Abstract
Background
Tumor suppression of Transforming Growth Factor (TGF-β) signaling pathway requires an adaptor protein, Embryonic Liver Fodrin (ELF). Disruption of ELF expression resulted in miscolocalization of Smad3 and Smad4, then disruption of TGF-β signaling. However, the prognostic significance of ELF for hepatocellular carcinoma (HCC) hasn’t been clarified. This study aimed to investigate whether measuring both TGF-β1 and ELF provides a more powerful predictor for HCC prognosis than either marker alone.
Methods
TGF-β1 and ELF protein were detected by immunohistochemistry. The relationship between TGF-β1/ELF expression and patients’ clinicopathologic factors was analyzed. The association between TGF-β1/ELF expression and disease-free survival and overall survival was analyzed by Kaplan-Meier curves, the log-rank test, and Multivariate Cox regression analyses.
Results
The expression of TGF-β1 in HCC tissues was significantly higher than that in normal liver tissues. Conversely, the expression of ELF in HCC tissues declined markedly. ELF protein was correlated with HBsAg, tumor size, tumor number, TNM and recurrence. Data also indicated a significant negative correlation between ELF and TGF-β1. Patients with high TGF-β1 expression or/and low ELF expression appeared to have a poor postoperative disease-free survival and overall survival compared with those with low TGF-β1 expression or/and high ELF expression. Furthermore, the predictive range of ELF combined with TGF-β1 was more sensitive than that of either one alone.
Conclusions
TGF-β1 and ELF protein are potential and reliable biomarkers for predicting prognosis in HCC patients after hepatic resection. Our current study has demonstrated that the prognostic accuracy of testing can be enhanced by their combination.
Keywords: Transforming growth factor, Embryonic liver fodrin, Hepatocellular carcinoma, Prognosis, Biomarkers
Background
Hepatocellular cancer (HCC) is one of the most common, aggressive malignancies, the third leading cause of cancer-related deaths worldwide (World Health Organization Report, 2006) [1-3]. Although surgical resection, percutaneous ablation and liver transplantation are considered as the curative treatments for HCC, the long-term prognosis of patients undergoing potentially curative treatments is still poor. Fully 60% to 70% of patients develop recurrence or metastasis within 5 years after resection [4,5]. It is therefore a very important and urgent task to find an effective biomarker to identify patients with a high risk of recurrence or metastases, and provide personalized therapy according to the predicted risk of recurrence.
The transforming growth factor β (TGF-β) signaling pathway is known to play an important role in multiple cellular processes, including cell growth, differentiation, adhesion, migration, apoptosis, extracellular matrix formation and immunosuppressant [6-9]. TGF-β signals are conveyed from type I and type II transmembrane serine/threonine kinase receptors to the intracellular mediators-Smad2 and Smad3, which further complex with Smad4, translocate to the nucleus and bind to Smad-binding elements (SBE) in target gene promoters, thereby activating its targets, such as p21, p15, p16, p27 [10-14]. TGF-β is particularly active as a profound tumor suppressor by prohibiting cell cycle progression and arresting cells in early G1 phase. However, misregulation of TGF-β signaling promotes tumor growth and invasion, evasion of immune surveillance, and cancer cell dissemination and metastasis [11-14]. In HCC tissues, the overexpression of TGF-β1 was found and correlated with carcinogenesis, progression, and prognosis of HCC, while normal hepatocytes had not any TGF-β1 staining [15]. In our previous study, we found hepatocarcinogenesis could be closely related to the low expression of Smad4 and phosphorylated Smad2, and the high expression of TGF-β1 and Smad7 in advanced stage of liver cirrhosis [16].
Embryonic Liver Fodrin (ELF), also named as β2-spectrin (β2SP), first isolated from foregut endodermal stem cell libraries, functions as a Smad3/4 adaptor protein, plays critical roles in the proper control of Smad access to activating receptors involved in regulation of TGF-β signaling [17-19]. Interestingly, ELF is a key suppressor of tumorigenesis [20,21]. Disruption of ELF expression by gene knockout was found to result in miscolocalization of Smad3 and Smad4, and disruption of TGF-β signaling [22]. About half of mice with heterozygous deletion of ELF developed hepatocellular carcinoma, and 90% of ELF+/−/Smad4+/− mice developed gastric cancer and other gastrointestinal cancers [23,24]. Loss of ELF may play a role in the malignant transformation of hepatic progenitor/stem cells [22]. However, the prognostic value of ELF for HCC is not well-known. Testing the combination of TGF-β1 and ELF as a predictor for HCC prognosis is also merits study.
In the present study, we examined the pattern of expression of TGF-β1 and ELF in HCC tumor tissues and normal tissues. Together with the known function, it is therefore of interest to investigate that TGF-β1 and ELF protein are potential and reliable biomarker for predicting prognosis in HCC patients after hepatic resection, and prognostic accuracy of testing can be enhanced by their combination in the patients with HCC.
Methods
Patients and tissue samples
A total of 84 adult patients with HCC who underwent hepatic resection in the Department of Hepatobiliary Surgery, First Affiliated Hospital of Sun Yat-sen University between June 2007 and October 2009, were enrolled in this study, including 68 males and 16 females with an average age of 48 years (range 23 to 75 years). Written informed consent was obtained from all patients, and the study was conducted in accordance with the protocol approved by the Declaration of Helsinki and the guidelines of the Ethics Review Committee of First Affiliated Hospital of Sun Yat-sen University. In addition, normal liver tissues were collected from patients with cavernous hemangioma of liver or patients with intrahepatic stones.
The diagnosis of HCC met the criteria of the American Association for the study of Liver Disease [25]. The volume of liver resection and the surgical procedures were decided by tumor size, tumor location, and liver functional reserve based on a multidisciplinary team meeting every week. Tumor stages were classified according to the tumor-node-metastasis (TNM) system of the International Union Against Cancer by the American Joint Committee [26]. The histologic grade of tumor was assigned according to the Edmondson Steiner grading system [27]. Fresh HCC tissues and HCC adjacent tissues were collected within 30 minutes after resection. These tissues were fixed with 10% formalin and then embedded in paraffin.
