Tongcheng pig using gene-specific primers (Table 1). This fragment was inserted downstream of the Renilla luciferase open reading frame in the psiCheck-2 vector (Promega, USA) using NotI and XhoI restriction sites. The mutant SFRP1 3′-UTR sequence, which had a 7 bp deletion in the binding site, was cloned by bridge PCR and inserted into the final destination vectors to construct the mutated vector. To construct the SFRP1 over-expression vector, a 247 bp fragment containing the coding sequence of SFRP1 was cloned into the NheI and XhoI restriction sites of the psiCHECK-2 vector to replace the Renilla coding sequence, resulting in SFRP1-CDS-3′-UTR-psiCHECK-2. All the PCR products were confirmed by direct sequencing.
Cell culture and dual luciferase reporter assay
Porcine iliac endothelial cells (PIECs; obtained from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, P.R. China) were cultured in Dulbecco’s modified Eagle's medium with high glucose (Gibco, Invitrogen, Carlsbad, CA, USA), supplemented with 10% fetal bovine serum (Gibco), 1% glutamine, and 1% penicillin/streptomycin (Gibco). The cells were incubated at 37°C in 5% CO2. Chemically synthesized miRNA-1/206 or the negative control duplexes (Gene Pharma, Shanghai, China) were transfected into the PIECs in combination with a luciferase reporter containing wild-type or mutant SFRP1 3′UTR using Lipofectamine 2000 reagent(Gibco) in 24-well plates. Each transfection was performed in triplicate. At 48 h after transfection, all the cells were harvested. Renilla and Firefly luciferase activities were measured with the Dual Luciferase Assay System (Promega, Madison, WI, USA) in a TD-20/20 luminometer (Turner Biosystems, Sunnyvale, CA, USA). The Renilla luciferase signal was normalized to the Firefly luciferase signal. The normalized Renilla luciferase activity was compared with the control, miRNA-1/206 and the mutant groups using Student’s t-test (p < 0.05) with SPSS 15.0 software.
Western-blot analysis
PIECs, transfected with the indicated plasmids, were harvested at 48 h after transfection, and washed twice in phosphate-buffered saline. The total protein was extracted using M-PER Mammalian Protein Extraction Reagent (Thermo Scientific, USA). Protein concentrations were measured with a Pierce BCA Protein Assay Kit (Thermo Scientific, USA) according to the manufacturer’s instructions. Western-blot analysis was performed as follows: 20 μg of protein per lane was resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). The PVDF membranes were probed with primary antibodies (Santa Cruz Biotech., Santa Cruz, CA, USA) at a 1:1,000 dilution and with β-actin (Santa Cruz Biotech., Santa Cruz, CA, USA) at a 1:5,000 dilution as a control. The membranes were further incubated with an horseradish-peroxidase-conjugated secondary antibody (Zymed, San Diego, CA, USA) at a 1:10,000 dilution.
Results
Target prediction of miRNA-1/206 and bioinformatic analysis
The candidate targets of miRNA-1/206 were predicted using TargetScan and PicTar programs. 258 candidate targets were predicted by both programs. To explore the biological function of the candidate genes, a DAVID functional annotation analysis was performed using the thresholds EASE adjusted to p < 0.05. The Gene Ontology (GO) analysis revealed that these targets were significantly enriched in the regulation of transcription, positive regulation of macromolecule metabolic processes, positive regulation of nitrogen compound metabolic processes, positive regulation of cellular biosynthetic processes and other biological processes (Figure 1). KEGG pathway analysis showed that these targets were significantly enriched in four pathways (p <0.05), which were SNARE interactions in vesicular transport, the Wnt signaling pathway, small cell lung cancer and the Neurotrophin signaling pathway (Table 2). Seven putative targets, including CCND1, SFRP1, CCND2, PPP2R5A, NFAT5, DAAM1 and FZD7, participated in the Wnt signaling pathway. Therefore, the SFRP1 gene, which was predicted to be a target of miRNA-1/206 and was involved in the Wnt signaling pathway, was selected for further study.
Figure 1.

Top 10 GO biological process terms significantly enriched in for target genes of miRNA-1/206. GO analysis was conducted with a DAVID functional annotation program.
Table 2.
