Abstract
Metanephric adenoma (MA) is a rare benign renal tumor comprised of a neoplastic proliferation of primitive metanephric tubular cells. A previous study identified BRAF V600E mutations in approximately 90% of MA and found that similar BRAF exon 15 mutations are exceedingly rare in other common renal tumors, including renal cell carcinoma (RCC) and oncocytoma. A recent follow-up study has validated mutation-specific immunohistochemistry (IHC) for detection of BRAF V600E mutations in a small cohort of MA. Here, we extend these findings to a larger, independent cohort of MA, demonstrating an overall 88% sensitivity and 100% specificity for BRAF V600E IHC. In addition, we report two cases of MA with novel BRAF exon 15 mutations, including a V600D missense mutation and a compound V600D and K601L missense mutation. Finally, we evaluate BRAF V600E IHC in a large TMA cohort of common renal tumors and find no significant expression in several RCC subtypes. These data support a role for BRAF V600E IHC in diagnostically challenging cases of MA and expand the spectrum of BRAF exon 15 mutations in this uncommon but unique renal neoplasm.
Keywords: BRAF, metanephric adenoma, immunohistochemistry, V600E, Sanger sequencing
Introduction
Metanephric adenoma (MA) is a benign, often asymptomatic and incidentally identified renal tumor comprised of primitive metanephric tubular cells1-3. Patients commonly present in the fifth to sixth decade of life, and there is a slight female preponderance (F:M = approximately 2:1). Grossly, MA is usually a solid, solitary, unilateral mass that lacks a true fibrous capsule. On average, these tumors measure 5.5 cm in greatest dimension, however, they may be very small (less than 0.5 cm) or large (greater than 10 cm). Microscopically, MA usually appears solid at low magnification, although areas of cystic degeneration are sometimes present1-3. On higher magnification, these tumors consist of primitive metanephric tubular epithelial cells with scant cytoplasm and monotonous round to oval nuclei with fine chromatin and without nucleoli; these cells are usually arranged in small acinar structures, although areas with papillary, tubular, and glomeruloid growth patterns may be identified. Generally, there is minimal admixed paucicellular or hyalinized stroma, but cases may also demonstrate regressive changes, including dense sclerosis, dystrophic calcification, and/or psammomatous calcification. Mitotic activity is usually very low or absent in MA, and necrosis is very rare.
In the majority of cases, MA can be diagnosed on routine hematoxylin and eosin (H&E) stain; however, the differential diagnosis includes the solid variant of papillary renal cell carcinoma (RCC), as well as epithelial predominant Wilms tumor (WT) – both of which require definitive treatment1-3. In challenging cases, particularly core needle biopsy specimens, immunohistochemistry (IHC) may be helpful3-5. MA usually express WT1 and CD57 and can demonstrate focal CK7 expression in areas with elongated tubules3, 6.
A recent study by Choueiri et al. has begun to shed light on the molecular underpinnings of this renal tumor, demonstrating that approximately 90% of MA harbor BRAF V600E mutations7. BRAF encodes a serine/threonine-specific protein kinase upstream of the MAPK/ERK signaling pathway, and somatic activating BRAF mutations have been identified in a wide variety of common human malignancies, including melanoma, papillary thyroid carcinoma, and colonic adenocarcinoma8, 9. BRAF exon 15 mutations, including the V600E missense mutation, are frequently detected in a range of benign and malignant human tumors10-15, however, BRAF mutations in common non-MA renal tumors (i.e., RCC, oncocytoma, WT) are either very infrequent (less than 1%) or absent7, 16, 17. A mutation-specific antibody against the BRAF V600E protein product has been recently validated for IHC detection of BRAF V600E mutations in situ and utilized successfully in a variety of human tumor types10, 18-26. In a follow-up study to their work on BRAF V600E mutations in MA, Pinto et al. demonstrated that six of six (100%) MA cases, including five with confirmed BRAF V600E mutations, exhibited BRAF V600E expression by mutation-specific IHC27. In contrast, less than 1% of cases from a large TMA cohort of common renal tumors demonstrated BRAF V600E expression.
In this study, we extend the findings reported in Pinto et al. to a larger, independent cohort of MA and report two cases of BRAF V600E-negative MA with novel BRAF exon 15 mutations. Finally, we evaluate BRAF V600E protein expression by mutation-specific IHC in a large TMA cohort of common renal tumors.
Materials and Methods
Identification of MA cases
This study was approved by the Institutional Review Board at the University of Michigan Medical School. A comprehensive retrospective search of the University of Michigan Health System (UMHS) pathology records database was performed to identify all available MA cases between 1985 and 2014, and a total of eleven such cases were available for the purposes of this study. H&E stained slides from all cases were reviewed by two study pathologists (A.M.U. and R.M), and representative formalin-fixed, paraffin-embedded (FFPE) tissue blocks were selected for BRAF V600E IHC and BRAF exon 15 sequencing.
Renal tumor tissue microarray (TMA) construction
A TMA representing common renal tumor types from eighty-six unique patients was constructed with FFPE tissue from partial or total nephrectomy specimens retrieved from the UMHS pathology specimen archive. This TMA included specimens from various renal tumor types, including: chromophobe RCC (n = 26); oncocytoma (n = 20); papillary RCC (n = 20); clear cell RCC (n = 16); RCC, unclassified (n = 2); clear cell papillary RCC (n = 1); and, Xp11 translocation-associated RCC (n = 1). The tumor samples were represented on this TMA in at least triplicate cores, and samples of benign renal parenchyma from ten patients served as internal controls.