Immunohistochemical analysis
The techniques have been described previously [16]. The sections were incubated with pre-diluted primary Rabbit polyclonal anti-ELF antibody (ab72239, Abcam, USA) at a dilution of 1:100, with Rabbit monoclonal anti-TGF-β1 antibody (Y369, Bioworld, USA) at dilution of 1:100, at 4°C overnight. Negative controls were treated the same way, omitting the primary antibodies.
Evaluation of immunohistochemical staining
The immunohistochemical staining in the tissue was scored independently by 2 pathologists blinded to the clinical data, by applying a semiquantitative immunoreactivity score (IRS) reported elsewhere [28-30]. Category A documented the intensity of immunostaining as 0–3 (0, negative; 1, weak; 2, moderate; 3, strong). Category B documented the percentage of immunoreactive cells as 0 (less than 5%),1 (6%–25%), 2 (26%–50%), 3 (51%–75%), and 4 (76%–100%). Multiplication of category A and B resulted in an IRS ranging from 0 to 12 for each tumor or nontumor. Sections with a total score of 0 or 1 or 2 were defined as negative (−), score of 3 or 4 were defined as weakly positive (+), score of 6 or 8 were defined as moderately positive (++), score of 9 or 12 were defined as strongly positive (+++). For categorical analyses, the immunoreactivity was graded as low level (total score < =4) or high level (total score >4).
Follow-up
The postoperative patients were followed up once a month during the first half year post-operatively and every 3 months thereafter. Serum AFP level and abdominal ultrasonography were done routinely during the postoperative review. Computed tomography (CT) was performed every 3 to 6 months together with chest radiographic examination. The endpoint of study was December 2013. Survival time was calculated from the date of surgery to the date of death or to the last follow-up. Date of death was obtained from patient records or patients’ families through follow-up telephone calls. Date of death for each case was double verified by local civil affairs department and public security department. The median follow-up period was 39 months (range 3 to 81 months).
Recurrence or metastasis was detected by imaging examination such as ultrasonography, contrast-enhanced ultrasonography, CT, magnetic resonance imaging (MRI), hepatic arterial angiography, or positron emission tomography -CT (PET-CT). Isolated increases in serum AFP were not regarded as recurrent events. Once tumor recurrence was verified, patients received the appropriate further treatments, including repeat liver resection, radiofrequency ablation, percutaneous ethanol injection, chemoembolization, and/or molecular targeting therapy by sorafenib.
Statistical analysis
Statistical analyses were carried out using the SPSS v 13. 0 software (Chicago, IL, USA). The Wilcoxon W rank sum test and chi-square test was used to compare qualitative variables. Spearman correlation was used to investigate the correlation between ELF and TGF-β1 expression. Survival curves were calculated using the Kaplan-Meier method and were compared by a log-rank test, illustrated by survival plots. The Cox proportional hazards model was used to determine the independent risk factors associated with prognosis. P < 0.05 was considered statistically significant.
Results
The low expression of ELF and the high expression of TGF-β1 in HCC tissues
Using immunohistochemical staining, we examine expression of ELF and TGF-β1 on 20 normal liver tissues, 84 HCC samples and adjacent tissues. All normal liver tissues expressed high level of ELF (20/20). In HCC adjacent tissues, there was a 77.4% high expression rate for ELF (65/84). However, the ELF high expression rate declined to 47.6% (40/84) in HCC tissues. There was significant difference among the groups examined (P < 0.001) (Table 1, Figure 1A, B). On the contrary, the expression rate of TGF-β1 in HCC tissues (59.5%, 50/84) was significantly higher than that in the normal liver tissues (0, 0/20, P < 0.001), but not in HCC adjacent tissues (46.4%, 39/84, P = 0.089, Table 2, Figure 1A, C). These results suggested that there was the low expression of ELF and high expression of TGF-β1 in HCC tissues.
Table 1.
The expression of ELF in HCC
| Group | n | Expression of ELF | |
|---|---|---|---|
| High | Low | ||
| Normal liver tissues | 20 | 20(100.0%) | 0(0.0%) |
| Adjacent tissues* | 84 | 65(77.4%) | 19(22.6%) |
| HCC tissues*# | 84 | 40(47.6%) | 44(52.4%) |
*compared with Normal liver tissues, P < 0.001 (by chi-square test).
#compared with Adjacent tissues, P < 0.001 (by chi-square test).
Figure 1.
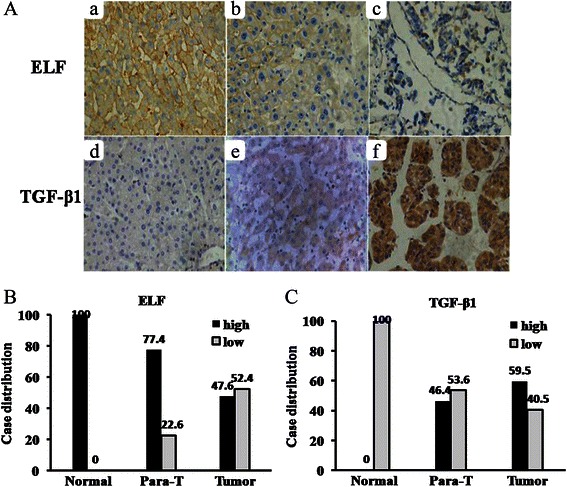
Expression of ELF and TGF-β1 protein. (A) Immunohistochemical staining in different tissues is shown. Normal liver tissues (Aa and Ad), HCC adjacent tissues (Ab and Ae), HCC tissues (Ac and Af) (original magnification × 400). (B) and (C) Case distribution of ELF/TGF-β1 expression in normal liver tissues (Normal), HCC adjacent tissues (Para-T) and HCC tissue (Tumor).
Table 2.
The expression of TGF-β1 in HCC
| Group | n | Expression of TGF-β1 | |
|---|---|---|---|
| High | Low | ||
| Normal liver tissues | 20 | 0(0.0%) | 20(100.0%) |
| Adjacent tissues* | 84 | 39(46.4%) | 45(53.6%) |
| HCC tissues* | 84 | 50(59.5%) | 34(40.5%) |
*compared with Normal liver tissues, P < 0.001 (by chi-square test).
