SUMMARY
The mechanisms by which transcription factor haploinsufficiency alters the epigenetic and transcriptional landscape in human cells to cause disease are unknown. Here, we utilized human induced pluripotent stem cell (iPSC)-derived endothelial cells (ECs) to show that heterozygous nonsense mutations in NOTCH1 that cause aortic valve calcification disrupt the epigenetic architecture resulting in derepression of latent pro-osteogenic and -inflammatory gene networks. Hemodynamic shear stress, which protects valves from calcification in vivo, activated anti-osteogenic and anti-inflammatory networks in NOTCH1+/+, but not NOTCH1+/−, iPSC-derived ECs. NOTCH1 haploinsufficiency altered H3K27ac at NOTCH1-bound enhancers, dysregulating downstream transcription of over 1000 genes involved in osteogenesis, inflammation, and oxidative stress. Computational predictions of the disrupted NOTCH1-dependent gene network revealed regulatory nodes that when modulated restored the network toward the wild-type state. Our results highlight how alterations in transcription factor dosage affect gene networks leading to human disease and reveal nodes for potential therapeutic intervention.
INTRODUCTION
Human disease is often caused by genetic variants that quantitatively affect dosage of the encoded gene product, particularly those involving major regulatory factors. The use of induced pluripotent stem cells (iPSCs) has facilitated the understanding of many human diseases, but it remains unclear how reduction in dosage of transcriptional regulators selectively affects the transcription of target genes, alters the epigenetic landscape, and perturbs gene networks resulting in disease. The ability to model haploinsufficiency of a transcription factor (TF) in human iPSCs combined with integration of broad “-omic” data may reveal mechanisms underlying dose-sensitivity of regulatory proteins and novel targets for intervention.
We previously reported two families with heterozygous non-sense mutations in the membrane-bound TF, NOTCH1 (N1), which led to a congenital defect of the aortic valve known as bicuspid aortic valve (BAV) and severe aortic valve calcification in adults (Garg et al., 2005). Calcific aortic valve disease (CAVD) is the third leading cause of adult heart disease and is responsible for over 100,000 valve transplants annually in the United States alone (Garg et al., 2005). BAV, which occurs in 1–2% of the population and involves the formation of two valve leaflets rather than the normal three leaflets, is a major risk factor for early valve calcification, although the mechanism for the calcification is unknown (Go et al., 2014). Recent studies identified N1 mutations in additional familial cases of BAV and CAVD as well as approximately 4% of sporadic cases, underscoring the importance of N1 in this disease (Foffa et al., 2013; Mohamed et al., 2006).
Hemodynamic shear stress protects against aortic valve calcification in adults, similar to shear-induced protection against atherosclerosis and vascular calcification. Accordingly, the first region of the valve to calcify is the aortic side that experiences less laminar shear stress than the ventricular side (Weinberg et al., 2010). Shear stress activates signaling through the N1 transmembrane receptor in endothelial cells (ECs) in vitro, and NOTCH signaling in vivo is greater on the ventricular side of the aortic valve (Combs and Yutzey, 2009; Masumura et al., 2009). Furthermore, in mice, EC-specific deletion of the Notch ligand Jagged1 leads to valve malformations and aortic valve calcification (Hofmann et al., 2012). These findings suggest that N1 signaling in the endothelium is uniquely positioned to mediate the anti-calcific response to shear stress within the valve.
Here, we utilized human iPSC-derived ECs to show that heterozygous nonsense mutations in N1 disrupt the epigenetic architecture resulting in derepression of latent pro-osteogenic and -inflammatory gene networks. Hemodynamic shear stress activated anti-osteogenic and anti-inflammatory networks in N1+/+, but not N1+/−, iPSC-derived ECs. N1 haploinsufficiency altered H3K27ac at N1-bound enhancers, dysregulating downstream transcription of over 1000 genes involved in osteogenesis, inflammation, and oxidative stress. Computational predictions of the disrupted N1-dependent gene network revealed regulatory nodes that when modulated restored the network toward the wild-type (WT) state. Our results highlight how alterations in TF dosage affect gene networks leading to human disease and reveal nodes for potential therapeutic intervention.
RESULTS
Transcriptional States in EC Differentiation and Response to Shear Stress
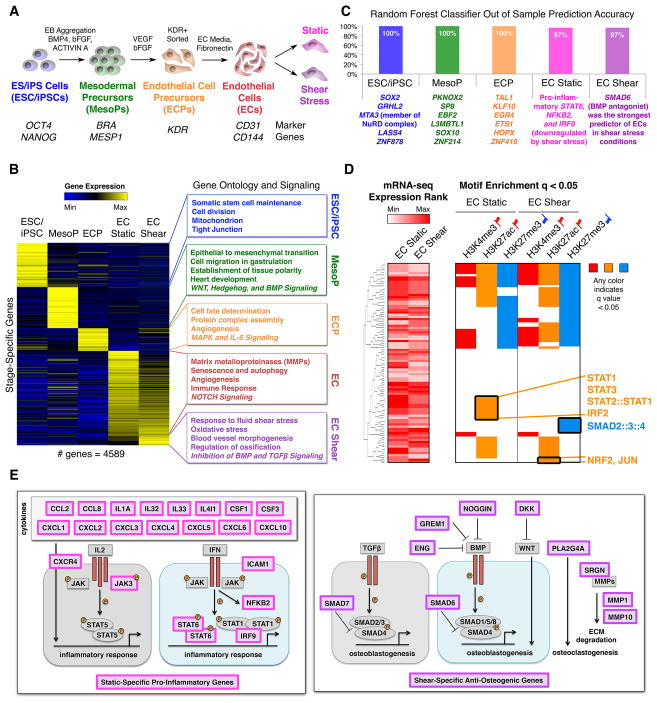
To investigate the consequences of N1 heterozygosity in ECs, we first needed to describe the normal transcriptional and epigenetic state of human ECs during differentiation and under static and fluid shear stress conditions. We therefore differentiated 2 human embryonic stem cell (ESC) lines (H7, H9) and 3 human iPSC lines into ECs using a protocol previously developed in our lab (Figure 1A) (White et al., 2012). We collected cells at key stages of EC differentiation: undifferentiated pluripotent cells, mesodermal precursors (MesoPs), EC precursors (ECPs), and ECs that we exposed to either static or laminar shear stress conditions to model the effects of hemodynamic shear stress on the ventricular side of the aortic valve (Figure 1A). We only conducted experiments on ECPs and ECs that were 70–100% pure for their respective markers by FACS (Figure S1A–B).
Figure 1. Transcriptional Mechanisms in EC Differentiation and Response to Shear Stress.
(A) Stages of EC differentiation analyzed.
(B) Unique signature of EC differentiation stages by RNA-seq. Stage-unique genes were expressed most highly at the given stage and significantly upregulated relative to immediately preceding or following stages. p < 0.05 by negative binomial test with false discovery rate (FDR) correction.
(C) Top stage-predictive TFs identified by random forest classifier.
(D) Left: Expression of TFs whose motifs were tested in the corresponding rows on the right. Right: Motif enrichment within activating or repressive chromatin marks in ECs exposed to static or shear stress conditions suggesting activated or repressed signaling pathways. Any color indicates significant motif enrichment (q < 0.05) by motif Diverge with FDR correction while white indicates non-significance. Red up flags: activating marks; blue down flags: repressive marks.
(E) Left: Diagram of static-specific pro-inflammatory genes (pink). Right: Diagram of shear-specific anti-osteogenic genes (violet).
We first identified the unique signature of key stages of EC differentiation using RNA-seq data from each aforementioned cell population (Figure 1B and Table S1–2). As expected, genes related to cell division and stem cell maintenance defined pluripotent cells while genes involved in WNT, HEDGEHOG, and BMP signaling were enriched in MesoPs. By the ECP stage, genes involved in angiogenesis and MAPK signaling were upregulated, indicating the commencement of EC specification. NOTCH signaling was a unique feature of the final shear-responsive EC stage. ECs also showed upregulation of matrix metalloproteinases (MMPs), which are involved in degrading extracellular matrix (ECM) (Vu and Werb, 2000).
Genes upregulated in shear stress conditions were involved in antagonizing pro-osteogenic BMP and TGFβ signaling pathways and included TFs such as SMAD6 and SMAD7 and secreted factors such as GREM1 (Figure 1B) (Bragdon et al., 2011). The secreted factors may provide a mechanism for ECs exposed to shear stress to prevent the calcification of underlying valve interstitial cells (VICs). Additionally, shear stress-upregulated genes involved factors that increase the resistance to oxidative damage, including NQO1 and TXNRD1 (Gorrini et al., 2013). Using a random forest machine learning approach to identify stage-predictive TFs from the RNA-seq data, we found that static conditions were predicted by expression of genes encoding inflammatory proteins including STAT6, NFKB2, and IRF9, all of which were downregulated by shear stress, while the TF most predictive of shear stress conditions was SMAD6, which inhibits BMP signaling (Figure 1C and S1C) (Hervas-Stubbs et al., 2011).