BRAF V600E IHC
Whole sections were obtained from MA FFPE tissue blocks for BRAF V600E IHC (clone VE1; pre-dilute; 790-4855; Ventana Medical Systems, Tucson, AZ), which was performed using a BenchMark ULTRA automated stainer and the ultraView Universal DAB Detection Kit (Ventana Medical Systems) by the CLIA-certified clinical IHC laboratory of the Department of Pathology at UMHS. This anti-BRAF V600E antibody is a mouse monoclonal antibody generated against a synthetic peptide representing the mutated BRAF V600E amino acid sequence (from amino acid 596 to 606; GLATEKSRWSG)18, 20. Whole sections from a melanoma case with a confirmed BRAF V600E mutation were included as batch positive controls for BRAF V600E IHC; consistent with previously published data (as well as practical experience in the UMHS clinical IHC laboratory), the batch positive control demonstrated diffuse, moderate to strong cytoplasmic staining in melanoma cells but negative or weak staining in adjacent non-neoplastic tissue10, 18-27. BRAF V600E IHC results for all MA cases were reviewed and scored independently by two study pathologists (A.M.U. and R.M.), who were blinded to both the general and specific BRAF exon 15 genomic sequencing results; only cases with diffuse, moderate to strong cytoplasmic staining in tumor cells were recorded as BRAF V600E IHC positive. For assessment of the renal tumor TMA, only cases with cores showing diffuse, moderate to strong cytoplasmic staining in tumor cells were recorded as BRAF V600E IHC positive.
BRAF exon 15 genomic sequencing
For each MA case, tumor genomic DNA was isolated from FFPE tissue blocks using the QIAamp DNA FFPE Tissue Kit (Qiagen, Venlo, Netherlands), according to the manufacturer's protocol. Commercially available male human genomic DNA (G1471; Promega, Madison, WI) served as a wild type BRAF exon 15 control, while genomic DNA isolated from SK-MEL-5 cells (HTB-70; ATCC, Manassas, VA) using the DNeasy Blood & Tissue Kit (Qiagen) served as a positive BRAF V600E control. BRAF exon 15 was PCR amplified from 50 nanograms of genomic DNA using flanking sequence-specific primers (forward: 5′-M13-TTTGTGAATACTGGGAACTATGAAA-3′; and, reverse: 5′-TCATCCTAACACATTTCAAGCC-3′) and HotStarTaq DNA polymerase (Qiagen). The PCR conditions were as follows: initial denaturation at 95°C for 15 minutes; 45 cycles of 94°C for 60 seconds, 50°C for 60 seconds, and 72°C for 60 seconds; and, final extension at 72°C for 7 minutes. After confirming the expected amplicon size by standard agarose gel electrophoresis, all PCR products were incubated with ExoSAP-IT (Affymetrix, Santa Clara, CA) prior to bi-directional Sanger sequencing by the University of Michigan DNA Sequencing Core using the M13 forward and BRAF exon 15 PCR reverse primers. The resulting chromatograms were analyzed with Sequencher software, version 4.5 (Gene Codes, Ann Arbor, MI), and compared to reference sequence for BRAF exon 15 (NM_004333.4).
BRAF Exon 15 subcloning
BRAF exon 15 PCR products from samples with complex somatic aberrations were resolved by standard gel electrophoresis, and the excised DNA fragments were purified using the S.N.A.P. Gel Purification Kit (Life Technologies, Waltham, MA). The purified PCR products were subcloned into the pCR4-TOPO vector using the TOPO TA Cloning Kit (Life Technologies) and transformed into Mach1 cells (Life Technologies) under kanamycin selection. Plasmid DNA was isolated from ten independent bacterial colonies and subjected to Sanger sequencing with M13 forward or reverse primers by the University of Michigan Sequencing Core. The resulting chromatograms were analyzed as described above.
Results
Clinicopathologic features of a retrospective MA cohort
Eleven cases of MA from the UMHS pathology records database were available for this study (Table 1). These patients included six women and five men, and the age at diagnosis ranged from 16 to 84 years (median = 45 years). Six of the tumors arose in the right kidney, while four developed in the left kidney; one MA occurred within the left side of a horseshoe kidney. One patient had polycythemia at the time of presentation, consistent with prior estimates of the incidence in patients with MA1. Eight of the cases were from partial or total nephrectomy specimens, while the other three were from core needle biopsies. The maximum tumor dimension ranged from 1.3 to 5.1 cm (median = 2.7 cm). These tumors demonstrated the typical histomorphologic spectrum of MA, including varying degrees of hyalinization, psammomatous calcification, and papillary, tubular, or glomeruloid growth patterns (see Table 1 for details).
Table 1.