Correlation between TGF-β1/ELF expression and 16 clinico-pathologic characteristics in HCC
In order to further understand the prognostic value of TGF-β1/ELF expression for HCC after resection, the relationships between the expression of these proteins and 16 clinico-pathologic characteristics, such as age, gender, HCC family, HBsAg, ALT, AFP, cirrhosis, ascites, PVTT, tumor size, tumor number, tumor differentiation, tumor encapsulation, TNM stage, recurrence and complication, were analyzed. The expression level of ELF was negatively correlated with HBsAg (P =0.04), tumor size (P = 0.010), tumor number (P = 0.001), TNM stage (P = 0.027) and recurrence (P < 0.001). As predicted, TGF-β1 expression was positively associated with the tumor size (P = 0.001), tumor number (P = 0.003), TNM stage (P = 0.002) and recurrence (P < 0.001), too (Table 3). In addition, we found the significant negative correlation between ELF and TGF-β1 expression patterns by using Spearman correlation (r = −0.271, P = 0.013, Table 4).
Table 3.
Correlation between the clinicopathological characteristics and expression of ELF and TGF-β1 in the 84 HCC patients
| Variables | Cases | ELF expression | P value | TGF-β1 expression | P value | ||
|---|---|---|---|---|---|---|---|
| Low | High | Low | High | ||||
| Age(yrs) | |||||||
| > = 60 | 16 | 7(43.8%) | 9(56.2%) | 0.442 | 9(54.4%) | 7(54.4%) | 0.325 |
| <60 | 68 | 37(54.4%) | 31(45.6%) | 29(42.6%) | 39(57.4%) | ||
| Sex | |||||||
| Male | 68 | 37(54.4%) | 31(45.6%) | 0.44 | 27(39.7%) | 41(60.3%) | 0.77 |
| Female | 16 | 7(43.8%) | 9(56.2%) | 7(43.8%) | 9(56.2%) | ||
| HCC family history | |||||||
| Yes | 6 | 2(33.3%) | 4(66.7%) | 0.83 | 3(50.0%) | 3(50.0%) | 0.95 |
| No | 78 | 42(53.8%) | 36(46.2%) | 31(39.7%) | 47(60.3%) | ||
| HbsAg | |||||||
| Positive | 72 | 41(56.9%) | 31(43.1%) | 0.04 | 29(40.3%) | 43(59.7%) | 0.93 |
| Negative | 12 | 3(25.0%) | 9(75.0%) | 5(41.7%) | 7(58.3%) | ||
| ALT(U/L) | |||||||
| ≥80 | 9 | 3(33.3%) | 6(66.7%) | 0.39 | 1(11.1%) | 8(88.9%) | 0.12 |
| <80 | 75 | 41(54.7%) | 34(45.3%) | 33(44.0%) | 42(56.0%) | ||
| PLT(×109) | |||||||
| >100 | 74 | 41(55.4%) | 33(44.6%) | 0.24 | 28(37.8%) | 46(62.2%) | 0.32 |
| ≤100 | 10 | 3(30.0%) | 7(70.0%) | 6(60.0%) | 4(40.0%) | ||
| Cirrhosis | |||||||
| Yes | 64 | 32(50.0%) | 32(50.0%) | 0.43 | 27(42.2%) | 37(57.8%) | 0.57 |
| No | 20 | 12(60.0%) | 8(40.0%) | 7(35.0%) | 13(65.0%) | ||
| AFP(ug/L) | |||||||
| ≥20 | 48 | 28(58.3%) | 20(41.7%) | 0.21 | 19(39.6%) | 29(60.4%) | 0.85 |
| <20 | 36 | 16(44.4%) | 20(55.6%) | 15(41.7%) | 21(58.3%) | ||
| Tumor size (cm) | |||||||
| ≥5 | 50 | 32(64.0%) | 18(36.0%) | 0.01 | 13(26.0%) | 37(74.0%) | 0.001 |
| <5 | 34 | 12(35.3%) | 22(64.7%) | 21(61.8%) | 13(38.2%) | ||
| Tumor number | |||||||
| Single | 62 | 26(41.9%) | 36(58.1%) | 0.001 | 31(50.0%) | 31(50.0%) | 0.003 |
| Multiple | 22 | 18(81.8%) | 4(18.2%) | 3(13.6%) | 19(86.4%) | ||
| Differentiation | |||||||
| I-II | 62 | 31(50.0%) | 31(50.0%) | 0.46 | 27(43.5%) | 35(56.5%) | 0.34 |
| III-IV | 22 | 13(59.1%) | 9(40.9%) | 7(31.8%) | 15(68.2%) | ||
| TNM stage | |||||||
| I-II | 55 | 24(43.6%) | 31(56.4%) | 0.03 | 29(52.7%) | 26(47.3%) | 0.002 |
| III-IV | 29 | 20(69.0%) | 9(31.0%) | 5(17.2%) | 24(82.8%) | ||
| PVTT | |||||||
| Yes | 11 | 8(72.7%) | 3(27.3%) | 0.15 | 2(18.2%) | 9(81.8%) | 0.11 |
| No | 73 | 36(49.3%) | 37(50.7%) | 32(43.8%) | 41(56.2%) | ||
| Tumor encapsulation | |||||||
| Complete | 64 | 30(46.9%) | 34(53.1%) | 0.07 | 29(45.3%) | 35(54.7%) | 0.11 |
| None | 20 | 14(70.0%) | 6(30.0%) | 5(25.0%) | 15(75.0%) | ||
| Recurrence | |||||||
| Yes | 56 | 39(69.6%) | 17(30.4%) | <0.001 | 12(21.4%) | 44(78.6%) | <0.001 |
| No | 28 | 5(17.9%) | 23(82.1%) | 22(78.6%) | 6(21.4%) | ||
| Complication | |||||||
| No | 73 | 41(56.2%) | 32(43.8%) | 0.07 | 31(42.5%) | 42(57.5%) | 0.34 |
| Yes | 11 | 3(27.3%) | 8(72.7%) | 3(27.3%) | 8(72.7%) | ||
AFP, Alpha-fetoprotein; HBsAg, hepatitis B surface antigen; PLT, platelet; PVTT, portal vein tumor thrombi.