To investigate whether the anti-inflammatory and anti-osteogenic effects of shear stress were mediated by changes in genome occupancy of these key TFs, we tested the distribution of their putative occupancy sites based on motif analysis within active enhancers or repressed regions as identified by H3K27ac and H3K27me3 ChIP-seq, respectively, in human iPSC-derived ECs (Figure 1D). In static conditions, we found a unique overrepresentation of pro-inflammatory STAT and IRF motifs in H3K27ac-marked enhancers. By contrast, H3K27ac enhancers in the shear stress condition showed a unique overrepresentation of anti-inflammatory and antioxidant NRF2 motifs and TGFβ-inhibitory JUN motifs (Dennler et al., 2000; Gorrini et al., 2013). H3K27me3-marked repressive regions in shear stress conditions showed a unique overrepresentation of SMAD2/3/4 motifs, suggesting repression of pro-osteogenic TGFβ signaling in shear stress conditions. Thus, shear stress may protect against calcification by antagonizing pro-osteogenic BMP and TGFβ signaling pathways and repressing pro-inflammatory STAT and IRF signaling pathways at the transcriptional and epigenetic level (Figure 1E).
Dynamic Chromatin States Correlated with Distinct Transcriptional Patterns
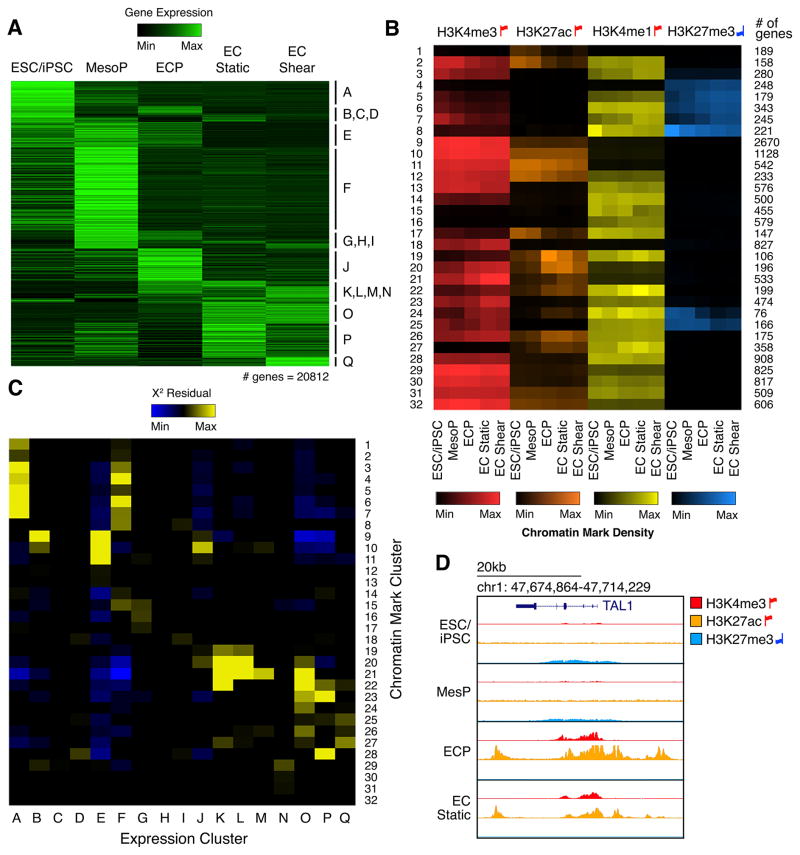
To understand the epigenetic changes occurring near the transcriptional start sites (TSSs) of dynamically expressed genes, we performed ChIP-seq for the H3K4me3 active promoter mark, H3K27ac active enhancer mark, H3K4me1 poised/active mark, and H3K27me3 repressive mark across EC differentiation (Rada-Iglesias et al., 2010; Wamstad et al., 2012) (Figure 2A–B and Table S3). Dynamic changes in histone modifications at promoters fell into distinct clusters. To test whether dynamic histone modifications correlated with distinct transcriptional patterns to distinguish functional groups of co-expressed genes, we compiled the number of genes shared between each chromatin and expression cluster and determined statistical enrichment (Figure 2C). Most expression clusters correlated with multiple chromatin clusters. For example, expression cluster F contained genes highly transcribed at the MesoP stage, which were enriched in chromatin clusters 3–8 and 14–15. Previous studies have shown that genes important for early development often correlate with a chromatin modification pattern similar to that of chromatin cluster 8, which includes a high level of repressive H3K27me3 at the pluripotent stage that is then relieved to allow expression during early development (Rada-Iglesias et al., 2010). Indeed, we found that there was a significant enrichment in cluster F of genes annotated as developmental within chromatin cluster 8 compared to genes annotated as non-developmental (Figure S1D). This indicated that chromatin cluster 8 identified a specific functional subset of genes in expression cluster F.
Figure 2. Correlation of Dynamic Chromatin Patterns with Transcriptional Transitions.
(A) Hierarchical clustering of mRNA expression.
(B) Hierarchical clustering of genes based on enrichment of histone modifications within 1 kb of the TSS. Color indicates mean enrichment for each gene cluster. Red up flags: activating marks; blue down flags: repressive marks.
(C) The overlap of genes within expression clusters (horizontal axis) and chromatin clusters (vertical axis). Color represents X2 residuals (any yellow indicates significant overlap between genes in the corresponding expression and chromatin cluster).
(D) Histone modification enrichment around TAL1 (Cluster J) during EC differentiation.
Conversely, some expression clusters correlated strongly with a single chromatin cluster. For example, expression cluster J predominantly clustered with chromatin cluster 10. While genes expressed in J showed very specific upregulation at the ECP stage, chromatin cluster 10 was characterized by stable H3K4me3 and H3K27ac activating marks throughout differentiation. All other expression clusters correlating with this non-dynamic chromatin cluster included genes upregulated at the ECP stage. This pattern was upheld even when evaluating chromatin marks up to 15 kilobases (kb) from gene TSSs.
The aforementioned random forest approach to discern stage-predictive TFs identified several ECP-predictive TFs that may confer the specificity of cluster J genes, including SCL1/TAL1, an important regulator of ECPs and vasculogenesis in addition to hematopoiesis (Drake et al., 1997; Liao et al., 1998; Van Handel et al., 2012) (Figure 1C). Unlike other cluster J genes that showed consistently active promoters throughout differentiation, H3K4me3 and H3K27ac marked the TAL1 promoter most strongly at the ECP stage (Figure 2D), when TAL1 drives critical fate decisions (Van Handel et al., 2012). Additionally, repressive H3K27me3 marked the TAL1 promoter at prior stages, suggesting its repression may prevent premature activation of downstream ECP gene networks. TAL1 gene expression was elevated in ECPs, but TAL1 DNA-binding motifs were not enriched in any stage-specific enhancers throughout EC differentiation. Thus, genes expressed at the pivotal ECP specification stage maintain an epigenetic state primed for transcriptional activation throughout differentiation and may rely on the expression of a discrete set of regulators such as TAL1 to confer temporal specificity. Only 11 other cluster J genes shared TAL1’s dynamic chromatin pattern (chromatin cluster 24). These included GATA2, a known cofactor of EC regulator ETV2 (Shi et al., 2014), as well as genes with no previously known role in EC differentiation such as C16orf74 that may represent novel ECP regulators. Together, these ECP-enriched genes and their targets activate genes critical for downstream endothelial development such as ANGPT2, a known TAL1 target gene that is required for postnatal angiogenesis (Deleuze et al., 2012), maintaining a high level of activation in ECs under both static and shear stress conditions (Figure S1E).
We next investigated whether chromatin clusters could discern functional groups of genes involved in the response to shear stress. Two groups of expression clusters contained static or shear stress-specific gene expression: K-L and P-Q. While K-L was enriched for cytoskeletal genes, P-Q was enriched for genes involved in SMAD signaling and blood vessel development. We focused on expression clusters P and Q to test whether the associated chromatin clusters could distinguish functional gene groups important for the anti-calcific effects of shear stress (Table S4). GO term enrichment in individual expression-chromatin cluster intersections showed that static-specific cluster intersections P22 and P25 contained immune process and interleukin signaling factors, respectively. However, when intersected with shear-specific genes, the same chromatin clusters, 22 and 25, were associated with TGFβ signaling antagonism (including ENG and INHBA) and bone mineralization (including inhibitory GREM1) (Bragdon et al., 2011; Guo et al., 2004). Additional shear-specific cluster intersection Q24 contained activin-binding genes such as FSTL3, an inhibitor of TGFβ signaling (Bragdon et al., 2011). Thus, chromatin clusters were able to distinguish immune process-related genes active in the static condition and antagonism of pro-osteogenic TGFβ signaling in shear stress conditions.