Clinicopathologic features of a retrospective MA cohort.
| Gender | Age (years) | Specimen Type | Lat | Size (cm) | Histologic Features | BRAF V600E IHC | BRAF exon 15 mutation(s) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | T | G | C | H | PC | Ne | S | |||||||
| M | 16 | Nep | L | 4.4 | Fo | Fo | Fo | N | N | NFo | N | N | Pos | V600E |
| F | 32 | Nep | R | 1.3 | NFo | NFo | Fo | N | Fo | NFo | Fo | N | Neg | V600D |
| M | 32 | Nep | L | 3.2 | NFo | Fo | Fo | N | NFo | E | N | Fo | Neg | Wild type |
| F | 41 | Nep | R | 3.6 | Fo | Fo | Fo | N | F | Fo | N | N | Pos | V600E |
| F | 43 | Nep | R | 5.1 | Fo | Fo | Foo | N | E | E | N | N | Neg | V600E |
| F | 45 | Nep | L | 2.0 | NFo | NFo | Fo | Fo | NFo | E | N | N | Pos | V600E |
| F | 55 | Nep | L | 2.3 | Fo | N | N | Fo | Fo | E | N | N | Pos | V600E |
| M | 60 | CNB | L | 2.7 | N | Fo | Fo | N | N | N | N | N | Pos | V600E |
| M | 62 | CNB | R | 1.8 | N | Fo | N | N | N | N | N | N | Neg | V600D, K601L |
| F | 74 | CNB | L | 2.5 | N | N | Fo | N | N | N | N | N | Pos | V600E |
| M | 84 | Nep | R | 3.4 | Fo | Fo | Fo | Fo | E | N | N | Fo | Pos | V600E |
M = male, F = female, Nep = partial or total nephrectomy, CNB = core needle biopsy, Lat = laterality, L =left, R = right, P = papillary, T = tubular, G = glomeruloid, C = cystic, H = hyalinization, PC = psammomatous calcification, Ne = necrosis, S = sclerosis, N = no, Fo = focal, NFo = non-focal, E = extensive, IHC = immunohistochemistry, Pos = positive, Neg = negative
BRAF V600E IHC in a retrospective cohort of MA
Given the reported high frequency of BRAF V600E mutations in MA7, as well as the high sensitivity and specificity of mutation-specific IHC for detecting these mutations in a wide variety of neoplasms (including MA)10, 18, 19, 21, 22, 24-27, we sought to examine BRAF V600E IHC in our retrospective MA cohort. Seven (64%) of the cases in our cohort demonstrated diffuse, moderate to strong cytoplasmic BRAF V600E IHC staining (Figure 1), while the remaining four cases showed negative to weak cytoplasmic BRAF V600E IHC staining (Figure 2). Of note, in the vast majority of cases, the adjacent benign renal parenchyma showed variable weak cytoplasmic and weak to moderate nuclear staining. Overall, the tumor areas in the IHC positive cases demonstrated unequivocal, diffuse moderate to strong cytoplasmic expression of mutated BRAF protein.
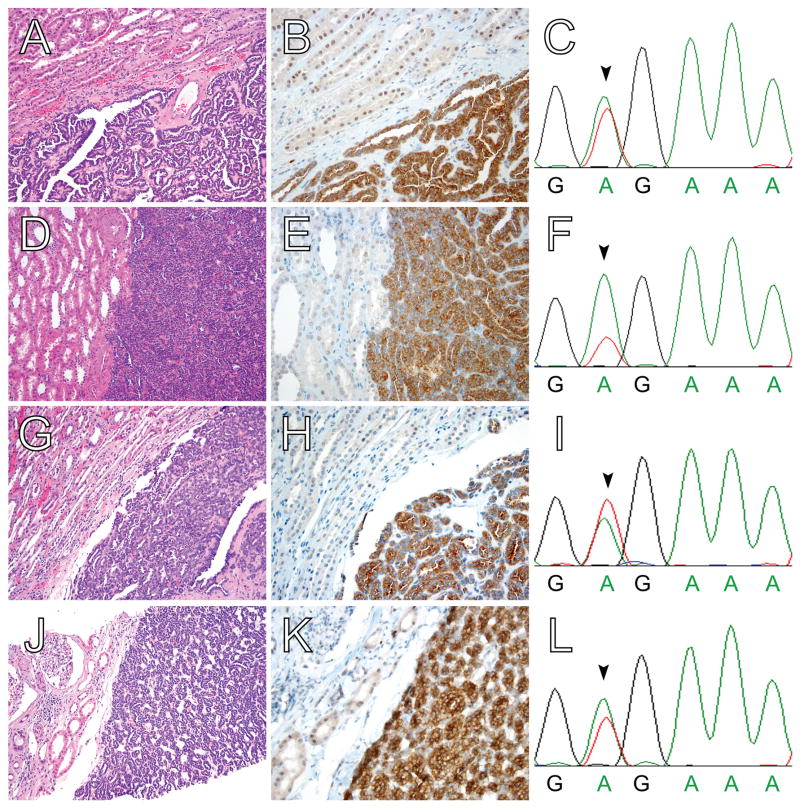
Figure 1. BRAF V600E protein expression in MA, as detected by IHC with a mutation-specific antibody, is highly correlated with the presence of BRAF V600E mutation by Sanger sequencing.