Table 4.
The correlationship between ELF and TGF-β1 in HCC
| ELF | TGF-β1 | r | P value | ||
|---|---|---|---|---|---|
| +++ | ++ | - ~ + | |||
| +++ | 7 | 4 | 12 | −0.271 | 0.013 |
| ++ | 4 | 1 | 12 | ||
| - ~ + | 20 | 14 | 10 | ||
Independent prognostic factors of HCC
To further identify the risk factors linked to postoperative Disease Free Survival (DFS) and Overall Survival (OS), ELF, TGF-β1 and 16 clinicopathologic factors were evaluated by univariate analysis and the Cox regression model. The univariate analysis showed that the significant prognostic factors for DFS of HCC were tumor number, portal vein tumor thrombus (PVTT), tumor encapsulation, TNM stage, ELF expression, and TGF-β1 expression. Similarly, the analysis showed that the significant factors for OS of HCC were tumor number, PVTT, tumor size, resection margin, tumor differentiation, TNM stage, ELF expression, and TGF-β1 expression (all P < 0.05). Using the Cox regression multivariate analysis, we found that PVTT, ELF expression, and TGF-β1 expression were the significant independent related factors for DFS (all P < 0.05), in addition, tumor differentiation (P = 0.029), PVTT (P = 0.011), ELF expression (P = 0.042) and TGF-β1 expression (P < 0.001) were the significant independent related factors for OS (Tables 5 and 6).
Table 5.
Prognostic factors for DFS and OS by univariate analysis
| Variables | n | DFS | P | OS | P | ||||
|---|---|---|---|---|---|---|---|---|---|
| 1-yr | 3-yrs | 5-yrs | 1-yr | 3-yrs | 5-yrs | ||||
| Sex | |||||||||
| Male | 68 | 54.4% | 36.8% | 30.7% | 0.53 | 79.4% | 50.0% | 41.2% | 0.48 |
| Female | 16 | 56.3% | 37.5% | 37.5% | 87.5% | 56.3% | 50.0% | ||
| Age(yrs) | |||||||||
| <60 | 68 | 45.6% | 32.4% | 29.3% | 0.15 | 82.4% | 48.5% | 39.7% | 0.39 |
| ≥60 | 16 | 62.5% | 56.3% | 42.2% | 75.0% | 62.5% | 56.3% | ||
| HCC family history | |||||||||
| Yes | 6 | 50.0% | 33.3% | 16.7% | 0.57 | 83.3% | 50.0% | 50.0% | 0.63 |
| No | 78 | 48.7% | 37.2% | 33.3% | 79.5% | 51.3% | 42.3% | ||
| PLT(×109) | |||||||||
| <100 | 10 | 80.0% | 60.0% | 60.0% | 0.07 | 100.0% | 80.0% | 70.0% | 0.08 |
| ≥100 | 74 | 44.6% | 33.8% | 28.2% | 78.4% | 47.3% | 39.2% | ||
| HBsAg | |||||||||
| Positive | 72 | 47.2% | 38.9% | 33.3% | 0.90 | 79.2% | 50.0% | 44.4% | 0.88 |
| Negative | 12 | 58.3% | 25.0% | 25.0% | 91.7% | 58.3% | 33.3% | ||
| AFP(μg/L) | |||||||||
| <20 | 36 | 52.8% | 38.9% | 38.9% | 0.34 | 83.3% | 50.0% | 44.4% | 0.75 |
| ≥20 | 48 | 45.8% | 35.4% | 26.7% | 79.2% | 52.1% | 41.7% | ||
| Ascites | |||||||||
| No | 68 | 52.9% | 39.7% | 34.1% | 0.14 | 83.8% | 51.5% | 44.1% | 0.55 |
| Yes | 16 | 31.3% | 25.0% | 25.0% | 68.8% | 50.0% | 37.5% | ||
| Cirrhosis | |||||||||
| No | 24 | 45.0% | 35.0% | 30.0% | 0.78 | 95.0% | 60.0% | 45.0% | 0.49 |
| Yes | 60 | 50.0% | 37.5% | 32.9% | 76.6% | 48.4% | 42.2% | ||
| Tumor number | |||||||||
| Single | 62 | 59.7% | 43.5% | 36.9% | 0.003 | 85.5% | 61.3% | 51.6% | <0.001 |
| Multiple | 22 | 18.2% | 18.2% | 18.2% | 68.2% | 22.7% | 18.2% | ||
| PVTT | |||||||||
| No | 73 | 54.8% | 41.1% | 35.4% | <0.001 | 87.7% | 56.2% | 47.9% | <0.001 |
| Yes | 11 | 9.1% | 9.1% | 9.1% | 36.4% | 18.2% | 9.1% | ||
| Tumor size (cm) | |||||||||
| <5 | 34 | 64.7% | 47.1% | 39.2% | 0.05 | 97.1% | 67.6% | 52.9% | 0.04 |
| ≥5 | 50 | 38.0% | 30.0% | 28.0% | 70.0% | 40.0% | 36.0% | ||
| Tumor encapsulation | |||||||||
| None | 20 | 30.0% | 25.0% | 15.0% | 0.01 | 60.0% | 45.0% | 30.0% | 0.08 |
| Complete | 64 | 54.7% | 40.6% | 37.7% | 87.5% | 53.1% | 46.9% | ||
| Resection margin | |||||||||
| <2 cm | 45 | 40.0% | 26.7% | 24.4% | 0.07 | 80.0% | 40.0% | 28.9% | 0.01 |
| ≥2 cm | 39 | 59.0% | 48.7% | 39.6% | 82.1% | 64.1% | 59.0% | ||
| Complication | |||||||||
| No | 73 | 49.3% | 38.4% | 33.1% | 0.37 | 84.9% | 53.4% | 45.2% | 0.10 |
| Yes | 11 | 45.5% | 27.3% | 27.3% | 54.5% | 36.4% | 27.3% | ||
| Tumor differetiation | |||||||||
| I-II | 62 | 54.8% | 41.9% | 35.1% | 0.16 | 85.5% | 56.5% | 48.4% | 0.04 |
| III-IV | 22 | 31.8% | 22.7% | 22.7% | 68.2% | 36.4% | 27.3% | ||
| TNM stage | |||||||||
| I-II | 55 | 60.0% | 43.6% | 38.0% | 0.01 | 90.9% | 60% | 52.7% | 0.001 |
| III-IV | 29 | 27.6% | 24.1% | 20.7% | 62.1% | 34.5% | 24.1% | ||
| ELF expression | |||||||||
| Low | 44 | 25.0% | 15.9% | 10.2% | <0.001 | 72.7% | 31.8% | 23.7% | <0.001 |
| High | 40 | 75.0% | 60.0% | 57.5% | 90.0% | 72.5% | 65.0% | ||
| TGFβ1 expression | |||||||||
| Low | 34 | 79.4% | 73.5% | 62.0% | <0.001 | 94.1% | 85.3% | 76.5% | <0.001 |
| High | 50 | 28.0% | 12.0% | 12.0% | 72.0% | 28.0% | 20.0% | ||
AFP, Alpha-fetoprotein; HBsAg, hepatitis B surface antigen; PLT, platelet; PVTT, portal vein tumor thrombi.
