Summary
Bacteria use rapid contraction of a long sheath of the Type VI secretion system (T6SS) to deliver effectors into a target cell. Here we present an atomic resolution structure of a native contracted Vibrio cholerae sheath determined by cryo-electron microscopy. The sheath subunits, composed of tightly interacting proteins VipA and VipB, assemble into a six-start helix. The helix is stabilized by a core domain assembled from four β-strands donated by one VipA and two VipB molecules. The fold of inner and middle layers is conserved between T6SS and phage sheaths. However, the structure of the outer layer is distinct and suggests a mechanism of interaction of the bacterial sheath with an accessory ATPase, ClpV, that facilitates multiple rounds of effector delivery. Our results provide a mechanistic insight into assembly of contractile nanomachines that bacteria and phages use to translocate macromolecules across membranes.
Introduction
Several critical components of the T6SS are structurally and functionally related to components of contractile tails of bacteriophages. Secreted VgrG and PAAR proteins form a complex similar to phage spike, secreted Hcp is a structural homolog of a phage tube protein, and TssE (type six secretion E) is a homolog of T4 phage baseplate protein gp25 (Leiman et al., 2009; Pukatzki et al., 2007; Shneider et al., 2013). VipA and VipB (TssB and TssC) proteins were shown to form a cog-wheel like tubular structure in V. cholerae (Bönemann et al., 2009) that was noticed to resemble T4 phage gp18 polysheath (Leiman et al., 2009). The VipA/VipB sheath assembles around an inner Hcp tube and is attached to a structure called a baseplate that spans the bacterial membranes (Basler et al., 2012; Brunet et al., 2014; Zoued et al., 2013). Importantly, VipA/VipB sheath was shown to form a long contractile organelle in V. cholerae (Basler et al., 2012; Kapitein et al., 2013) and in E. coli (Brunet et al., 2013), suggesting that sheath contraction powers the secretion. In vivo, the contracted sheath is specifically recognized by the ClpV ATPase, which disassembles the sheath by unfolding VipB from the N-terminus (Basler and Mekalanos, 2012; Kapitein et al., 2013; Pietrosiuk et al., 2011). Even though sheath contraction has been implicated in powering protein translocation across a membrane for phages, pyocins and T6SS (Leiman and Shneider, 2012), a mechanistic understanding of this process is currently limited, mostly due to the lack of a high-resolution structure of a native and fully assembled sheath.
There is no high resolution information available for the T6SS sheath but several crystal structures are available for fragments of phage sheath proteins (Aksyuk et al., 2009a, 2011). However, a major limitation of these atomic structures is that monomeric proteins were used for crystallization and thus, in principle, cannot reveal atomic details of inter-subunit interactions in a native fully assembled sheath polymer. The structure of the T4 phage sheath polymer was so far solved only at low resolution using electron microscopy (Kostyuchenko et al., 2005; Leiman et al., 2004), again not providing the necessary details to fully understand sheath assembly.
Native T6SS sheath has only been isolated from V. cholerae in a contracted form (Basler et al., 2012). Even though the sheath was isolated without the inner Hcp tube, Hcp and other components of T6SS were shown to be necessary for sheath assembly (Basler et al., 2012; Brunet et al., 2014; Kapitein et al., 2013). Indeed, in contrast to a long and regular T6SS sheath that can be isolated from T6SS-positive V. cholerae (Basler et al., 2012), VipA/VipB from P. aeruginosa and V. cholerae heterologously expressed in E. coli only form short tubes (Bönemann et al., 2009; Kube et al., 2014; Lossi et al., 2013); electron microscopy of these tubes provided low resolution density maps (Kube et al., 2014; Lossi et al., 2013). Nonetheless, a recent ~ 6 Å resolution structure of V. cholerae sheath provided insights into a possible mechanism of ClpV specific disassembly of the contracted sheath (Kube et al., 2014).
Due to recent advances in direct electron detection cameras and software tools (Egelman, 2010; Faruqi et al., 2003; Li et al., 2013; Lu et al., 2014), it is now possible to obtain density maps with a resolution that allows de novo building of atomic models (Kühlbrandt, 2014). These technical advances allowed for directly generating atomic models of the subunit of the mitochondrial ribosome (Amunts et al., 2014) or the ribosome-Sec61 complex (Voorhees et al., 2014) and provided fundamentally insights into mechanisms of those macromolecular machines. Here, we used the state-of-the-art electron microscopic approaches and the Rosetta density-guided structural modeling methods to reveal the structure of the contracted VipA/VipB sheath from V. cholerae in atomic detail.
Results and Discussion
Atomic structure of the VipA/VipB protomer
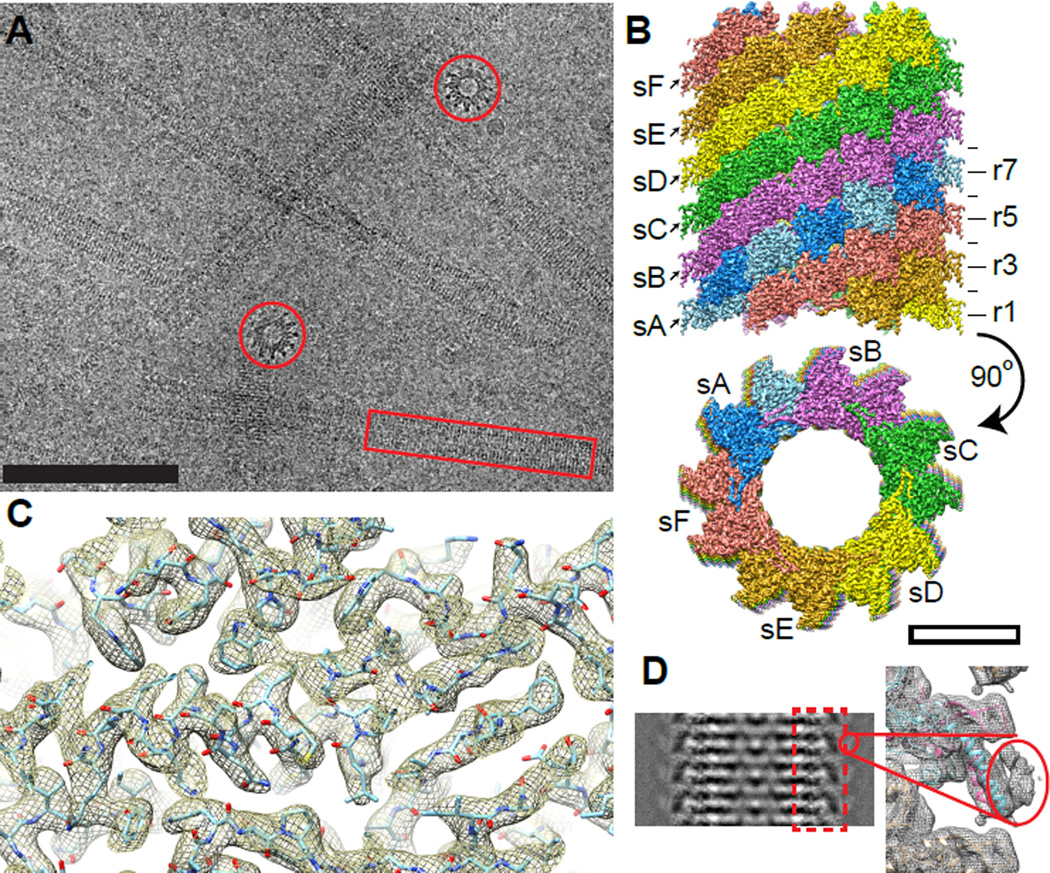
We purified the native contracted sheath from Vibrio cholerae and imaged it by cryo-electron microscopy (Figure 1A). Fourier transforms of recorded images showed Thon rings up to ~3 Å with layer lines in single micrographs up to a resolution of 5 Å (Figure S1A). Helical reconstruction was performed by the iterative helical real space reconstruction (IHRSR) method (Egelman, 2000) with the final helical parameters being a 21.8 Å axial rise, 29.4° rotation and a C6 rotational symmetry about the helical axis (Figures 1B, S1B, C and Movie S1). Helical parameters and an overall shape of the sheath are similar to the previously reported structure (Kube et al., 2014), however, our approach allowed us to obtain a resolution of ~3.5–4.0 Å, which improved up to ~3.2 Å for the inner and middle layers of the sheath (Figure S1D). Most of the amino acid side chains and some oxygen atoms in the backbone were resolved in the most ordered parts of the structure (Figure 1C and Movie S1).
