Summary
A conserved feature of the midblastula transition (MBT) is a requirement for a functional DNA replication checkpoint to coordinate cell cycle remodeling and zygotic genome activation (ZGA). We have investigated what triggers this checkpoint during Drosophila embryogenesis. We find that the magnitude of the checkpoint scales with the quantity of transcriptionally engaged DNA. Measuring RNA Polymerase II (Pol II) binding at 20-minute intervals over the course of ZGA reveals that the checkpoint coincides with widespread de novo recruitment of Pol II that precedes and does not require a functional checkpoint. This recruitment drives slowing or stalling of DNA replication at transcriptionally engaged loci. Reducing Pol II recruitment in zelda mutants both reduces replication stalling and bypasses the requirement for a functional checkpoint. This suggests a model where the checkpoint functions as a feedback mechanism to remodel the cell cycle in response to nascent ZGA.
Introduction
Embryogenesis initiates with a period of cellular proliferation with minimal changes in cellular differentiation and functional specialization (O'Farrell et al., 2004; Tadros and Lipshitz, 2009). In embryos from Drosophila, Xenopus, and Zebrafish, cellular proliferation occurs with an abbreviated cell cycle consisting of sequential S- and M-phases without intervening gap phases. Concurrently, constrained transcriptional activity suppresses zygotic patterning in response to maternal cell fate determinants. Upon reaching a precise nucleo:cytoplasmic (N:C) ratio, embryos undergo coordinated cell cycle remodeling and large scale zygotic gene activation (ZGA), and enter a period of cell fate specification and morphogenesis with reduced cellular proliferation. This remodeling of cell cycle behavior and transcription accompanies a genetic transition from maternal to zygotic control of development collectively termed the midblastula transition (MBT). Although the temporal control of MBT timing via the N:C ratio is precise and reproducible within species, little is known about how the nuclear content is measured, and how the resultant N:C ratio regulates the cell cycle and ZGA.
One attractive candidate for a ‘sensor’ is the DNA replication checkpoint, whose activity is necessary for cell cycle remodeling and for maintaining ZGA (Brodsky et al., 2000; Conn et al., 2004; Crest et al., 2007; Di Talia et al., 2013; Fogarty et al., 1994; Sibon et al., 1999; Sibon et al., 1997). In Drosophila embryos, for example, rapid early mitoses are followed by a gradual checkpoint-mediated lengthening of the final pre-MBT cell cycles. The effect of this checkpoint is most obvious in Drosophila at nuclear cycle 13 (NC13), when it is required to extend interphase from 12 to 19 minutes. Drosophila mutants for two checkpoint kinases, ataxia telangiectasia and Rad3 related (mei-41/ATR) and checkpoint kinase 1 (grapes/chk1), fail to trigger a checkpoint at NC13 and prematurely enter mitosis prior to completion of S-phase, resulting in catastrophic DNA damage and ultimately death (Fogarty et al., 1994; Sibon et al., 1999; Sibon et al., 1997). The cues that activate the DNA damage response at the MBT are not known. It has been proposed that replication factors become limiting as embryos approach the N:C ratio, thus causing replication stress and triggering the checkpoint (Dasso and Newport, 1990; Sibon et al., 1999). Some support for this model comes from Xenopus embryos, where overexpression of a subset of replication factors will increase the number of pre-MBT mitoses from 12-14 to 13-15 (Collart et al., 2013).
To understand further the workings of the MBT clock in Drosophila, we have investigated the molecular mechanism that activates the MBT replication checkpoint. Rather than focusing on additional mitoses and other post-MBT (NC14) phenotypes associated with checkpoint defects, we directly test how checkpoint activity scales with the dosage of zygotic DNA at NC13. We find a non-equivalence of genomic DNA for triggering the checkpoint that correlates with the relative quantity of transcriptionally engaged DNA. By use of time-resolved ChIP-seq analysis of RNA Pol II occupancy over the course of ZGA, we determine that checkpoint activation at NC13 likewise correlates with the induction of large-scale de novo binding of Pol II to thousands of promoters. We find evidence that DNA replication slowing or stalling at NC13 co-localizes with and depends upon RNA Pol II activity. Pol II is recruited to chromatin normally in mei-41 mutant embryos, and reducing Pol II occupancy suppresses the mei-41 mitotic catastrophe. Thus, we propose that the primary effector downstream of the N:C ratio for timing the MBT is the initial establishment of transcriptional competence at the onset of large scale ZGA.
Results
Non-equivalence of genomic DNA for triggering the MBT replication checkpoint
Following fertilization, Drosophila embryos undergo 13 rapid metasynchronous syncytial mitoses, gradually lengthening the cell cycle period from an initial period of eight minutes prior to nuclear cycle 10 (NC10) to approximately 19 minutes at NC13 [(Foe and Alberts, 1983), and Figure 1A]. The characteristic lengthening of NC13 corresponds to a lengthening of S-phase (Shermoen et al., 2010), and therefore serves as a read-out for the magnitude of an induced DNA replication checkpoint. Compared with a 12.6 ± 0.16 minute wild type NC12, the NC13 is lengthened in wild type embryos by 53 ± 3%, whereas NC13 is only 4 ± 3% longer in mei-41, and 12.7 ± 5% longer in grp (Figure 1A). This genetic requirement for a replication checkpoint indicates that NC13 embryos may be subject to a new source of replication stress. Unlike in other organisms, this replication stress does not seem to be related directly to replication capacity (Collart et al., 2013; Dasso and Newport, 1990; Sibon et al., 1999), as reducing levels of the 180 kDa subunit of DNA Polymerase α (Brodsky et al., 2000; LaRocque et al., 2007), or the noncatalytic subunit of Cdc7 kinase Dbf4/chiffon has as no impact on NC13 duration (Figure S1).
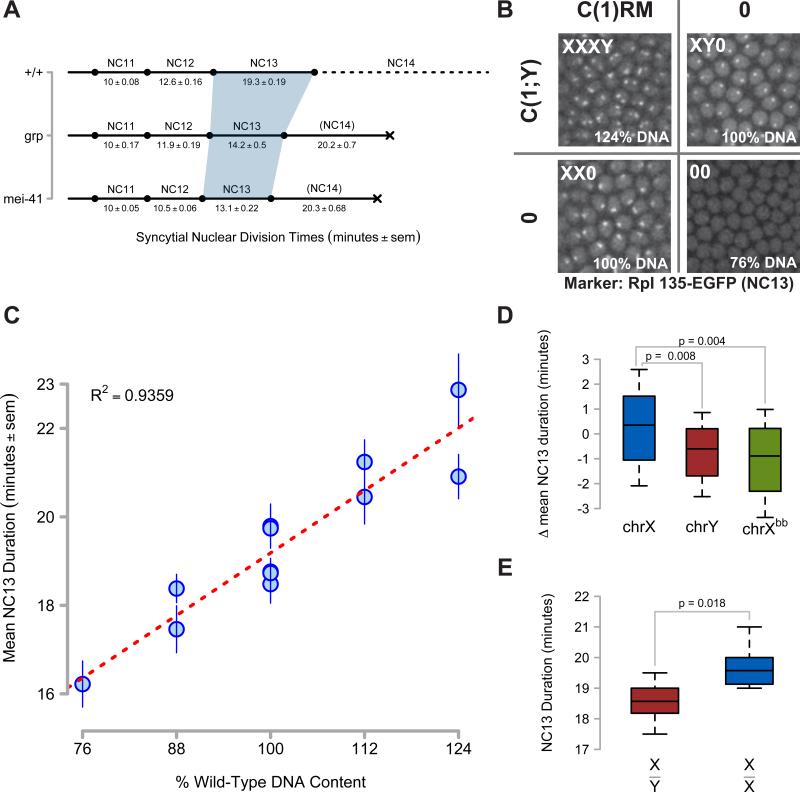
Figure 1. Non-Equivalence of Zygotic DNA for MBT Checkpoint Activation.
(A) Timelines of syncytial cell cycle times for wild type (+/+), grp1, and mei-41D3/29D embryos were measured by time-lapse confocal microscopy of H2Av-GFP or RFP. The shaded region highlights NC13. Lethality is signified by a black X.