Significantly enriched KEGG pathways of target genes
| Category | Term | Count | % | p-value | Genes |
|---|---|---|---|---|---|
| KEGG_PATHWAY | hsa04130:SNARE interactions in vesicular transport | 4 | 1.55 | 0.023 | BET1, VAMP4, VAMP2, SNAP25 |
| KEGG_PATHWAY | hsa04310:Wnt signaling pathway | 7 | 2.71 | 0.034 | CCND1, SFRP1 , CCND2, FZD7, PPP2R5A, NFAT5, DAAM1, |
| KEGG_PATHWAY | hsa05222:Small cell lung cancer | 5 | 1.94 | 0.046 | CCND1, PIAS3, CDK6, RARB, FN1 |
| KEGG_PATHWAY | hsa04722:Neurotrophin signaling pathway | 6 | 2.23 | 0.049 | BDNF, YWHAZ, RAP1A, RAP1B, NGFR, CALM2 |
SFRP1 is involved in the Wnt signaling pathway and selected for further study.
Sequence analysis of the porcine SFRP1 gene
The SFRP1 protein sequences (Accession ID: XM_003359868.3) were retrieved from the GenBank database. First, the amino acid sequence of porcine SFRP1 was compared with those of the human, mouse and rat, and the alignment results showed that the protein sequence of the pig had a six amino acid depletion. The porcine SFRP1 polypeptide exhibited 96.09%, 94.26% and 93.31% similarity with the human, mouse and rat homologs, respectively (Figure 2). Based on the phylogenetic tree analysis, the protein sequence of the pig was closely related to that of cow, while the rat and mouse formed another closely related group, and these four species formed a group with the humans (Figure 3A). SMART software was used to predict SFRP1 protein domains. The precursor protein of SFRP1 contained a Frizzled domain and a Netrin C-terminal domain(Figure 3B). The sites of SFRP1 protein localization had a 44.4% possibility of being extracellular, a 22.2% possibility of being cytoplasmic and a 22.2% possibility of being in the endoplasmic reticulum. Additionally, the theoretical isoelectric point (pI) and molecular weight (Mw) of SFRP1 were 9.05 and 34.79 KDa, respectively.
Figure 2.
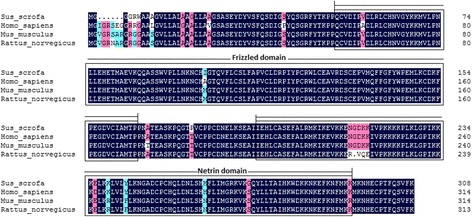
Comparison of porcine SFRP1 sequence (GenBank accession ID: XP_003359916.2) with those of human (GenBank accession ID: NP_003003.3), mouse (GenBank accession ID: NP_038862.2) and rat (GenBank accession no. NP_001263641.1) sequence. Shading shows identical and similar amino acid residues among the four species. Common structural domains are indicated by boxes including a Frizzled domain and a Netrin C-terminal domain.
Figure 3.
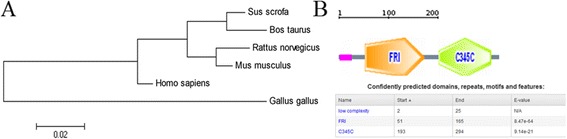
Phylogenetic tree and domains of porcine SFRP1 protein. (A) Phylogenetic tree of SFRP1 from different species. The GenBank accession numbers of those sequences are as follows: pig, XP_003359916.2; human, NP_003003.3; cattle,NP_776885.1; rat, NP_001263641.1; mouse, NP_038862.2, and chicken, NP_989884.1. (B) Porcine SFRP1 protein domains. The porcine SFRP1 precursor protein contains a Frizzled domain and a Netrin C-terminal domain.
Distribution of porcine SFRP1 mRNA and miRNA-206 in tissues
The expression of SFRP1 and miRNA-206 was measured in seven tissues from adult Tongcheng pigs. The SFRP1 gene was highly expressed in the kidney, liver, lung, spleen and small intestine, moderately expressed in the heart, and weakly expressed in the longissimus dorsi muscle (Figure 4A). However, miRNA-206 was abundantly expressed in the longissimus dorsi muscle and heart, and was weakly expressed in the other tissues (Figure 4B).
Figure 4.
