Abstract
Messenger RNA degradation represents a critical regulated step in gene expression. While the major pathways in turnover have been identified, accounting for disparate half-lives has been elusive. We show that codon optimality is one feature that contributes greatly to mRNA stability. Genome-wide RNA decay analysis revealed that stable mRNAs are enriched in codons designated optimal, whereas unstable mRNAs contain predominately non-optimal codons. Substitution of optimal codons with synonymous, non-optimal codons results in dramatic mRNA destabilization, while the converse substitution significantly increases stability. Further, we demonstrate that codon optimality impacts ribosome translocation, connecting the processes of translation elongation and decay through codon optimality. Finally, we show that optimal codon content accounts for the similar stabilities observed in mRNAs encoding proteins with coordinated physiological function. This work demonstrates that codon optimization exists as an mechanism to finely tune levels of mRNAs, and ultimately, proteins.
INTRODUCTION
Messenger RNA (mRNA) degradation plays a critical role in regulating transcript levels in the cell and is a major control point for modulating gene expression. Degradation of most mRNAs in Saccharomyces cerevisiae is initiated by removal of the 3′ poly(A) tail (deadenylation), followed by cleavage of the 5′ 7mGpppN cap (decapping) and exonucleolytic degradation of the mRNA body in a 5′-3′ direction (Coller and Parker, 2004; Ghosh and Jacobson, 2010). Despite being targeted by a common decay pathway, turnover rates for individual yeast mRNAs differ dramatically with half-lives ranging from <1 minute to 60 minutes or greater (Coller and Parker, 2004). RNA features that influence transcript stability have long been sought, and some sequence and/or structural elements located within 5′ and 3′ untranslated regions (UTRs) have been implicated in contributing to the decay of a subset of mRNAs (Lee and Lykke-Andersen, 2013; Muhlrad and Parker, 1992; Geisberg et al., 2014). However, these features regulate mRNA stability predominantly in a transcript-specific manner through binding of regulatory factors and cannot account for the wide variation in half-lives observed across the entire transcriptome (Geisberg et al., 2014). Therefore, it seems likely that additional and more general features which act to modulate transcript stability could exist within mRNAs.
We have previously shown that inclusion of a cluster of rare arginine codons within the open reading frame (ORF) of a reporter mRNA dramatically enhanced its turnover (Hu et al., 2009; Sweet et al., 2012). The mRNA destabilization caused by rare codons was manifest in the enhancement of both deadenylation and decapping of the transcript. This effect was not dependent on RNA surveillance pathways such as No-Go, Nonsense-Mediated, or Non-Stop Decay (Shoemaker and Green, 2012). The links between rare codons and enhanced mRNA turnover rates of reporter mRNAs are consistent with earlier observations for several endogenous transcripts in yeast (Caponigro et al., 1993; Hoekema et al., 1987).
The rare codons used in our previous studies belong to a general class of codons defined as non-optimal (Pechmann and Frydman, 2013; Reis et al., 2004). Conceptually, codon optimality is a scale that reflects the balance between the supply of charged tRNA molecules in the cytoplasmic pool and the demand of tRNA usage by translating ribosomes, representing a measure of translation efficiency. Critically, optimal codons are postulated to be decoded faster and more accurately by the ribosome than non-optimal codons (Akashi, 1994; Drummond and Wilke, 2008), which are hypothesized to slow translation elongation (Novoa and Ribas de Pouplana, 2012; Tuller et al., 2010). Therefore, codon optimality is hypothesized to play an important role in modulation of translation elongation rates and the kinetics of protein synthesis (Kri Ko et al., 2014; Novoa and Ribas de Pouplana, 2012; Pechmann and Frydman, 2013; Reis et al., 2004). In this work, we present four lines of evidence in support of the finding that codon optimality has a broad and powerful influence on mRNA stability in yeast cells. First, global analysis of RNA decay rates reveals that mRNA half-life correlates with optimal codon content. Many stable mRNAs demonstrate a strong preference towards the inclusion of optimal codons within their coding regions, while many unstable mRNAs harbor non-optimal codons. Second, we demonstrate that substitution of optimal codons with synonymous, non-optimal codons results in a dramatic destabilization of the mRNA and that the converse replacement leads to a significant increase in mRNA stability. Third, we experimentally demonstrate an impact of codon optimality on ribosome translocation indicating that the effect on mRNA decay occurs through modulation of mRNA translation elongation. These findings indicate that transcript-specific translation elongation rate, as dictated by codon usage, is an important determinant of mRNA stability. Fourth, we observe tightly coordinated optimal codon content in genes encoding proteins with common physiological function. We hypothesize that this finding explains the previously observed similarity in mRNA decay rates for these gene families. Taken together, our data suggest that there is evolutionary pressure on protein coding regions to coordinate gene expression at the level of protein synthesis and mRNA decay.