(A,D,G,J) H&E, (B,E,H,K) BRAF V600E IHC, and (C,F,I,L) BRAF exon 15 genomic sequencing of four MA cases. All four cases show the typical histomorphologic features of MA and demonstrate diffuse, moderate to strong cytoplasmic BRAF V600E staining. Adjacent non-neoplastic kidney parenchyma shows negative to weak cytoplasmic staining and weak to moderate nuclear staining. In each case, Sanger sequencing of BRAF exon 15 reveals a corresponding thymidine to adenine substitution at codon 600 (arrowheads in C,F,I,L); this substitution results in the missense BRAF V600E mutation. Sanger sequencing confirmed the presence of BRAF V600E mutations in the remaining three MA cases with diffuse, moderate to strong cytoplasmic BRAF V600E staining (data not shown). No other BRAF exon 15 mutations were identified in these seven cases. Magnification: (A,D,G,J) 20X; and, (B,E,H,K) 40X.
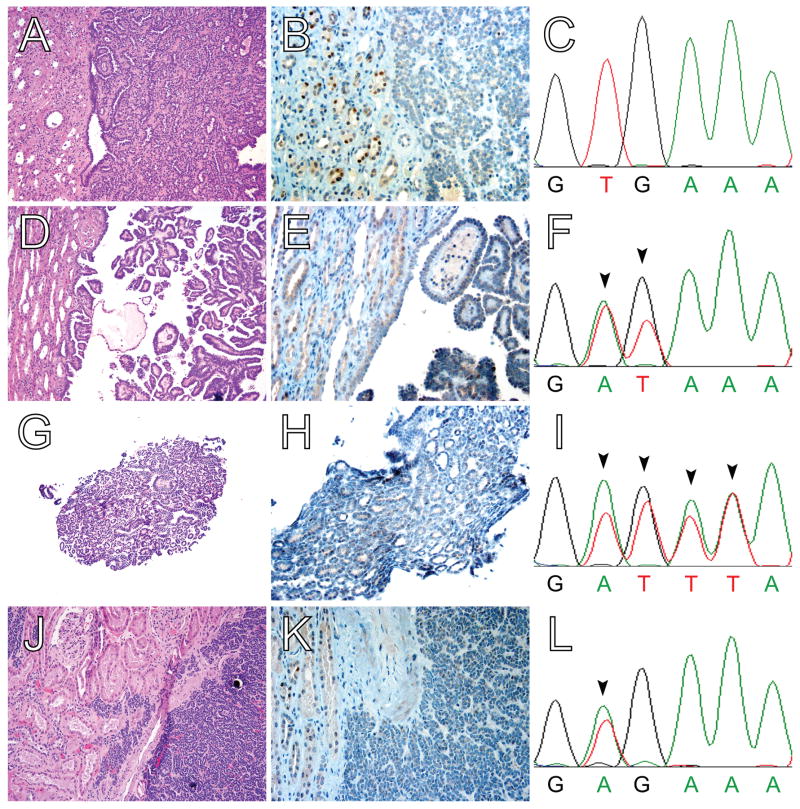
Figure 2. Novel BRAF exon 15 mutations in BRAF V600E-negative MA.
(A,D,G,J) H&E, (B,E,H,K) BRAF V600E IHC, and (C,F,I,L) BRAF exon 15 genomic sequencing of four MA cases. All four cases show the typical histomorphologic features of MA but demonstrate negative to weak cytoplasmic and/or nuclear staining. Adjacent non-neoplastic kidney parenchyma shows negative to weak cytoplasmic staining and weak to moderate nuclear staining. For one case, Sanger sequencing of BRAF exon 15 revealed no mutations (wild type sequence at codons 600 and 601 shown in panel C). Two additional cases of V600E-negative MA showed novel BRAF exon 15 mutations by Sanger sequencing. For one case, compound substitution of adenine and thymidine at codon 600 for thymidine and guanine (arrowheads in F) results in the missense BRAF V600D mutation; for the other case, a complex compound substitution and/or indel at codons 600 and 601 (arrowheads in I) results in the compound missense BRAF mutations V600D and K601L. (These novel BRAF exon 15 mutations were subsequently confirmed by Sanger sequencing of the subcloned PCR product.) Finally, for one case, Sanger sequencing demonstrated a thymidine to adenine substitution at codon 600 (arrowhead in L), corresponding to a BRAF V600E missense mutation; this is the only instance of discordance between V600E mutation-specific immunohistochemistry and Sanger sequencing. Magnification: (A,D,G,J) 20X; and, (B,E,H,K) 40X.
Concordance between BRAF V600E mutation status by mutation-specific IHC and Sanger sequencing of BRAF exon 15
As determined by mutation-specific IHC, the frequency of BRAF V600E mutations in our retrospective MA cohort was lower than previously reported (64% versus 89%)7. To further investigate this apparent difference, we interrogated our cohort for V600E mutations via Sanger sequencing of BRAF exon 15. Genomic DNA harvested from the SK-MEL-5 melanoma cell line (which carries the BRAF V600E mutation) and commercially available wild type male human genomic DNA were utilized as positive and negative controls, respectively (data not shown). All seven cases with diffuse, moderate to strong cytoplasmic BRAF V600E IHC staining harbored a thymidine to adenine substitution at codon 600 (i.e., GTG → GAG), which corresponds to the BRAF V600E missense mutation (Figure 1). In addition, for one of the remaining four cases with negative to weak cytoplasmic BRAF V600E IHC staining, Sanger sequencing demonstrated a BRAF V600E mutation (Figure 2J-L). All other cases were negative for BRAF V600E mutations, and no other BRAF exon 15 mutations were detected in tumors harboring BRAF V600E mutations. Therefore, overall eight (73%) of the cases in our retrospective MA cohort contain BRAF V600E mutations, and the overall sensitivity and specificity for mutation-specific IHC was 88% and 100%, respectively.