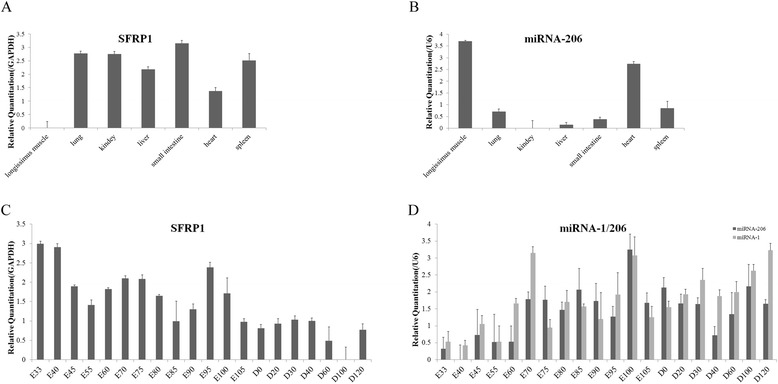
Relative expression of SFRP1 and miRNA-206 in the different tissues and in skeletal muscle during development of Tongcheng pigs. (A) Tissue distribution of SFRP1 in the adult Tongcheng pig. (B) Tissue distribution of miRNA-206 in the adult Tongcheng pig. (C) Relative expression of SFRP1 mRNA and (D) miRNA-1/206 in porcine skeletal muscle from Tongcheng pigs at different developmental stages (20 stages). miRNA-1 expression in Tongcheng pigs is cited from Tang et al. [35]. The values are the average (±SE) levels of SFRP1 mRNA and miRNA-1/206 from three independent experiments normalized to GAPDH and U6, respectively. In each group, the lowest expression value was arbitrarily set to 0 by log10 transformation to evaluate the relative expression levels.
Developmental expression of porcine SFRP1 mRNA and miRNA-206
We collected the longissimus dorsi muscle from 20 prenatal and postnatal developmental stages of Tongcheng pigs. Quantitative real-time PCR indicated that SFRP1 was highly expressed at the E33 stage, and was then down-regulated from E33 to E55. It was then up-regulated from E55 to E70 and down-regulated from E70 to E85. Subsequently, it was up-regulated from the E85 to E95, and then down-regulated again from E95 to D0. The expression of SFRP1 was maintained at a stable low level in postnatal skeletal muscle. In postnatal myogenesis, SFRP1 was consistently expressed from days 0 to 40. Subsequently, it decreased and reached a minimum at D100 (Figure 4C). The expression of miRNA-206 was weak in skeletal muscle in the early embryonic stages, but then remained stable with high levels of expression in the remaining prenatal stages, although some fluctuations occurred. The expression level of miRNA-206 reached a peak at E100. In postnatal muscle, miRNA-206 was down-regulated from D0 to D40 and was up-regulated from D40 to D100 (Figure 4D).
Co-expression analysis of SFRP1 and microRNA-1/206
Tang et al. have explored the spatial and dynamic expression of miRNA-1 in the Tongcheng pig (Figure 4D) [35]. According to their study and this study, miRNA-1 and miRNA-206 were abundantly expressed in skeletal muscle and weakly expressed in other tissues. In contrast, SFRP1 was expressed at very low levels in adult skeletal muscle. The co-expression analysis revealed that the SFPR1 mRNA was significantly negatively correlated with miRNA-1 (Pearson’s RSFRP1/miRNA-1 = −0.928, p value = 0.003) (Figure 5A) and miRNA-206 (Pearson’s RSFRP1/miRNA-1 = −0.922, p value = 0.003) (Figure 5B) at the mRNA level in different tissues adult tissues.
Figure 5.
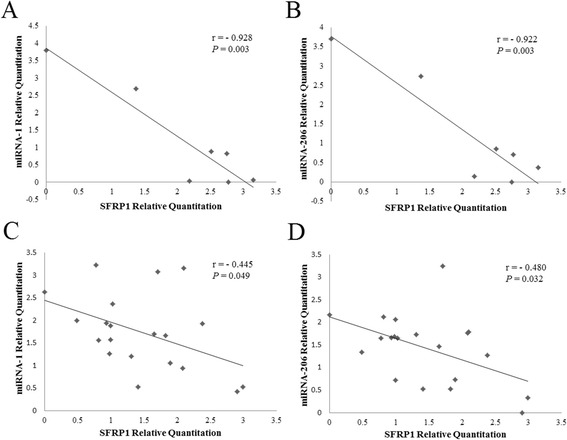
Correlation analyses of miRNA-1/206 and SFRP1 expression. (A) Pearson’s correlation between SFRP1 and miRNA-1 expression in different tissues. (B) Pearson’s correlation between SFRP1 and miRNA-206 expression in different tissues. (C) Pearson’s correlation between SFRP1 and miRNA-1 expression during skeletal muscle development. (D) Pearson’s correlation between SFRP1 and miRNA-206 expression during skeletal muscle development.