RESULTS
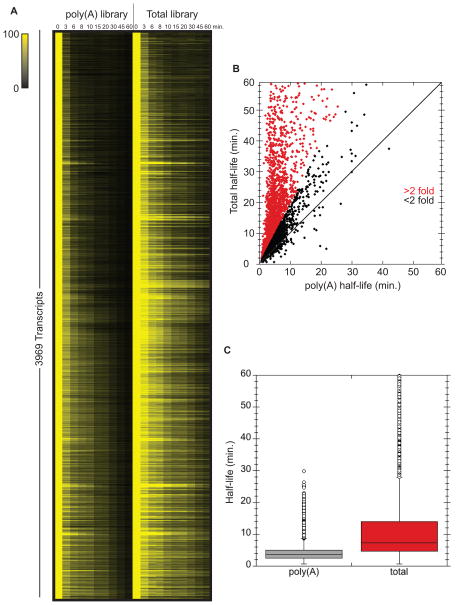
Measuring global mRNA decay rates using methods that either enrich for polyA+ RNA from total RNA samples and/or synthesize complementary DNA (cDNA) using oligonucleotides annealed to the poly(A) tail may fail to capture important information for several reasons. Although it is firmly established that deadenylation is the rate limiting step in mRNA turnover, we and others have observed that specific mRNAs persist in cells as “stable” deadenylated species (Hu et al., 2009; Muhlrad et al., 1995). For such transcripts, decapping and subsequent decay is delayed and decapping becomes the rate defining step for mRNA degradation. Moreover, some mRNAs may contain structures that impede poly(A) tail function (Geisberg et al., 2014). Lastly, since the process of deadenylation converts an mRNA species from one that can be efficiently captured by oligo dT to one that cannot, the overall level of information gained may vary with the level of poly(A) enrichment achieved in the protocol used. With this in mind, we sought to determine how prevalent these phenomena are on a transcriptome-wide level. For this purpose, we performed a time course after inactivation of RNA polymerase II (Nonet et al., 1987). At each time point, libraries were prepared from either oligo-dT selected mRNAs or rRNA-depleted whole cell RNA and subjected to Illumina sequencing (see experimental procedures). This approach allowed us to compare poly(A) half-lives (oligo dT) with total mRNA decay rates (rRNA depleted; Figure 1A). Remarkably, the vast majority (92%) of transcripts for which we could confidently calculate half-lives (3969) had longer half-lives when the rRNA depleted libraries were analyzed relative to the half-lives determined from poly(A) selected libraries (Fig. 1B and C). It is important to note that not all of these transcripts exist as deadenylated RNAs since mRNAs with short poly(A) tails will not bind oligo dT. These data indicate that mRNA half-lives determined by oligo dT selection give highly skewed values. For example, the ADH1 mRNA had a calculated half-life of 4.2 minutes when determined from poly(A) selected RNA and a 31.7 minute half-life when determined from rRNA depleted RNA (see Table S1 for complete list).
Figure 1. Half-lives calculated from poly(A)+ vs total mRNA differ significantly.
RNA-seq was performed on poly(A)+ and total RNA libraries prepared from rpb1-1 transcriptional shut-off experiments across a 60 minute time course. (A) All mRNAs with reliable half-lives in both libraries are plotted visually. Color intensity represents normalized mRNA remaining (time 0 is set to 100% for each mRNA). (B) Half-life of each mRNA plotted as calculated from total mRNA sequencing against the poly(A) sequencing. Data points with a >2 fold difference are highlighted in red. (C) Overview of the distribution of half-lives for both libraries. See also Table S1.
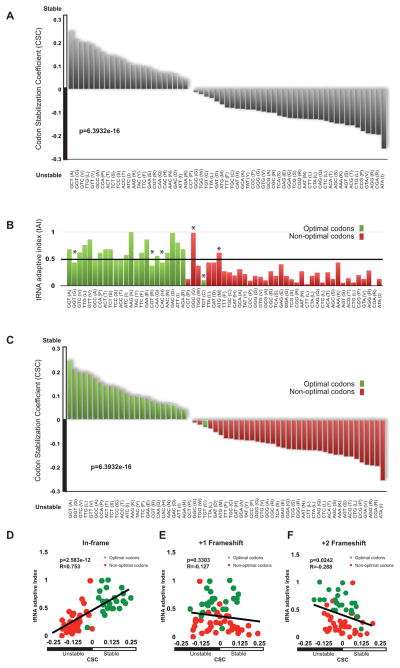
With this data in hand we attempted to identify sequence motifs that might dictate stability or instability, without success. Following up on previous observations that inclusion of ten consecutive rare codons in the open reading frames of an otherwise stable mRNA caused a dramatic decrease in stability (Hu et al., 2009; Sweet et al., 2012), we inspected our transcriptome-wide mRNA half-life data to determine whether codon content within ORFs could affect mRNA stability. To do so, we determined if mRNAs enriched in any individual codon demonstrated greater or lesser stability. We defined mRNAs as stable if they have a half-life greater than 2-fold longer than the average (~20 min), and unstable if they have a half-life less than half of the average (~5 min). For each codon, we calculated a correlation between the frequency of occurrence of that codon in mRNAs and the stabilities of the mRNAs. Occurrences of a codon were compared to the half-life for each mRNA and a Pearson correlation calculation was used to generate an R-value (graphically represented for sample codons in Fig. S1E). We refer to this metric as the Codon occurrence to mRNA Stability Correlation coefficient (CSC). The CSC values for all codons were then compared to each other (Fig. 2A). Strikingly, it was observed that some codons preferentially occurred in stable mRNAs while others occurred preferentially in unstable mRNAs (overall p-value = 1.496e-14, permutation p-value < 10−4). For example, the GCT alanine codon was highly enriched in stable transcripts as defined by our RNA-seq analysis, while its synonymous codons, GCG and GCA were preferentially present in unstable transcripts (Fig. 2A). Approximately one-third of all codon triplets were over-represented in stable mRNAs, while the remaining two-thirds appeared to predominate in unstable mRNAs. As a consequence of the large dataset and significance of the observed correlation, these data strongly suggest that codon usage influences mRNA degradation rates.
Figure 2. Codon composition correlates with stability.
(A) The Codon occurrence to mRNA Stability Correlation coefficient (CSC) plotted for each codon as calculated from the total RNA data set. The CSC is the R-value of the correlation between the occurrences of that codon and the half-lives of mRNA. Overall p-value is 6.3932e-16, permutation p-value is < 10−4. (B) tRNA Adaptability Index values for each codon plotted in the same order as (A). Codon optimality as defined in Pechmann and Frydman (2013) is color coded, using green for optimal codons and red for non-optimal codons. Codons designated with an asterisk (*) were called optimal or non-optimal according to additional criteria discussed therein. (C) The Codon occurrence to mRNA Stability Correlation coefficient (CSC) plotted for each codon as in (A), but optimality information presented in (B) is added by color-coding. Green color represents optimal codons and red represents non-optimal. (D) tRNA Adaptive Index values plotted vs CSC when ORFs are considered in-frame. Green indicates optimal codons, red indicates non-optimal codons (R = .7255, p-value is p-value = 2.075e-09, permutation p-value < 10−4) (E) tRNA Adaptive Index values plotted vs CSC when ORFs are frameshifted by one nucleotide. Green indicates optimal codons, red indicates non-optimal codons. (F) tRNA Adaptive Index values plotted vs CSC when ORFs are frameshifted by two nucleotides. Green indicates optimal codons, red indicates non-optimal codons. See also Figure S1.