(B) Representative confocal images (2500μm2) of nucleolar RNA Pol I GFP expression in NC13 embryos produced from a cross between C(1)RM/0 ; RpI135-EGFP/+ and C(1;Y)1/0 adults. First chromosome dosage is indicated in the upper left of each panel, and the corresponding amount of zygotic genomic DNA is indicated in the bottom left. NC13 nucleolar morphology in XY0 embryos is punctate, whereas it is barbell-shaped in XX0 embryos (See Supplemental Information). No nucleolar RpI135 EGFP is detected at NC13 in 00 embryos.
(C) NC13 times were measured for embryos with zygotic DNA dosage between 76% and 124% (see Experimental Procedures). Mean NC13 times ± sem for N ≥ 11 embryos per genotype are plotted as a function of zygotic genomic DNA content. Linear regression is represented as a red line.
(D) Box plots showing deviations from mean NC13 time for genotypes differing in chrX (N = 74), chrY (N = 40), or rDNA dosage (Xbb, N = 41). Brackets indicate the results of two-tailed t-tests.
(E) NC13 times for male (X/Y, N = 11) and female (X/X, N = 12) embryos produced from w; His2Av-RFP x w, HbP2>GFPnls /Y; + adults. Box plots show the distribution of NC13 times for each genotype. Brackets indicate the results of a two-tailed t-test.
See also Figure S1.
To test whether the checkpoint scales with the N:C ratio, we measured the correlation of NC13 time with the overall quantity of zygotic genomic DNA. We generated embryos containing between 76% and 124% DNA content by varying the dosage of chromosomes X and Y (chrX, chrY) using compound chromosome stocks (See Experimental Procedures and Figure 1B). The duration of NC13 positively correlates with zygotic DNA content (Figure 1C), but we observe two notable and informative discrepancies. First, the mean duration of NC13 for chrX+ genotypes is 1.1 ± 0.5 minutes longer than chrY+ genotypes of otherwise equivalent DNA content (Figure 1D). We observe a similar discrepancy between male and female embryos in an otherwise wild-type His2Av-RFP stock, where X/Y embryos complete NC13 in 18.7 ± 0.33 minutes whereas X/X embryos complete NC13 in 19.7 ± 0.20 minutes, a difference of 1.0 ± 0.5 minutes (Figure 1E). The character of the chrX DNA also influences the mean duration of NC13. Embryos with a wild-type X have NC13 duration that is 1.3 ± 0.6 minutes longer than those with an X lacking rDNA repeats (Figure 1D). Although small, these differences are significant. If NC13 duration depended solely on absolute DNA content, based on the linear fit of NC13 times to DNA content (Figure 1C, red dashed line), shortening the cycle by one minute would require reducing DNA dosage by 8.3%, or approximately 70% of the first chromosome. We conclude that not all DNA sequences are equivalent for triggering the replication checkpoint at the MBT. One major difference between chrX and chrY is the degree of transcriptionally active tracts of euchromatic DNA, with the X consisting of approximately 50% euchromatin and 50% heterochromatin in contrast to the 100% heterochromatic Y. In addition, highly transcribed rDNA repeats also modulate the magnitude of the checkpoint (Figure 1D). Therefore, we set out to test the alternative model that the checkpoint scales with the degree of transcriptionally engaged genomic DNA.
Large-scale recruitment of poised RNA Pol II distinguishes NC13 from NC12
To characterize transcriptional competency in the early embryo, we developed a method for performing ChIP-seq on small numbers of precisely staged embryos to measure the dynamics of RNA Pol II occupancy during ZGA. We carefully optimized sample preparation to generate high-quality measurements of RNA Pol II occupancy using 100 to 200 embryos collected during single interphases (NC12 or 13), or for three timepoints within NC14 interphase (Early, Middle, and Late, approximately 0-15’, 15-35’, and 35-60’ after NC14, see Extended Experimental Procedures), with an average interval of 18 minutes between timepoints. This approach extends previous analyses of Pol II binding during ZGA (Chen et al., 2013) allowing the dynamics of RNA Pol II recruitment to be reconstructed at each cycle over the course of the MBT (Figure 2).
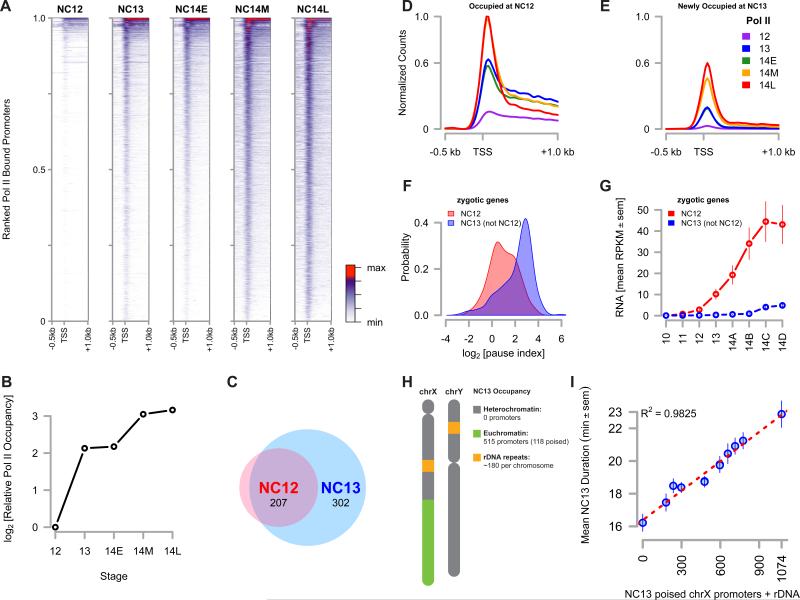
Figure 2. Large Scale Recruitment of Poised RNA Pol II at NC13.
(A) Promoter-proximal RNA Pol II (CTD pSer5) was plotted for timepoints spanning the MBT. Significantly enriched promoters are ranked from the top to the bottom of the y-axis by high to low mean intensity over the entire timecourse. The x-axis spans −0.5 kb to +1.0 kb and the TSS is noted. The colorbar is at the right hand margin.
(B) The sum of normalized Pol II CPM values for each gene in the Drosophila genome was calculated for each timepoint and plotted as an estimate of total Pol II occupancy over the course of MBT.
(C) The number of purely zygotic genes present at either NC12 or NC13 was determined and plotted as a venn diagram.
(D and E) Mean distributions of Pol II over promoters occupied at NC12 (D) versus promoters newly occupied at NC13 (E) are plotted per timepoint. The y-axis for both plots represents Pol II counts normalized to the maximum count value in both data sets. The maximum count value for genes newly occupied at NC13 is 0.6 and is noted on both axes.
(F) A kernel density estimate was plotted for the set of pause indices for each gene in both the ‘bound at NC12’ set (red) or the ‘newly bound at NC13’ set (blue).
(G) RPKM values for purely zygotic genes in the ‘bound at NC12’ (red) and the ‘newly bound at NC13’ (blue) sets were extracted from (Lott et al., 2011) and averaged. Mean RPKM values ± sem are plotted from NC10 through NC14.
(H) The schematic representation of chrX and Y showing relative quantities of heterochromatic and euchromatic sequences on each. The observed number of promoters occupied and poised at NC13 is annotated on the right.
(I) NC13 cell cycle time data for different X-Y chromosome combinations from Figure 1E is re-scaled and plotted according to the sum of poised chrX promoters plus rDNA repeats. Data are represented as mean ± sem with a linear regression (red line).
See also Table S1
At the outset of NC12, a small cohort of pre-MBT transcribed genes is occupied by initiating RNA Pol II [CTD pSer5] (‘Pol II’ hereafter, Figure 2A and B, and Table S1). Between NC12 and NC13, there is a 5.4 fold increase in the number of promoters occupied by Pol II. During NC13, initiating Pol II is significantly enriched within 1 kb of 2988 promoters, in contrast to NC12, when only 550 promoters are significantly bound. This trend matches the overall quantity of Pol II binding over this time, where between NC12 and NC13, there is a 4.4 fold change in the total Pol II occupancy within the genome (Figure 2B). To characterize this increase in Pol II occupancy between NC12 and NC13, we calculated the mean Pol II distribution over genes within two classes of NC13 peaks: those bound at NC13 and also at NC12, versus those newly occupied at NC13 (Figure 2C). The mean Pol II distribution for genes occupied at NC12 is initially uniformly distributed throughout the gene body, whereas Pol II is largely found concentrated near the transcription start site (TSS) for genes newly bound at NC13 (Figure 2D,E).