Figure 1. Cryo-EM structure of the T6SS sheath.
(A) A representative low dose cryo-EM micrograph with side (red box) and top (red circle) views of the sheath. Scale bar - 100 nm; (B) Assembly of the protomers into a six start helix; s - individual strands, r - horizontal rings. Scale bar - 10 nm. (C) An example of the atomic model fitted into the protein density. (D) Left: a class average of the sheath showing a density on the surface; right: protein density filtered to low resolution showing density of the VipB N-terminus and VipA C-terminus. See also Figure S1 and Movie S1.
Even though the resolution of our protein density decreased for the outer surface layer, we were able to de novo trace residues 2 to 126 (out of 168) of VipA and residues 61 to 492 of VipB (Figures 2A, B and S2A–E). The VipA C-terminus and the VipB N-terminus were clearly localized to a less ordered layer on the surface of the sheath as shown in class averages of sheath images (Figure 1D and S1E, F). To further improve the geometry of the side chains, the model was subject to Rosetta density-guided all-atom refinement using a physically realistic energy function (Song et al., 2013 and DiMaio et al., in press). An atomic model built into an independently generated EM map of lower resolution had a Cα RMSD to the original atomic model of 0.34 Å (see Experiential Procedures) suggesting that model building is highly reliable. Analysis of the conservation and coevolution of VipA/VipB protein residues shows that the conserved residues are generally facing the inner part of the protomer, variable residues are exposed on the protomer surface and distances between most coevolving residues are within 10 Å (Figure S4, Table S1).
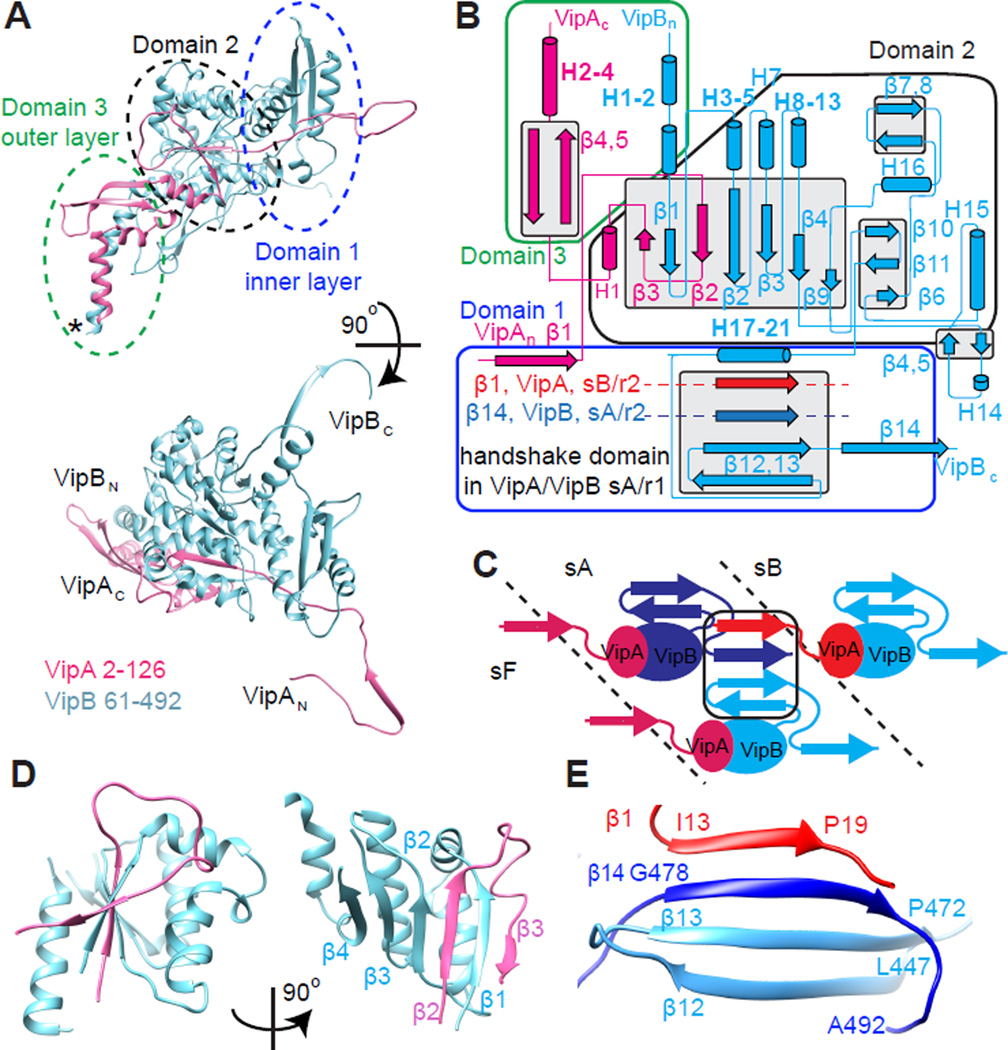
Figure 2. Atomic model of the sheath protomer.
(A) An atomic model for VipA (pink) and VipB (blue) with the outlined domains. Domain 3 contains untraced residues predicted to form 5 α-helices (location marked with *). (B) Interactions of secondary structure elements in the protomer; (C) scheme of the handshake domain assembly by three protomers of VipA/VipB; (D) Two views of domain 2 - six β-strands surrounded by 5 α-helices stabilizing the interaction between VipA and VipB in the protomer; (E) A “handshake domain” in the domain 1 connecting four β-strands: β12 and β13 of VipB in strand A, ring 1 (light blue) with β14 of VipB in strand A, ring 2 (blue) and β1 of VipA in strand B, ring 2 (red). See also Figure S2 and Movie S1.
The atomic model allowed us to calculate energies of protein-protein interactions using PISA (Krissinel and Henrick, 2007) and understand how the sheath structure is stabilized in its contracted form. The strongest intermolecular interaction was calculated between VipA and VipB to form a heterodimeric sheath protomer with 1:1 stoichiometry (Table 1, Figures 2A, B and S2F). Two β-strands of VipA and four β-strands of VipB intertwine forming the middle layer of the sheath (domain 2, Figure 2D). VipA further binds to one side of VipB, forming 35 hydrogen bonds and 14 salt bridges. The total interfacial area for this interaction is 3,493 Å2 and ΔG = −54.8 kcal/mol/protomer represents over half of all the interaction energy within the assembled sheath (Table 1).
Table 1.
Energy of interactions between VipA/VipB in the sheath assembly.
| Interface area, Å2 |
Number of H-H bonds |
Number of salt bridges |
ΔG kcal/mol |
% of interaction energy |
|
|---|---|---|---|---|---|
| VipA-B main interface | 3493 | 35 | 14 | −54.8 | 57 |
| VipB-B interaction in the strand | 2444 | 33 | 14 | −19.3 | 20 |
| VipA-B between the strands | 1143 | 7 | 2 | −14.7 | 15 |
| VipA-B in the horizontal ring | 634 | 11 | 4 | −4.5 | 5 |
| VipB-B vertical interaction | 454 | 5 | 2 | −3.0 | 3 |
Intermolecular interactions
Resolution limitations of the previous study (Kube et al., 2014) led to an imprecise segmentation of a sheath subunit from the low resolution density map (Figure S1G). Here we show that in the native sheath, subunits assemble by forming a unique set of interactions in the innermost layer of the sheath. A domain with four β-strands is assembled from two anti-parallel β-strands (β12 and β13) of one VipB molecule, one parallel β-strand (β14) of a second VipB on the same six-start helical strand, and one parallel β-strand (β1) of a VipA molecule from a neighboring strand in the six-start helix (Figure 2B, C, E). The interaction surface between VipB and VipB covers ~2,444 Å2, represents about 20% of the total interaction energy, and stabilizes the protomers within the strand. Finally, the interface area between VipA and VipB is 1,143 Å2 and contributes approximately 14.7 kcal/mol/protomer energy to the stabilization of the individual strands within the six-start helix. Together, these previously unrecognized interactions represent an energy of 34 kcal/mol/protomer and are the major contributors to sheath stability.