Novel BRAF exon 15 mutations in V600E-negative MA
For the three remaining cases with negative to weak cytoplasmic BRAF V600E IHC staining and without detectable BRAF V600E mutations by Sanger sequencing, BRAF exon 15 was closely examined for novel, non-V600E mutations. For one case, compound two base substitution of adenine and thymidine at codon 600 for thymidine and guanine results in the missense BRAF V600D mutation (Figure 2D-F), and for another case, a complex compound substitution and/or indel at codons 600 and 601 results in the compound missense BRAF mutations V600D and K601L (Figure 2G-I). These novel BRAF exon 15 mutations were subsequently confirmed by Sanger sequencing of the subcloned PCR product (data not shown). No BRAF exon 15 mutations were identified in the third case (Figure 2A-C).
BRAF V600E IHC in a large TMA cohort of common renal tumors
Finally, previously published data suggests that BRAF V600E mutations are rare in other common renal tumors7, 17, 27. Investigation of the available provisional The Cancer Genome Atlas (TCGA; www.cbioportal.org) renal cancer datasets showed no BRAF codon 600 mutations in either chromophobe RCC (n = 66) or clear cell RCC (n = 523), and only one case of papillary RCC (n = 168) harbored a BRAF V600E mutation; no other BRAF exon 15 mutations were identified in this large RCC sequencing cohort. In addition, a recent study examined BRAF V600E IHC in a large TMA cohort of renal tumors, including clear cell RCC, papillary RCC, chromophobe RCC, and oncocytoma, and found that less than 1% of these tumors showed positive staining with this mutation-specific antibody27. We sought to independently extend these findings in a large TMA cohort of common renal tumors from our institution. BRAF V600E IHC was performed on a TMA comprising tumor samples from eighty-six unique patients (see Materials and Methods). Consistent with the rarity of documented BRAF V600E mutations in common renal tumors, none (0%) of the 84 tumors available for evaluation demonstrated diffuse, moderate to strong cytoplasmic staining (Figure 3). Importantly, representative cores of benign renal parenchyma from a subset of the TMA tumor cases demonstrated variable weak cytoplasmic and weak to moderate nuclear staining – similar to the aforementioned adjacent benign renal parenchyma in the MA cases. This reproducible (albeit non-specific) background staining serves as a positive internal control for BRAF V600E IHC and, thereby, supports the experimental validity of negative BRAF V600E IHC in this TMA cohort of common renal tumors.
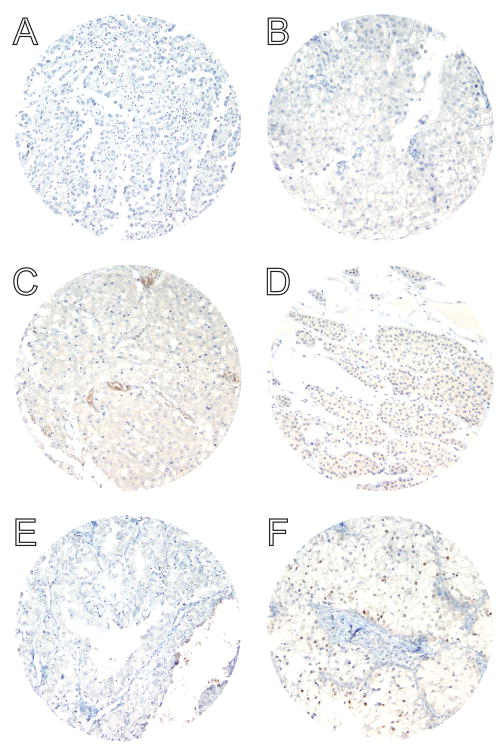
Figure 3. BRAF V600E protein is not detected by a mutation-specific antibody in a large cohort of common non-MA renal tumors.
BRAF V600E IHC of representative TMA cores of (A) type I papillary RCC, (B) chromophobe RCC, (C) eosinophilic variant of chromophobe RCC, (D) oncocytoma, (E) type II papillary RCC, and (F) clear cell RCC. Variable weak cytoplasmic and/or moderate nuclear staining is present (see clear cell RCC in panel F), similar to background non-neoplastic renal parenchyma; however, no moderate or strong cytoplasmic staining is identified in any of the eighty-four non-MA renal tumors analyzed. These results are in accord with prior sequencing data and IHC studies showing only very rare BRAF V600E mutations in RCC7, 27. Magnification: 20X.
Discussion
In this study, we examined BRAF V600E protein expression by mutation-specific IHC in a large cohort of MA. BRAF V600E IHC showed high sensitivity and specificity (88% and 100%, respectively) for the detection of BRAF V600E mutations (as assayed by Sanger sequencing) in this MA cohort. In addition, we report three cases of BRAF V600E-negative MA, including two with novel BRAF exon 15 mutations: one with a BRAF V600D mutation; and, one with compound BRAF V600D and K601L mutations. Finally, we showed that BRAF V600E protein expression by mutation-specific IHC was essentially absent in a large TMA cohort of common renal tumors.