Table 6.
Prognostic factors for disease-free and overall survival by the multivariate Cox proportional hazards regression model
| Variables | DFS | OS | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Tumor differentiation | 0.498 | 0.266-0.932 | 0.029 | |||
| PVTT | 0.405 | 0.199-0.824 | 0.013 | 0.398 | 0.195-0.812 | 0.011 |
| ELF expression | 2.135 | 1.115-4.088 | 0.022 | 1.989 | 1.024-3.862 | 0.042 |
| TGFβ1 expression | 0.219 | 0.099-0.486 | <0.001 | 0.210 | 0.093-0.474 | <0.001 |
HR, hazard ratio; CI, confidence interval; PVTT, portal vein tumor thrombi.
Low expression of ELF and high expression of TGF-β1 predict HCC patients’ poor prognosis
Firstly, we divided 84 patients with HCC into 2 groups according to their ELF expression profiles: the low-expression group (n = 44) and the high-expression group (n = 40). Using the Kaplan-Meier method to analyze patients’ survival, we found that the 1-, 3- and 5-year DFS rates of the high-expression ELF group were remarkably higher than the low-expression group (75.0%, 60.0% and 57.5% vs 25.0%, 15.9% and 10.2%, respectively, P < 0.001) (Figure 2A), while the 1-, 3- and 5-year OS rates of the high-expression ELF group were significantly higher than those of the low-expression group (90.0%, 72.5% and 65.0% vs 72.7%, 31.8% and 23.7%, respectively, P < 0.001) (Figure 2B). Our findings therefore indicated that ELF expression levels were positively correlated with patients’ DFS and OS.
Figure 2.

Kaplan-Meier curves are shown for time to disease recurrence (A) and overall survival (B) among patients with high or low intratumoral ELF expression.
Similarly, Two groups were divided from 84 HCC patients according to their TGF-β1 expression profiles: the low-expression group (n = 34) and the high-expression group (n = 50). We observed that the 1-, 3- and 5-year DFS rates of the low-expression TGF-β1 group were markedly higher than the high-expression group (79.4%, 73.5% and 62.0% vs 28.0%, 12.0% and 12.0%, respectively, P < 0.001) (Figure 3A). Also, the 1-, 3- and 5-year OS rates of the low-expression TGF-β1 group were significantly higher than those of the high-expression group (94.1%, 85.3% and 76.5% vs 72.0%, 28.0% and 20.0%, respectively, P < 0.001) (Figure 3B). These data suggested that TGF-β1 expression levels were negatively correlated with patients’ DFS and OS.
Figure 3.
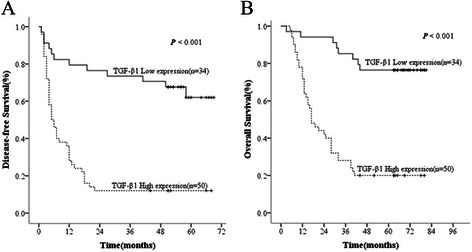
Kaplan-Meier curves are shown for time to disease recurrence (A) and overall survival (B) among patients with high or low intratumoral TGF-β1 expression.
The combination of TGF-β1 and ELF exhibits the improved prognostic accuracy for HCC
To analyze the prognostic value of combining TGF-β1 and ELF levels for HCC, we divided patients into the following four groups, such as: TGF-β1 high expression- ELF high expression group, TGF-β1 low expression- ELF high expression group, TGF-β1 high expression - ELF low expression group, TGF-β1 low expression- ELF low expression group. The data showed that the TGF-β1 low expression- ELF high expression group had the best DFS and OS rates, TGF-β1 low expression- ELF low expression group was the second best, the next was TGF-β1 high expression- ELF high expression group, whereas TGF-β1 high expression- ELF low expression group had the worst prognosis.
The 1-, 3- and 5-year DFS rates of TGF-β1 low expression- ELF high expression group (87.5%, 79.2% and 75.0%) were significantly higher than those of TGF-β1 high expression- ELF high expression group (56.3%, 31.3% and 31.3%, P = 0.003) and TGF-β1 high expression- ELF low expression group (26.5%, 2.9% and 2.9%, P < 0.001). The 1-, 3- and 5-year OS rates of TGF-β1 low expression- ELF high expression group (95.8%, 91.7% and 83.3%) were also significantly higher than those of TGF-β1 high expression- ELF high expression group (81.3%, 43.8% and 37.5%, P = 0.001) and TGF-β1 high expression- ELF low expression group (67.6%, 20.6% and 11.8%, P < 0.001) (Figure 4A and B).
Figure 4.