The distribution of Pol II at genes newly bound at NC13 resembles that of stalled or poised Pol II, previously been shown to be established over the course of the MBT (Chen et al., 2013). To estimate the degree of poising, we calculated a ‘pause index’, in which higher pause indices indicate a greater probability of a gene being poised (Zeitlinger et al., 2007). Indeed, the mean pause index for the set of NC12-bound zygotic genes is significantly different than that of the set of promoters newly bound at NC13 (Figure 2D, 1.00 versus 2.14, p << 0.01, two-tailed t-test). To confirm that Pol II is largely recruited in a poised status at NC13, we extracted RNA expression profiles from a published dataset for zygotic genes in each class of promoters (Lott et al., 2011). Of the 550 promoters bound by Pol II at NC12, 233 lack a significant maternal contribution and can be classified as ‘zygotic only’. Similarly, of the 2988 NC13 promoters, 509 are ‘zygotic only’, and 302 of these are not present in the set of NC12-bound promoters (Figure 2B). Poly-A mRNA expression from the set of NC12-bound zygotic genes begins at or around NC12 and steadily increases over the duration of NC14 (Figure 2C, “NC12”). Little or no new poly-A mRNA expression is detected until late in NC14 for the set of promoters newly bound at NC13 (Figure 2C, “NC13 (not NC12)”), consistent with de novo recruitment of Pol II at NC13 directly into the poised status. We conclude from these experiments that the major qualitative distinction in Pol II characteristics between NC12 and NC13 is the large-scale recruitment of Pol II to previously unbound genes and the subsequent establishment of transcriptional poising.
Importantly, the duration of NC13 correlates better with the number of transcriptionally engaged promoters than with bulk DNA alone. At NC13, Pol II occupies 515 promoters within chrX euchromatin, of which 118 are poised (pause index ≥ 2, Figure 2H). We recovered zero bound promoters on either chrX heterochromatin, or on chrY. In addition, we estimated an average of 180 rDNA repeats per X and Y based on previously published measurements (Long and Dawid, 1980). Re-scaling the x-axis of Figure 1E with our estimate of poised chrX promoters plus rDNA repeats at NC13 yields a better correlation between the measured NC13 times than DNA content alone (Figure 2i). These results suggest that some property of transcriptionally engaged chromatin architecture presents an unforeseen challenge to the DNA replication machinery, such that its abrupt establishment at NC13 triggers a replication checkpoint.
A functional replication checkpoint is not necessary for ZGA
We next revisited the question of whether a functional checkpoint is necessary for ZGA. First, we compared the temporal mRNA expression patterns of two zygotic genes in the NC12-bound class whose expression ultimately requires a functional checkpoint: runt and sry-α (Sibon et al., 1997). Precisely staged single-embryo QPCR in wild-type and grp mutant embryos shows that eliminating the checkpoint has no measurable effect on expression until 30 minutes post NC14 (Figure 3A). The reduction in runt and sry-α mRNA expression corresponds with a precocious catastrophic mitosis in NC14 (Figure 3B and 1A). Both runt and sry-α initiate normal expression in the absence of the checkpoint. Likewise, by immunostaining for initiating RNA Pol II (CTD pSer5) in syncytial blastoderm stage embryos, no gross difference in Pol II distribution or intensity is observed between wild-type and checkpoint mutant embryos (Figure 3C). These observations indicate that the initial phase of ZGA proceeds normally in checkpoint mutant embryos.
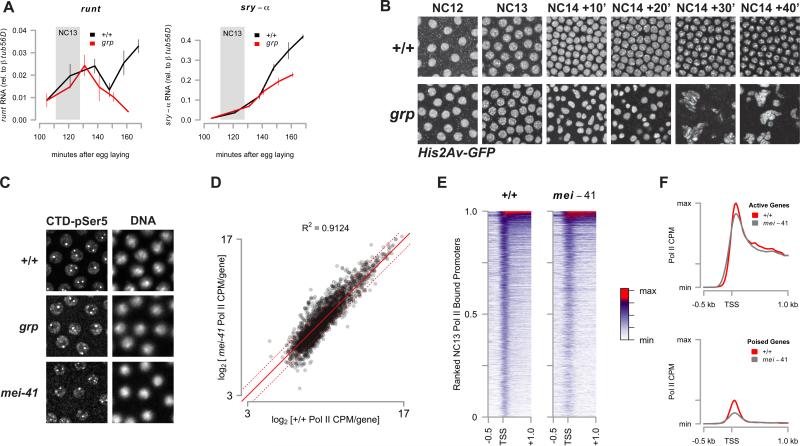
Figure 3. A Functional Replication Checkpoint is Not Necessary for Zygotic Gene Activation.
(A) Quantitative RT-PCR was performed on random-primed cDNA from precisely staged single w; His2Av-GFP (+/+, black) or trans-heterozygous w; grp1/209; His2Av-GFP/+ (grp, red) embryos (N = 3 per timepoint). Mean expression of runt or sry-α mRNA ± sem is quantified relative to expression of β-tubulin 56D mRNA. The period corresponding to NC13 in wild type embryos is highlighted (grey box).
(B) Representative time lapse confocal images (2500 μm2) are shown of His2Av-GFP in wild type (+/+, top) and grp1 mutant embryos (bottom) corresponding to the time points in Figure 3A.
(C) Staining of RNA Pol II (CTD pSer5) (left) and DNA (DAPI, right) in wild type (+/+, top), grp1 (middle), and mei-41D3/29D (bottom).
(D) log2[Pol II CPM] values for genes in the set of NC13-bound promoters were plotted for both wild-type and for mei-41D3 NC13 stage embryos. The solid red line indicates no change between samples and the dotted red lines indicate ±2-fold changes.
(E) Promoter-proximal Pol II counts for both wild-type (+/+) and mei-41D3 were plotted as in Figure 2A.
(F) Mean promoter-proximal Pol II counts for the set of ‘active’ (upper panel) or ‘poised’ (lower panel) genes in the wild-type (+/+) or mei-41D3 datasets are plotted. The y-axis is identical between plots and is scaled to the maximal value plotted.
See also Table S2
To confirm this observation, we compared the genome-wide distribution of Pol II in NC13-staged wild-type and mei-41 mutant embryos. The gene-by-gene distribution of Pol II intensities for both wild-type and mei-41 mutants is highly correlated (Figure 3D, 3E, and Table S2). Notably, the TSS-centered peak of Pol II in mei-41 is broader and more diffuse (Figure 3E, 3F). Although the mean promoter-proximal distribution of Pol II between wild-type and mei-41 is largely unchanged in the set of active genes (Figure 3F, top), the peak corresponding to poised Pol II is reduced by 31% relative to wild-type in mei-41 in the set of poised genes (Figure 3F, bottom). The overall effect of this reduction is small. Summing normalized Pol II counts over all bound genes, genome-wide Pol II occupancy in mei-41 is 92% of wild-type. This effect of mei-41 on poised loci could reflect either a feedback mechanism between mei-41 and poised Pol II, or could stem from the shorter cell cycle time of NC13 in mei-41 mutants, if establishment of poising is sensitive to interphase length as is seen with activation of a subset of zygotic loci (Edgar and Schubiger, 1986). We conclude that embryos lacking a functional replication checkpoint initiate this early phase of large scale ZGA normally.
DNA replication stalls at active and poised promoters
The initial event sensed by the replication checkpoint machinery is the formation of tracts of single-stranded DNA at sites of replication stress. These exposed sites of single-stranded DNA are rapidly bound by the conserved Replication Protein A (Rpa) complex that consists of three subunits (RpA-70, Rpa2, and Rpa3, in Drosophila), which recruits ATR to sites of stress (Zou and Elledge, 2003). In addition, Rpa also functions as a DNA elongation factor [reviewed in (Wold, 1997)]. Therefore, as an independent approach to studying the MBT replication checkpoint, we developed a fluorescent RpA-70 reporter in order to measure both optically and by ChIP-seq the magnitude and genomic distribution of sites of stalled DNA replication in the NC13 embryo. To test whether this reporter responds to induced replication stress, we depleted dNTPs from wild type embryos during NC12 (Figure 4B), triggering a temporary Chk1-dependent replication checkpoint [(Fasulo et al., 2012), and Figure 4C]. Under conditions of induced replication stress, RpA-70 forms intense foci within nuclei that gradually increase in both number and intensity over the course of the lengthened NC12 interphase.