During skeletal muscle development, SFPR1 had a higher expression level in the prenatal stages than in the postnatal stages. However, in contrast to SFRP1, miRNA-1/206 exhibited a relatively higher level of expression in postnatal muscle compared with prenatal muscle. SFRP1 mRNA was significantly negatively correlated with miRNA-1/206 [Pearson’s RSFRP1/miRNA-1 = −0.445, pvalue = 0.032 (Figure 5C); Pearson’s RSFRP1/miRNA-206 = −0.480, p value = 0.049(Figure 5D)]. These results indicated that SFRP1 expression might be regulated by miR-1/206 in pigs.
SFRP1 is a putative target of microRNA-1/206
The prediction from the miRNA-mRNA profiles and bioinformatics suggested that SFRP1 was potentially a miRNA-1/206 target in pigs. In the 3′-UTR region of SFRP1 mRNA, a putative binding site was identified (7 bp conserved homology) (Figure 6A). The seed sequence of miRNA-1/206 and the target-binding site between SFRP1 and miRNA-1/ 206 were highly conserved across mammals (Figure 7).
Figure 6.

SFRP1 3′-UTR has miR-1/206 target binding sites. (A) SFRP1 was predicted as a target of microRNA-1/206. (B) Schematic of the predicted miRNA-1 and miRNA-206 binding sites (underlined) in the 3′ UTR of SFRP1. The binding site region was deleted in the mutant 3′ -UTR reporters.
Figure 7.
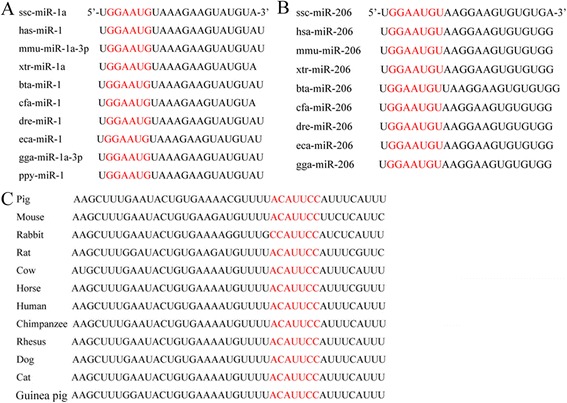
Predicted miRNA-1 and miRNA-206 binding sites (highlighted in red) in the 3′-UTR of SFRP1 showing species conservation. (A) Seed sequence of miRNA-1 showing species conservation. (B) Seed sequence of miRNA-206 showing species conservation. (C) Binding sites (highlighted in red) in the 3′-UTR of SFRP1 showing species conservation.
To validate whether SFRP1 was directly targeted by miRNA-1/206 in pigs, we constructed the psiCheck2-SFRP1-3′-UTR , a luciferase reporter vector. Subsequently, miRNA-1and miRNA-206 mimics and a normal control (NC) were co-transfected into PIECs, and luciferase activity was detected. The miRNA-1 mimic-transfected group exhibited 68.29% less luciferase activity compared with the NC group (p < 0.01) and the miRNA-206 mimic-transfected group exhibited 71.25% less luciferase activity compared with the control (p < 0.01) (Figure 8). To further validate the specific target site, the binding region of the SFRP1 3′-UTR was mutated by bridge PCR (Figure 6B). The luciferase activity of the psiCHECK-2-SFRP1–3′-UTR (mut) was not significantly decreased by both the miRNA-1 and miRNA-206 mimics (12.39% of control for the miRNA-1 mimic-transfected group and 16.76% of control for the miRNA-206 mimic-transfected group) (p >0.05) (Figure 8).
Figure 8.
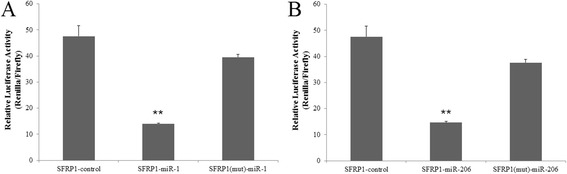
Validating SFRP1 as a positive target for miRNA-1 and miRNA-206. Cotransfection of porcine pre-miRNA-1 (A) and pre-miRNA-206 (B) or control and porcine SFRP1 UTR-derived psiCHECK-2 construct or mutant in PIEC cells. Renilla activity at 48 h post-transfection shows a significant decrease in normalized values compared with the control and mutant. Three replicates were performed for each group. **Indicates a p-value of less than 0.01 in Student’s t-test.