For decades, a large body of literature has hypothesized that some codons may be translated more efficiently than others. Reis et al. (2004) laid out a measure of how efficiently a codon would be translated, and termed it the tRNA Adaptive Index (tAI). This metric is meant to reflect the efficiency of tRNA usage by the ribosome. The term codon optimality has been introduced in an attempt to define the differential recognition of codons by the translational apparatus (Zhou et al., 2009; Pechmann and Frydman 2013). Frydman and colleagues generally defined any codon with a tAI above 0.47 as optimal and any codon with a tAI below 0.47 as non-optimal (Pechmann and Frydman 2013; Fig. 2B). Their final designation of codons also takes into account the over- and under-representation of certain codons in the genome, known as codon bias (Fig. 2B; marked with *; Zhou et al., 2009; Pechmann and Frydman 2013). As such, codon optimality is somewhat reflected in genomic codon usage (Fig. S1A); however, commonly occurring codons can be optimal or non-optimal, while uncommon codons can also be optimal or non-optimal (Fig. S1B). Strikingly, codons associated with stable or unstable mRNAs nearly perfectly mirrored their assignment as optimal or non-optimal, respectively (Fig. 2C). Direct comparison between our CSC metric and tAI revealed very good overall agreement between these values (Fig. 2D; R = 0.753, p-value = 2.583e-12, permutation p-value < 10−4). Importantly, the relationship between optimal codon content and mRNA half-life is independent of the method used to determine half-life. We repeated our analysis of codon usage vs. mRNA half-life using mRNA decay rates obtained by Miller et al., (2011). In contrast to our own, these data were obtained with a steady state approach calculation using metabolic labelling that minimally perturbs the cell and is completely distinct from our method (Miller et al. 2011). Both datasets show a similar and striking correlation between optimal codon content and mRNA decay rate (Fig. S1C and D).
To determine if the codon optimality correlation was possibly masking other features that might actually be determining mRNA half-life (e.g. sequence content, GC percentage, or secondary structure), we reanalyzed our data after computationally introducing +1 and +2 frameshifts. In the analysis of these frameshifted ORFs, the correlation between codon content and stability completely disappears, thus eliminating other variables as determinative (Fig. 2E; R = −0.127, p-value = 0.3303, permutation p-value = 0.8847 and Fig. 2F; R= −0.288, p-value = 0.0242, permutation p-value = 0.0012).
Stable and unstable mRNAs demonstrate different optimal codon content
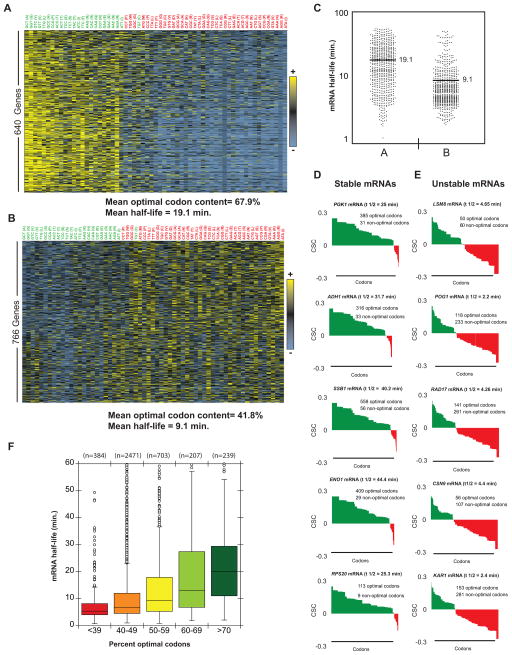
As shown above, computational analysis of our global mRNA stability data revealed a relationship between codon occurrence and mRNA half-life. These data indicate that either particular codons alter stability or overall codon content within an mRNA works collectively on stability. To evaluate the relationship between optimal codon content and decay rate on the level of individual transcripts, codon usage was mapped across all individual transcripts (Fig. S2). Cluster analysis revealed that different mRNAs are biased towards using different types of codons. The overall result is not surprising, as codon bias has been well studied (Gustafsson et al., 2004); however, the pattern of codon usage demonstrates that certain classes of mRNAs predominately use either optimal or non-optimal codons (Fig. 3A and B; overrepresented codons in yellow, underrepresented codons in blue) and that this usage correlates with the overall transcript stability (Fig. 3C). Closer inspection of several stable mRNAs revealed that these transcripts were not enriched in any particular codon, but an overwhelming proportion (>80%) of codons fell into the category of optimal (Fig. 3D). By contrast, individual unstable mRNAs were found to be enriched (60% or greater) in non-optimal codons (Fig. 3E). These analyses demonstrated that in this set of mRNAs, the stable mRNAs are biased towards harboring predominately optimal codons and the unstable mRNAs are enriched in non-optimal codons, though the specific codon identities vary between individual transcripts.
Figure 3. Multiple codons are enriched in stable and unstable mRNA classes.