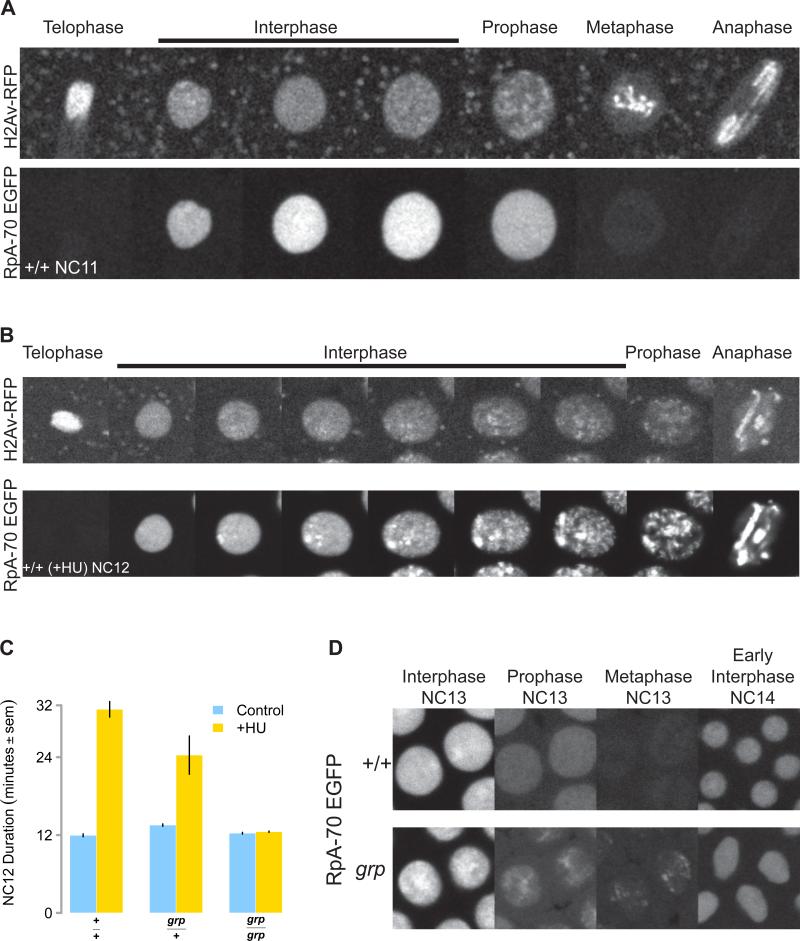
Figure 4. RpA-70 EGFP Marks Sites of Stalled Replication.
(A) RpA-70 EGFP uniformly localizes to interphase nuclei before NC13. An RpA-70 EGFP; H2Av RFP embryo was imaged by confocal microscopy. Successive representative images of a single NC11 stage nucleus are shown at the cell cycle stages indicated on top.
(B) RpA-70 EGFP and H2Av RFP as visualized in a HU-treated embryo by time-lapse confocal microscopy. Successive representative images of a single NC12 nucleus are shown at the cell cycle stages indicated on top.
(C) Wild type (+/+), grp/+, and grp mutant embryos were treated with HU and total NC12 duration was measured by time lapse confocal microscopy.
(D) Wild type (+/+) and grp mutant embryos expressing RpA-70 EGFP were visualized by time-lapse confocal microscopy. Successive representative images of two nuclei per genotype are shown at the cell cycle stages indicated.
During early syncytial blastoderm interphases (NC10-12), RpA-70 EGFP is uniformly distributed within the nucleoplasm beginning after nuclear envelope formation (Figure 4A). Upon entry into mitotic prophase, RpA-70 is rapidly exported into the cytoplasm, and is undetectable on undamaged chromatin throughout mitosis (Figure 4A). The first deviation from this pattern is observed between 9 and 15 minutes into NC13 when weak and diffuse foci of RpA-70 EGFP are observed in wild type embryo. These foci do not persist on chromatin by the time nuclei enter mitosis 13 (Figure 4D, +/+). In contrast, grp mutant embryos (and mei-41, data not shown) form more intense RpA-70 EGFP foci during NC13 (Figure 4D, grp Interphase NC13) that are still visible on condensing chromatin when the nuclei enter their premature mitosis (Figure 4D, grp Prophase and Metaphase NC13). These results confirm that in wild type embryos sites of stalled DNA replication arise by NC13 and are resolved prior to mitosis by activation of the replication checkpoint.
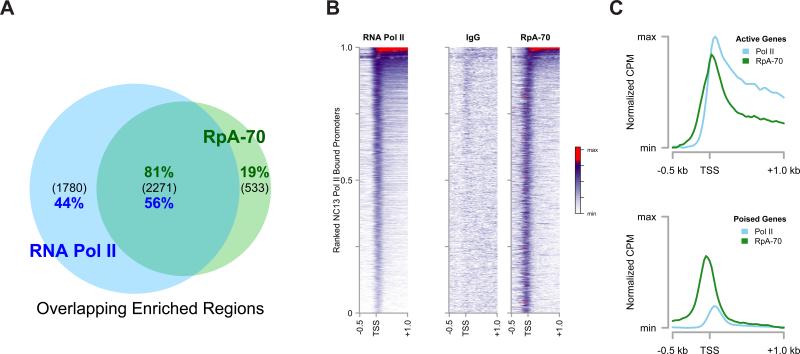
To map the genomic distribution of these RpA-70 foci, we performed ChIP-seq for both RpA-70 and Pol II on NC13 RpA-70 EGFP embryos in parallel, and sites of RpA-70 enrichment were calculated as for the Pol II experiments above. We expected that peaks of RpA-70 above background would correspond to sites of stalled DNA replication (see Discussion). Of the 2804 peaks of significant enrichment of RpA-70 over input DNA, 81% (2271) of these peaks overlap with those in the set of Pol II peaks, suggesting that the majority of RpA-70 enrichment over background localizes in the vicinity of transcription units (Figure 5A). The gene-by-gene distribution of promoter-proximal Pol II or RpA-70 in the set of overlapping peaks reveals similar distributions of both proteins (Figure 5B). On average, the distribution of RpA-70 near genes is displaced ~100 bp upstream of Pol II peaks, overlapping with TSSs (Figure 5B and C). The intensity of RpA-70 is relatively constant over the set of co-enriched promoters, whereas Pol II occupies a wider range of intensities. These results strongly support a correlation between transcriptionally engaged promoters and sites of stalled DNA replication within NC13 chromatin.
Figure 5. RpA-70 EGFP Co-Localizes with RNA Pol II in NC13 Embryos.
(A) Genomic regions significantly enriched by ChIP-seq for RpA-70 or Pol II were identified and intersected to yield a set of co-enriched regions. The total number of peaks in each group is indicated in parenthesis and the percentage of the total for each ChIP is shown (green: RpA-70, blue: Pol II).
(B and C) Promoter-by-promoter view of Pol II and RpA-70 localization on NC13 chromatin is shown (B) compared with a control ChIP (IgG). The mean distribution of Pol II and RpA-70 over actively expressed (C, top) and poised (C, bottom) is plotted. To facilitate comparison, mean CPM values per set of co-enriched promoters for each ChIP were calculated and are presented normalized to the maximal value per ChIP and are then floored to the minimum value (Normalized CPM).
Elimination of Zelda activity reduces RpA-70 binding to Zelda-dependent target genes
The transcription factor zelda (zld) is necessary for the expression of a broad set of early zygotic genes, and is regarded as a master regulator of ZGA (Harrison et al., 2011; Li et al., 2014; Liang et al., 2008; Nien et al., 2011). We reasoned that Pol II binding to NC13 promoters in zld embryos should be largely reduced, and we therefore tested whether reducing Pol II at promoters would have a corresponding effect on the binding of RpA-70.