T6SS and phage sheaths evolved from a common ancestor
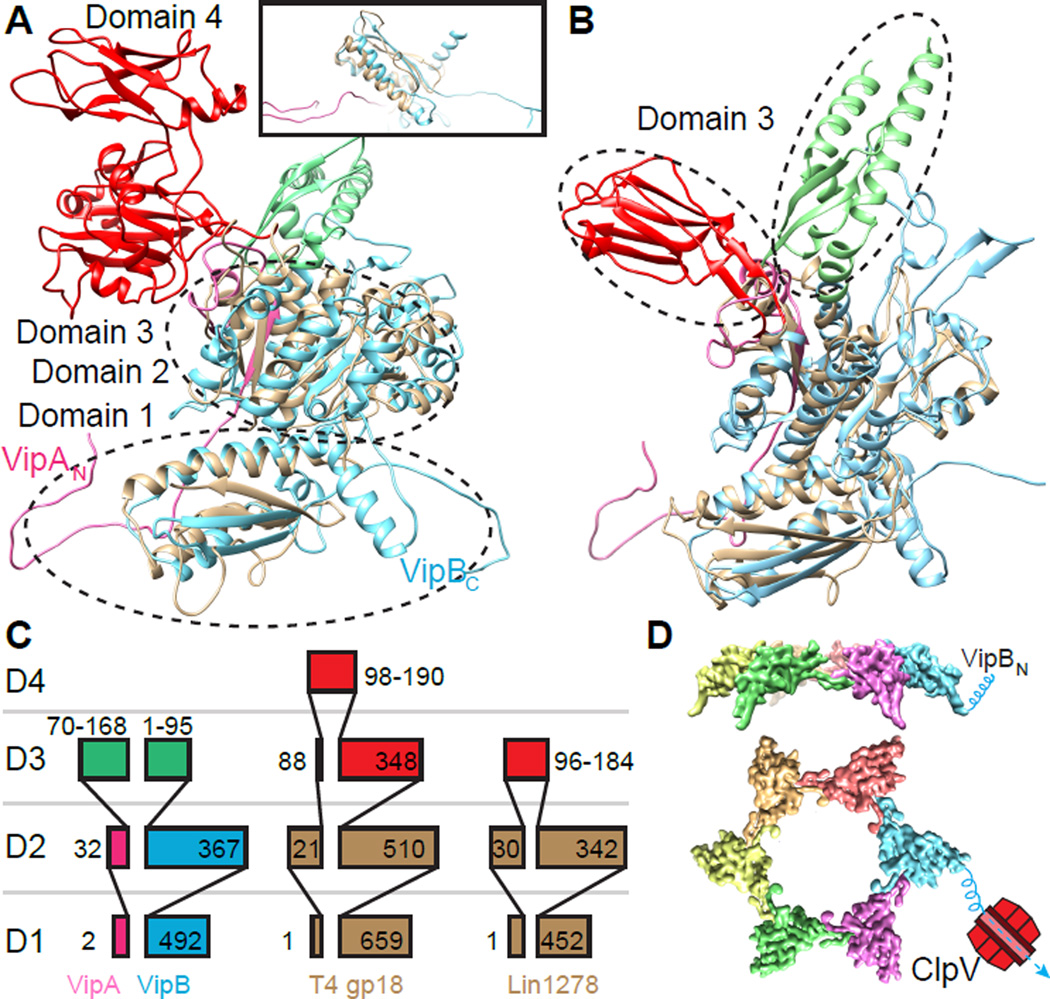
To understand the evolution of T6SS sheath we performed a structural alignment between VipA/VipB and a model of T4 phage sheath protein gp18 (Aksyuk et al., 2009a; Fokine et al., 2013) and a crystal structure of Listeria innocua phage sheath protein Lin1278 (Aksyuk et al., 2011). In contrast to sequence based alignments that only detect homology between VipB and phage sheath proteins, we show that domains 1 and 2, composed of both VipA and VipB, are highly conserved and the outer domains 3 and 4 are divergent (Figures 3 and S3). Domain 1 of T6SS sheath and the domain 1 of a model of gp18 or a crystal structure of Lin1278 align with RMSD of 2.7 Å and 2.2 Å, respectively. RMSDs between the domain 2 of the T6SS sheath and the crystal structures of the domain 2 of gp18 or Lin1278 are 3.7 Å and 2.8 Å, respectively.
Figure 3. Structural homology between the T6SS and phage sheaths.
(A, B) Structural alignment of VipA/VipB (pink/blue) with (A) model of full length T4 phage sheath gp18 (PDB: 3J2N), the inset shows a side view from the sheath lumen, (B) L. innocua phage sheath Lin1278 (PDB: 3LML). Structurally homologous domains 1 and 2 of phage sheaths are shown in brown; divergent domains 3 are shown in red for phage tails and in green for VipA/VipB. (C) A scheme depicting domain organization of VipA/VipB, gp18 and Lin1278 (partially adapted from (Leiman and Shneider, 2012)). (D) one ring of protomers showing N-terminus of VipB exposed to the outer surface of the sheath making it accessible to be disassembled by ClpV. See also Figure S3.
Interestingly, the architecture of domain 1 differs between phage and T6SS. In both phage sheath proteins, the first two β-strands have the same orientation as in the T6SS sheath but the third β-strand has an opposite orientation, and the fourth β-strand is missing (Figure 3A, B). Since the phage sheath structures were solved for monomers and not for fully assembled polymers, it is tempting to speculate that in a fully assembled phage sheath the corresponding handshake domain has the same architecture as in the native T6SS sheath and connects subunits and strands in the same manner as in T6SS.
The fundamental difference between phage and T6SS sheath is that phage sheath is used only once while T6SS sheath is recycled in vivo by ClpV (Basler and Mekalanos, 2012; Kapitein et al., 2013). Moreover, phages act in an extracellular space while the T6SS sheaths are functioning in bacterial cytoplasm. Here we show that the major difference between phage and T6SS sheaths is in the outer layer, which is not only structurally different but also positioned differently on the sheath surface. In the case of the T4 phage sheath, the domains 3 and 4 are inserted between β1 and H3 of VipB in the domain 2 (Figure 3A, B, S3). On the other hand, the T6SS sheath has its domain 3 inserted between H1 of VipA and H2 of VipB (Figure 3A, B, S3). This leads to a major difference in the angle between domain 3 and domain 2 compared to phage sheath architecture. Furthermore, the outermost layer of the phage sheaths are formed mostly by β-strands (Aksyuk et al., 2009a, 2011), whereas the T6SS sheath outer layer is predicted to be composed of 5 α-helices (Figure S2A–B).
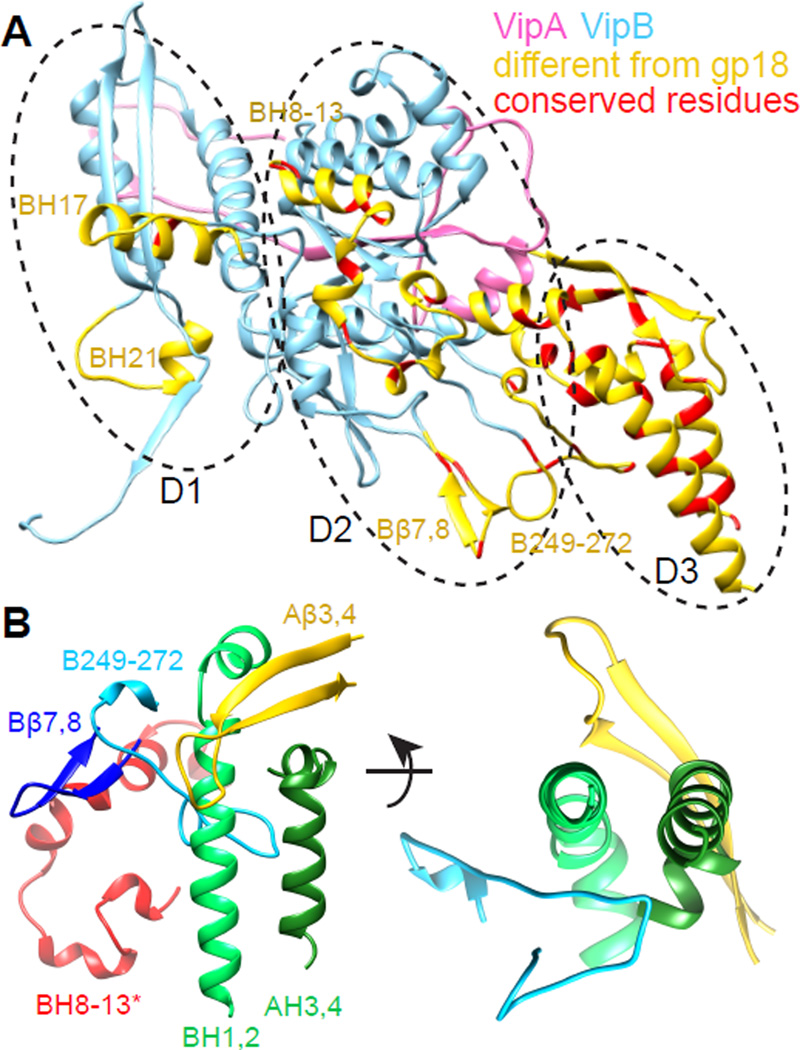
Even though the overall fold of domains 1 and 2 of phage and T6SS is conserved, the T6SS sheath has several potentially functional insertions compared to phage sheath (Figure 4A). The VipA/VipB protomer has two weakly conserved extra helices in the domain 1: VipB H17 and VipB H21. VipB H17 (aa 374–386) interacts with a loop of the next VipA in a strand (aa 18–24, originating from the handshake domain). A weakly conserved loop and a short VipB H21 interact with a loop (aa 412–415) close to VipB H19.