(A) Heat map of a class of relatively stable mRNAs with similar codon usage. Each column represents the usage of a single codon, with each row representing one mRNA. Yellow indicates above average usage of that codon, blue represents below average usage. See Fig. S4 for full heat map. (B) As (A), but showing a relatively unstable class of mRNAs. (C) Dot plot showing the distribution of half-lives in the mRNA classes shown in (A, B). (D) Codon optimality diagrams in selected stable mRNAs. Genes are broken down and plotted as individual codons. Codons are presented in order of optimality rather than in their natural order. Higher bars represent more optimal codons (CSC on y-axis). Green indicates optimal codons, red indicates non-optimal codons. (E) Codon optimality diagrams in selected unstable mRNAs, as in (D). (F) Box plot of mRNAs half-lives separated into optimality groups. Half of the data fall within the boxed section, with the whiskers representing the rest of the data. Data points falling further than 1.5 fold the interquartile distance are considered outliers. See also Figure S2.
Extending this analysis to the level of the whole transcriptome, a correlation between optimal codon content and mRNA stability was observed when the proportion of optimal codons within an mRNA was evaluated by percentiles. Specifically, mRNAs with less than 40% optimal codons were typically found to be unstable, with a median half-life of 5.4 minutes. In contrast, mRNAs with 70% optimal codon content or greater were found to be stable, with a median half-life of 17.8 minutes (Fig. 3F).
Optimal codon content directly influences mRNA decay rate
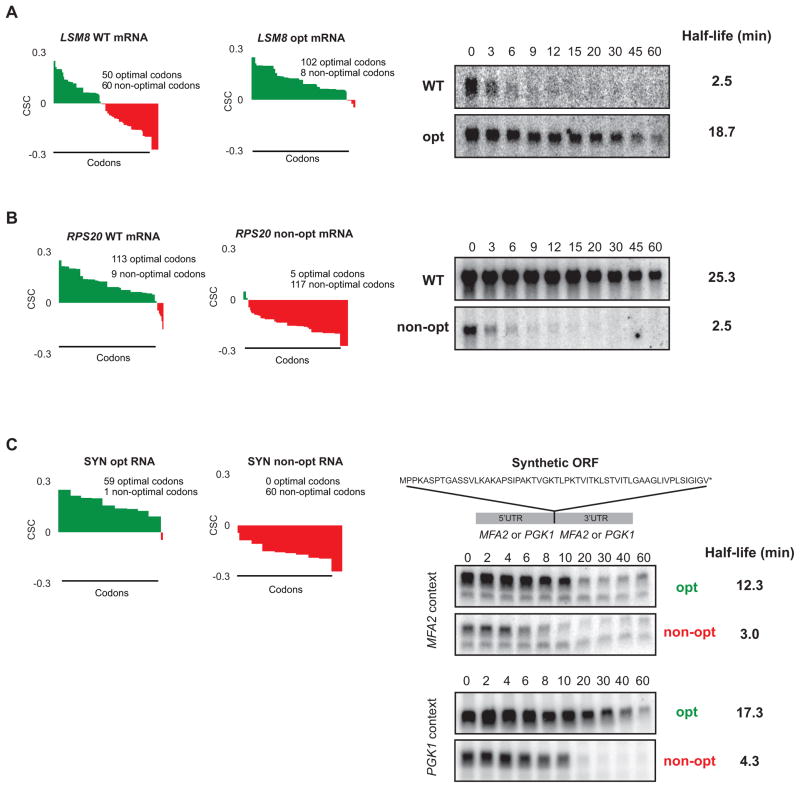
To experimentally validate the relationship observed in the computational analysis, we evaluated the effects on stability of altering the percentage of optimal codons within an mRNA. We modified the codon content of the unstable LSM8 mRNA (half-life = 4.65 min) by making synonymous optimal substitutions in 52 of its 60 non-optimal codons. Similarly, we replaced the majority of optimal codons (108 of 113) within the coding region of the stable RPS20 mRNA (half-life = 25.3 min) with synonymous, non-optimal codons. This methodology ensured that the polypeptides encoded by these sequences were unchanged from the native form. Moreover, the substitutions were selected to avoid significantly altering the GC content of the coding region or introducing any predicted RNA secondary structure (data not shown). Northern blot analysis of these mRNAs after transcriptional inhibition revealed that alteration of the codons within these two transcripts resulted in dramatic changes in their stability. Specifically, the half-life of LSM8 mRNA was increased greater than 7-fold as a consequence of the conversion of non-optimal codons into synonymous optimal codons in its ORF (half-life = 18.7 min; Fig. 4A). In contrast, substitution of non-optimal for optimal codons within the stable RPS20 mRNA resulted in a sharp (10 fold) reduction in its stability (half-life = 2.5 min; Fig. 4B). These data demonstrate that identity of codons within an mRNA can strongly influence stability, and that optimal codon content contributes significantly to determining the rate of mRNA decay in vivo.
Figure 4. Stability of mRNAs can be controlled by altering codon optimality.
(A) Codon optimality diagram of LSM8 (as Fig. 3E), a naturally non-optimal mRNA shown. LSM8 OPT is a synonymously substituted version of LSM8 engineered for higher optimality. Northern blots of rpb1-1 shut-off experiments are shown on the right with half-life of both reporters. Quantitation is normalized to SCR1 loading controls not shown. (B) As (A), except a naturally optimal mRNA, RPS20 (as in Fig. 3D), has been engineered for lower optimality as RPS20 non-opt. Northern blots of rpb1-1 shut-off experiments are shown on the right with half-life of both messages. Quantitation is normalized to SCR1 loading controls not shown. (C) Codon optimality diagrams showing a synthetic mRNA (SYN) encoding the polypeptide shown. Peptide is artificially engineered and has no similarity to any known proteins. SYN opt and non-opt were both inserted into flanking regions from a stable transcript (PGK1) and unstable transcript (MFA2). Northern blots on the right show GAL shut-off experiments demonstrating stability of the SYN mRNA in context of the MFA2 and PGK1 flanking sequences. Quantitation is normalized to SCR1 loading controls not shown. See also Figure S3.