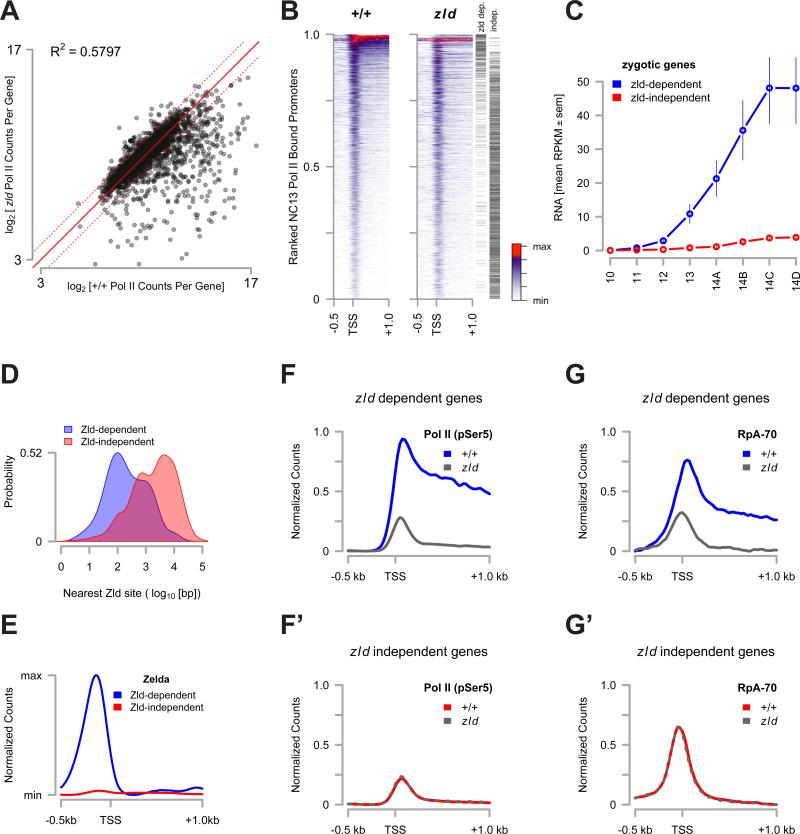
We performed ChIP-seq for Pol II and RpA-70 as above, comparing wild-type and zld mutant embryos. We estimate that overall Pol II binding in zld mutant embryos is 51% of wild-type, summing normalized Pol II counts over all NC13-bound genes. The set of genes bound by Pol II in wild-type NC13 embryos can be subdivided into at least two subclasses: zld-dependent and zld-independent. zld-dependent genes were defined as the set of genes where Pol II binding was reproducibly reduced by greater than 2-fold in zld embryos at an adjusted p-value of < 0.01 (Figure 6A, and Table S3). The set of zld-independent genes was defined as the set of promoters whose Pol II binding was unaffected (i.e., the rounded fold-change between zld and wild-type equals zero at an adjusted p-value of <0.01, Figure 6A).
Figure 6. Loss of Pol II binding reduces RpA-70 binding to transcribed regions.
(A) log2[Pol II CPM] values for genes in the set of NC13-bound promoters were plotted for both wild-type and for zld294 NC13 stage embryos. The solid red line indicates no change between samples and the dotted red lines indicate 2-fold changes in either direction.
(B) Promoter-proximal Pol II counts for both wild-type (+/+) and zld294 were plotted as in Figure 2A. The position of each promoter in the classes of ‘zelda dependent’ or ‘zelda independent’ loci are marked by a black hashmark on the right margin.
(C) RPKM values for genes in the ‘zelda dependent’ (blue) and the ‘zelda independent’ (red) sets were extracted from (Lott et al., 2011) and averaged. Mean RPKM values ± sem are plotted from NC10 through NC14.
(D) Kernel density estimates for distances between a known Zelda protein binding site (from (Harrison et al., 2011)) and TSSs in the ‘zelda dependent’ (blue) and ‘zelda independent’ (red) classes (p<<0.01, Wilcoxon rank sum test).
(E) The promoter proximal distribution of Zelda protein (from (Harrison et al., 2011)) for genes in the ‘zelda dependent’ (blue) and ‘zelda independent’ (red) sets was plotted.
(F and F’) The NC13 promoter proximal distribution of Pol II for zelda dependent loci (F) was plotted for wild type (blue) and zelda (grey) embryos. (F’) shows the distribution of Pol II at zelda independent loci for wild type (red) and zelda (grey dashed) embryos.
(G and G’) The NC13 promoter proximal distribution of RpA-70 for zelda dependent loci (G) was plotted for wild type (blue) and zelda (grey) embryos. (G’) shows the distribution of RpA-70 at zelda independent loci for wild type (red) and zelda (grey dashed) embryos.
See also Table S3
Of the set of 2988 genes with significant Pol II binding in wild-type NC13 embryos, 435 genes (15%) are zld-dependent, of which 266 (61%) fall into the set of genes bound by Pol II at NC12 (Figure 2). In contrast, 1793 genes (60%) are zld-independent, of which 1671 (93%) are members of the set of genes newly bound at NC13 (Figure 2). zld largely affects a subset of the actively expressed genes, consisting of loci with the highest average Pol II count distributions, with little or no effect on the establishment of Pol II binding at the TSS of genes with lower Pol II distributions (Figure 6B), which largely fall into the poised class. Consistent with previous reports (Harrison et al., 2011; Liang et al., 2008), the zld-dependent class consists of zygotic genes that are highly expressed early in development (Figure 6C) and are found within 792 ± 89 bp of a Zld binding site (Figure 6D,E). In contrast, zld-independent genes generally show little or no zygotic expression before the end of NC14 (Figure 6C), and are farther from a mapped Zld binding site (5495 ± 210 bp, Figure 6D,E). Correspondingly, zld is necessary for Pol II binding to the zld-dependent class of genes (Figure 6F), whereas zld does not affect Pol II recruitment in the zld-independent class (Figure 6F’).
We next asked whether reducing Pol II occupancy alters the occupancy of RpA-70 at the zld-dependent class of promoters. We compared the distribution of RpA-70 between wild-type and zld embryos within zld-dependent and –independent promoters (Figure 6G,G’). Reduced Pol II occupancy at zld-dependent promoters in zld mutants corresponds to a reduction in occupancy by RpA-70 (Figure 6G), whereas no change of RpA-70 binding is seen at zld-independent promoters (Figure 6G’). This result strongly supports our model that sites of transcriptional activity serve as roadblocks to DNA replication in NC13 embryos.
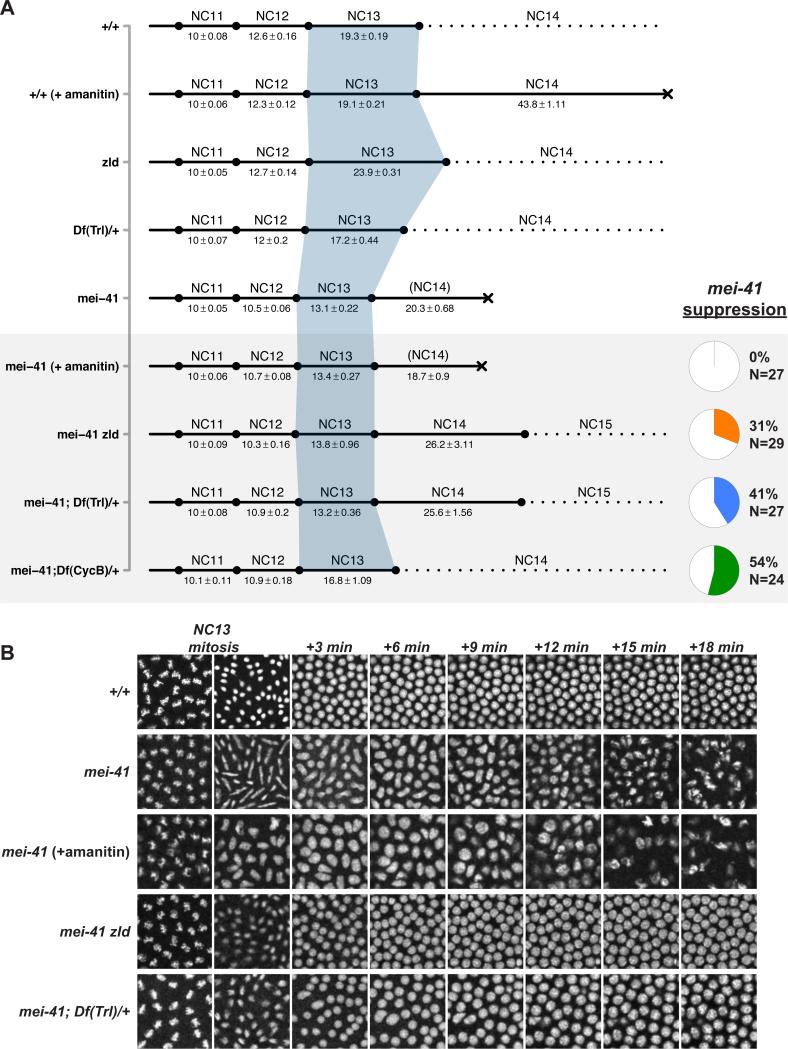
To test this model, we predicted that reducing total Pol II occupancy at NC13 would suppress the mitotic catastrophe in mei-41 mutant embryos. Indeed, embryos from mei-41 zld double-mutant mothers complete the syncytial mitotic divisions without catastrophe in 31% of cases following a short (13.8 ± 0.96 minute) NC13 (Figure 7A,B). In contrast, blocking Pol II transcription with α-amanitin fails to suppress the mei-41 mitotic catastrophe (Figure 7A,B). In the short timescales relevant to this experiment, α-amanitin functions by inhibiting the translocation of RNA Pol II along DNA (Gong et al., 2004), and does not affect either recruitment of Pol II or initialization of the transcription complex at the TSS [for example, (Li et al., 1996)]. These results are consistent with a model in which a feature of ZGA upstream of entry into transcriptional elongation drives DNA replication stalling at NC13.