Figure 4. Divergence of the T6SS sheath from the phage sheath.
(A) VipA/VipB protomer (pink/blue) with the additional insertions compared to phage sheath Lin1278 marked in yellow; residues with sequence conservation over 70% are marked in red. (B) Interaction network in the outer domain of VipA/VipB. BH 8–13* are part of VipB in the neighboring subunit. See also Figure S4.
As hypothesized previously (Basler and Mekalanos, 2012; Kapitein et al., 2013; Kube et al., 2014), during the sheath contraction the VipB N-terminus is likely exposed on the sheath surface to allow disassembly by ClpV. Although an atomic model of an extended T6SS sheath is not available yet, it is likely that the N-terminus of VipB is not accessible for binding by ClpV in the extended state to prevent disassembly of the extended sheath. We show that domain 3 is exposed on the surface of the contracted sheath, aligning the domains 3 from the neighboring strands on top of one another. This is in agreement with the recently proposed model (Kube et al., 2014), however, here we show that two helices from VipAC and three from VipBN are exposed on the surface. This indeed makes the VipB N-terminus fully accessible for disassembly by ClpV (Figure 3D) as suggested previously (Kube et al., 2014) but raises a possibility that VipA is involved in properly positioning VipB on the sheath surface. Furthermore, our atomic model suggests that precise positioning of domain 3 could be stabilized by interactions of three T6SS specific insertions into the surface of VipB in domain 2: short helices H8–H13, a loop V246-N276 and an outward facing hairpin β7–β8. These insertions appear to form a network of hydrophobic interactions with the domain 3 at the outer surface of the sheath (Figure 4B). Hairpin β7–β8 forms an interaction with the H8–H13 of the VipB in the neighboring strand and with the loop VipB246-276. Loop VipB246-276 interacts with the two long helices VipA H4 and VipB H1 of the domain 3 from the inside while the other hairpin VipA β3–β4 stabilizes them from the outside. The two long helices are further stabilized by a helix-helix interaction with the conserved interfaces (Figure 4A).
Attachment of the sheath to the baseplate
Whole cell cryo-electron tomography provided only a low resolution structure of the sheath (Basler et al., 2012) and therefore it is not possible directly from those data to orient the VipA/VipB structure relative to a baseplate in the bacterial cell wall. However, considering the degree of structural similarity between T6SS and phage, it is likely that VipA and VipB are oriented relative to the baseplate in the same way as gp18 in T4 phage (Aksyuk et al., 2009a). In Figure 3A, and all other similar top views, the baseplate would be located behind the plane of view; on all side views, like the inset of Figure 3A, the baseplate would be located on the bottom. This orientation of the VipA/VipB protomer suggests that two β-strands per subunit are free to bind to an unknown T6SS component in the baseplate (Figure 3A). A natural interacting partner for those two β-strands would be a structure similar to an “empty” 2-β-stranded handshake domain organized in a hexameric ring similarly to an actual sheath ring.
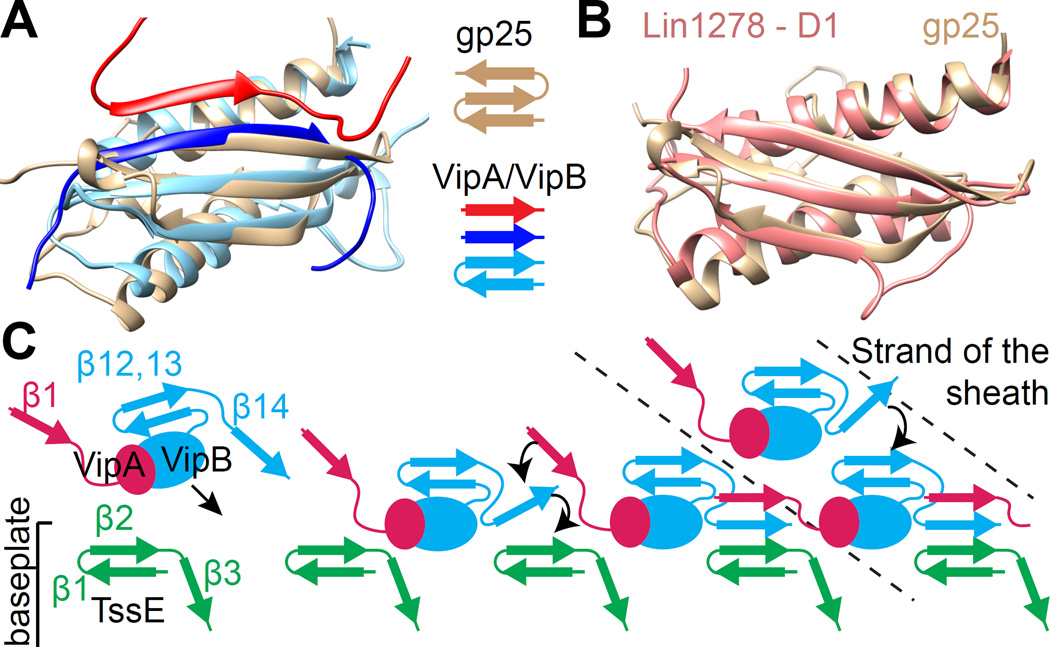
A search for structural homologs of the T6SS sheath revealed that protein NP_952040.1 from Geobacter sulfurreducens (PDB: 2IA7), a homolog of the T4 phage baseplate protein gp25, aligns with the T6SS sheath domain 1 with an RMSD of 2.7 Å (Figure 5A). As noted previously (Leiman and Shneider, 2012), phage sheath domain 1 has a fold that is similar to that of gp25-like protein (Figure 5B). Importantly, gp25 is positioned near the sheath in the T4 phage baseplate (Aksyuk et al., 2009b).
Figure 5. Evolutionary conservation of the handshake interactions and sheath assembly initiation.
(A) Structural alignment of the gp25-like phage protein from G. sulfurreducens (brown, PDB: 2IA7) and a T6SS sheath handshake domain containing VipB (light blue), VipB from the next subunit on the same strand (blue) and VipA from the next strand (red). (B) Alignment of the gp25-like phage protein from G. sulfurreducens (brown) and domain 1 of phage sheath Lin1278 (red). (C) A model for sheath assembly initiation and polymerization as viewed from inside the tube: Recruitment of VipA/VipB protomers (through their free β-strand) to the baseplate protein TssE (providing 2 β-strands); establishment of the full 4-β-stranded handshake domain starting with VipB, followed by VipA; Recruitment of additional VipA/VipB protomers to the newly formed ring. See also Figure S5.
In a fully assembled handshake domain of T6SS sheath the orientation of the third β-strand (counting from the lumen of the sheath) is parallel to the second β-strand but antiparallel in crystal structures of gp25 and its homolog (Figures 5A and S5). We detected significantly coevolving, and thus potentially interacting, residues only between the first two β-strands of gp25 (Figure S5). This suggests that similarly to the sheath handshake domain, only two β-strands of gp25 are present in a native assembly. The third β-strand of the gp25 could flip out of the domain and interact with yet another component of the baseplate. Therefore, gp25 could accept two additional β-strands from interacting proteins in a similar mechanism to the mechanism of sheath subunit interaction.