Then, we determined whether SFRP1 was affected by miRNA-1/206 at the protein level. The overexpression vector of SFRP1 was constructed and transfected into PIECs, and the quantitative real-time PCR (qPCR) results showed that the expression of SFRP1 was increased approximately 16-fold compared with the NC group. This result indicated that the SFRP1 overexpression vector was successful (Figure 9A). The porcine SFRP1-CDS-3′-UTR-psiCHECK-2 vector was constructed, and it was co-transfected with miRNA-1 and miRNA-206 mimics in PIECs. The Western blot results showed that the protein level of SFRP1 in the groups containing miRNA-1/206 mimics was decreased compared with the NC group (Figure 9B). These results suggested that the SFRP1 gene was a target of miRNA-1/206.
Figure 9.
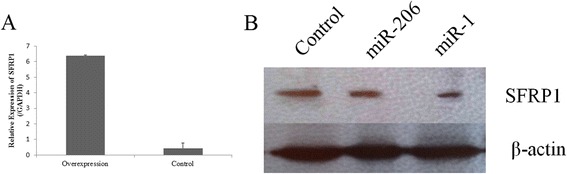
The miRNA-1 and miRNA-206 regulate SFRP1 at the protein level. (A) Overexpression of porcine SFRP1 in PIEC cells. Histogram indicates overexpression of SFRP1 48 h after transfection. (B) miRNA-1 and miRNA-206 down-regulated the SFRP1 at the protein level. The expression of SFRP1 was normalized against β-actin.
Discussion
We predicted the targets of miRNA-1/206 with the TargetScan and PicTar programs. Among the putative targets, many genes had been validated by previous studies, such as HDAC4 [29], GJA1, KCNJ2 [36], Pola1 [28], and Met [37]. However, all these results were based on myoblast C2C12 cells in vitro. Few reports have considered miRNA-1/206 targets during skeletal muscle development in vivo, particularly in pigs. To discover potential targets of miRNA-1/206 during swine myogenesis, we conducted GO and KEGG pathway analyses of targets based on the prediction data. The results suggested that these targets were significantly enriched in the Wnt signaling pathway (p < 0.05). Wnts signaling proteins are secreted proteins that function in differentiation, embryonic development and cell proliferation [38]. The Wnt pathways play an important role in the formation of muscle fibers during prenatal [39] and postnatal myogenesis with the activation of stem cells in the adult muscles [40,41]. SFRP1, a secreted antagonist of the Wnt-Frizzled pathway, was predicted to be a target of miRNA-1/206 and to participate in the Wnt signaling pathway. The expression of SFRP1 mRNA was up-regulated in muscle regeneration [42] and in skeletal muscle after denervation [43]. Additionally, an impairment of the Wnt-Frizzled pathway via SFRP1 over-expression controlled proliferation and neovascularization after muscle ischemia [13]. A comparison of the amino acid sequence of pig SFRP1 with those of human, rat and mouse demonstrated remarkably high similarity across species, and the frizzled domain and netrin C-terminal domain were especially highly conserved.
Previous studies reported SFRP1 expression at the mRNA level in various tissues, such as the brain, kidney and heart, and that the transcript was present both in the adult and during embryogenesis [7,44]. Moreover, SFRP1 was weakly expressed in matured skeletal muscles [45]. The qPCR results for SFRP1 in various adult porcine tissues were mostly consistent with these reports. These results indicated that SFRP1 had almost no antagonistic effect in adult skeletal muscle. Postnatal muscle growth is largely determined by the total number of fibers, which is determined by two major waves of fiber generation before birth: primary muscle fiber formation at 35–60 dpc and assemblage of secondary muscle fibers at 54–90 dpc [46-48]. The SFRP1 gene was down-regulated in skeletal muscle from E33 to E55 in Tongcheng pigs, indicating that SFRP1 was primarily involved in the formation of primary muscle fibers. We also found that the expression of SFRP1 was higher in embryonic skeletal muscle compared with postnatal skeletal muscle. miRNA-206, meanwhile, was abundantly expressed in skeletal muscle and heart, and up-regulated from pre- to postnatal-stage skeletal muscle. These results demonstrated that miR-206 plays a key role in skeletal muscle development in pigs. SFRP1 primarily affected skeletal muscle development in embryonic stages. Wnts signaling contributes to the overall process of myogenesis by activating myogenic regulatory factor genes such as Myf5 and MyoD [49]. miRNA-1/206 promoted skeletal muscle satellite cell proliferation and differentiation [27] and SFRP1 might inhibit myoblast differentiation [12]. The temporal expression patterns of miRNA-1/206 and SFRP1 in Tongcheng pigs were consistent with these previous findings.