To further examine the relationship between optimal codon content and mRNA stability, we generated two synthetic open reading frames which encode identical 59 amino acid polypeptides but differ in the optimality at each codon (SYN reporters; Fig. S3A, B, and C). We introduced the synthetic ORFs into a reporter bearing the 5′ and 3′ UTRs of MFA2, a well-studied mRNA which is rapidly degraded in the cell (half-life = 3.0 min), a phenomenon shown to be mediated, in part, by elements encoded within its 3′ UTR (LaGrandeur and Parker, 1999; Muhlrad and Parker, 1992). We also introduced the synthetic ORFs into a reporter with the 5′ and 3′ UTRs of PGK1, a well-characterized and stable mRNA (half-life = 25 min; Muhlrad et al., 1995). When stability of the four reporter mRNAs was measured by transcriptional shut-off analysis, the transcripts encoding the optimal SYN ORF were found to be significantly more stable (~4-fold) than their counterparts bearing the non-optimal codons (Fig. 4C). Importantly, degradation of both the optimally and non-optimally encoded SYN reporter mRNAs was determined to occur through the deadenylation-dependent decapping pathway used to degrade the majority of endogenous mRNAs in yeast, and was not mediated by any of the three pathways known to target aberrant mRNA (Fig. S3G and H). High-resolution northern analysis of the decay of these mRNAs confirmed that the rates of both deadenylation and decapping, the regulated steps in the normal decay pathway, were affected as a consequence of changes in codon composition within the reporter ORFs (Fig. S3D, E, and F). These data demonstrate that optimal codon content is a critical determinant of mRNA stability influencing both the rate of deadenylation and decapping during turnover of the mRNA independently of 5′ and 3′ UTRs, which can act in parallel to stabilize or destabilize the mRNA.
Optimal codon content influences translational efficiency
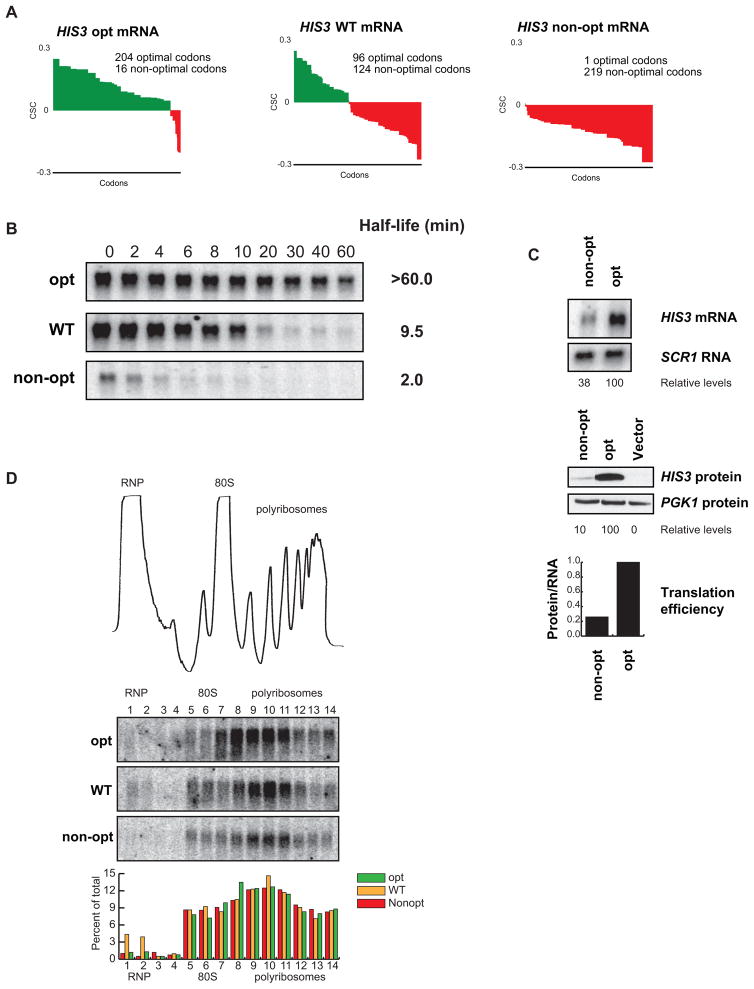
To evaluate the influence of codon optimality on mRNA translation efficiency in vivo, we generated three new reporters that differ in optimal codon content but do not differ in amino acid sequence. Specifically, we engineered the ORF of the HIS3 gene to contain either all optimal (HIS3 opt) or all non-optimal codons (HIS3 non-opt), with the wild-type HIS3 gene providing an intermediate point at 43% optimal codons (Fig. 5A). The HIS3 gene was chosen because it has a relatively long ORF (220 amino acids) compared to our other synonymous mutation constructs, allowing us to effectively monitor ribosome association by sucrose density gradients (see below). We then determined the mRNA decay rate of the three HIS3 constructs by transcriptional shutoff analysis using an rpb1-1 strain. Consistent with our previous results, it was observed that changing optimal codon content produced a dramatic effect on mRNA half-life (Fig. 5B). Notably, the effect on HIS3 mRNA decay matched the percent of optimal codons used. The half-life of the optimal construct (half-life >60 min) was much greater that of the WT construct (half-life = 9.5 min) whose half-life was markedly greater than the non-optimal construct (half-life = 2.0 min). Thus, we can achieve a full range of mRNA half-lives in yeast without altering protein sequence or flanking sequences by changing optimal codon content.
Figure 5. Optimality can affect translation and stability of an mRNA without changes in ribosome association.
(A) Codon optimality diagram of HIS3, a transcript with an intermediate half-life, as well as versions engineered with synonymous substitutions to contain higher and lower percent optimal codons, HIS3 opt and HIS3 non-opt respectively. (B) Northern blots of rpb1-1 shut-off experiments are shown with half-lives of all three messages. Quantitation is normalized to SCR1 loading controls not shown. (C) Northern and western blots for steady state concentrations of the optimal and non-optimal versions of HIS3. Loading controls and quantitation are shown below. Translational efficiency is calculated as relative protein levels divided by relative mRNA levels and plotted at the bottom. (D) A trace of sucrose density gradient analysis, along with northern blot analysis of the gradient fractions. The blots show location of the three HIS3 reporters within the gradient. Quantitation for each fraction is shown below.