Interestingly, T6SS component TssE was suggested to be a homolog of gp25 (Leiman et al., 2009; Lossi et al., 2011), co-purifies with the T6SS sheath in V. cholerae (Basler et al., 2012), and is important for sheath assembly (Basler et al., 2012; Kapitein et al., 2013). We therefore speculate that the TssE protein could be the part of the T6SS baseplate that accepts VipA-β1 and VipB-β14 strands of the first sheath ring and thus initiates the sheath assembly and also anchors the sheath to the baseplate (Figure 5C). Moreover, TssG and TssK were shown to copurify with sheath in V. cholerae (Basler et al., 2012) and VipB was shown to interact with TssK in E. coli (Zoued et al., 2013), suggesting that additional proteins are likely involved in attaching the sheath to the baseplate as well. A stable attachment of a contractile sheath to a baseplate is likely crucial for generation of the force needed to deliver substrates across target cell membranes. The sheath has to bind to the baseplate as strongly as individual sheath rings bind together otherwise the sheath would likely detach from the baseplate during a rapid contraction.
Interactions in the handshake domain are critical for T6SS sheath assembly and dynamics
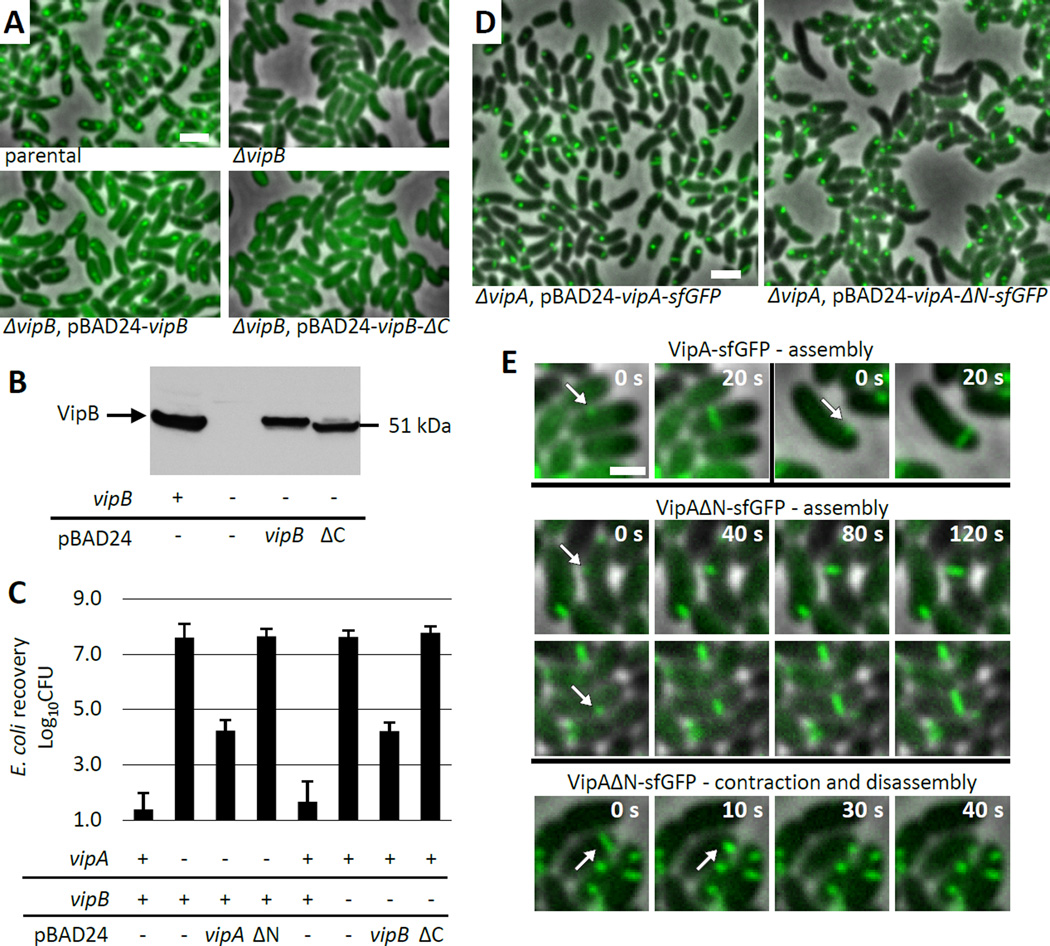
Our structural data indicate that interactions between β-strands in domain 1 are important for initiation of sheath polymerization, extension and potentially also for sheath contraction. To test this, we generated truncated versions of VipA and VipB lacking β1 and β14, respectively. In a background of a fully functional VipA-msfGFP chromosomal fusion we show that deletion of vipB abolishes sheath assembly and target cell killing (Figure 6A, C, Movie S2). As shown in Figure 6A–C and Movie S2, sheath assembly and target cell killing can be restored by a wild type level of expression of a full length VipB from a plasmid but not by a similar level of expression of a mutant lacking β14 (VipB-ΔC). This indicates that a connection between the sheath protomers on the same strand is essential for sheath assembly and T6SS function.
Figure 6. Handshake domain integrity is important for sheath dynamics.
(A) Sheath assembly was detected by fluorescence microscopy. Parental strain - V. cholerae with VipA-msfGFP fusion encoded in the native locus. Deletion of vipB gene was complemented by expression of either wt vipB or vipB lacking C-terminal β-strand (VipB-ΔC) from pBAD24 plasmid. 15×10 µm fields of cells are shown. Bar is 1 µm. See also Supplemental Movie S2. (B) Expression of VipB or VipB-ΔC was detected in the indicated strains prepared as for the imaging shown in (A) by western-blotting using VipB specific antibody. (C) Level of E. coli killing on a plate was measured for indicated strains after 3 h of incubation at 10:1 ratio. Presence or absence of vipA or vipB on the chromosome is indicated by “−” or “+”, respectively. Complementation was from pBAD24 plasmid carrying indicated genes. ΔN – vipA lacking N-terminal β-strand; ΔC – vipB lacking C-terminal β-strand. Data represented as mean +/− standard deviation. (D) Sheath assembly was detected by fluorescence microscopy. Parental strain - V. cholerae vipA−. Deletion of vipA gene was complemented by expression of either wt vipA or vipA lacking N-terminal β-strand (VipA-ΔN) from pBAD24 plasmid. 20×20 µm field of cells shown. Bar is 1 µm. See also Supplemental Movie S3. (E) Dynamics of sheath assembly for wild type VipA (2 examples, top) and VipA lacking N-terminal β-strand (VipA-ΔN) (2 examples, middle). An example of sheath contraction and disassembly shown for VipA-ΔN (bottom).
To assess the role of β1 strand of VipA we compared dynamics of a full length VipA-sfGFP expressed in vipA deletion background with dynamics of β1 strand deletion mutant (VipA-ΔN). As shown on Figure 6D and Movie S3, the wild type sheaths rapidly assemble and contract in almost all cells during 5 min. Sheaths with disrupted domain 1 are capable of assembling into structures resembling extended wild type sheaths but exhibit very little dynamics (Figure 6D, E, Movie S3). On average, we observe only one assembly event per approximately 500 cells over 5 minutes. Furthermore, the time of sheath assembly is increased for the VipA-ΔN sheath to about two minutes (Figure 6E) while the most of wild type sheaths fully assemble in 20 to 40 seconds under the same conditions. This clearly indicates that a fully assembled handshake domain is critical for efficient sheath assembly initiation and the fast assembly rate of the T6SS sheath. Interestingly, even though we inspected sheath dynamics in ~50,000 cells over 5 minutes, we identified only 5 examples of unambiguous sheath contraction and disassembly (one example is given in Figure 6E). This suggests that the ability to contract is preserved to some degree, but raises the possibility that domain 1 is involved in triggering sheath contraction in vivo. Alternatively, the rate of sheath assembly may play a role in triggering sheath contraction. Target cell killing in vipA deletion background was restored by expression of VipA-sfGFP but not by expression of VipA-ΔN-sfGFP mutant (Figure 6C) suggesting that mere ability to assemble sheaths is not sufficient for T6SS-dependent killing.
Concluding remarks
The unusual four-strand β-sheet handshake domain assembled from three molecules invites comparisons with other protein polymers. In most protein filaments that have been intensively studied, such as F-actin (von der Ecken et al., 2014; Galkin et al., 2014), microtubules (Alushin et al., 2014), bacterial flagellar filaments (Yonekura et al., 2003) or Type IV pili (Craig et al., 2006), subunits can be treated as compact and the assemblies are held together by the exclusion of solvent at the buried interfaces (Miller et al., 1987). In contrast to these, Type I pili from bacteria have a polymerization mechanism that involves an N-terminal extension of one subunit that becomes a β-strand within a β-sheet of an adjacent subunit (Waksman and Hultgren, 2009). This β-sheet formed by two subunits gives a remarkable stability to the filaments, and allows Type I pili to resist very large shear forces (Castelain et al., 2011; Miller et al., 2006). We expect that this architecture allows the sheath to transfer a large force and remain intact during its rapid contraction.