The mRNA-miRNA co-expression correlation analysis was reported to identify the putative targets of miRNA [50,51], and this method could improve the positive rate for identifying the mRNA target genes of miRNA. miRNA-1 and miRNA-206 were abundant in the postnatal stages and were at low levels in the prenatal stages of muscle development, while SFRP1 exhibited an opposite expression patterns. Correlation analysis revealed that the SFPR1 was significantly negatively correlated with miRNA-1/206 (p < 0.05), and these results indicate that SFRP1 is potentially regulated by miRNA-1/206.
The interaction between SFRP1 and miRNA-1/206 in pigs has not been previously reported. This study demonstrated that SFRP1 expression was regulated by miRNA-1/206. The SFRP1 3′-UTR sequence around the miRNA-1/206 target sites and the seed sequence of mature miRNA-1/206 are well conserved in mammals, which suggests that the target region is important in SFRP1 regulation and that the regulation of SFRP1 by miRNA-1/206 may also exist in other species. Similar results were observed in other miRNA studies [30,52].
The luciferase activity of psiCHECK-2 containing the SFRP1 3′-UTR sequence was significantly decreased by co-transfection with miRNA-1/206 mimics (p < 0.05). However, with the SFRP1 3′-UTR mutant sequence, activity was not significantly decreased (p > 0.05). These results indicated that the target binding site was specific and unique in pigs, and that miRNA-1/206 might repress SFRP1 expression by degrading the mRNA transcripts. We propose that SFRP1 is regulated by miRNA-1/206, is >involved in the proliferation of muscle cells, and affects prenatal skeletal muscle development. Moreover, we explored the interactions between SFRP1 and miRNA-1/206 at the protein level, and Western-blot analysis confirmed that SFRP1 was significantly down-regulated by miRNA-1/206.
Conclusions
In summary, we predicted the target genes of miRNA-1/206 and performed a functional annotation of the target genes. We performed a molecular characterization analysis of the porcine SFRP1 gene,, which was one of the predicted targets for miRNA-1/206. We also explored the spatial-temporal profile of SFRP1 mRNA and miRNA-206 in adult tissues and in skeletal muscle tissues during development in Tongcheng pigs. The results indicated that SFRP1 was primarily involved in prenatal skeletal muscle development. Finally, we verified that porcine SFRP1 was a target of miRNA-1/206 using dual luciferase and Western blot assay. These results increase our understanding of the biological functions of SFRP1 and miRNA-1/206 in skeletal muscle development.
Acknowledgments
The authors are grateful to Dr. Jianhua Cao from Huazhong Agricultural University for providing the psiCheck-2 vector and to Sanping Xu in the Tongcheng Animal Husbandry Bureau in Hubei Province for sample collection. This work was supported by the National Key Project (2014ZX08009-001), the National Basic Research Program of China (2015CB943101, 2012CB124706), the National Natural Science Foundation of China (31372295, 31330074) and the Agricultural Science and Technology Innovation Program (ASTIP-IAS05).
Abbreviations
- SFRP1
Secreted frizzled-related protein 1
- MRFs
Myogenic regulatory factors
- PIECs
Porcine iliac endothelial cells
- HDAC4
Histone deacetylase 4
- Pola1
the largest subunit of DNA polymerase α
- miRNA
microRNA
- NC
Normal control
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- qPCR
quantitative real-time PCR
Footnotes
Yalan Yang and Wei Sun contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YY, WS, CL and ZT conceived and designed the experiments. YY, WS and RW performed the experiments. YY and RZ analyzed data. YY and WS drafted the manuscript. KL and ZT critically revised the manuscript. All authors contributed to revision of the manuscript and approved the final version.
Contributor Information
Yalan Yang, Email: yangyalan1988@126.com.
Wei Sun, Email: 25092596@qq.com.
Ruiqi Wang, Email: rachelwangbio@gmail.com.
Chuzhao Lei, Email: leichuzhao1118@126.com.
Rong Zhou, Email: 85391032@qq.com.
Zhonglin Tang, Email: tangzhonglin@caas.cn.
Kui Li, Email: likui@caas.cn.