We hypothesized that codon optimality should influence translation elongation. We tested this hypothesis using two approaches. First, we monitored the protein output from the HIS3 optimal construct vs. the HIS3 non-optimal construct by western blot, and then normalized the protein expression to the mRNA levels, as determined by northern blot. We observed that the non-optimal construct had four-fold less protein output than the optimal construct (Fig. 5C). Second, we evaluated the ribosome density on the HIS3 mRNA constructs. Ribosome density was monitored using sucrose gradients, followed by fractionation and northern blotting of the isolated fractions (Hu et al., 2009). Critically, it was observed that the ribosome occupancy was nearly identical for all three HIS3 reporter mRNAs (Fig. 5D). Thus, we propose that a four-fold decrease in protein output, in conjunction with nearly identical localization within a polyribosome, suggests a decrease in ribosome translocation rate on the non-optimal construct as compared to the optimal.
Optimal codon content impacts ribosome translocation
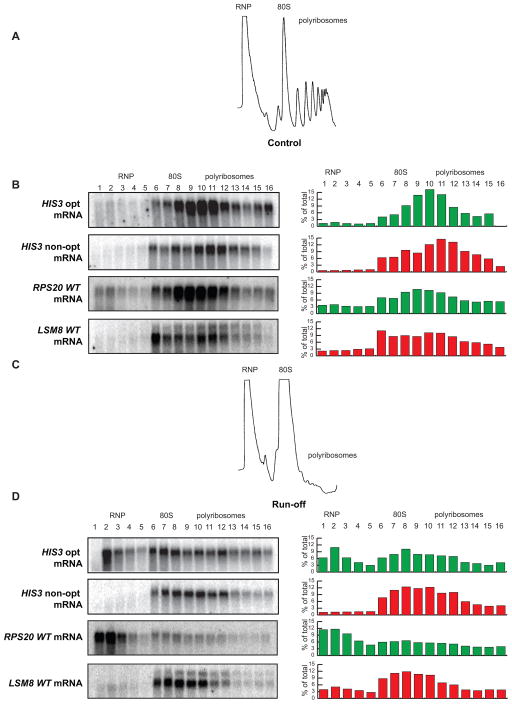
To directly determine whether ribosomes translocate slower on mRNAs containing non-optimal codons vs. optimal codons, we monitored ribosomal run-off of these two reporters. To do this, we blocked translational initiation by depriving cells of glucose for 10 minutes. Glucose deprivation results in rapid inhibition of translational initiation and thus bulk polyribosomes are lost by run-off (Coller and Parker 2005; Fig. 6A vs. C). To monitor ribosomal run-off, we extracted mRNA-ribosome complexes before and after glucose deprivation, separated the material with a sucrose gradient, collected fractions, and monitored the presence of the HIS3 mRNAs in each fraction by northern analysis. Importantly, under normal conditions the ribosome occupancy of the HIS3 opt and non-opt constructs was determined to be similar (Fig. 6B); however, upon induction of ribosome run-off, a large fraction of the optimal construct mRNA relocated to the top of the gradient in the ribosome-free area, while the HIS3 non-opt mRNA remained largely associated with polyribosomes (Fig. 6D). We extended this analysis to two endogenous mRNA transcripts that differ dramatically in codon optimality, LSM8 (45% optimal codons) and RSP20 (92% optimal codons). Notably, the endogenous LSM8 mRNA was retained on polyribosomes following inhibition of translational initiation, while the RPS20 mRNA dissociated efficiently. We propose that the difference in retention is due to more efficient ribosome translocation on messages with high optimal codon content. Thus, the retention of the mRNAs bearing predominantly non-optimal codons in polyribosomal fractions indicates that codon optimality can impact the rate of ribosome translocation directly.
Figure 6. Optimal and non-optimal transcripts are retained differently on polysomes.
(A) Representative A260 trace of sucrose density gradient analysis demonstrating normal distribution into RNP, 80S, and polyribosome fractions. (B) Distribution of the optimal and non-optimal HIS3 reporters and the RPS20 and LSM8 mRNAs in the sucrose density gradients under normal conditions showing localization primarily in the polyribosome fractions. (C) Representative A260 trace of sucrose density gradient analysis under run-off conditions, showing collapse of the polyribosome fractions. (D) Distribution of the optimal and non-optimal HIS3 reporters and the RPS20 and LSM8 mRNAs under run-off conditions, demonstrating differential relocation.