Experimental Procedures
Bacterial strains and DNA manipulations
V. cholerae 2740-80 parental and ΔvipA strains and the pBAD24-VipA-sfGFP plasmid were described previously (Basler et al., 2012). pBAD24-VipA-ΔN-sfGFP plasmid was created by replacing vipA gene in pBAD24-VipA-sfGFP plasmid with a gene lacking codons encoding 23 N-terminal amino acids using standard methods. V. cholerae 2740-80 vipA-msfGFP strain was created by replacing vipA on the chromosome with vipA-msfGFP fusion by sacB-mediated allelic exchange using the pWM91 suicide plasmid as described previously (Basler and Mekalanos, 2012; Basler et al., 2012; Bina and Mekalanos, 2001; Metcalf et al., 1996). msfGFP differs from previously used sfGFP by Val 206 to Lys substitution, which was previously described to reduce dimerization of GFP (Zacharias et al., 2002). Comparison of VipA-msfGFP to VipA-sfGFP expressed from pBAD24 plasmid in ΔvipA strain revealed no difference in dynamics of the fusion proteins (data not shown). The linker between VipA and msfGFP was 3xAla 3xGly as used previously on pBAD24 plasmid (Basler et al., 2012). To limit effects of the fusion gene on the downstream genes in the T6SS locus we added last 21 bp of vipA at the end of vipA-msfGFP. V. cholerae 2740-80 vipA-msfGFP ΔvipB strain was created by replacing vipB with a gene encoding “MMSTTEKGRLDQA” peptide (first seven and last six residues of vipB fused in frame) by allelic exchange as described above and was done previously (Basler et al., 2012). Standard techniques were used to clone a PCR amplified vipB or the first 477 codons of vipB to pBAD24 plasmid (Guzman et al., 1995) to generate pBAD24-VipB and pBAD24-VipB-ΔC plasmids, respectively. All PCR generated products were verified by sequencing. Plasmids were transformed to V. cholerae by electroporation. Gentamicin resistant E. coli MG1655 strain was used in bacterial killing assays. Strain list provided as supplementary Table S3.
Antibiotic concentrations used were streptomycin (100 µg/ml), ampicillin (200 µg/ml), gentamicin (15 µg/ml). Luria-Bertani (LB) broth was used for all growth conditions. Liquid cultures were grown aerobically at 37 °C.
Fluorescence microscopy
Procedures similar to those described previously (Basler et al., 2012) were used to detect fluorescence signal in V. cholerae. Overnight cultures of V. cholerae carrying pBAD24-vipA-sfGFP, pBAD24-vipA-ΔN-sfGFP, pBAD24-vipB, or pBAD24-vipB-ΔC were washed by LB and diluted 50× into fresh LB supplemented with ampicillin, streptomycin and 0.003% arabinose for VipA and 0.006% arabinose for VipB and cultivated for 2.5 – 3.0 hours to optical density at 600nm (OD) of about 0.8 – 1.2. Strains without plasmid were grown similarly but no ampicillin and arabinose was added. Cells from 100 µL of the culture were re-suspended in 5 – 10 µL of fresh LB (to OD ~10), spotted on a thin pad of 1% agarose in LB, and covered with a glass cover slip. Cells were immediately imaged at room temperature using an objective heated to 37 °C. Microscope configuration similar to the one described previously (Basler et al., 2013) was used: Nikon Ti-E inverted motorized microscope with Perfect Focus System and Plan Apo 100× Oil Ph3 DM (NA 1.4) objective lens. SPECTRA × light engine (Lumencore), ET-GFP (Chroma #49002) filter set was used to excite and filter fluorescence. sCMOS camera pco.edge 4.2 (PCO, Germany) (pixel-size 65 nm) and VisiView software (Visitron Systems, Germany) was used to record images. Fiji (Schindelin et al., 2012) was used for all image analysis and manipulations as described previously (Basler et al., 2013). Contrast on compared sets of images was adjusted equally. All imaging experiments were performed with three biological replicates.
Bacterial killing assay
V. cholerae 2740-80 strains and E. coli MG1655 strain were incubated overnight at 37 °C in LB supplemented with appropriate antibiotics. Cultures were washed in fresh LB, diluted 100× and grown to OD 0.8–1.2 in presence of appropriate antibiotics and 0.01% arabinose for strains with pBAD24 plasmids. Cells were washed and mixed at final OD of ~10 in 10:1 ratio (V. cholerae to E. coli) as specified and 5 µL of the mixture was spotted on a dry LB agar plate containing 0.01% arabinose but no antibiotics. After 3 hours bacterial spots were cut out and the cells were re-suspended in 0.5 mL LB. The cellular suspension was serially diluted (1:10) in LB and 5 µL of the suspensions were spotted on selective plates (gentamicin for E. coli, and streptomycin for V. cholerae). Colonies were counted after ~16 h incubation at 30 °C. Three biological replicates were analyzed.
VipA/VipB sheath purification
Sheath was purified similarly to a previous method (Basler et al., 2012). An overnight culture of flgG in-frame deletion mutant of the parental V. cholerae 2740-80 strain (Basler et al., 2012) was diluted 1:200 into 1000 mL of fresh LB and then shaken at 37 °C for 2.5–3.0 hours to reach an OD of 1.0–1.5. Cells were cooled on ice, centrifuged for 10 min 7000×g and lysed in 50 mL lysis buffer (150 mM NaCl, 20 mM Tris, pH 8, lysozyme 200 µg/mL, DNAse I 50 µg/mL, 5 mM EDTA, 0.1% SDS, 0.5% Triton X-100). Cell lysis was complete after 5–10 minutes incubation at 37 °C. To activate DNAse to cleave DNA, MgCl2 was added to 10mM final concentration and after 2–5min incubation at 37 °C EDTA was added to reach 15 mM final concentration. Cell debris was removed by centrifugation for 20 min at 10.000×g. Cleared lysates were subjected to ultraspeed centrifugation at 150,000×g for 1 h at 4 °C. Pellets were re-suspended in 1 mL in TND buffer (150mM NaCl, 20mM Tris, pH 8, 0.5% Triton-100, 0.1% SDS) and insoluble material was removed by centrifugation for 1min at 10.000×g. Supernatant was diluted to 50 mL in TND buffer and subjected to ultraspeed centrifugation at 150,000×g for 1 h at 4 °C. Pellet was washed with 2mL of phosphate buffered saline (PBS) and resuspended in 2 mL PBS. Insoluble material was removed by centrifugation for 1min at 10.000×g. Supernatant was diluted to 50mL by PBS and subjected to ultraspeed centrifugation at 150,000×g for 1 h at 4 °C. Pellet was washed by 2mL of PBS, resuspended in 1 mL PBS and insoluble material was removed by centrifugation for 1min at 10.000×g. Supernatant contained pure sheath. Purity was assessed by Coomassie stained gel and protein concentration was measured by standard approaches.
Peptide specific antibodies
Antigen purified rabbit polyclonal antibodies raised against VipB peptide QENPPADVRSRRPL were obtained commercially (GenScript, USA). Specificity of the antibodies was tested on V. cholerae strains expressing or lacking vipB.
Cell fractionation and immunoblot analysis
Cells from 250 µL culture prepared for imaging as described above were re-suspended in 100 µL PBS and subjected to sonication (20 cycles, 100 % amplitude, 0.5 s cycle) (UIS215V Hielscher Ultrasonics GmbH, Germany). Then 10 µL of 10 % SDS was added and the sample was sonicated as before. Samples were incubated for 10 min at 95°C, centrifuged and 17 µl were mixed with 7 µl 4× NuPAGE LDS Sample Buffer (Life Technologies) and 2 µl 1 M DTT. Samples were heated again for 10 min at 72°C, centrifuged and loaded on 4–12 % pre-cast polyacrylamide gels (Life Technologies) and transferred to nitrocellulose membrane (Amersham Biosciences, UK). Membrane was blocked by 5 % milk in Tris buffered saline (pH 7.4) containing Tween 0.1 % (TBST), incubated with primary peptide antibody for 16 hours at 4 °C, washed with TBST, incubated for 1.5 h with horseradish peroxidase labeled anti-rabbit antibody (Jackson Lab), washed with TBST, and peroxidase was detected by LumiGLO Chemiluminescent Substrate (Cell Signaling Technology, Inc., USA).