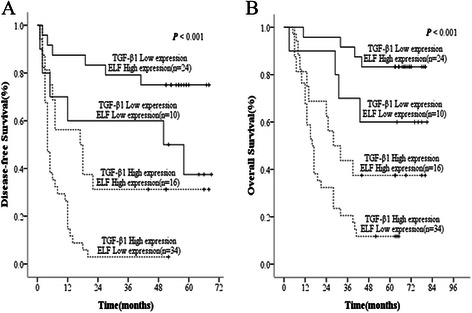
The combination of ELF and TGF-β1 was found to enhance prognostic accuracy for HCC. Disease-free survival curves (A) and overall survival curves (B).
Furthermore, we found that the 1-, 3- and 5-year DFS rates of TGF-β1 high expression-ELF low expression (26.5%, 2.9% and 2.9%) were remarkably lower than TGF-β1 high expression- ELF high expression (56.3%, 31.3% and 31.3%, P = 0.002) and TGF-β1 low expression-ELF low expression (60.0%, 60.0% and 37.5%, P = 0.002). Also, the 1-, 3- and 5-year OS rates of TGF-β1 high expression-ELF low expression (67.6%, 20.6% and 11.8%) were markedly lower than TGF-β1 low expression-ELF low expression group (90.0%, 70.0% and 60.0%, P = 0.003). However, there was no significant difference of OS rates between TGF-β1 high expression-ELF low expression and TGF-β1 high expression- ELF high expression (67.6%, 20.6% and 11.8% vs 81.3%, 43.8% and 37.5%, respectively, P = 0.058). We also found no significant difference of DFS and OS rates between TGF-β1 low expression-ELF high expression group and TGF-β1 low expression- ELF low expression group, or between TGF-β1 low expression-ELF low expression group and TGF-β1 high expression-ELF high expression group (Figure 4A and B). Collecting, the results indicated that the combination of TGF-β1 elevation and ELF reduction in HCC tissues appears to be predictive of the poorest prognosis.
Discussion
In the past few decades, great efforts have been made to explore the molecular mechanism of HCC to identify biomarkers for prediction and to develop effective treatments. In this study, we focused on investigating the prognostic significance of TGF-β1 and ELF, in particular their combination, for HCC. Our first finding showed that the TGF-β1 protein was upregulated in human HCC tissues and no normal liver tissues with strong cytoplasmic TGF-β1 protein immunostaining. The results were consistent with our previous study that the low-expression of TGF-β1 in normal rat liver tissues and the high-expression of TGF-β1 in rat HCC tissues [16]. Like others reports [31,32]. We also found the positive correlation between TGF-β1 and several clinicopathological characteristics: tumor size, tumor number, TNM stage and recurrence. A shorter post-operative survival of HCC patients with high level of TGF-β1 had been documented in this study. The 1-, 3- and 5-year DFS rates and OS rates of HCC patients with high level of TGF-β1 were markedly lower than the low-expression group.
Why do the functions of TGF-β switch from tumor suppression to tumor promotion? Mishra L et al. indicated that proper control of TGF-β signaling tumor suppressor function requires an additional adaptor protein, ELF. Research from that group indicated that disruption of ELF expression results in miscolocalization of Smad3 and Smad4, then disruption of TGF-β signaling, allowing normal cells to escape from the regulation of proliferation in carcinogenesis [21,33-36]. However, it was not reported if ELF expression level correlated with survival of HCC patients.
It is therefore of interest to investigate the expression and clinical significance of ELF in patients with HCC. We found that ELF was lost or underexpressed in the majority of HCC tissues, and that a high level of ELF expression predicted a favorable DFS rate and OS rate for HCC patients. Our data showed that the expression of ELF negatively correlated with HbsAg, tumor size, tumor number, TNM and recurrence. The 1-, 3- and 5-year DFS rates of HCC patients with the high level of ELF expression were remarkably higher than those of HCC patinets with the low levels. Similarly, the 1-, 3- and 5-year OS rates of HCC patients with the high level of ELF expression were significantly higher than those of HCC patients with the low levels. These data were consistent with previous studies, which showed that significant ELF reduction was found in HCC, gastric cancer and lung cancer [33-36].
Further, we studied the correlation between ELF and TGF-β1 in HCC patients, and demonstrated their significant negative correlation. Then we used univariate analysis and the Cox regression mode to study the role of ELF and TGF-β1 on HCC, finding that the expression of ELF and TGF-β1 were both significant and independent prognostic factors for DFS or OS of HCC. These data further verified that ELF and TGF-β1 were important and promising candidate tumor biomarker for predicting the prognosis of patients with HCC, and we hypothesized if combination of ELF and TGF-β1 could give us a more sensitive way to predict HCC patients’ outcome.
It is widely understood that a combination of multiple markers might yield more information for predicting clinical outcome of HCC patients [37]. Elevation of TGF-β1 or reduction of ELF in HCC tissues appears to be predictive of a poor prognosis. The combination of TGF-β1 and ELF expression were therefore used as a predictor of clinical outcome. The results indicated that their combination has a better prognostic value compared with either one alone. For example, those patients with low ELF expression and high TGF-β1 expression had the poorest OS and DFS rates, whereas those patients with high ELF expression and low TGF-β1 expression had the most favorable OS and DFS rates. The second best prognosis belonged to these patients with low ELF expression and low TGF-β1 expression. In addition, we found that high level of ELF could partially rescue TGF-β1 related tumor promotion, but TGF-β1still was the more important factor for prognosis of patient with HCC.