Isogenic iPSC-Derived ECs Model N1 Haploinsufficiency
We sought to understand how N1 heterozygosity perturbs the normal EC gene expression and epigenetic state to cause CAVD. We derived and characterized iPSCs from the fibroblasts of three individuals from two families affected with CAVD due to heterozygous nonsense mutations in N1 (Figure 3A, S2A–E, and S3). Additionally, we derived and characterized iPSCs from a related individual who was N1+/+ and unaffected by CAVD. As unrelated controls, we used two established ESC lines (H7, H9) and two previously established iPSC lines.
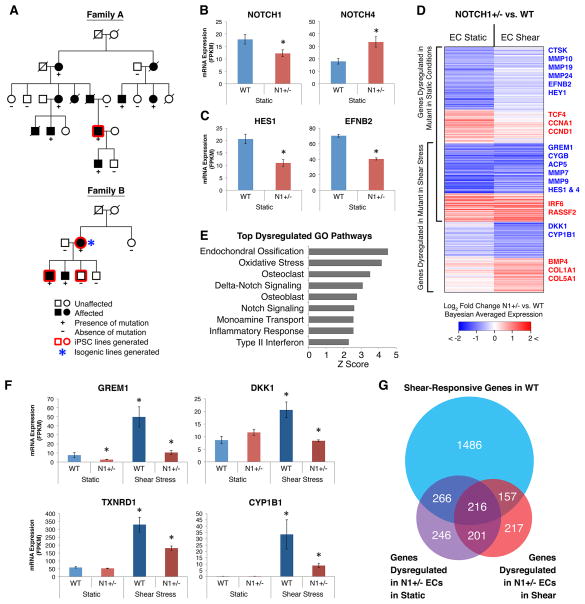
Figure 3. Gene Networks Dysregulated in N1 Haploinsufficient Isogenic iPSC-derived ECs.
(A) Pedigrees of two families affected with congenital heart disease and valve calcification due to N1 mutations. Squares, males; circles, females.
(B) mRNA expression of N1 and compensatory upregulation of NOTCH4.
(C) mRNA expression of canonical N1 targets HES1 and EFNB2.
(D) Log2 fold change in mRNA expression in N1+/− vs. WT ECs in static and shear stress conditions of 1303 genes significantly dysregulated in N1+/− ECs.
(E) Top GO pathways enriched among genes dysregulated in N1+/− ECs.
(F) Examples of anti-osteogenic (GREM1, DKK1), antioxidant (TXNRD1), and anti-atherogenic (CYP1B1) shear-responsive genes not properly activated in N1+/− ECs.
(G) Overlap of statistically significant gene sets.
In (B–G): WT n = 3, N1+/− n = 2 (isogenic ECs); error bars represent standard error; *p < 0.05 by negative binomial test with FDR correction. See also Figures S2–S5 and Table S5–6.
To derive isogenic control lines, we corrected the N1 mutation using TALENs (Figure 3A and S4A–D). We detected no off-target effects by Southern blot for the donor DNA and compared multiple corrected N1+/+ clones to multiple TALEN-targeted but uncorrected N1+/− clones to control for any effects of the TALEN-targeting process. We differentiated control and mutant iPSCs into ECs and exposed them to either static or shear stress conditions to model the anti-calcific effects of shear stress.
Using isogenic cell lines, we found that N1 mRNA levels were reduced by 30–40% in the N1+/− ECs by RNA-seq (Figure 3B). Additionally, 70–100% of N1 mRNA present in N1+/− ECs was transcribed from the WT allele, suggesting that the mutant mRNA largely undergoes nonsense mediated decay (Figure S5D). This indicated that the iPSC-derived ECs were effectively modeling a decreased dosage of N1. N1+/− ECs showed increased levels of NOTCH4 mRNA, encoding the other major NOTCH protein in ECs, reflecting a possible compensatory response. Although N1+/− ECs did not exhibit altered differentiation capacity, their transcriptome clustered separately from isogenic N1+/+ ECs (Figures S4C and S5A). Furthermore, canonical N1 targets, including HES1 and EFNB2, were downregulated in N1+/− ECs, indicating N1+/− ECs were haploinsufficient for N1 activity, at least at some targets (Figure 3C).
Gene Networks Dysregulated Due to N1 Haploinsuffiency
RNA-seq of iPSC-derived ECs in static conditions revealed that 929 mRNAs were dysregulated in N1+/− ECs, while in shear stress conditions 791 mRNAs were altered (with ~half being dysregulated in both conditions) (Figure 3D and 3G and Table S5). GO analysis showed that NOTCH and DELTA-NOTCH signaling were among the top pathways dysregulated in N1 haploinsufficiency (shear and/or static conditions), indicating that the N1+/− iPSC-derived ECs were indeed modeling a defect in N1 activity (Figure 3E and Table S6). Furthermore, top dysregulated GO pathways included endochondral ossification and inflammatory response, two pathways thought to play a major role in CAVD. In addition, 180 small non-coding RNAs (ncRNAs) were significantly dysregulated in N1+/− ECs, and these were enriched for miRNAs and CDBox RNAs (Figure S5B–C). Surprisingly, no ncRNAs were significantly dysregulated under shear stress, indicating that shear stress-induced N1 signaling may be sufficient to restore ncRNA networks in N1+/− ECs to their WT state, similar to the shear stress-induced restoration of a subset of dysregulated mRNAs. ncRNAs altered in N1+/− ECs reflected similar processes as observed for dysregulated mRNAs. Anti-osteogenic (e.g., miR-20a, 26a, 30e, and 106a) and anti-atherogenic miRNAs (e.g., miR-126) were downregulated in N1+/− ECs, while pro-osteogenic (e.g., miR-30d) and pro-atherogenic (e.g., miR-663) miRNAs were upregulated (Goettsch et al., 2013; Li et al., 2013; Schober et al., 2014).
When we tested whether unique features of the shear stress condition were dysregulated in N1 heterozygosity, we found that N1+/− ECs did not properly activate anti-calcific genes normally induced by shear stress (Figure 3D and F). In total, 30% of shear-responsive genes in WT ECs were dysregulated in N1+/− ECs and were thus N1-dependent (Figure 3G). N1+/− ECs showed downregulation of shear-specific antagonists of pro-osteogenic BMP and WNT pathways, including genes producing secreted proteins GREM1 and DKK, respectively (Van Handel et al., 2012). Furthermore, shear-exposed N1+/− ECs failed to upregulate anti-atherogenic factors such as CYP1B1 and showed aberrant upregulation of pro-inflammatory genes, including IRF6 (Conway et al., 2009; Kwa et al., 2014). In both static and shear stress conditions, N1+/− ECs showed an increase in cell cycle genes such as CDC20 and CDCA2 (with additional genes such as CCNA1 and CCND1 activated in static conditions). Furthermore, we observed downregulation of PDE3A and upregulation of PDE2A, which would be predicted to alter EC permeability and promote inflammatory cell infiltration (Surapisitchat et al., 2007). Finally, when exposed to shear stress, N1+/− ECs did not appropriately upregulate genes involved in the oxidative stress response such as CYGB and TXNRD1 (Li et al., 2007) and showed downregulation of MMPs such as MMP7 and 9 (with additional MMPs 10, 19, and 24 downregulated in static conditions) (Figure S5E). Overall, N1 haploinsufficient ECs could not mediate the normal anti-calcific response induced by shear stress and showed aberrant upregulation of pro-osteogenic and inflammatory signaling.
Epigenetic Dysregulation Correlated with Pro-Calcific Gene Expression in N1+/− ECs
To determine whether decreased dosage of N1 perturbed the epigenetic state of genes dysregulated in N1+/− ECs, we performed genome-wide ChIP-seq for H3K4me3, H3K27ac, H3K4me1, and H3K27me3 in WT or N1+/− patient-specific iPSC-derived ECs under static and shear stress conditions. We determined the density of the proximal promoter mark H3K4me3 within 3 kb of TSSs of genes dysregulated in N1+/− ECs. Given that H3K27ac, H3K4me1, and H3K27me3 mark both proximal and distal regulatory domains, we determined the density of these marks within 15 kb of TSSs of dysregulated genes.
When we clustered dysregulated genes based on their expression and epigenetic state in N1+/− ECs compared to WT ECs, the resulting clusters defined functional groups of genes involved in distinct aspects of CAVD (Figure 4A). For example, cluster I contained genes involved in ECM and pro-osteogenic WNT signaling, which showed increased transcription in both static and shear stress conditions without much change in any of the four evaluated chromatin marks. However, cluster J distinguished genes involved in pro-osteogenic BMP signaling whose transcriptional upregulation was accompanied by increased H3K4me3 and H3K27ac activating marks in both static and shear stress conditions as well as decreased H3K27me3 repressive marks in the static condition. Thus, functional groups of genes shared common epigenetic mechanisms associated with transcriptional dysregulation in N1+/− ECs.