Cryo-electron microscopy
Sample was placed on holey carbon grids (Quantifoil GmbH, Germany) and plunge-frozen into liquid ethane cooled down to LN2 temperature using a Vitrobot MK4 (FEI Corp, The Netherlands). Frozen grids were stored in LN2 and directly observed in a Titan Krios (FEI Corp, The Netherlands) operated at 300 kV and quipped with a K2 Summit direct electron detector (Gatan, Pleasanton, CA). All images were acquired in a single two-day session at a defocus range of 0.5 – 1.5 µm. Images were recorded in dose fractionation mode, with a dose rate 3–4 e−/pix/second, exposures per image sub-frames between 1 and 1.5 e−/ Å2 and a cumulative dose for the entire image series of 30 e−/ Å2. The final pixel size for the resulting 4k images was 1.0 Å/pix at the sample level.
Image processing and 3D reconstruction
Alignment for beam induced movement was performed by 2dx_automator (Scherer et al., 2014) that provides on-the-fly drift-correction based on the algorithm implemented by Li et al. (Li et al., 2013). 4k images recorded as movie-data in ‘counting mode’ were drift-corrected with the algorithm by Li et al (Li et al., 2013). The quality of the images drastically improved after drift correction especially at high resolution (Figure S1A). Drift on the order of 10 Å could be fixed and resulted in Thon rings up to 3–3.5 Å. All recorded frames up to 30 e−/ Å2 were used, and no weighting was performed. From the recorded ~ 250 images the best 77 were selected based on ice thickness and the quality of the Thon rings. CTF determination was performed by CTFFIND3 (Mindell and Grigorieff, 2003). This led to exclusion of one image, due to a poor fit between the theoretical and observed Thon ring pattern. The images were then multiplied by the estimated CTF in SPIDER to both correct phases and to improve the SNR. Filaments were boxed using the e2helixboxer function within EMAN2 (Tang et al., 2007), using a box width of 600 pixels for the initial alignment and 384 pixels for the final reconstruction. The SPIDER software package (Frank et al., 1996) was used for most subsequent steps. From the long boxes, overlapping segments were cut that were 600 pixels long with a shift of 30 pixels between boxes, where the shift (yielding 95% overlap) was chosen to be ~1.5 times the axial rise per subunit. A total of 10,203 segments were obtained. The segments were then padded to 600 × 600 pixels and decimated to 200 × 200 pixels size (3 Å/pix) for initial alignments and reconstruction using IHRSR (Egelman, 2000). Once these were reconstructed, the original images were subsequently decimated to 300 × 300 pixels for further processing that included out-of-plane tilts. Finally, the initial boxes were windowed to 384 × 384 pixels for several cycles of IHRSR with 1.0 Å/pix until convergence. At the end of each iteration helical symmetry with a rise of 21.8 Å and a rotation of 29.4 degrees with C6 symmetry was applied. Class averages for Figures 1D and S1F were generated using Spring (Desfosses et al., 2014).
To test reproducibility of the atomic model building, a completely independent EM-density map was generated starting from the initial micrographs followed by independent particle picking using the e2helixboxer function within EMAN2 (Tang et al., 2007). Square boxes of 400 Å length (1 Å/pix) were picked with a step of 30 Å. Iterative real space helical reconstruction (Egelman, 2000) was performed with Spring (Desfosses et al., 2014) starting with a featureless cylinder as an initial model. At the end of each iteration C6 symmetry was applied to the reference. All segments were processed as one dataset, resolution estimated by Fourier Shell Correlation between the half-datasets was 4.3 Å (FSC = 0.5).
Atomic model building
Model building was done de novo, with initial models of a single subunit built first, and then the system was refined in a symmetrical complex with all the interacting subunits present. A model of a single-subunit VipA/VipB was manually built in Coot (Emsley et al., 2010) guided by an initial partial model from Buccaneer (Cowtan, 2006), which placed a total of 513 residues into the density map. In parallel, automated model building was carried out independently using a newly developed approach (Wang et al., in press). The automated method uses sequence-derived backbone conformations with side-chain density-fit to accurately assign sequence into density maps. Starting with a map segmented to containing a single subunit, the computational method was able to place 466 residues into the density.
The two independently derived models showed reasonably good agreement: 394 residues were assigned in both models with a Cα RMSD of 1.05 Å. However, there were parts of the protein assigned in each model that were unassigned in the other. Thus, to build and refine the final model, we used RosettaCM (Song et al., 2013), a comparative modeling protocol that assembles protein structures by recombining portions of several models; in this case, the inputs were the two independently traced models. RosettaCM was guided by experimental density data, with agreement to the density map as an additional score term while building and refining models. A total of 1000 models were generated, and a best model was selected based on the all-atom energy plus the “fit to density” energy.
Using this model, a final refinement step was carried out in the context of the symmetrical assembly, improving model geometry and relieving clashes at the symmetric interfaces (DiMaio et al., in press). The final model shows very good agreement to the density, with 504 of 558 traced residues matching the map with real-space correlations of 0.60 or greater (using density_tools in Rosetta), and very good model geometry, with 0.36% Ramachandran outliers, 0% rotamer outliers, a Molprobity clash score of 2.15, and an overall Molprobity score of 1.38 (Chen et al., 2010).
To test for overfitting during model building we uniformly perturbed the final model and refined it against the independently generated EM map. A long refinement cycle (1000 cycles of backbone rebuilding) was used to ensure the refined model is unbiased from the model fit to the original reconstruction. The resulting model had 0.34 Å Cα RMSd to the original model.
Atomic B-factors were capped to 600 for heavy atoms and to 720 for H atoms. Methionine in position 1 of VipA was not included in the model due to a lack of EM density and evidence from mass spectrometry analysis of isolated sheath (data not shown) that it is not present on the N-terminus.
Molecular analysis
Interaction energy was calculated using PISA (Krissinel and Henrick, 2007). Secondary structure prediction for the Figures S2A,B was performed by Jnet (Cole et al., 2008). Structural alignments were performed by RaptorX (Wang et al., 2013), the RMSD presented in the text are calculated from these alignments. Structural homologs were found using PDB Structure Navigator http://pdbj.org/strucnavi.
Evolutionary constraints
Evolutionary constraints were generated by the Gremlin server (http://gremlin.bakerlab.org/) (Ovchinnikov et al., 2014) or FreeContact software (Kaján et al., 2014). All reliable constraints with scores over 1.5 are listed in Table S1. The distance in 3D was measured between the weighted centers of mass of the contacting residues. The distance was also estimated between the contacting residues in the neighboring protomers and in case the inter-protomer distance was less than intra-protomer distance the inter-protomer distance was used in Table S1. This was implemented using Matlab (Mathworks).
Coloring of the EM maps was done with Dynamo package for electron tomographic image processing (Castaño-Díez et al., 2012). The visualization of atomic models, evolutionary constraints, and rendering of the Movie S1 was performed in UCSF Chimera (Pettersen et al., 2004).
Supplementary Material
Acknowledgements
We thank Mihai Ionescu for an excellent technical assistance in preparation of the sheath sample. We thank Kenneth Goldie and Bill Anderson for expert assistance with cryo-EM; Venkat Dandey for support with software; Grant support: NIH EB001567 (to EHE), Swiss systems biology initiative SystemsX.ch grant CINA, NIH R01GM092802 (RYW and DB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
The EM map was deposited to EMDB (http://www.emdatabank.org/) with an accession number EMD-2699. Electron micrographs used for processing were deposited to EMPIAR (http://www.ebi.ac.uk/pdbe/emdb/empiar/) with an accession number EMPIAR-10019. Atomic coordinates were deposited to PDB (www.rcsb.org) with an accession number XXX.
Author contributions
MK performed cryo-EM analysis, built a manual atomic model with TM, interpreted the data, and wrote the manuscript. RW, FD, DB performed automated model building and refinement, contributed to data interpretation. MBr generated new plasmids and strains, imaged sheath dynamics, performed bacterial killing assays, analyzed protein levels by western-blot, and contributed to data analysis and interpretation. SS contributed to data collection and processing. TM built a manual atomic model with MK. HS supervised data collection and contributed essential analytical tools. EE performed cryo-EM image analysis and made critical contributions to data interpretation. MBa analyzed and interpreted the data, and wrote the manuscript. All authors participated in writing the manuscript.