Precision in gene expression is achieved through coordination of optimal codon content
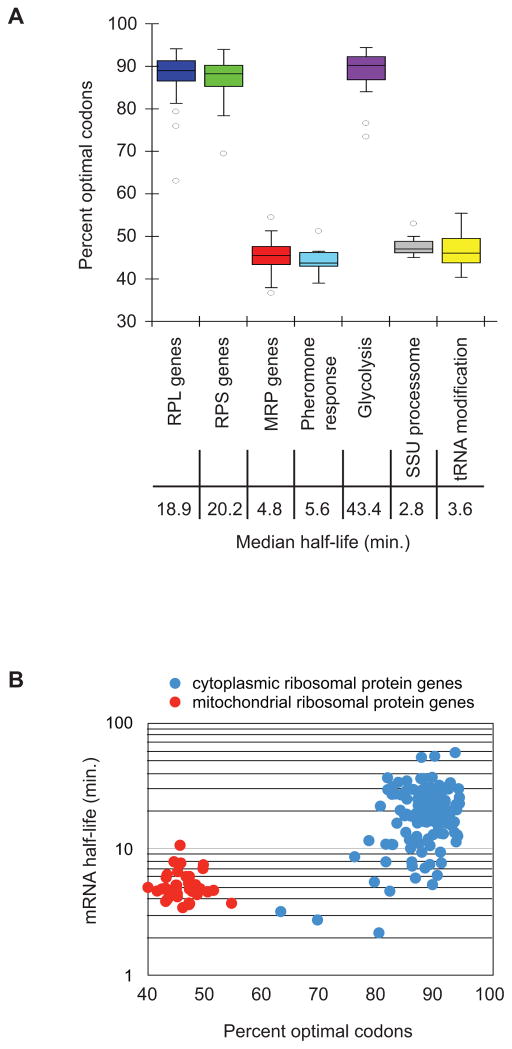
A previous analysis of mRNA stability in yeast revealed that the decay rates of some mRNAs encoding proteins that function in the same pathway or are part of the same complex were similar. Turnover of individual mRNAs appears to be based on the physiological function and cellular requirement of the proteins they encode (Wang et al., 2002). We hypothesized that modulation of optimal codon content may provide the mechanism for the cell to coordinate the metabolism of transcripts expressing proteins of common function. We assessed codon usage for genes whose protein products function in common pathways and/or complexes. We observed that mRNAs encoding the enzymes involved in glycolysis (n=10) had a similar and extraordinarily high proportion of optimal codons (mean=86%; Fig. 7A). These transcripts were determined to be stable both previously and in our genome-wide analysis (median half-life=43.4 min; Wang et al., 2002). In contrast, mRNAs encoding polypeptides involved in pheromone response in yeast cells (n = 14) were all unstable (median half-life=5.6 min; Wang et al., 2002) and harbored an average of only 43% optimal codons (Fig. 7A). Our analysis revealed that other groups of transcripts behave similarly. The stable large and small cytosolic ribosomal subunit protein mRNAs (n=70 and 54, respectively; median half-life=18.9 min and 20.2 min, respectively) demonstrated an average optimal codon content of 89% and 88% respectively, but mRNAs that encode ribosomal proteins functioning in the mitochondria are unstable (n=42; median half-life=4.8 min), consistent with the observation that they have 45% optimal codon content. (Fig. 7A and B). Other families of genes that have similar decay rates include those whose protein products are involved in ribosomal processing, tRNA modification, the TCA cycle, RNA processing, and components of the translational machinery (Fig. 7 and data not shown). These data provide evidence that transcripts expressing proteins of related function are coordinated at the level of optimal codon content as well as decay rate, suggesting that these genes may have evolved specific codon contents as a mechanism to facilitate precise synchronization of expression based on their function in the cell.
Figure 7. Functionally related genes display similar optimality.
(A) Groups of genes whose protein products have related functions are plotted to show their optimality. Half of the data fall within the boxed section, with the whiskers representing the rest of the data. Data points falling further than 1.5 fold the interquartile distance are considered outliers. Represented gene groups are: 70 RPL (large ribosomal subunit proteins) genes, 54 RPS (small ribosomal subunit proteins) genes, 42 MRP (mitochondrial ribosomal proteins) genes, 14 pheromone response genes, 10 glycolysis enzymes, 15 SSU (small subunit processosome) genes, 12 tRNA processing genes. (B) Breakdown of two groups to show relationship between optimal codon content and half-life within the groups. mRNA half-life for each protein in the cytoplasmic ribosome and the mitochondrial ribosome is plotted against the optimal codon content of that mRNA.
DISCUSSION
We have provided several lines of evidence indicating that codon optimality is a major determinant of mRNA stability in budding yeast. First, bioinformatic analysis demonstrates a strong correlation between the percentage of optimal codons and mRNA half-life. For example, mRNAs with less than 40% optimal codons have a median half-life of 5.3 min. while mRNAs with greater than 70% optimal codons have a median half-life of 20.1 min. The conclusions emerging from the bioinformatics were verified experimentally, showing that changing optimal codons to non-optimal destabilized otherwise stable mRNAs while changing non-optimal codons to optimal stabilized otherwise unstable mRNAs. Most importantly, we provide evidence that optimal and non-optimal codons exert their effects by modulating translation elongation rates.
Several ribosomal profiling studies have failed to detect codon-specific differences in the translation of optimal and non-optimal codons (Ingolia et al., 2009; Qian et al., 2012; Charneski et al., 2013). Nevertheless, we observe striking differences in ribosome clearance when mRNAs encoding the same polypeptide are comprised of optimal or non-optimal codons. These differences may reflect the additive effects of many small ribosome hesitations at non-optimal codons. Such hesitations would be imperceptible in ribosomal profiling analyses. Alternatively, the overall codon composition of an ORF could set a uniform translational elongation rate across the ORF. If this were true, no change in rate at individual codons would be detected by ribosome profiling.
It is important to note that while codon content is clearly a major determinant of mRNA stability, it does not predict half-lives of all mRNAs. For example, mRNAs for several histone components, such as HHF2 and HHT1, contain 85% optimal codons, but yet are very unstable with half-lives of 2.4 and 3.5 minutes, respectively. The half-lives of such mRNAs could be dictated by their ability to initiate translation efficiently (or inefficiently) and/or by elements in 5′ or 3′ UTRs. Numerous examples of each have been described (Goldstrohm et al., 2007; Olivas and Parker, 2000). It is also possible that features within the ORFs might explain some of the outliers (e.g. distribution of optimal and non-optimal codons).
Because of the effects of optimal codon content on translational elongation rates, it is most likely that some factors(s) monitor these rates while mRNAs are engaged with ribosomes. Indeed, we have previously shown that slowing of ribosomal movement by insertion of rare codons promotes mRNA decay (Hu et al., 2009; Sweet et al., 2012). A prime candidate for a monitoring factor is the DEAD-box RNA helicase DHH1, an integral component of the mRNA decay machinery (Presnyak and Coller, 2013) that has been shown to act as an activator of decapping through its role in promoting translational repression (Coller and Parker, 2005; Sweet et al., 2012). Further studies will be needed to determine the mechanism by which translation elongation rate influences mRNA decay.