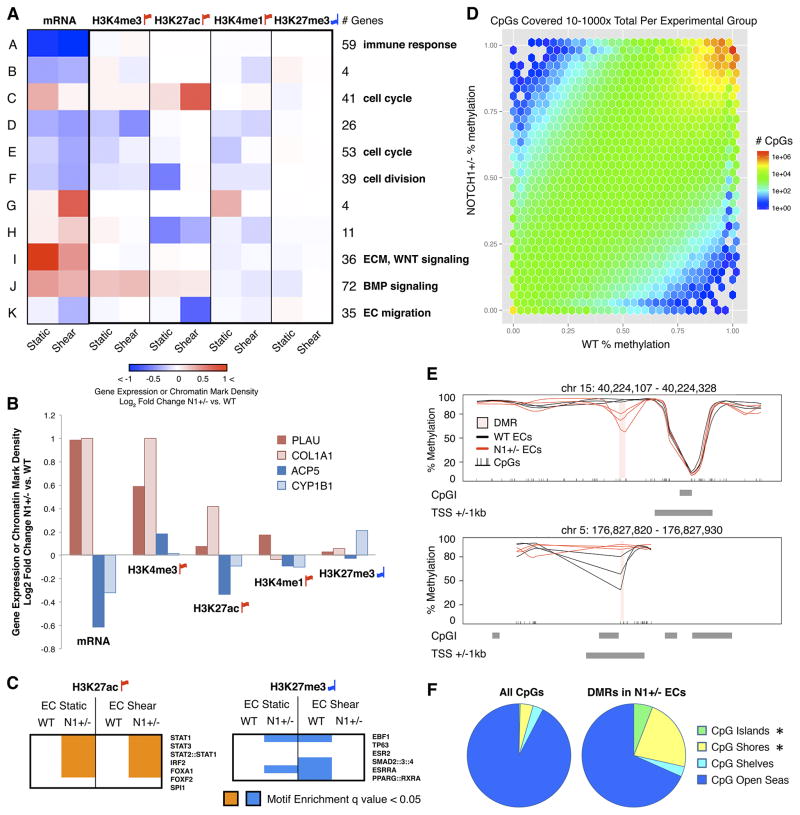
Figure 4. Epigenetic Dysregulation in N1+/− ECs.
(A) Hierarchical clustering of genes based on log2 fold change of expression and enrichment of histone modifications within 3 kb (H3K4me3) or 15 kb (H3K27ac, H3K4me1, H3K27me3) of the TSS in N1+/− vs. WT ECs. Enriched GO pathways within each cluster are shown on the right.
(B) Mean log2 fold change in N1+/− vs. WT static ECs of mRNA expression and histone modifications as in (A) of individual pro-osteogenic (PLAU, COL1A1), osteoclast (ACP5), and anti-atherogenic (CYP1B1) genes.
(C) TF motif enrichment in N1+/− vs. WT chromatin marks in static or shear stress conditions. Motifs tested were drawn from unique clusters identified in Figure 1D.
(D) Relative mean DNA methylation of CpGs in N1+/− (vertical axis) vs. WT (horizontal axis) ECs in static conditions. Plot includes only CpGs covered 10–1000x total between three biological replicates per experimental group.
(E) Examples of the 248 DMRs identified in N1+/− vs. WT ECs.
(F) Distribution of DMRs or all CpGs relative to CpGIs. Shores are < 2 kb flanking CpGIs; shelves are < 2kb flanking outwards from shores; open seas are > 4 kb flanking CpGIs. *p <0.05 by X2 test with Bonferroni correction.
In (A–C): WT n = 5, N1+/− n = 3 (patient-specific ECs). Red up flags: activating marks; blue down flags: repressive marks. In (D–F): WT n = 3, N1+/− n = 3 (patient-specific ECs). See also Figures S6 and Table S7.
Focusing on individual genes revealed that histone modification changes correlated with pro-calcific gene dysregulation in N1+/− ECs (Figure 4B). Pro-osteogenic genes involved in endochondral ossification (e.g., PLAU) and osteoblast regulation (e.g., COL1A1) showed upregulated mRNA expression associated with increased activating marks H3K4me3, H3K27ac, and H3K4me1 (Engelholm et al., 2001). Conversely, transcriptionally downregulated anti-calcific factors including anti-atherogenic genes such as CYP1B1 and osteoclast genes such as ACP5 showed decreased H3K27ac activating marks as well as increased repressive H3K27me3 marks (Alatalo et al., 2000).
We next investigated whether epigenetic dysregulation in N1+/− ECs reflected changes in the distribution of key TF motifs within active and repressed regions (Figure 4C). Epigenetic studies in WT ECs above showed that pro-inflammatory STAT and IRF motifs were enriched in H3K27ac-marked enhancers only in static conditions. Notably, STAT and IRF motifs were significantly enriched in H3K27ac-marked activation sites in N1+/− as compared to WT ECs during both static and shear stress conditions (Figure 4C). This suggests that in N1+/− ECs there is an even greater increase in STAT and IRF signaling in static conditions and that these pro-inflammatory signals are not appropriately downregulated in response to shear stress. Conversely, H3K27me3-marked repressive domains in WT ECs were enriched for SMAD2/3/4 motifs in shear stress conditions, while H3K27me3 sites in N1+/− ECs were depleted for SMAD2/3/4 motifs in shear stress, suggesting a de-repression of pro-osteogenic TGFβ signaling as a consequence of N1 haploinsufficiency. Furthermore, in both static and shear stress conditions, H3K27ac sites in N1+/− ECs were enriched for RUNX1 motifs compared to WT ECs, likely indicating a progression to early chondrogenic signaling given RUNX1’s role in chondroblasts and bone formation (Smith et al., 2005; Yamashiro et al., 2004). In sum, N1+/− ECs revealed a shift of activated chromatin marks towards putative pro-inflammatory and osteogenic regulatory domains and a depletion of repressive marks from putative pro-osteogenic enhancers.
To understand whether N1+/− ECs underwent more permanent silencing of regulatory regions that may in principle be related to disease, we evaluated the DNA methylation landscape in WT and N1+/− ECs by whole-genome bisulfite sequencing. We concentrated on the static condition as preliminary studies showed negligible change in methylation status in response to shear stress assays, which occurred over only 24 hours with little cell division to allow for methylation turnover. Bisulfite sequencing revealed 248 differentially methylated regions (DMRs) in N1+/− ECs compared to WT ECs (Figures 4D and E). Nearly half of these DMRs were novel regions not identified in previous efforts to capture the dynamic DNA methylation landscape essential for normal development (Ziller et al., 2013). This suggests that dysregulation of DNA methylation in disease settings may partially occur in regulatory domains otherwise stable throughout development.
DMRs due to N1 haploinsufficiency were significantly enriched for CpG islands (CpGIs) and shores (+/− 2 kb from CpGIs) and depleted for CpG open seas (> 4 kb from CpGIs) (Figure 4F). The largest enrichment of DNA methylation changes occured at CpG shore regions, suggesting that these regions might be the least stable in the disease state. In many regions, changes in DNA methylation were accompanied with chromatin mark dysregulation. Regions hypermethylated in N1+/− ECs lost H3K4me3 or H3K27ac activating marks present in WT ECs while regions hypomethylated in N1+/− ECs gained H3K4me3 or H3K27ac marks (Figure S6A). The extensive conversion of these DMRs between the silenced and activated state suggests robust changes in the epigenetic landscape of N1+/− ECs.
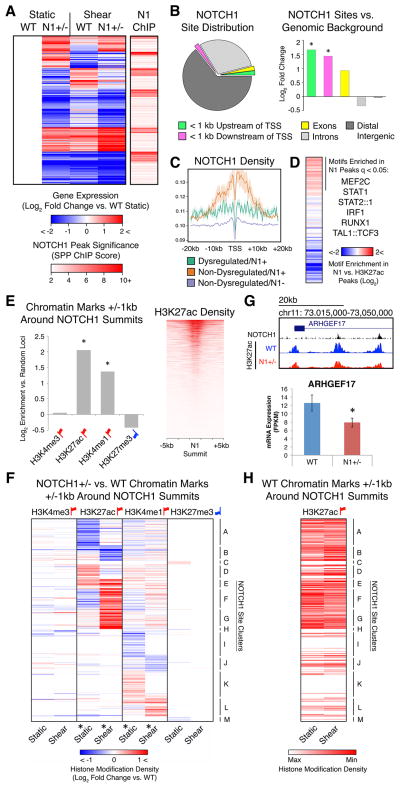
N1 Binding Sites Showed Dysregulation in H3K27ac Chromatin Marks in N1+/− ECs
To discern the aspects of transcriptional and epigenetic dysregulation in N1+/− ECs directly associated with N1 genome occupancy, we performed endogenous N1 ChIP-seq in primary human aortic ECs (HAECs). We found that 414 of the 1303 genes dysregulated in N1+/− ECs showed N1 binding in proximity to their TSSs (within 20 kb) as determined by peaks called in the N1 ChIP-seq (Figure 5A). Overall, dysregulated genes were significantly more likely to be found in proximity to a N1 peak than non-dysregulated genes (p < 0.05). Genes with the most significant N1 ChIP peaks near their TSSs were most often downregulated in N1+/− ECs, which is consistent with N1 acting as a transcriptional activator. However, many putative direct targets were also upregulated in N1 heterozygosity, indicating that N1 might also directly repress pro-calcific genes in WT ECs.