References
- 1.von Maltzahn J, Chang NC, Bentzinger CF, Rudnicki MA. Wnt signaling in myogenesis. Trends Cell Biol. 2012;22(11):602–9. doi: 10.1016/j.tcb.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cisternas P, Henriquez JP, Brandan E, Inestrosa NC. Wnt signaling in skeletal muscle dynamics: myogenesis, neuromuscular synapse and fibrosis. Mol Neurobiol. 2014;49(1):574–89. doi: 10.1007/s12035-013-8540-5. [DOI] [PubMed] [Google Scholar]
- 3.Ridgeway AG, Petropoulos H, Wilton S, Skerjanc IS. Wnt signaling regulates the function of MyoD and myogenin. J Biol Chem. 2000;275(42):32398–405. doi: 10.1074/jbc.M004349200. [DOI] [PubMed] [Google Scholar]
- 4.Rudnicki MA, Le Grand F, McKinnell I, Kuang S. The molecular regulation of muscle stem cell function. Cold Spring Harb Symp Quant Biol. 2008;73:323–31. doi: 10.1101/sqb.2008.73.064. [DOI] [PubMed] [Google Scholar]
- 5.Vertino AM, Taylor-Jones JM, Longo KA, Bearden ED, Lane TF, McGehee RE, Jr, et al. Wnt10b deficiency promotes coexpression of myogenic and adipogenic programs in myoblasts. Mol Biol Cell. 2005;16(4):2039–48. doi: 10.1091/mbc.E04-08-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otto A, Schmidt C, Luke G, Allen S, Valasek P, Muntoni F, et al. Canonical Wnt signalling induces satellite-cell proliferation during adult skeletal muscle regeneration. J Cell Sci. 2008;121(Pt 17):2939–50. doi: 10.1242/jcs.026534. [DOI] [PubMed] [Google Scholar]
- 7.Rattner A, Hsieh JC, Smallwood PM, Gilbert DJ, Copeland NG, Jenkins NA, et al. A family of secreted proteins contains homology to the cysteine-rich ligand-binding domain of frizzled receptors. Proc Natl Acad Sci U S A. 1997;94(7):2859–63. doi: 10.1073/pnas.94.7.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Q, D'Amore PA, Sokol SY. Functional and biochemical interactions of Wnts with FrzA, a secreted Wnt antagonist. Development. 1998;125(23):4767–76. doi: 10.1242/dev.125.23.4767. [DOI] [PubMed] [Google Scholar]
- 9.Jones SE, Jomary C. Secreted Frizzled-related proteins: searching for relationships and patterns. Bioessays. 2002;24(9):811–20. doi: 10.1002/bies.10136. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Stump RJ, Lovicu FJ, McAvoy JW. Expression of Frizzleds and secreted frizzled-related proteins (Sfrps) during mammalian lens development. Int J Dev Biol. 2004;48(8–9):867–77. doi: 10.1387/ijdb.041882yc. [DOI] [PubMed] [Google Scholar]
- 11.Lavergne E, Hendaoui I, Coulouarn C, Ribault C, Leseur J, Eliat PA, et al. Blocking Wnt signaling by SFRP-like molecules inhibits in vivo cell proliferation and tumor growth in cells carrying active beta-catenin. Oncogene. 2010;30(4):423–33. doi: 10.1038/onc.2010.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Descamps S, Arzouk H, Bacou F, Bernardi H, Fedon Y, Gay S, et al. Inhibition of myoblast differentiation by Sfrp1 and Sfrp2. Cell Tissue Res. 2008;332(2):299–306. doi: 10.1007/s00441-008-0574-z. [DOI] [PubMed] [Google Scholar]
- 13.Ezan J, Leroux L, Barandon L, Dufourcq P, Jaspard B, Moreau C, et al. FrzA/sFRP-1, a secreted antagonist of the Wnt-Frizzled pathway, controls vascular cell proliferation in vitro and in vivo. Cardiovasc Res. 2004;63(4):731–8. doi: 10.1016/j.cardiores.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 16.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301(5631):336–8. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 18.Ren J, Huang HJ, Gong Y, Yue S, Tang LM, Cheng SY. MicroRNA-206 suppresses gastric cancer cell growth and metastasis. Cell Biosci. 2014;4:26. doi: 10.1186/2045-3701-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 20.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102(39):13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krutzfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab. 2006;4(1):9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11(4):441–50. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 23.McCarthy JJ. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim Biophys Acta. 2008;1779(11):682–91. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol. 2007;102(1):306–13. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- 25.Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, et al. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120(Pt 17):3045–52. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- 26.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11(3):241–7. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao X, et al. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190(5):867–79. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174(5):677–87. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228–33. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou X, Tang Z, Liu H, Wang N, Ju H, Li K. Discovery of MicroRNAs associated with myogenesis by deep sequencing of serial developmental skeletal muscles in pigs. PLoS One. 2012;7(12):e52123. doi: 10.1371/journal.pone.0052123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ai J, Zhang R, Li Y, Pu J, Lu Y, Jiao J, et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun. 2010;391(1):73–7. doi: 10.1016/j.bbrc.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Tang Z, Liang R, Zhao S, Wang R, Huang R, Li K. CNN3 Is Regulated by microRNA-1 during Muscle Development in Pigs. Int J Biol Sci. 2014;10(4):377–85. doi: 10.7150/ijbs.8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13(4):486–91. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 37.Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, et al. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119(8):2366–78. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11(24):3286–305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 39.van Amerongen R, Berns A. Knockout mouse models to study Wnt signal transduction. Trends Genet. 2006;22(12):678–89. doi: 10.1016/j.tig.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 40.Tsivitse S. Notch and Wnt signaling, physiological stimuli and postnatal myogenesis. Int J Biol Sci. 2010;6(3):268–81. doi: 10.7150/ijbs.6.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steelman CA, Recknor JC, Nettleton D, Reecy JM. Transcriptional profiling of myostatin-knockout mice implicates Wnt signaling in postnatal skeletal muscle growth and hypertrophy. FASEB J. 2006;20(3):580–2. doi: 10.1096/fj.05-5125fje. [DOI] [PubMed] [Google Scholar]
- 42.Zhao P, Hoffman EP. Embryonic myogenesis pathways in muscle regeneration. Dev Dyn. 2004;229(2):380–92. doi: 10.1002/dvdy.10457. [DOI] [PubMed] [Google Scholar]
- 43.Svensson A, Norrby M, Libelius R, Tagerud S. Secreted frizzled related protein 1 (Sfrp1) and Wnt signaling in innervated and denervated skeletal muscle. J Mol Histol. 2008;39(3):329–37. doi: 10.1007/s10735-008-9169-y. [DOI] [PubMed] [Google Scholar]
- 44.Finch PW, He X, Kelley MJ, Uren A, Schaudies RP, Popescu NC, et al. Purification and molecular cloning of a secreted, Frizzled-related antagonist of Wnt action. Proc Natl Acad Sci U S A. 1997;94(13):6770–5. doi: 10.1073/pnas.94.13.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melkonyan HS, Chang WC, Shapiro JP, Mahadevappa M, Fitzpatrick PA, Kiefer MC, et al. SARPs: a family of secreted apoptosis-related proteins. Proc Natl Acad Sci U S A. 1997;94(25):13636–41. doi: 10.1073/pnas.94.25.13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Picard B, Lefaucheur L, Berri C, Duclos MJ. Muscle fibre ontogenesis in farm animal species. Reprod Nutr Dev. 2002;42(5):415–31. doi: 10.1051/rnd:2002035. [DOI] [PubMed] [Google Scholar]
- 47.Lefaucheur L, Edom F, Ecolan P, Butler-Browne GS. Pattern of muscle fiber type formation in the pig. Dev Dyn. 1995;203(1):27–41. doi: 10.1002/aja.1002030104. [DOI] [PubMed] [Google Scholar]
- 48.Wigmore PM, Stickland NC. Muscle development in large and small pig fetuses. J Anat. 1983;137(Pt 2):235–45. [PMC free article] [PubMed] [Google Scholar]
- 49.Cossu G, Borello U. Wnt signaling and the activation of myogenesis in mammals. EMBO J. 1999;18(24):6867–72. doi: 10.1093/emboj/18.24.6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagalla S, Shaw C, Kong X, Kondkar AA, Edelstein LC, Ma L, et al. Platelet microRNA-mRNA coexpression profiles correlate with platelet reactivity. Blood. 2011;117(19):5189–97. doi: 10.1182/blood-2010-09-299719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ponsuksili S, Du Y, Hadlich F, Siengdee P, Murani E, Schwerin M, et al. Correlated mRNAs and miRNAs from co-expression and regulatory networks affect porcine muscle and finally meat properties. BMC Genomics. 2013;14:533. doi: 10.1186/1471-2164-14-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao S, Zhang J, Hou X, Zan L, Wang N, Tang Z, et al. OLFML3 expression is decreased during prenatal muscle development and regulated by microRNA-155 in pigs. Int J Biol Sci. 2012;8(4):459–69. doi: 10.7150/ijbs.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]