Figure 7. Reduced Pol II recruitment suppresses mei-41 lethality.
(A) Syncytial cell cycle times for the indicated genotypes/treatments were measured by time-lapse confocal microscopy of H2Av-GFP. Time is represented in minutes ± sem. Lethality is represented by a black X. The grey box highlights conditions tested for suppression of mei-41 lethality. Data for suppressing individuals only are shown for the final three genotypes. The pie charts at bottom right indicate the frequency of mei-41 suppression for the associated genotypes. Wild-type and mei-41 data is reproduced from Figure 1. N = 27 (+/+ (+amanitin)), N = 30 (zld germline clones), N = 20 (Df(Trl)/+).
(B) Representative images (2500μm2) from time-lapse recordings from (A) are shown in 3-minute intervals beginning at metaphase 13 through 18 minutes into NC14. Note the absence of defective NC13 mitosis in mei-41 zld and mei-41; Df(Trl)/+ and subsequent wild-type nuclear morphology compared with mei-41 alone or mei-41 (+amanitin).
Next, we compared mei-41 suppression by zld with heterozygosity for Cyclin B, previously reported to suppress MBT replication checkpoint defects (Sibon et al., 1999). Heterozygosity for Cyclin B (Df(CycB)/+) suppresses mei-41 in 54% of cases. Unlike zld, CycB-dependent suppression is accompanied by a significantly lengthened (16.8 ± 1.09) NC13 time (Figure 7A, and Sibon et al., 1999). Together, these results suggest two mechanisms for suppressing a requirement for a functional DNA replication checkpoint at the MBT, either by reducing the source of replication stalling (e.g., by reducing ZGA via factors such as zld), or by providing enough time to complete DNA replication (e.g., by reducing CycB).
To test this prediction, we examined mei-41 embryos heterozygously deficient for the transcription factor Trithroax-like/GAGA Factor known to be required for establishment of poised Pol II at heat-shock promoters and for embryonic transcriptional regulation (Bhat et al., 1996; Shopland et al., 1995). Embryos from mei-41; Df(3L)ED4543/+ (Df(Trl)/+) mothers complete the syncytial divisions without mitotic catastrophe in 41% of cases following a short (13.2 ± 0.36 minute) NC13 (Figure 7A,B), ultimately yielding hatching larvae (13%, N = 199). Similar hatch rates are obtained using both an overlapping deficiency (Df(3L)fz-M21) and an allele of Trl (81.1, data not shown). Additionally, Df(Trl)/+ embryos otherwise wild type for mei-41 demonstrate a moderately shortened NC13 time (17.2 ± 0.44 minutes) compared with wild type embryos (Figure 7A). Taken together, we conclude that the initial phases of ZGA trigger the MBT replication checkpoint, and that the conflict between ZGA and DNA replication can be mitigated by reducing transcriptional initiation without a corresponding effect on cell cycle duration.
Discussion
On the basis of five central observations, we conclude that the MBT replication checkpoint is activated in response to the de novo recruitment of Pol II to chromatin at NC13. First, zygotic DNA differs in its capacity to trigger the checkpoint, correlating not with total DNA content, but rather with the quantity of transcriptionally engaged loci. Second, checkpoint activation coincides with large-scale de novo recruitment of Pol II throughout the genome. Third, sites of replication stalling as measured by RpA-70 localize to transcribed regions of the genome. Fourth, reduced Pol II occupancy in zelda germline clones results in a local reduction of the genomic occupancy of RpA-70 at zelda targets. Finally, reducing total Pol II occupancy at NC13 suppresses the lethality associated with defects in the replication checkpoint. Our results therefore suggest a simple model for the coordination of zygotic genome activation and cell cycle remodeling downstream of N:C ratio measurement.
Central to the concept of the MBT are the timing mechanisms that coordinate changes in maternal/zygotic RNA expression and cell cycle behavior. In our model, the MBT replication checkpoint coordinates ZGA with cell cycle remodeling, responding to large-scale transcriptional engagement to initiate changes in the maternal cell cycle. In this sense, the replication checkpoint is an indirect ‘sensor’ of the N:C ratio, responding instead to a proxy of nuclear content in the form of the fraction of the zygotic genome engaged in transcription. We therefore propose that cell cycle remodeling at the Drosophila MBT is zygotically driven by a two-step mechanism. First, the replication checkpoint, in response to de novo Pol II recruitment, drives Chk1 dependent down-regulation of Cdc25 catalytic activity (Edgar and Datar, 1996; Peng et al., 1997), leading to attenuation of Cdk1 kinase activity and transient cell cycle lengthening. Next, several zygotic genes drive the specific proteolytic degradation of the Cdc25 homolog Twine during NC14 (Di Talia et al., 2013; Farrell and O'Farrell, 2013). The resultant down-regulation of Cdk1 activity leads to the acquisition of several hallmarks of the zygotic cell cycle including a G2 phase, and early and late replicating chromatin domains (Farrell et al., 2012). In this model, cell cycle remodeling is initiated by checkpoint-dependent regulation of catalytic levels but ultimately completed and stabilized by zygotic gene activity.
Our model therefore predicts that characterizing the control of Pol II recruitment to chromatin will elucidate how the N:C ratio timer ultimately drives cell cycle remodeling. For at least a subset of zygotic genes, the onset of transcription correlates with the duration of interphase (Edgar and Schubiger, 1986). Interphase length is itself controlled by Cyclin/Cdk activity, and Cyclin dosage is gradually titrated by increasing nuclear content, resulting in a gradual checkpoint-independent lengthening of the syncytial cell cycle (Edgar et al., 1994; Ji et al., 2004). Therefore, N:C ratio dependent ZGA could be activated once interphase time advances beyond a critical length. In addition, one or more uncharacterized N:C-ratio independent timers drive maternal mRNA clearance and activation of the class of time- or stage-dependent zygotic transcripts (Benoit et al., 2009; Lu et al., 2009; Tadros et al., 2003). Since expression of both N:C ratio dependent and independent classes of zygotic transcripts is prevented by blocking translation before syncytial blastoderm stages (Edgar and Schubiger, 1986), one possible mechanism for timing events independently of the N:C-ratio is regulated translation of essential factors such as smaug and zelda (Benoit et al., 2009; Harrison et al., 2010). Indeed, the class of zld-dependent genes is enriched for the class of time/stage-dependent zygotic genes (Lu et al., 2009), supporting the idea that zelda drives N:C ratio independent ZGA. These observations support the emerging idea that ZGA is driven not by any one discrete mechanism, but rather by a collection of different, yet synchronized, systems.