Figure 5. Transcriptional and Epigenetic Dysregulation Directly Associated with N1 Genome Occupancy.
(A) Left: K means clustering of putative direct N1 targets defined as genes significantly dysregulated in N1+/− ECs with N1 ChIP peaks within 20 kb of the TSS. Right: Significance of N1 peaks within 20 kb of the TSS of 414 putative direct N1 targets.
(B) Left: Distribution of N1 peaks. Right: Log2 fold change of proportion of N1 peaks vs. genomic background in indicated regions. *p < 0.05 by X2 test with Bonferroni correction.
(C) N1 density around the TSS of genes dysregulated in N1 haploinsufficiency with N1 peaks within 20 kb (green), non-dysregulated genes with N1 peaks within 20 kb (orange), or non-dysregulated genes without N1 peaks within 20 kb (blue).
(D) Motifs significantly enriched (q < 0.05 by motif Diverge with FDR correction) within 25 bps of N1 peak summits compared to H3K27ac peaks in ECs in static conditions.
(E) Left: Log2 fold change of overlap of chromatin marks in WT ECs with 1 kb around N1 summits vs. random non-gap genomic loci. *p < 0.05 by X2 test with Bonferroni correction. Right: H3K27ac density near N1 summits.
(F) Hierarchical clustering based on log2 fold change of N1+/− vs. WT histone modification density within 1 kb of N1 summits. *p < 0.05 by KS test with Bonferroni correction (histone modification dysregulation around N1 summits vs. random non-gap genomic loci).
(G) Top: N1 peaks and WT or N1+/− H3K27ac near ARHGEF17. Bottom: Mean mRNA expression of ARHGEF17. Error bars represent standard error; *p < 0.05 by negative binomial test with FDR correction.
(H) Relative H3K27ac density within 1 kb of N1 summits ordered as in (F) in WT ECs in static or shear stress conditions.
In (A–H): Gene expression: WT n = 3, N1+/− n = 2 (isogenic iPSC-derived ECs). Chromatin marks: WT n = 5, N1+/− n = 3 (patient-specific iPSC-derived ECs). N1 genome occupancy: WT n = 1 (union of 3 technical replicates) (primary HAECs). Red up flags: activating marks; blue down flags: repressive marks. See also Figure S6.
Most N1 binding events occurred in distal intergenic regions, but binding was most significantly enriched in promoter regions within 1 kb from TSSs compared to genomic background (Figure 5B). Putative direct N1 targets whose expression was unchanged by N1 haploinsufficiency on average showed greater enrichment of N1 binding near the TSS than targets dysregulated in N1+/− ECs (Figure 5C). Sites bound with lower levels of N1 may be more sensitive to a reduction in N1 dose, as they may be closer to a critical occupancy threshold required for gene activation. To determine motifs enriched in N1 binding sites, we compared motifs in genomic regions +/− 25 base pairs (bps) from N1 peak summits to motifs found in general open chromatin regions within ECs as determined by H3K27ac ChIP-seq (Figure 5D). Only 17% of N1 peaks contained the motif bound by its canonical DNA-binding partner CSL, suggesting that N1 may target DNA through alternate binding partners in ECs. However, N1-bound sites were enriched for motifs including RUNX1 motifs, as previously seen in T-lymphoblastic leukemia cells (Wang et al., 2011), as well as IRF and STAT motifs. Since RUNX1, IRF, and STAT motifs were all enriched in enhancers active specifically in N1+/− ECs, N1 complex binding may sufficiently compete with the binding of these TFs in the WT, but not mutant, state to prevent activation of related pro-osteogenic and pro-inflammatory signaling pathways.
We next investigated whether histone modifications near direct N1 binding sites were altered in ECs with decreased dosage of N1. In WT ECs, genomic regions +/− 1 kb from N1 peak summits were significantly enriched for H3K27ac and H3K4me1 and depleted, though not significantly, for H3K27me3 as defined by ChIP-seq (Figure 5E). In contrast, genomic regions +/− 1 kb from N1 peak summits showed significant alterations in H3K27ac and to a lesser degree in H3K4me1 in N1+/− ECs in both static and shear stress conditions compared to WT ECs (Figure 5F and S6B). N1+/− ECs showed increased H3K27ac at some sites and decreased H3K27ac at other sites, suggesting that N1 binding has a locus-specific effect on H3K27ac. For example, decreased H3K27ac surrounding two N1 binding sites within the gene body of the previously undescribed target ARHGEF17 correlated with its significant transcriptional downregulation in static conditions (Figure 5G).
We clustered N1 binding sites by the change in H3K27ac in N1 haploinsufficiency compared to WT, which revealed that the effect of shear stress on H3K27ac at N1 binding sites was reduced in N1+− ECs (Figure 5F and H). Within N1-bound sites where shear stress normally increased acetylation (Cluster B), mutant ECs showed less acetylation in shear stress than WT ECs. Conversely, sites with decreased acetylation in shear stress (Clusters E–G) showed incomplete deacetylation in mutant ECs as compared to WT ECs. This dampening of shear stress effects was less pronounced in distal H3K27ac loci devoid of N1 binding sites (Figure S6C). However, the pattern was present in H3K27ac loci proximal (within 1 kb) to TSSs and distinguished functional groups of genes (Figure S6D and Table S7). For example, promoters of genes encoding inhibitors of pro-osteogenic WNT signaling (e.g., HBP1, TLE) (Sampson et al., 2001; Wu et al., 2014) normally experienced increased H3K27ac in response to shear stress, but showed lower levels of acetylation in N1+/− ECs as compared to WT ECs in shear stress conditions. Since almost all N1 binding sites showing H3K27ac dysregulation were distal from gene TSSs, these data suggest that N1 binding may mediate the effect of shear stress on H3K27ac specifically at distal enhancers.
Manipulating Dysregulated Regulatory Nodes Restores Expression of N1 Downstream Targets Towards WT Levels
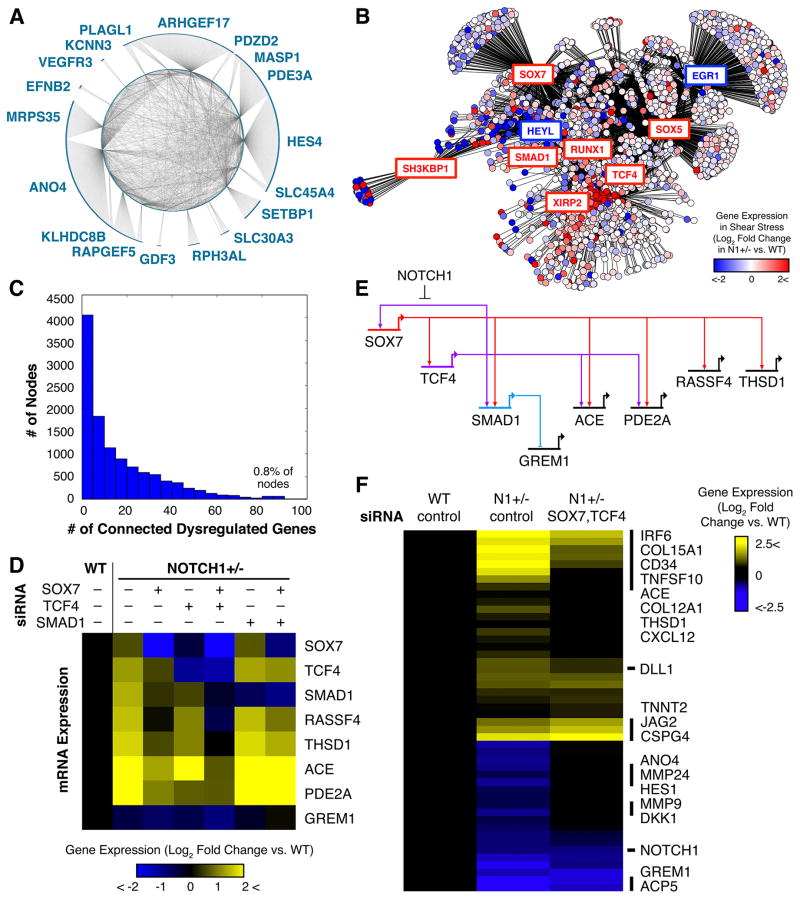
We sought to harness our knowledge of the transcriptional and epigenetic dysregulation caused by N1 haploinsufficiency to identify putative regulatory nodes within the gene network downstream of N1 that might serve as therapeutic targets. We employed a network inference algorithm to predict network connections using RNA-seq data of WT and N1+/− ECs in static and shear stress conditions (Margolin et al., 2006). With N1 as a network hub, we predicted several genes to be directly connected to N1 (Figure 6A). These included the canonical targets EPHNB2 and HES4 as well as ARHGEF17, which we described above as a direct N1 target dysregulated in N1+/− ECs. Genes with putative direct connections to N1 were themselves highly interconnected, suggesting that these genes might co-regulate one another to provide additional network stability. Each of these genes further branched out to connect to particular sets of genes, such as HES4, which connected to many genes strongly dysregulated in N1+/− ECs, and ARHGEF17, which connected to genes that were nearly all highly shear-responsive (Figure S7A–B).