Conclusions
Our study determined that loss or reduction of ELF and elevation of TGF-β1 was correlated with disease progression and metastasis in patients with HCC. And the most interesting finding was that the predictive range of ELF levels combined with TGF-β1 expression was more sensitive than that of either ELF or TGF-β1 alone with regard to OS and cumulative disease recurrence in patients with HCC. From a diagnostic viewpoint, our results suggest that the detection of tumor ELF alone or the combined evaluation of ELF/ TGF-β1 levels could be used as a new prognostic marker in patients with HCC. However, the exact mechanisms of ELF and TGF-β1 expression regulation and function in HCC should been elucidated further. In the future, ELF might be used as potentially powerful target for treatment of HCC through enhancing the tumor suppression of TGF-β pathway.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (NO. 81201918), Science and Technology Project of Guangdong Province (No.2012B031800099), Doctorial Fellowship of Higher Education of China (NO.200805581172). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- HCC
Hepatocellular cancer
- TGF-β
The transforming growth factor β
- SBE
Smad-binding elements
- ELF
Embryonic Liver Fodrin
- β2SP
β2-spectrin
- TNM
Tumor-node-metastasis
- IRS
Immunoreactivity score
- CT
Computed tomography
- MRI
Magnetic resonance imaging
- PET-CT
Positron emission tomography -CT
- DFS
Disease Free Survival
- OS
Overall Survival
- AFP
Alpha-fetoprotein
- HBsAg
Hepatitis B surface antigen
- PLT
Platelet
- PVTT
Portal vein tumor thrombi. HR, hazard ratio
- CI
Confidence interval
Footnotes
Fei Ji and Shun-Jun Fu contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
FJ, SJF and YPH were the main authors of the manuscript. They were involved in the conception, design and coordination of the study as well as in data analysis, interpretation of results and drafting the manuscript. YPH was in charge of all experimental procedures. SLS, LJZ, QHC, SQL, BGP, and LJL participated in the experimental procedures and revised critically the content of the manuscript. All authors contributed to the interpretation of data and critically revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Fei Ji, Email: jifei1215@163.com.
Shun-Jun Fu, Email: fsj103@163.com.
Shun-Li Shen, Email: shunlishen@163.com.
Long-Juan Zhang, Email: zhlongj@mail.sysu.edu.cn.
Qing-Hua Cao, Email: bpigeon@163.com.
Shao-Qiang Li, Email: Lisq@medmail.com.cn.
Bao-Gang Peng, Email: pengbaogang@medmail.com.cn.
Li-Jian Liang, Email: dr_lianglj@163.com.
Yun-Peng Hua, Email: hyp0427@163.com.
References
- 1.EI-serag HB, Rudolph KL. Hepatocellular carcinoma:epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Tang ZY. Hepatocellular carcinoma-cause, treatment, and metastasis. World J Gastroenterol. 2001;7:445–54. doi: 10.3748/wjg.v7.i4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization: The world health report 2006. http://www.who.int/whr/2006/en/
- 4.Ng KK, Lo CM, Liu CL, Poon RT, Chan SC, Fan ST. Survival analysis of patients with transplantable recurrent hepatocellular carcinoma: implications for salvage liver transplant. Arch Surg. 2008;143:68–74. doi: 10.1001/archsurg.2007.15. [DOI] [PubMed] [Google Scholar]
- 5.Kim DY, Paik YH, Ahn SH, Youn YJ, Choi JW, Kim JK, et al. PIVKA-II is a useful tumor marker for recurrent hepatocellular carcinoma after surgical resection. Oncology. 2007;72:52–7. doi: 10.1159/000111707. [DOI] [PubMed] [Google Scholar]
- 6.Zarzynska JM. Two Faces of TGF-Beta1 in Breast Cancer. Mediators Inflamm. 2014;2014:141747. doi: 10.1155/2014/141747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heldin CH, Landström M, Moustakas A. Mechanism of TGF-β signaling to growth arrest, apoptosis, and epithelialmesenchymal transition. Curr Opin Cell Biol. 2009;21:166–76. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 8.Ikushima H, Miyazono K. Biology of transforming growth factor-β signaling. Curr Pharm Biotechnol. 2011;12:2099–107. doi: 10.2174/138920111798808419. [DOI] [PubMed] [Google Scholar]
- 9.Moses H, Barcellos-Hoff MH. TGF-β biology in mammary development and breast cancer. Cold Spring Harb Perspect Biol. 2011;3:a003277. doi: 10.1101/cshperspect.a003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parvani JG, Taylor MA, Schiemann WP. Noncanonical TGF-β signaling during mammary tumorigenesis. J Mammary Gland Biol Neoplasia. 2011;16:127–46. doi: 10.1007/s10911-011-9207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katz LH, Li Y, Chen JS, Muñoz NM, Majumdar A, Chen J, et al. Targeting TGF-β signaling in cancer. Expert Opin Ther Targets. 2013;17:743–60. doi: 10.1517/14728222.2013.782287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bierie B, Moses HL. Transforming growth factor beta (TGF-β) and inflammation in cancer. Cytokine Growth Factor Rev. 2010;21:49–59. doi: 10.1016/j.cytogfr.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kajdaniuk D, Marek B, Borgiel-Marek H, Kos-Kudła B. Transforming growth factorβ1 (TGFβ1) in physiology and pathology. Endokrynol Pol. 2013;64:384–96. doi: 10.5603/EP.2013.0022. [DOI] [PubMed] [Google Scholar]
- 14.Zu X, Zhang Q, Gao R, Liu J, Zhong J, Wen G. Transforming growth factor-β signaling in tumor initiation, progression and therapy in breast cancer: an update. Cell Tissue Res. 2012;347:73–84. doi: 10.1007/s00441-011-1225-3. [DOI] [PubMed] [Google Scholar]
- 15.Malaguarnera G, Giordano M, Paladina I, Berretta M, Cappellani A, Malaguarnera M. Serum markers of hepatocellular carcinoma. Dig Dis Sci. 2010;55:2744–55. doi: 10.1007/s10620-010-1184-7. [DOI] [PubMed] [Google Scholar]