Our results are in concert with previously published data regarding BRAF V600E mutations and BRAF V600E protein expression in MA7, 27. One additional case of MA with a confirmed BRAF V600E mutation has been reported in the literature28. Thus, together with the cases reported in this study, thirty-five of forty-one (85%) published genotyped MA harbor BRAF V600E. When the novel BRAF mutations identified in our study are included, overall thirty-seven of forty-one (90%) published genotyped MA contain BRAF exon 15 mutations. Similarly, of the published MA cases analyzed for BRAF V600E protein expression by mutation-specific IHC, thirteen of seventeen (76%) showed the diffuse, moderate to strong cytoplasmic staining characteristic of tumors with BRAF V600E mutations10, 18-27. The diagnosis of MA is often straightforward on routine H&E stains, however, some cases present challenging diagnostic dilemmas – particularly differentiating MA from the solid variant of papillary RCC4. In addition, abdominal radiographic imaging has increased the number of incidentally identified small renal masses, many of which are amenable to core needle biopsy diagnosis29; however, the diagnosis of MA on core needle biopsy may be challenging because of limited material available for evaluation. In these cases, ancillary studies (especially IHC) may be helpful3-5. MA characteristically express WT1 and CD57 and are frequently negative for CK7 and AMACR expression6. The value of BRAF V600E IHC in diagnostically challenging cases has not been specifically examined, however given the relatively low frequency of documented BRAF V600E mutations and/or BRAF V600E expression in other common renal tumors7, 16, 17, 27, positive IHC staining for BRAF V600E supports the diagnosis of MA. At the same time, however, it is also important to recognize that the absence of BRAF V600E staining does not exclude the possibility of MA.
This study is the first to document non-V600E BRAF exon 15 mutations in MA. In the only other published large MA sequencing study, a small pilot cohort of seven tumors was initially analyzed for oncogenic mutations by mass spectrometric genotyping with the OncoMap platform, which identified recurrent BRAF V600E mutations7; an additional twenty-two cases were then analyzed for specific hotspot BRAF mutations (including BRAF V600D) by multiple base extension (MBE) chemistry and secondarily validated by pyrosequencing with the BRAF Pyro® kit (Qiagen) in a CLIA-certified laboratory. Interestingly, in contrast to our study, no non-V600E BRAF exon 15 mutations were identified. In addition to one case without detectable BRAF exon 15 mutations, we identified two cases with novel BRAF exon 15 mutations: one with a BRAF V600D mutation; and, one with compound BRAF V600D and K601L mutations. While the sample size of our cohort is too small to draw definitive conclusions, there was no obvious correlation between BRAF genotype and specific clinicopathologic factors (i.e., age, histomorphologic features; see Table 1).
BRAF V600D mutations are uncommon but have been identified in variety of neoplasms, including melanoma, cholangiocarcinoma, and Langerhans cell histiocytosis9, 30-32. Similar to the more common V600E mutation, a variety of in vitro assays have demonstrated sensitivity of cultured cells lines harboring BRAF V600D mutations to the BRAF inhibitors vemurafenib and dabrafenib33-35. These findings suggest that BRAF V600D mutations may be functionally similar to V600E mutations in the tumorigenesis of MA – probably downstream activation of the MAPK/ERK signaling pathway. The significance of the novel BRAF K601L mutation identified in our study is unclear. Although codon 601 is a hotspot for BRAF exon 15 mutations, according to the Catalogue of Somatic Mutations In Cancer (COSMIC) database, this particular mutation has only been reported in one other case – a lung adenocarcinoma36. Sanger sequencing of the subcloned PCR product showed that the compound mutation (V600D/K601L) in this case likely arose as a single frame-preserving indel event, and given the concomitant V600D mutation, the K601L mutation may be of little additional functional significance. Overall, four of forty-one (10%) published genotyped MA do not harbor BRAF exon 15 mutations. Increasingly, recurrent somatic mutations in other signaling pathways genes have been described in BRAF V600E-negative neoplasms10, 37. In addition, a candidate tumor suppressor gene in MA has been mapped to a small region on chromosome 2p38, and deletion of a portion of chromosome 2p has been documented in a BRAF V600E-positive MA case28. These findings suggest a need for additional comprehensive sequencing of MA – both cases with and without BRAF exon 15 mutations.
Finally, MA is thought to represent one end of a spectrum of benign metanephric neoplasms that includes metanephric adenofibroma (MAF) and metanephric stromal tumor (MST). Furthermore, this group of benign metanephric neoplasms has been hypothetically postulated as the possible hyperdifferentiated, benign counterpart to WT – a morphologically heterogeneous group of tumors that may represent multiple distinct genetic entities. With this in mind, it is interesting to note the documented lack of BRAF mutations in WT16. Future studies should rigorously examine WT, MAF, and MST for BRAF exon 15 mutations. It will also be important to examine these tumors for BRAF V600E protein expression by mutation-specific IHC.
Acknowledgments
We would like to thank Tina Fields and the staff of the Department of Pathology clinical IHC laboratory at UMHS for technical assistance.