One important question for future investigation will be to define the features of ZGA that trigger the MBT replication checkpoint. Our work suggests that the trigger of the checkpoint is upstream of entry into productive transcriptional elongation. Importantly, two mutants that confer premature ZGA have a corresponding premature activation of the MBT replication checkpoint (Pérez-Montero et al., 2013; Sung et al., 2013). A mutant of the early embryonic linker histone BigH1, for example, causes early ZGA in the presence of widespread DNA damage (Pérez-Montero et al., 2013). Similarly, a mutation of the large subunit of RNA Polymerase II (RpII215X161) also causes premature ZGA, triggering an early replication checkpoint and cell cycle pause (Sung et al., 2013). Although the precise nature of these mutant phenotypes is not yet clear, it is possible that they result from increased accessibility of Pol II to pre-MBT chromatin. These phenotypes are consistent with a model where the MBT replication checkpoint scales with zygotic transcriptional engagement.
It is also important to note that we have not determined whether the form of the trigger is actual replication stress, or rather stress-independent recruitment of the Rpa complex to promoters. It is possible that Pol II occupancy represents a previously unseen ‘roadblock’ to the DNA replisome (Azvolinsky et al., 2009), which can lead to replication stress [reviewed in (Bermejo et al., 2012)]. However, evidence from other model systems supports a stress-independent pathway. In budding yeast, Rpa binds to promoters and actively transcribed genes independently of the DNA replisome (Sikorski et al., 2011). We show that RpA-70 binds to both active and poised promoters at NC13, and further evidence suggests that Rpa could be recruited as part of the Pol II complex itself (Maldonado et al., 1996) or even function as an essential component of poised chromatin architecture (Fujimoto et al., 2012). In the latter example, interaction with the Rpa complex is necessary for HSF1 binding and for pre-loading of RNA Pol II at heat-shock promoters by recruiting the histone chaperone FACT (Fujimoto et al., 2012). Although proteomic screens have not identified an HSF-Rpa interaction in Drosophila, Rpa does appear to interact physically with GAGA-binding proteins Pipsqueak and Trithorax-like (Guruharsha et al., 2011), the latter of which we show to interact genetically with mei-41 (Figure 7). Therefore, it remains possible that the mechanism driving engagement of Pol II itself involves large-scale recruitment of the Rpa complex to chromatin, thus mimicking a signal of replication stress to activate the checkpoint.
Experimental Procedures Summary
Complete Experimental Procedures are included in the Supplement
Measurement of NC13 Duration
Three different crosses were used to generate embryos with 76% - 124% DNA content: [C(1)RM/0 ; RpI135 EGFP/+ × C(1;Y)1/0] , [w ; RpI135 EGFP/+ × C(1;Y)1/0], and [C(1)DX/Y ; RpI135 EGFP/+ × C(1;Y)1/0]. Genotypes were scored by counting the number of RpI135 EGFP foci at NC13 (e.g., as in Figure 1B) and by scoring NC14 nullo-X phenotypes of 0/0 and Y/0 embryos. Wild type male and female embryos were distinguished by scoring zygotic expression of a paternally supplied X-linked GFP transgene (= X/X).
Up to 15 embryos were dechorionated and affixed to a glass coverslip, overlaid with halocarbon oil, and simultaneously imaged by laser scanning confocal microscopy at a 30 second frame rate per embryo. Cell cycles times were scored as the duration between successive anaphases. To control for day-to-day fluctuations in room temperature, cell cycle times were normalized by setting the mean NC11 time to 10 minutes and scaling NC12 and 13 accordingly based on the mean NC11 time of all embryos on the slide. The NC13 times reported in Figure 1 are based on time-lapse recordings from 182 embryos. (N= 11 [X/Y, Y/0 and 0/0], N=12 [X/X], N= 13 [XY/0], N= 15 [XX/XY], N= 17 [XY/Y], N= 18 [XX/0], N= 20 [XXbb/0 and X/0], and N = 21 [XX/Y and XXbb/XY])
Scoring mei-41 suppression
Embryos from mei-4129D f zld294 FRT19A; H2Av-GFP/+ germline clones were used to score zld suppression of mei-41. mei-41D3/29D; Df(3L)ED4543/H2Av-GFP was the maternal genotype for scoring Trl suppression and mei-41D3/29D; Df(2R)59AB/+; H2Av-GFP/+ was the maternal genotype for scoring CycB suppression. Embryos were imaged as described above. The mei-41 phenotype was scored as ‘suppressed’ if greater than 75% of imaged nuclei successfully completed mitosis 13 (and 14 in cases of extra divisions) without evidence of anaphase bridging and if wild type blastoderm morphology was maintained during post-MBT cell cycle pause.
Chromatin Immunoprecipitation
Embryos were crosslinked for 15 minutes in a solution of 2 ml PBS + 0.5% Triton X-100 overlaid with 6 ml Heptane and 180 μl 20% fresh paraformaldehyde. Interphase embryos of specific stages were sorted under an epifluorescent dissection microscope on the basis of nuclear density by means of the RpA-70 EGFP transgene. Subsets of NC14 embryos were collected by measurement of nuclear elongation on a compound microscope with 20x objective. ChIP was performed essentially as described in (Blythe et al., 2009), with modifications noted in the Supplemental Information.
Sequencing and Analysis
Single-end sequencing of barcoded libraries was performed by the Lewis Sigler Institute for Integrative Genomics Sequencing Core Facility (Princeton) on an Illumina HiSeq 2500 with read length of 67 bp. Libraries were prepared with the NEBNext ChIP-seq library prep mastermix kit (NEB) according to the manufacturer's instructions. All data reflect the mean of two independent biological replicates.
Sequences were mapped to the Drosophila genome (dm3) using default settings on Bowtie (Langmead et al., 2009). Regions of significant enrichment were determined using Zinba (Rashid et al., 2011), differential binding was determined using edgeR (Robinson et al., 2010), and all other analysis was performed using the GenomicRanges package in R [(Lawrence et al., 2013), http://www.R-project.org/]. Sequences and peaks mapping to chrU and Uextra were not considered. Regions of enrichment were mapped to a modified Ensembl transcript database by identifying peaks within 1 kb of an annotated TSS, excluding transcripts less than 125 bp in length. The mean CPM values for 25 bp windows across the length of the genome were calculated and used to determine additional comparisons described in the text.
Supplementary Material
Acknowledgments
We thank the Bloomington Drosophila Stock Center for fly stocks. We thank C. Hannon, A. Roknabadi, and B. Pelham-Webb for help with experiments, all members of the Wieschaus and Schupbach labs, especially S. Little and B. He for lively discussion. We thank R. Kadzik, P. Klein, T. Schupbach, J. Lieb, M. Harrison, J. Zeitlinger, T. Orr-Weaver, W. Sullivan, and P. O'Farrell for helpful comments, and C. Rushlow, P. Schedl, and J. Sekelsky for fly stocks. We are indebted to the staff of the Sequencing Core Facility of the Lewis Sigler Institute and to L. Parsons for support for the ChIP-seq studies. We thank S. Di Talia for comments on the manuscript. This work was supported in part by Grant Number 5R37HD15587 from NICHD to E.F.W. and Ruth Kirschstein NRSA Postdoctoral Fellowship 1F32HD072653 from NICHD to S.A.B.. E.F.W. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions:
S.A.B. designed and performed all experiments and analysis. E.F.W. discussed experimental results and performed difficult crosses. S.A.B. and E.F.W. wrote the manuscript.
The GEO accession number for the ChIP-seq data is GSE62925
The authors declare no conflicts of interest.