Figure 6. Manipulation of Dysregulated Regulatory Nodes to Restore the EC Gene Network.
(A) Putative regulatory nodes directly connected to N1 in the predicted network and their interconnections (p < 0.05).
(B) Predicted gene regulatory network in ECs (p < 0.05) with each circle representing a gene and color indicating log2 fold change of N1+/− vs. WT expression in shear stress conditions. Boxed genes are putative dysregulated regulatory nodes with red and blue boxes indicating up-or downregulated genes, respectively.
(C) Histogram of number of nodes with different numbers of connected dysregulated genes. A small number of master regulators may control the majority of dysregulated genes.
(D) Effect of control, SOX7, TCF4, and/or SMAD1 siRNA on N1+/− EC mRNA expression of indicated genes as detected by QPCR.
(E) Gene regulatory subcircuit assembled based on perturbation results and network prediction.
(F) Effect of combined SOX7 and TCF4 siRNA on restoring N1+/− vs. WT expression of 48 genes dysregulated in N1 haploinsufficiency. N = 2.
See also Figure S7.
When we expanded our network prediction to include all dysregulated genes as hubs, we obtained a gene network with scale-free properties illustrating the predicted connections between genes downstream of N1 (Figure 6B). While most genes were connected to few dysregulated targets, several genes encoding transcriptional regulators were connected to a large portion of the genes dysregulated in N1 haploinsufficiency, suggesting that these putative regulatory nodes may control the majority of altered genes (Figure 6B and C). Among the nodes connected to the most dysregulated genes were SOX7, the WNT signaling effector TCF4, and the BMP signaling effector SMAD1, all upregulated in N1+/− ECs.
Using siRNAs, we corrected the aberrant upregulation of SOX7, TCF4, or SMAD1, alone or in combination, in an effort to restore the normal EC gene network. We monitored the effects of these perturbations on the initial putative targets (RASSF4, THSD1, ACE, PDE2A, and GREM1) selected based on connections predicted in our inferred gene network (Figure 6D). When we intervened solely with SOX7 siRNA, both TCF4 and SMAD1 were restored towards their WT expression levels, indicating that SOX7 is upstream of these predicted nodes. In addition, the expression of all other putative targets except GREM1 was partially to fully restored to WT levels. Intervening with TCF4 siRNA also reduced the aberrant upregulation of SOX7, demonstrating a positive feedback loop between SOX7 and TCF4. SMAD1 also shifted towards its WT expression whether through direct regulation by TCF4 or indirect regulation through SOX7. Knockdown of TCF4 also partially restored the expression of most putative downstream targets tested (RASSF4, THSD1, PDE2A), albeit to a lesser degree than knockdown of SOX7. However, treating N1+/− ECs with a combination of SOX7 and TCF4 siRNA had even more dramatic effects in restoring the WT expression of all putative targets excluding GREM1. In contrast, knocking down SMAD1 restored GREM1 expression towards WT levels without affecting SOX7, TCF4, or any of the other putative targets. Ultimately, based on our perturbation experiments and network inference, we were able to identify a gene network subcircuit involving nodes SOX7, TCF4, and SMAD1 that controls downstream gene dysregulation in N1+/− ECs (Figure 6E). N1 is responsible for repressing this subcircuit from activation in WT ECs while decreased N1 levels in N1+/− ECs are insufficient to prevent its activation.
To determine whether intervening with the SOX7 and TCF4 regulatory nodes had more widespread effects on restoring the gene network downstream of N1, we expanded our panel to include 48 genes dysregulated in N1+/− ECs selected based on our inferred gene network and RNA-seq data. We found that the combination of SOX7 and TCF4 siRNA restored the expression of the majority of these genes towards the WT state. N1 expression was unaffected, but expression of its canonical downstream target HES1 was restored towards WT, suggesting possible repair of the pathways downstream of N1 signaling. The siRNA treatment alleviated the downregulation of genes encoding MMPs (MMP24, MMP9), which may serve to degrade ECM in the valve, and secreted anti-osteogenic factors such as the WNT inhibitor DKK1 that may help prevent calcification in underlying VICs. Additionally, the treatment reduced upregulation of pro-inflammatory genes such as IRF6 and cytokine CXCL12 as well as collagens COL15A1 and COL12A1 that may contribute to calcification. Overall, correcting the aberrant upregulation of regulatory nodes SOX7 and TCF4 using siRNA was able to restore expression of genes within the network dysregulated in N1+/− ECs towards the WT state.
DISCUSSION
We have defined the mechanisms critical for normal EC differentiation and response to shear stress, determined how these mechanisms are perturbed in N1 haploinsufficient cells, and intervened at key regulatory nodes to restore N1+/− ECs towards their WT state. Overall, iPSC-based modeling of human N1 mutations allowed a rigorous interrogation of the gene networks disrupted in ECs from patients with CAVD to reveal novel targets for intervention. Moreover, the findings here demonstrate mechanisms by which dose-reduction of a TF can alter the epigenetics and transcriptome in a human disease model.
Transcriptional and Epigenetic Mechanisms Governing EC Differentiation and Response to Shear Stress
Among the more interesting findings from transcriptional and epigenetic profiling during EC differentiation was that shear stress induced a highly coordinated suppression of pro-osteogenic and inflammatory signaling in ECs that may be critical to protect against calcification events. Shear-dependent histone modifications correlated with upregulation of anti-osteogenic genes, including those encoding secreted BMP and WNT antagonists that may represent a paracrine signaling method for shear-exposed ECs to prevent the calcification of neighboring tissue. Synchronously, shear stress downregulated pro-inflammatory cytokines, reduced STAT and IRF signaling effectors characteristic of ECs in static conditions, and shifted activating histone modifications towards anti-inflammatory and antioxidant motifs. It is interesting to consider that ECs throughout the vasculature may function to repress pro-osteogenic events in response to laminar shear stress, given the propensity for calcification at sites of vascular bifurcation experiencing turbulent blood flow.
Transcriptional Consequences of N1 Haploinsufficiency in iPSC-Based Modeling of CAVD
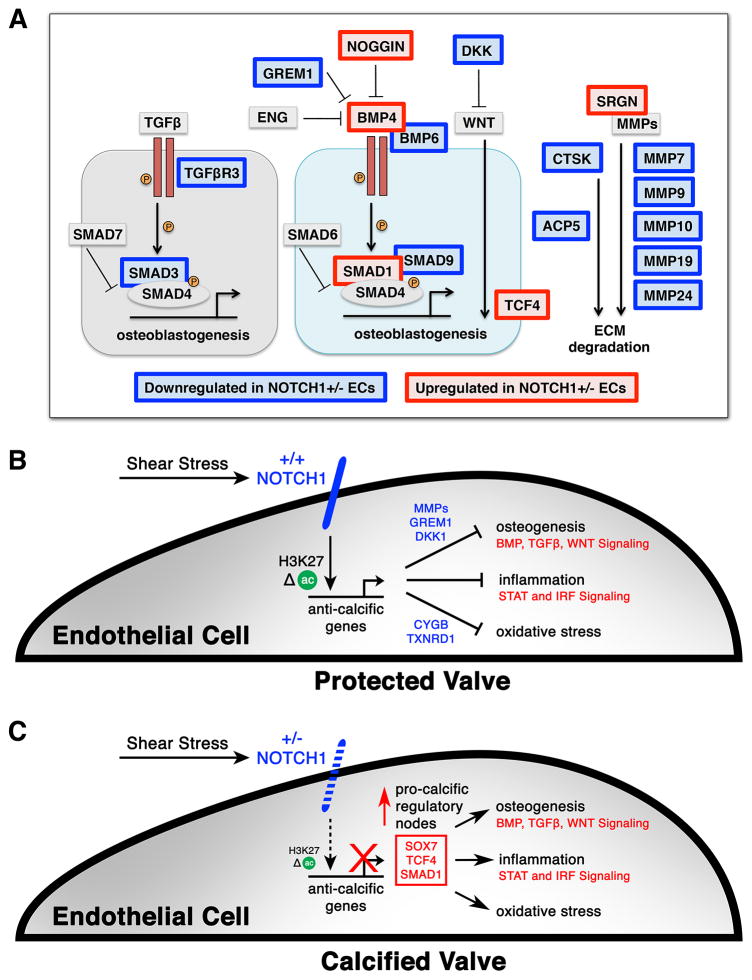
iPSC-based modeling of human N1 mutations in CAVD revealed that N1 haploinsufficiency disrupts the appropriate EC response to shear stress (Figure 7A–C). In contrast to WT ECs, shear-exposed N1+/− ECs failed to upregulate anti-osteogenic factors, including secreted BMP and WNT antagonists that may be critical for preventing calcification of underlying VICs. They instead overexpressed pro-osteogenic genes such as BMP4, suggestive of an osteoblast-like switch, highlighting the importance of the N1-dependent response to shear stress in maintaining the cell fate of valve ECs. Conversion of valve ECs into osteoblast-like cells has been reported in disease states (Hofmann et al., 2012), consistent with N1 functioning to repress this aberrant gene program. Interestingly, the secreted anti-osteogenic factor, Matrix Gla Protein (MGP), was very lowly expressed in iPSC-derived ECs despite its abundance in native murine valve tissue (Luo et al., 1997), making it difficult to determine if MGP was a shear-responsive N1 target in human cells. Nevertheless, the transcriptional disturbances in N1+/− ECs indicated a dysregulated inflammatory environment and vulnerability to oxidative stress that may fuel the progression of calcification in an aortic valve without the anti-calcific barriers normally erected by shear-exposed ECs.