Funding: R.M. and A.M.C. are supported by the Prostate Cancer Foundation and National Human Genome Research Institute (UM1HG006508). A.M.C. is also supported by the National Cancer Institute (P50CA069568 and U01CA111275), A. Alfred Taubman Medical Institute, American Cancer Society, Doris Duke Charitable Foundation, and Howard Hughes Medical Institute.
Footnotes
Disclosures: None.
References
- 1.Davis CJ, Jr, Barton JH, Sesterhenn IA, et al. Metanephric adenoma. Clinicopathological study of fifty patients. Am J Surg Pathol. 1995;19:1101–1114. doi: 10.1097/00000478-199510000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Jones EC, Pins M, Dickersin GR, et al. Metanephric adenoma of the kidney. A clinicopathological, immunohistochemical, flow cytometric, cytogenetic, and electron microscopic study of seven cases. Am J Surg Pathol. 1995;19:615–626. doi: 10.1097/00000478-199506000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Argani P. Metanephric neoplasms: the hyperdifferentiated, benign end of the Wilms tumor spectrum? Clin Lab Med. 2005;25:379–392. doi: 10.1016/j.cll.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Mantoan Padilha M, Billis A, Allende D, et al. Metanephric adenoma and solid variant of papillary renal cell carcinoma: common and distinctive features. Histopathology. 2013;62:941–953. doi: 10.1111/his.12106. [DOI] [PubMed] [Google Scholar]
- 5.Muir TE, Cheville JC, Lager DJ. Metanephric adenoma, nephrogenic rests, and Wilms' tumor: a histologic and immunophenotypic comparison. Am J Surg Pathol. 2001;25:1290–1296. doi: 10.1097/00000478-200110000-00010. [DOI] [PubMed] [Google Scholar]
- 6.Olgac S, Hutchinson B, Tickoo SK, et al. Alpha-methylacyl-CoA racemase as a marker in the differential diagnosis of metanephric adenoma. Mod Pathol. 2006;19:218–224. doi: 10.1038/modpathol.3800520. [DOI] [PubMed] [Google Scholar]
- 7.Choueiri TK, Cheville J, Palescandolo E, et al. BRAF mutations in metanephric adenoma of the kidney. Eur Urol. 2012;62:917–922. doi: 10.1016/j.eururo.2012.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahman MA, Salajegheh A, Smith RA, et al. B-Raf mutation: a key player in molecular biology of cancer. Exp Mol Pathol. 2013;95:336–342. doi: 10.1016/j.yexmp.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 10.Brown NA, Rolland DC, McHugh JB, et al. Activating FGFR2-RAS-BRAF Mutations in Ameloblastoma. Clin Cancer Res. doi: 10.1158/1078-0432.CCR-14-1069. 2014 July 3 Epub. [DOI] [PubMed] [Google Scholar]
- 11.Chappe C, Padovani L, Scavarda D, et al. Dysembryoplastic neuroepithelial tumors share with pleomorphic xanthoastrocytomas and gangliogliomas BRAF(V600E) mutation and expression. Brain Pathol. 2013;23:574–583. doi: 10.1111/bpa.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serrano C, Simonetti S, Hernandez-Losa J, et al. BRAF V600E and KRAS G12S mutations in peripheral nerve sheath tumours. Histopathology. 2013;62:499–504. doi: 10.1111/his.12021. [DOI] [PubMed] [Google Scholar]
- 13.Brastianos PK, Taylor-Weiner A, Manley PE, et al. Exome sequencing identifies BRAF mutations in papillary craniopharyngiomas. Nat Genet. 2014;46:161–165. doi: 10.1038/ng.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Badalian-Very G, Vergilio JA, Degar BA, et al. Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood. 2010;116:1919–1923. doi: 10.1182/blood-2010-04-279083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taube JM, Begum S, Shi C, et al. Benign nodal nevi frequently harbor the activating V600E BRAF mutation. Am J Surg Pathol. 2009;33:568–571. doi: 10.1097/PAS.0b013e31818a64fb. [DOI] [PubMed] [Google Scholar]
- 16.Miao J, Kusafuka T, Fukuzawa M. Hotspot mutations of BRAF gene are not associated with pediatric solid neoplasms. Oncol Rep. 2004;12:1269–1272. [PubMed] [Google Scholar]
- 17.Gattenlohner S, Etschmann B, Riedmiller H, et al. Lack of KRAS and BRAF mutation in renal cell carcinoma. Eur Urol. 2009;55:1490–1491. doi: 10.1016/j.eururo.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 18.Capper D, Preusser M, Habel A, et al. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011;122:11–19. doi: 10.1007/s00401-011-0841-z. [DOI] [PubMed] [Google Scholar]
- 19.Andrulis M, Penzel R, Weichert W, et al. Application of a BRAF V600E mutation-specific antibody for the diagnosis of hairy cell leukemia. Am J Surg Pathol. 2012;36:1796–1800. doi: 10.1097/PAS.0b013e3182549b50. [DOI] [PubMed] [Google Scholar]
- 20.Capper D, Berghoff AS, Magerle M, et al. Immunohistochemical testing of BRAF V600E status in 1,120 tumor tissue samples of patients with brain metastases. Acta Neuropathol. 2012;123:223–233. doi: 10.1007/s00401-011-0887-y. [DOI] [PubMed] [Google Scholar]