References
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit B, He CH, Zhang F, Votruba SM, Tadros W, Westwood JT, Smibert CA, Lipshitz HD, Theurkauf WE. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development. 2009;136:923–932. doi: 10.1242/dev.031815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Lai MS, Foiani M. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol Cell. 2012;45:710–718. doi: 10.1016/j.molcel.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Bhat KM, Farkas G, Karch F, Gyurkovics H, Gausz J, Schedl P. The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development. 1996;122:1113–1124. doi: 10.1242/dev.122.4.1113. [DOI] [PubMed] [Google Scholar]
- Blythe SA, Reid CD, Kessler DS, Klein PS. Chromatin immunoprecipitation in early Xenopus laevis embryos. Developmental Dynamics. 2009;238:1422–1432. doi: 10.1002/dvdy.21931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky MH, Sekelsky JJ, Tsang G, Hawley RS, Rubin GM. mus304 encodes a novel DNA damage checkpoint protein required during Drosophila development. Genes Dev. 2000;14:666–678. [PMC free article] [PubMed] [Google Scholar]
- Chen K, Johnston J, Shao W, Meier S, Staber C, Zeitlinger J. A global change in RNA polymerase II pausing during the Drosophila midblastula transition. Elife. 2013;2:e00861. doi: 10.7554/eLife.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart C, Allen GE, Bradshaw CR, Smith JC, Zegerman P. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science. 2013;341:893–896. doi: 10.1126/science.1241530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn CW, Lewellyn AL, Maller JL. The DNA damage checkpoint in embryonic cell cycles is dependent on the DNA-to-cytoplasmic ratio. Dev Cell. 2004;7:275–281. doi: 10.1016/j.devcel.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Crest J, Oxnard N, Ji J-Y, Schubiger G. Onset of the DNA replication checkpoint in the early Drosophila embryo. Genetics. 2007;175:567–584. doi: 10.1534/genetics.106.065219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso M, Newport JW. Completion of DNA replication is monitored by a feedback system that controls the initiation of mitosis in vitro: studies in Xenopus. Cell. 1990;61:811–823. doi: 10.1016/0092-8674(90)90191-g. [DOI] [PubMed] [Google Scholar]
- Di Talia S, She R, Blythe SA, Lu X, Zhang QF, Wieschaus EF. Posttranslational control of Cdc25 degradation terminates Drosophila's early cell-cycle program. Curr Biol. 2013;23:127–132. doi: 10.1016/j.cub.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Datar SA. Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila's early cell cycle program. Genes & Development. 1996;10:1966–1977. doi: 10.1101/gad.10.15.1966. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Schubiger G. Parameters controlling transcriptional activation during early Drosophila development. Cell. 1986;44:871–877. doi: 10.1016/0092-8674(86)90009-7. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Sprenger F, Duronio RJ, Leopold P, O'Farrell PH. Distinct molecular mechanism regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes Dev. 1994;8:440–452. doi: 10.1101/gad.8.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell JA, O'Farrell PH. Mechanism and regulation of Cdc25/Twine protein destruction in embryonic cell-cycle remodeling. Curr Biol. 2013;23:118–126. doi: 10.1016/j.cub.2012.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell JA, Shermoen AW, Yuan K, O'Farrell PH. Embryonic onset of late replication requires Cdc25 down-regulation. Genes & Development. 2012 doi: 10.1101/gad.186429.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasulo B, Koyama C, Yu KR, Homola EM, Hsieh TS, Campbell SD, Sullivan W. Chk1 and Wee1 kinases coordinate DNA replication, chromosome condensation, and anaphase entry. Mol Biol Cell. 2012;23:1047–1057. doi: 10.1091/mbc.E11-10-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- Fogarty P, Kalpin RF, Sullivan W. The Drosophila maternal-effect mutation grapes causes a metaphase arrest at nuclear cycle 13. Development. 1994;120:2131–2142. doi: 10.1242/dev.120.8.2131. [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Takaki E, Takii R, Tan K, Prakasam R, Hayashida N, Iemura S.-i., Natsume T, Nakai A. RPA assists HSF1 access to nucleosomal DNA by recruiting histone chaperone FACT. Mol Cell. 2012;48:182–194. doi: 10.1016/j.molcel.2012.07.026. [DOI] [PubMed] [Google Scholar]
- Gong XQ, Nedialkov YA, Burton ZF. Alpha-amanitin blocks translocation by human RNA polymerase II. J Biol Chem. 2004;279:27422–27427. doi: 10.1074/jbc.M402163200. [DOI] [PubMed] [Google Scholar]
- Guruharsha KG, Rual J-F, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, et al. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Botchan MR, Cline TW. Grainyhead and Zelda compete for binding to the promoters of the earliest-expressed Drosophila genes. Dev Biol. 2010;345:248–255. doi: 10.1016/j.ydbio.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Li X.-y, Kaplan T, Botchan MR, Eisen MB. Zelda Binding in the Early Drosophila melanogaster Embryo Marks Regions Subsequently Activated at the Maternal-to-Zygotic Transition. PLoS Genet. 2011;7:e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J-Y, Squirrell JM, Schubiger G. Both cyclin B levels and DNA-replication checkpoint control the early embryonic mitoses in Drosophila. Development. 2004;131:401–411. doi: 10.1242/dev.00944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRocque JR, Dougherty DL, Hussain SK, Sekelsky J. Reducing DNA polymerase alpha in the absence of Drosophila ATR leads to P53-dependent apoptosis and developmental defects. Genetics. 2007;176:1441–1451. doi: 10.1534/genetics.107.073635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Weber JA, Chen Y, Greenleaf AL, Gilmour DS. Analyses of promoter-proximal pausing by RNA polymerase II on the hsp70 heat shock gene promoter in a Drosophila nuclear extract. Mol Cell Biol. 1996;16:5433–5443. doi: 10.1128/mcb.16.10.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-y., Harrison MM, Villalta JE, Kaplan T, Eisen MB. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. Elife. 2014;3 doi: 10.7554/eLife.03737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E, Dawid I. Repeated genes in eukaryotes. Annual review of biochemistry. 1980 doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Lott SE, Villalta JE, Schroth GP, Luo S, Tonkin LA, Eisen MB. Noncanonical compensation of zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-seq. PLoS Biol. 2011;9:e1000590. doi: 10.1371/journal.pbio.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Li JM, Elemento O, Tavazoie S, Wieschaus EF. Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development. 2009;136:2101–2110. doi: 10.1242/dev.034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson CW, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- Nien C-Y, Liang H-L, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, Rushlow C. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 2011;7:e1002339. doi: 10.1371/journal.pgen.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell PH, Stumpff J, Su TT. Embryonic cleavage cycles: how is a mouse like a fly? Curr Biol. 2004;14:R35–45. doi: 10.1016/j.cub.2003.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- Pérez-Montero S, Carbonell A, Morán T, Vaquero A, Azorín F. The embryonic linker histone H1 variant of Drosophila, dBigH1, regulates zygotic genome activation. Dev Cell. 2013;26:578–590. doi: 10.1016/j.devcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- Rashid NU, Giresi PG, Ibrahim JG, Sun W, Lieb JD. ZINBA integrates local covariates with DNA-seq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome Biol. 2011;12:R67. doi: 10.1186/gb-2011-12-7-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen AW, McCleland ML, O'Farrell PH. Developmental control of late replication and S phase length. Curr Biol. 2010;20:2067–2077. doi: 10.1016/j.cub.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland LS, Hirayoshi K, Fernandes M, Lis JT. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes & Development. 1995;9:2756–2769. doi: 10.1101/gad.9.22.2756. [DOI] [PubMed] [Google Scholar]
- Sibon OC, Laurençon A, Hawley R, Theurkauf WE. The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition. Curr Biol. 1999;9:302–312. doi: 10.1016/s0960-9822(99)80138-9. [DOI] [PubMed] [Google Scholar]
- Sibon OC, Stevenson VA, Theurkauf WE. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature. 1997;388:93–97. doi: 10.1038/40439. [DOI] [PubMed] [Google Scholar]
- Sikorski TW, Ficarro SB, Holik J, Kim T, Rando OJ, Marto JA, Buratowski S. Sub1 and RPA associate with RNA polymerase II at different stages of transcription. Mol Cell. 2011;44:397–409. doi: 10.1016/j.molcel.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H.-w., Spangenberg S, Vogt N, Großhans J. Number of nuclear divisions in the Drosophila blastoderm controlled by onset of zygotic transcription. Curr Biol. 2013;23:133–138. doi: 10.1016/j.cub.2012.12.013. [DOI] [PubMed] [Google Scholar]
- Tadros W, Houston SA, Bashirullah A, Cooperstock RL, Semotok JL, Reed BH, Lipshitz HD. Regulation of maternal transcript destabilization during egg activation in Drosophila. Genetics. 2003;164:989–1001. doi: 10.1093/genetics/164.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong J-W, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.