Figure 7. Model of Mechanisms Regulating Pro-Calcific Events in N1 Haploinsufficient ECs.
(A) Diagram of osteogenic pathways dysregulated in N1 haploinsufficiency. Red indicates upregulation in N1+/− ECs and blue indicates downregulation in N1+/− ECs.
(B) Model of WT ECs. Shear stress activates N1 signaling in ECs, leading to epigenetic changes at N1-bound enhancers and transcriptional activation of anti-calcific gene programs that prevent osteogenesis, inflammation, and oxidative stress to protect the valve from calcification.
(C) Model of N1+/− ECs, which cannot mediate the proper response to shear stress, leading to epigenetic dysregulation at N1-responsive enhancers and aberrant upregulation of pro-calcific regulatory nodes.
Determining the pathways dysregulated in N1+/− ECs focuses therapeutic efforts on reinstating the proper EC response to shear stress and restoring the anti-osteogenic and anti-inflammatory barriers against calcification in the valve. Furthermore, defining the factors important for maintaining these barriers provides insight into alternative genes that may be mutated in CAVD patients without mutations in N1. Future work delineating whether mutations in various members of the network preventing CAVD lead to aortic valve disease may explain why only a portion of patients with BAV progress to valve calcification.
N1 Genome Occupancy and Epigenetic Dysregulation in N1+/− ECs
Determining the transcriptional and epigenetic consequences of N1 haploinsufficiency occurring directly at N1-bound sites provides insight into how TF dosage differentially affects targets leading to human disease. At baseline in WT ECs, gene targets dysregulated in N1+/− ECs had lower N1 occupancy proximal to their TSS compared to non-dysregulated targets. Thus, genes most sensitive to decreased dosage of N1 begin with lower levels of N1 binding at baseline, potentially placing them closer to the threshold at which binding becomes insufficient to affect transcription.
Consistent with previous reports of N1 recruitment of histone acetyltransferases (Yashiro-Ohtani et al., 2014), N1 binding sites showed the greatest changes in H3K27ac, compared to other epigenetic marks, and could not mount the proper epigenetic response to shear stress in N1+/− ECs. The changes in H3K27ac correlated with transcriptional dysregulation of putative direct targets such as ARHGEF17, which we found to be a major predicted regulatory node, indicating that WT levels of N1 binding are required to maintain the appropriate epigenetic state and transcriptional regulation of the most sensitive direct targets.
N1 haploinsufficiency had broad downstream effects on the epigenetic landscape in ECs. Epigenetic dysregulation extended to DNA methylation, indicating an extensive shift in the regulatory state of key domains in ECs. CpG shore regions were the most vulnerable to dysregulation, showing the largest enrichment of methylation changes in N1 haploinsufficient ECs. These data support the notion that CpG shores are the least stable in the disease state, consistent with previous reports of shores displaying the most methylation differences in the context of cancerous cells and specific tissue types (Irizarry et al., 2009). This ability to differentiate between distinct cell states may have important implications in diagnosis of disease. For example, DNA methylation may provide a useful marker for patients who are at risk for valve calcification.
Identification of Dysregulated Transcriptional Nodes
An incomplete understanding of the molecular mechanisms involved in the development of CAVD has hampered the design of effective therapies. By thoroughly interrogating the transcriptional and epigenetic consequences of N1 haploinsufficiency, we identified transcriptional nodes controlled by SOX7 and TCF4, where intervention impacted a large portion of the gene dysregulation in patient-specific ECs. The ability to exert a concerted influence on gene networks disrupted in cells from patients with CAVD by targeting discrete regulatory nodes has potential therapeutic implications. Using this iPSC-based disease model, we anticipate screening for small molecules that target central regulatory nodes to inhibit or delay the progression of calcification in patients at risk for CAVD.
The work presented here demonstrates the value of computationally integrating genome-wide transcriptome, DNA methylation, and histone modification data with gene network analyses to reveal the consequences of human disease-causing mutations. We believe this type of broad and comprehensive approach will serve as the foundation for rational drug design for many disorders in the coming years.
EXPERIMENTAL PROCEDURES
Experimental details can be found in Extended Experimental Procedures.
Supplementary Material
Acknowledgments
We are grateful to members of the Srivastava Lab and Gladstone/UCSF community for helpful scientific discussions. We thank the Gladstone Genomics, Histology, and Stem Cell Cores and UCSF ESC Targeting Core for their technical expertise. We thank G. Howard and B. Taylor for their editorial help and K. Ivey and C. Gifford for critical comments. This work was supported by NIH/NHLBI grants U01 HL098179 (B.G.B., K.S.P., D.S.) and U01 HL100406 (B.G.B., D.S.). D.S. was supported by the L.K. Whittier, William H. Younger Family, and Eugene Roddenberry Foundations and the California Institute for Regenerative Medicine. B.G.B. was supported by the Lawrence J. and Florence A. DeGeorge Charitable Trust/American Heart Association (AHA) Established Investigator Award and William H. Younger Family Foundation. K.S.P. was supported by a grant from NIH/NHLBI (HL089707). C.V.T. was supported by the AHA Predoctoral, UCSF Discovery, Roddenberry, and Winslow Fellowships and the UCSF Core Exploratory Award, Developmental and Stem Cell Biology Graduate Program (NIH T32HD007470), and Medical Scientist Training Program (NIH T32GM007618). M.P.W. was supported by the AHA Predoctoral and Winslow Fellowship, and M.L. was supported by a Sarnoff Fellowship.