References
- Aksyuk AA, Leiman PG, Kurochkina LP, Shneider MM, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 2009a;28:821–829. doi: 10.1038/emboj.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksyuk AA, Leiman PG, Shneider MM, Mesyanzhinov VV, Rossmann MG. The structure of gene product 6 of bacteriophage T4, the hinge-pin of the baseplate. Structure. 2009b;17:800–808. doi: 10.1016/j.str.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Aksyuk AA, Kurochkina LP, Fokine A, Forouhar F, Mesyanzhinov VV, Tong L, Rossmann MG. Structural conservation of the myoviridae phage tail sheath protein fold. Structure. 2011;19:1885–1894. doi: 10.1016/j.str.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, Nogales E. High-Resolution Microtubule Structures Reveal the Structural Transitions in αβ-Tubulin upon GTP Hydrolysis. Cell. 2014;157:1117–1129. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A, Brown A, Bai X, Llácer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SHW, Ramakrishnan V. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Mekalanos JJ. Type 6 secretion dynamics within and between bacterial cells. Science. 2012;337:815. doi: 10.1126/science.1222901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Ho BT, Mekalanos JJ. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell. 2013;152:884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina JE, Mekalanos JJ. Vibrio cholerae tolC is required for bile resistance and colonization. Infect. Immun. 2001;69:4681–4685. doi: 10.1128/IAI.69.7.4681-4685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bönemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 2009;28:315–325. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet YR, Espinosa L, Harchouni S, Mignot T, Cascales E. Imaging type VI secretion-mediated bacterial killing. Cell Rep. 2013;3:36–41. doi: 10.1016/j.celrep.2012.11.027. [DOI] [PubMed] [Google Scholar]
- Brunet YR, Henin J, Celia H, Cascales E. Type VI secretion and bacteriophage tail tubes share a common assembly pathway. EMBO Rep. 2014;15:315–321. doi: 10.1002/embr.201337936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño-Díez D, Kudryashev M, Arheit M, Stahlberg H. Dynamo: a flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. J. Struct. Biol. 2012;178:139–151. doi: 10.1016/j.jsb.2011.12.017. [DOI] [PubMed] [Google Scholar]
- Castelain M, Ehlers S, Klinth J, Lindberg S, Andersson M, Uhlin BE, Axner O. Fast uncoiling kinetics of F1C pili expressed by uropathogenic Escherichia coli are revealed on a single pilus level using force-measuring optical tweezers. Eur. Biophys. J. 2011;40:305–316. doi: 10.1007/s00249-010-0648-1. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Desfosses A, Ciuffa R, Gutsche I, Sachse C. SPRING - an image processing package for single-particle based helical reconstruction from electron cryomicrographs. J. Struct. Biol. 2014;185:15–26. doi: 10.1016/j.jsb.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Von der Ecken J, Müller M, Lehman W, Manstein DJ, Penczek PA, Raunser S. Structure of the F-actin--tropomyosin complex. Nature. 2014 doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- Egelman EH. Reconstruction of helical filaments and tubes. Meth. Enzymol. 2010;482:167–183. doi: 10.1016/S0076-6879(10)82006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faruqi AR, Cattermole DM, Henderson R, Mikulec B, Raeburn C. Evaluation of a hybrid pixel detector for electron microscopy. Ultramicroscopy. 2003;94:263–276. doi: 10.1016/s0304-3991(02)00336-4. [DOI] [PubMed] [Google Scholar]
- Fokine A, Zhang Z, Kanamaru S, Bowman VD, Aksyuk AA, Arisaka F, Rao VB, Rossmann MG. The molecular architecture of the bacteriophage T4 neck. J. Mol. Biol. 2013;425:1731–1744. doi: 10.1016/j.jmb.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Vos MR, Schröder GF, Egelman EH. Near-Atomic Resolution for One State of F-Actin. Structure. 2014 doi: 10.1016/j.str.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaján L, Hopf TA, Kalaš M, Marks DS, Rost B. FreeContact: fast and free software for protein contact prediction from residue co-evolution. BMC Bioinformatics. 2014;15:85. doi: 10.1186/1471-2105-15-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein N, Bonemann G, Pietrosiuk A, Seyffer F, Hausser I, Locker JK, Mogk A. ClpV recycles VipA/VipB tubules and prevents non-productive tubule formation to ensure efficient type VI protein secretion. Mol Microbiol. 2013;87:1013–1028. doi: 10.1111/mmi.12147. [DOI] [PubMed] [Google Scholar]
- Kostyuchenko VA, Chipman PR, Leiman PG, Arisaka F, Mesyanzhinov VV, Rossmann MG. The tail structure of bacteriophage T4 and its mechanism of contraction. Nat. Struct. Mol. Biol. 2005;12:810–813. doi: 10.1038/nsmb975. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Kube S, Kapitein N, Zimniak T, Herzog F, Mogk A, Wendler P. Structure of the VipA/B Type VI Secretion Complex Suggests a Contraction-State-Specific Recycling Mechanism. Cell Rep. 2014 doi: 10.1016/j.celrep.2014.05.034. [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W. Biochemistry. The resolution revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- Leiman PG, Shneider MM. Contractile tail machines of bacteriophages. Adv. Exp. Med. Biol. 2012;726:93–114. doi: 10.1007/978-1-4614-0980-9_5. [DOI] [PubMed] [Google Scholar]
- Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell. 2004;118:419–429. doi: 10.1016/j.cell.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossi NS, Dajani R, Freemont P, Filloux A. Structure-function analysis of HsiF, a gp25-like component of the type VI secretion system, in Pseudomonas aeruginosa. Microbiology (Reading, Engl.) 2011;157:3292–3305. doi: 10.1099/mic.0.051987-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossi NS, Manoli E, Förster A, Dajani R, Pape T, Freemont P, Filloux A. The HsiB1C1 (TssB-TssC) complex of the Pseudomonas aeruginosa type VI secretion system forms a bacteriophage tail sheathlike structure. J. Biol. Chem. 2013;288:7536–7548. doi: 10.1074/jbc.M112.439273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schröder GF, Fitzgerald KA, Wu H, Egelman EH. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf WW, Jiang W, Daniels LL, Kim SK, Haldimann A, Wanner BL. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- Miller E, Garcia T, Hultgren S, Oberhauser AF. The mechanical properties of E. coli type 1 pili measured by atomic force microscopy techniques. Biophys. J. 2006;91:3848–3856. doi: 10.1529/biophysj.106.088989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Lesk AM, Janin J, Chothia C. The accessible surface area and stability of oligomeric proteins. Nature. 1987;328:834–836. doi: 10.1038/328834a0. [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov S, Kamisetty H, Baker D. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. Elife. 2014;3:e02030. doi: 10.7554/eLife.02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pietrosiuk A, Lenherr ED, Falk S, Bönemann G, Kopp J, Zentgraf H, Sinning I, Mogk A. Molecular basis for the unique role of the AAA+ chaperone ClpV in type VI protein secretion. J. Biol. Chem. 2011;286:30010–30021. doi: 10.1074/jbc.M111.253377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S, Kowal J, Chami M, Dandey V, Arheit M, Ringler P, Stahlberg H. 2dx_automator: implementation of a semiautomatic high-throughput high-resolution cryo-electron crystallography pipeline. J. Struct. Biol. 2014;186:302–307. doi: 10.1016/j.jsb.2014.03.016. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, DiMaio F, Wang RY-R, Kim D, Miles C, Brunette T, Thompson J, Baker D. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Fernández IS, Scheres SHW, Hegde RS. Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell. 2014;157:1632–1643. doi: 10.1016/j.cell.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman G, Hultgren SJ. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat. Rev. Microbiol. 2009;7:765–774. doi: 10.1038/nrmicro2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ma J, Peng J, Xu J. Protein structure alignment beyond spatial proximity. Sci Rep. 2013;3:1448. doi: 10.1038/srep01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- Zoued A, Durand E, Bebeacua C, Brunet YR, Douzi B, Cambillau C, Cascales E, Journet L. TssK is a trimeric cytoplasmic protein interacting with components of both phage-like and membrane anchoring complexes of the type VI secretion system. J. Biol. Chem. 2013;288:27031–27041. doi: 10.1074/jbc.M113.499772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.