- 16.Hua YP, Li SQ, Lai JM, Liang LJ, Peng BG, Liang HZ, et al. Changes in TGF-β/Smads signaling pathway in rats with chemical hepatocarcinogenesis. J South Med Univ. 2008;28:1848–52. [PubMed] [Google Scholar]
- 17.Baek HJ, Lim SC, Kitisin K, Joqunoori W, Tang Y, Marshall MB, et al. Hepatocellular cancer arises from loss of transforming growth factor beta signaling adaptor protein embryonic liver fodrin through abnormal angiogenesis. Hepatology. 2008;48:1128–37. doi: 10.1002/hep.22460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thenappan A, Shukla V, Abdul Khalek FJ, Thenappan A, Shukla V, Abdul Khalek FJ, et al. Loss of transforming growth factor β adaptor protein β-2 spectrin leads to delayed liver regeneration in mice. Hepatology. 2011;53:1641–50. doi: 10.1002/hep.24111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Song Y, Tu W, He X, Lin J, Liu F. β-2 spectrin is involved in hepatocyte proliferation through the interaction of TGFβ/Smad and PI3K/AKT signaling. Liver Int. 2012;32:1103–11. doi: 10.1111/j.1478-3231.2012.02812.x. [DOI] [PubMed] [Google Scholar]
- 20.Tang Y, Katuri V, Srinivasan R, Foqt F, Redman R, Anand G, et al. Transforming growth factor-beta suppresses nonmetastatic colon cancer through Smad4 and adaptor protein ELF at an early stage of tumorigenesis. Cancer Res. 2005;65:4228–37. doi: 10.1158/0008-5472.CAN-04-4585. [DOI] [PubMed] [Google Scholar]
- 21.Mishra L, Katuri V, Evans S. The role of PRAJA and ELF in TGF-beta signaling and gastric cancer. Cancer Biol Ther. 2005;4:694–9. doi: 10.4161/cbt.4.7.2015. [DOI] [PubMed] [Google Scholar]
- 22.Tang Y, Kitisin K, Jogunoori W, Li C, Deng CX, Mueller SC, et al. Progenitor/ stem cells give rise to liver cancer due to aberrant TGF- beta and IL-6 signaling. Proc Natl Acad Sci U S A. 2008;105:2445–50. doi: 10.1073/pnas.0705395105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SS, Shetty K, Katuri V, Kitisin K, Baek HJ, Tang Y, et al. TGF-beta signaling pathway inactivation and cell cycle deregulation in the development of gastric cancer: role of the beta-spectrin, ELF. Biochem Biophys Res Commun. 2006;344:1216–23. doi: 10.1016/j.bbrc.2006.03.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitisin K, Ganesan N, Tang Y, Joqunoori W, Volpe EA, Kim SS, et al. Disruption of transforming growth factor-beta signaling through beta-spectrin ELF leads to hepatocellular cancer through cyclin D1 activation. Oncogene. 2007;26:7103–10. doi: 10.1038/sj.onc.1210513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020–2. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobin LH, Wittekind C. UICC (International Union against Cancer) TNM classification of malignant tumors. 6. New York: John Wiley; 2002. pp. 1–264. [Google Scholar]
- 27.Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462–503. doi: 10.1002/1097-0142(195405)7:3<462::AID-CNCR2820070308>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 28.Weichert W, Röske A, Gekeler V, Beckers T, Ebert MP, Pross M, et al. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: a retrospective analysis. Lancet Oncol. 2008;9:139–48. doi: 10.1016/S1470-2045(08)70004-4. [DOI] [PubMed] [Google Scholar]
- 29.Wu F, Liu SY, Tao YM, Ou DP, Fang F. Decreased expression of methyl methansulfonate and UV sensitive gene clone 81 is related to poor prognosis of patients with hepatocellular carcinoma. Cancer. 2008;112:2002–10. doi: 10.1002/cncr.23396. [DOI] [PubMed] [Google Scholar]
- 30.Wu F, Yang LY, Li YF, Ou DP, Chen DP, Fan C. Novel role for epidermal growth factor-like domain 7 in metastasis of human hepatocellular carcinoma. Hepatology. 2009;50:1839–50. doi: 10.1002/hep.23197. [DOI] [PubMed] [Google Scholar]
- 31.Tripsianis G, Papadopoulou E, Romanidis K, Katotomichelakis M, Anagnostopoulos K, Kontomanolis E, et al. Overall Survival and Clinicopathological Characteristics of Patients with Breast Cancer in Relation to the Expression Pattern of HER-2, IL-6, TNF-α and TGF-β1. Asian Pac J Cancer Prev. 2013;14:6813–20. doi: 10.7314/APJCP.2013.14.11.6813. [DOI] [PubMed] [Google Scholar]
- 32.de Kruijf EM, Dekker TJ, Hawinkels LJ, Putter H, Smit VT, Kroep JR, et al. The prognostic role of TGF-β signaling pathway in breast cancer patients. Ann Oncol. 2013;24:384–90. doi: 10.1093/annonc/mds333. [DOI] [PubMed] [Google Scholar]
- 33.Song S, Maru DM, Ajani JA, Chan CH, Honjo S, Lin HK, et al. Loss of TGF-β adaptor β2SP activates notch signaling and SOX9 expression in esophageal adenocarcinoma. Cancer Res. 2013;73:2159–69. doi: 10.1158/0008-5472.CAN-12-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baek HJ, Pishvaian MJ, Tang Y, Kim TH, Yang S, Zouhairi ME, et al. Transforming growth factor-β adaptor, β2-spectrin, modulates cyclin dependent kinase 4 to reduce development of hepatocellular cancer. Hepatology. 2011;53:1676–84. doi: 10.1002/hep.24128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katuri V, Tang Y, Marshall B, Rashid A, Jogunoori W, Volpe EA, et al. Inactivation of ELF/TGF-beta signaling in human gastrointestinal cancer. Oncogene. 2005;24:8012–24. doi: 10.1038/sj.onc.1208946. [DOI] [PubMed] [Google Scholar]
- 36.Baek HJ, Kim SS, da Silva FM, Volpe EA, Evans S, Mishra B, et al. Inactivation of TGF-beta signaling in lung cancer results in increased CDK4 activity that can be rescued by ELF. Biochem Biophys Res Commun. 2006;346:1150–7. doi: 10.1016/j.bbrc.2006.05.195. [DOI] [PubMed] [Google Scholar]
- 37.Li SX, Tang GS, Zhou DX, Pan YF, Tan YX, Zhang J, et al. Prognostic significance of cytoskeleton- associated membrane protein 4 and its palmitoyl acyltransferase DHHC2 in hepatocellular carcinoma. Cancer. 2014;120:1520–31. doi: 10.1002/cncr.28593. [DOI] [PubMed] [Google Scholar]