Footnotes
AUTHOR CONTRIBUTIONS
C.V.T. designed/performed experiments and designed/performed bioinformatic analyses on transcriptional, epigenetic, genome occupancy, and network inference studies. M.L. contributed to bioinformatic analyses on epigenetic data. M.P.W. supervised iPSC reprogramming and provided EC differentiation method. C.V.T. and L.L. performed EC differentiations and TALEN genome engineering. C.V.T. and D.H. performed ChIP assays. K.S.P supervised computational design, analyses, and interpretation and edited the manuscript. B.G.B. supervised epigenetic analyses and interpretation and edited the manuscript. D.S. designed experiments and analyses and supervised the work. C.V.T. and D.S. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alatalo SL, Halleen JM, Hentunen TA, Mönkkönen J, Väänänen HK. Rapid screening method for osteoclast differentiation in vitro that measures tartrate-resistant acid phosphatase 5b activity secreted into the culture medium. Clin Chem. 2000;46:1751–1754. [PubMed] [Google Scholar]
- Bragdon B, Moseychuk O, Saldanha S, King D, Julian J, Nohe A. Bone morphogenetic proteins: a critical review. Cell Sig. 2011;23:609–620. doi: 10.1016/j.cellsig.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Combs MD, Yutzey KE. Heart valve development: Regulatory networks in development and disease. Circ Res. 2009;105:408–421. doi: 10.1161/CIRCRESAHA.109.201566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway DE, Sakurai Y, Weiss D, Vega JD, Taylor WR, Jo H, Eskin SG, Marcus CB, McIntire LV. Expression of CYP1A1 and CYP1B1 in human endothelial cells: Regulation by fluid shear stress. Cardiovasc Res. 2009;81:669–677. doi: 10.1093/cvr/cvn360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleuze V, El-Hajj R, Chalhoub E, Dohet C, Pinet V, Couttet P, Mathieu D. Angiopoietin-2 is a direct transcriptional target of TAL1, LYL1, and LMO2 in endothelial cells. PLoS ONE. 2012;7:e40484. doi: 10.1371/journal.pone.0040484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennler S, Prunier C, Ferrand N, Gauthier J, Atfi A. c-Jun inhibits transforming growth factor beta -mediated transcription by repressing Smad3 transcriptional activity. J Biol Chem. 2000;275:28858–28865. doi: 10.1074/jbc.M910358199. [DOI] [PubMed] [Google Scholar]
- Drake CJ, Brandt SJ, Trusk TC, Little CD. TAL1/SCL is expressed in endothelial progenitor cells/angioblasts and defines a dorsal-to-ventral gradient of vasculogenesis. Dev Biol. 1997;192:17–30. doi: 10.1006/dbio.1997.8751. [DOI] [PubMed] [Google Scholar]
- Engelholm LH, Nielsen BS, Netzel-Arnett S, Solberg H, Chen XD, Garcia JML, Lopez-Otin C, Young MF, Birkedal-Hansen H, Danø K, et al. The urokinase plasminogen activator receptor–associated protein/endo180 is coexpressed with its interaction partners urokinase plasminogen activator receptor and matrix metalloprotease-13 during osteogenesis. Lab Invest. 2001;81:1403–1414. doi: 10.1038/labinvest.3780354. [DOI] [PubMed] [Google Scholar]
- Foffa I, Ait Alì L, Panesi P, Mariani M, Festa P, Botto N, Vecoli C, Andreassi MG. Sequencing of NOTCH1, GATA5, TGFBR1 and TGFBR2 genes in familial cases of bicuspid aortic valve. BMC Med Genet. 2013;14:44. doi: 10.1186/1471-2350-14-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, Dai S, Ford ES, Fox CS, Franco S, et al. Heart disease and stroke statistics--2014 update: A report from the American Heart Association. Circulation. 2014;129:e28–e292. doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goettsch C, Hutcheson JD, Aikawa E. MicroRNA in Cardiovascular Calcification: Focus on Targets and Extracellular Vesicle Delivery Mechanisms. Circ Res. 2013;112:1073–1084. doi: 10.1161/CIRCRESAHA.113.300937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- Guo B, Slevin M, Li C, Parameshwar S, Liu D, Kumar P, Bernabeu C, Kumar S. CD105 inhibits transforming growth factor-β-Smad3 signalling. Anticancer Res. 2004:1337–1346. [PubMed] [Google Scholar]
- Hervas-Stubbs S, Perez-Gracia JL, Rouzaut A, Sanmamed MF, Le Bon A, Melero I. Direct effects of type I interferons on cells of the immune system. Clin Cancer Res. 2011;17:2619–2627. doi: 10.1158/1078-0432.CCR-10-1114. [DOI] [PubMed] [Google Scholar]
- Hofmann JJ, Briot A, Enciso J, Zovein AC, Ren S, Zhang ZW, Radtke F, Simons M, Wang Y, Iruela-Arispe ML. Endothelial deletion of murine Jag1 leads to valve calcification and congenital heart defects associated with Alagille syndrome. Development. 2012;139:4449–4460. doi: 10.1242/dev.084871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwa MQ, Nguyen T, Huynh J, Ramnath D, De Nardo D, Lam PY, Reynolds EC, Hamilton JA, Sweet MJ, Scholz GM. Interferon regulatory factor 6 differentially regulates Toll-like receptor 2-dependent chemokine gene expression in epithelial cells. J Biol Chem. 2014;289:19758–19768. doi: 10.1074/jbc.M114.584540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Chen XQ, Li WJ, Yang YH, Wang JZ, Yu ACH. Cytoglobin up-regulated by hydrogen peroxide plays a protective role in oxidative stress. Neurochem Res. 2007;32:1375–1380. doi: 10.1007/s11064-007-9317-x. [DOI] [PubMed] [Google Scholar]
- Li H, Li T, Wang S, Wei J, Fan J, Li J, Han Q, Liao L, Shao C, Zhao RC. miR-17-5p and miR-106a are involved in the balance between osteogenic and adipogenic differentiation of adipose-derived mesenchymal stem cells. Stem Cell Res. 2013;10:313–324. doi: 10.1016/j.scr.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Liao EC, Paw BH, Oates AC, Pratt SJ, Postlethwait JH, Zon LI. SCL/Tal-1 transcription factor acts downstream of cloche to specify hematopoietic and vascular progenitors in zebrafish. Genes & Dev. 1998;12:621. doi: 10.1101/gad.12.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Ducy P, McKee MD, Pinero GJ, Loyer E, Behringer RR, Karsenty G. Spontaneous calcification of arteries and cartilage in mice lacking matrix GLA protein. Nature. 1997;386:78–81. doi: 10.1038/386078a0. [DOI] [PubMed] [Google Scholar]
- Masumura T, Yamamoto K, Shimizu N, Obi S, Ando J. Shear stress increases expression of the arterial endothelial marker ephrinB2 in murine ES cells via the VEGF-Notch signaling pathways. Arterioscler Thromb Vasc Biol. 2009;29:2125–2131. doi: 10.1161/ATVBAHA.109.193185. [DOI] [PubMed] [Google Scholar]
- Mohamed SA, Aherrahrou Z, Liptau H, Erasmi AW, Hagemann C, Wrobel S, Borzym K, Schunkert H, Sievers HH, Erdmann J. Novel missense mutations (p. T596M and p P1797H) in NOTCH1 in patients with bicuspid aortic valve. Biochem Biophys Res Commun. 2006;345:1460–1465. doi: 10.1016/j.bbrc.2006.05.046. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2010;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson EM, Haque ZK, Ku M, Tevosian S, Albanese C, Pestell R, Paulson KE, Yee AS. Negative regulation of the Wnt-beta-catenin pathway by the transcriptional repressor HBP1. EMBO J. 2001;20:4500–4511. doi: 10.1093/emboj/20.16.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Richard J, Zirbes KM, Gong W, Lin G, Kyba M, Thomson JA, Koyano-Nakagawa N, Garry DJ. Cooperative interaction of Etv2 and Gata2 regulates the development of endothelial and hematopoietic lineages. Dev Biol. 2014;389:208–218. doi: 10.1016/j.ydbio.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober A, Nazari-Jahantigh M, Wei Y, Bidzhekov K, Gremse F, Grommes J, Megens RTA, Heyll K, Noels H, Hristov M, et al. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat Med. 2014;20:368–376. doi: 10.1038/nm.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N, Dong Y, Lian JB, Pratap J, Kingsley PD, van Wijnen AJ, Stein JL, Schwarz EM, O’Keefe RJ, Stein GS, et al. Overlapping expression of Runx1(Cbfa2) and Runx2(Cbfa1) transcription factors supports cooperative induction of skeletal development. J Cell Physiol. 2005;283:133–143. doi: 10.1002/jcp.20210. [DOI] [PubMed] [Google Scholar]
- Surapisitchat J, Jeon KI, Yan C, Beavo JA. Differential regulation of endothelial cell permeability by cGMP via phosphodiesterases 2 and 3. Circ Res. 2007;101:811–818. doi: 10.1161/CIRCRESAHA.107.154229. [DOI] [PubMed] [Google Scholar]
- Van Handel B, Montel-Hagen A, Sasidharan R, Nakano H, Ferrari R, Boogerd CJ, Schredelseker J, Wang Y, Hunter S, Org T, et al. Scl Represses Cardiomyogenesis in Prospective Hemogenic Endothelium and Endocardium. Cell. 2012;150:590–605. doi: 10.1016/j.cell.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu TH, Werb Z. Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev. 2000;14:2123–2133. doi: 10.1101/gad.815400. [DOI] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, Pear WS, Schug J, Blacklow SC, Arnett KL, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci USA. 2011;108:14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg EJ, Mack PJ, Schoen FJ, García-Cardeña G, Kaazempur Mofrad MR. Hemodynamic environments from opposing sides of human aortic valve leaflets evoke distinct endothelial phenotypes in vitro. Cardiovasc Eng. 2010;10:5–11. doi: 10.1007/s10558-009-9089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MP, Rufaihah AJ, Liu L, Ghebremariam YT, Ivey KN, Cooke JP, Srivastava D. Limited gene expression variation in human embryonic stem cell and induced pluripotent stem cell derived endothelial cells. Stem Cells. 2012;31:92–103. doi: 10.1002/stem.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bowe DB, Sadlonova A, Whisenhunt TR, Hu Y, Rustgi AK, Nie Y, Paterson AJ, Yang X. O-GlcNAc transferase is critical for transducin-like enhancer of split (TLE)-mediated repression of canonical Wnt signaling. J Biol Chem. 2014;289:12168–12176. doi: 10.1074/jbc.M114.553859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro T, Wang XP, Li Z, Oya S, Aberg T, Fukunaga T, Kamioka H, Speck NA, Takano-Yamamoto T, Thesleff I. Possible roles of Runx1 and Sox9 in incipient intramembranous ossification. J Bone Miner Res. 2004;19:1671–1677. doi: 10.1359/JBMR.040801. [DOI] [PubMed] [Google Scholar]
- Yashiro-Ohtani Y, Wang H, Zang C, Arnett KL, Bailis W, Ho Y, Knoechel B, Lanauze C, Louis L, Forsyth KS, et al. Long-range enhancer activity determines Myc sensitivity to Notch inhibitors in T cell leukemia. Proc Natl Acad Sci USA. 2014;111:E4946–E4953. doi: 10.1073/pnas.1407079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LTY, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.