- 21.Koperek O, Kornauth C, Capper D, et al. Immunohistochemical detection of the BRAF V600E-mutated protein in papillary thyroid carcinoma. Am J Surg Pathol. 2012;36:844–850. doi: 10.1097/PAS.0b013e318246b527. [DOI] [PubMed] [Google Scholar]
- 22.Ilie M, Long E, Hofman V, et al. Diagnostic value of immunohistochemistry for the detection of the BRAFV600E mutation in primary lung adenocarcinoma Caucasian patients. Ann Oncol. 2013;24:742–748. doi: 10.1093/annonc/mds534. [DOI] [PubMed] [Google Scholar]
- 23.Long GV, Wilmott JS, Capper D, et al. Immunohistochemistry is highly sensitive and specific for the detection of V600E BRAF mutation in melanoma. Am J Surg Pathol. 2013;37:61–65. doi: 10.1097/PAS.0b013e31826485c0. [DOI] [PubMed] [Google Scholar]
- 24.Preusser M, Capper D, Berghoff AS, et al. Expression of BRAF V600E mutant protein in epithelial ovarian tumors. Appl Immunohistochem Mol Morphol. 2013;21:159–164. doi: 10.1097/PAI.0b013e31825d7402. [DOI] [PubMed] [Google Scholar]
- 25.Sinicrope FA, Smyrk TC, Tougeron D, et al. Mutation-specific antibody detects mutant BRAFV600E protein expression in human colon carcinomas. Cancer. 2013;119:2765–2770. doi: 10.1002/cncr.28133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roden AC, Hu X, Kip S, et al. BRAF V600E expression in Langerhans cell histiocytosis: clinical and immunohistochemical study on 25 pulmonary and 54 extrapulmonary cases. Am J Surg Pathol. 2014;38:548–551. doi: 10.1097/PAS.0000000000000129. [DOI] [PubMed] [Google Scholar]
- 27.Pinto A, Signoretti S, Hirsch MS, et al. Immunohistochemical staining for braf v600e supports the diagnosis of metanephric adenoma. Histopathology. 2014 Aug 11; doi: 10.1111/his.12509. Epub. [DOI] [PubMed] [Google Scholar]
- 28.Dadone B, Ambrosetti D, Carpentier X, et al. A renal metanephric adenoma showing both a 2p16e24 deletion and BRAF V600E mutation: a synergistic role for a tumor suppressor gene on chromosome 2p and BRAF activation? Cancer Genet. 2013;206:347–352. doi: 10.1016/j.cancergen.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Shah RB, Bakshi N, Hafez KS, et al. Image-guided biopsy in the evaluation of renal mass lesions in contemporary urological practice: indications, adequacy, clinical impact, and limitations of the pathological diagnosis. Hum Pathol. 2005;36:1309–1315. doi: 10.1016/j.humpath.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 30.Tannapfel A, Sommerer F, Benicke M, et al. Mutations of the BRAF gene in cholangiocarcinoma but not in hepatocellular carcinoma. Gut. 2003;52:706–712. doi: 10.1136/gut.52.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greaves WO, Verma S, Patel KP, et al. Frequency and spectrum of BRAF mutations in a retrospective, single-institution study of 1112 cases of melanoma. J Mol Diagn. 2013;15:220–226. doi: 10.1016/j.jmoldx.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kansal R, Quintanilla-Martinez L, Datta V, et al. Identification of the V600D mutation in Exon 15 of the BRAF oncogene in congenital, benign langerhans cell histiocytosis. Genes Chromosomes Cancer. 2013;52:99–106. doi: 10.1002/gcc.22010. [DOI] [PubMed] [Google Scholar]
- 33.Tap WD, Gong KW, Dering J, et al. Pharmacodynamic characterization of the efficacy signals due to selective BRAF inhibition with PLX4032 in malignant melanoma. Neoplasia. 2010;12:637–649. doi: 10.1593/neo.10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang H, Higgins B, Kolinsky K, et al. RG7204 (PLX4032), a selective BRAFV600E inhibitor, displays potent antitumor activity in preclinical melanoma models. Cancer Res. 2010;70:5518–5527. doi: 10.1158/0008-5472.CAN-10-0646. [DOI] [PubMed] [Google Scholar]
- 35.Gentilcore G, Madonna G, Mozzillo N, et al. Effect of dabrafenib on melanoma cell lines harbouring the BRAF(V600D/R) mutations. BMC Cancer. 2013;13:17. doi: 10.1186/1471-2407-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmid K, Oehl N, Wrba F, et al. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res. 2009;15:4554–4560. doi: 10.1158/1078-0432.CCR-09-0089. [DOI] [PubMed] [Google Scholar]
- 37.Brown NA, Furtado LV, Betz BL, et al. High prevalence of somatic MAP2K1 mutations in BRAF V600E-negative Langerhans cell histiocytosis. Blood. 2014;124:1655–1658. doi: 10.1182/blood-2014-05-577361. [DOI] [PubMed] [Google Scholar]
- 38.Pesti T, Sukosd F, Jones EC, et al. Mapping a tumor suppressor gene to chromosome 2p13 in metanephric adenoma by microsatellite allelotyping. Hum Pathol. 2001;32:101–104. doi: 10.1053/hupa.2001.21132. [DOI] [PubMed] [Google Scholar]