Precision and coordination of gene expression through codon optimality
Both long and short time scales provide important opportunities for the reassignment of codon optimality in the cell. In the short term, changes in cellular growth conditions and nutrient availability could significantly impact individual (or subsets of) charged tRNA levels. As a consequence of this reduction in supply, translation elongation rates of mRNAs enriched in the codons decoded by these tRNAs would be slowed and their levels decreased, due to enhanced turnover. In this way, codon optimality provides the cell not only with a general mechanism to hone mRNA levels, but also with a mechanism to sense environmental conditions and rapidly tailor global patterns of gene expression.
Long-term genetic changes can introduce synonymous mutations into protein coding genes that do not alter the amino acid sequence of the encoded polypeptide; however, such changes would impact mRNA and protein expression levels if the mutations significantly altered the proportion of optimal codons within the open reading frame of the mRNA. Thus, synonymous gene mutation can be envisioned as a method to evolve mRNA stability rates that are advantageous to the cell. We find that mRNAs encoding proteins that act together in similar pathways or are part of the same stoichiometric complexes, and which have been previously observed to decay at similar rates (Wang et al., 2002), encode nearly identical proportions of optimal codons (Fig. 7A). We suggest that codon optimality has been finely tuned for these gene sets as an elegant mechanism to ensure coordinated post-transcriptional regulation and parsimonious expression of proteins at the precise levels required by the cell. Interestingly, similar levels of optimal to non-optimal codons could ensure not only similarity of stability and translation rates for related mRNAs, but also coordination of response to changes in tRNA levels (e.g. nutrient availability, stress, cell type, etc.). Recent studies reveal that tRNA concentrations within the cell are not static but are constantly undergoing change, sometimes dramatically. For instance, large scale RNA profiling experiments have demonstrated that tRNA concentrations vary widely between proliferating and differentiating cells (Gingold et al., 2014). Based on our analysis, we would argue that significant alterations in tRNA concentrations could alter the mRNA expression profile within a cell by dynamically changing mRNA stability, even without any changes in transcription.
Ribosomes are the master gatekeepers, determining the downstream fate of both normal and aberrant mRNAs
As a final implication, our work suggests that co-translational mRNA surveillance by the ribosome is not only important to target aberrant mRNAs to rapid decay, but also to tune the degradation rates of normal mRNAs. In eukaryotes, aberrations in mRNAs lead to aberrant translation events such as premature termination, lack of translation termination, and ribosome stalling, which result in the accelerated turnover of the mRNA by the Nonsense-Mediated, Non-Stop, and No-Go Decay pathways, respectively (Shoemaker and Green, 2012). We find here that codon usage within normal mRNAs also influences translating ribosomes and can have profound effects on mRNA stability. Thus, the ribosome acts as the master sensor, helping to determine the fate of all mRNAs, both normal and aberrant, through modulation of its elongation and/or termination processes. The use of the ribosome as a sensor is ideal for protein-coding genes, whose primary function in the cell is to be translated. We suggest that a component of mRNA stability is built into all mRNAs as a function of codon composition. The elongation rate of translating ribosomes is communicated to the general decay machinery, which affects the rate of deadenylation and decapping. Individually, the identity of codons within an mRNA would be predicted to have a minute influence on overall ribosomal decoding; however, within the framework of an entire mRNA, we show that codon optimality can have profound effects on translation elongation and mRNA turnover. We therefore conclude that codon identity represents a general property of mRNAs and is a critical determinant of their stability.
Experimental Procedures
Yeast strains and growth conditions
The genotypes of all yeast strains used in this study are listed in Supplementary Table 2. Unless indicated, all strains are based on BY4741. Cells were grown in standard synthetic medium (pH 6.5) supplemented with appropriate amino acids and sugars. All cells were grown at 24 C and collected at mid-log phase (3 × 107 cells ml−1).
Plasmids and strain construction
The plasmids and oligonucleotides used in this study are listed in Supplementary Table 3 and 4 respectively. Reporter plasmids bearing native genes (LSM8, RPS20, HIS3 WT) were constructed by amplifying the native loci, adding restriction sites and several unique sites (to facilitate detection by northern probe) in the 3′ UTR by site-directed mutagenesis, and inserting the construct into an expression vector. The reporters with altered optimality (LSM8 opt, RPS20 non-opt, HIS3 opt & non-opt) were constructed by synthesizing the DNA in multiple pieces, annealing and amplifying them, and then subcloning into an expression vector. These reporter plasmids were transformed into an rpb1-1 yeast strain.
To construct the plasmids bearing the synthetic reporters, restriction sites were introduced into previously constructed plasmids bearing MFA2 and PGK1 under the control of a GAL1 UAS. The SYN ORFs were then synthesized and assembled as described for the altered reporters above. These reporters were transformed into a WT yeast strain.
Northern RNA analysis and sucrose density gradients
Northern RNA analysis of GAL-driven reporters and sucrose density gradients for polyribosome analysis was performed as previously described (Hu et al, 2009). For analysis of reporters in rpb1-1 was performed similarly to GAL, except cells were grown in media containing glucose and repression was achieved by shifting cells to 37C. Ribosomal run-off experiments were performed similarly to normal polyribosome analysis, except cells were resuspended in media lacking glucose for 10 minutes before harvesting (Coller and Parker, 2005).
RNA-seq
rpb1-1 mutant cells (Nonet et al, 1987) were grown to mid-log phase at 24°C and shifted to a non-permissive temperature of 37C. Aliquots were collected over 60 minutes. RNA was then extracted, external controls were added, and two sets libraries were prepared from each using the the Illumina TruSeq Stranded Total RNA and mRNA library prep kits. The libraries were quantitated using an Agilent Bioanalyzer and sequenced on an Illumina HiSeq2000 using paired-end 100 bp reads with an index read. Sequencing data and the processed data for each gene are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE57385.
Alignment and half-life calculation
Reads were aligned to the S. cerevisiae reference genome using bowtie (Langmead et al., 2009), with the unaligned reads then aligned to the sequences of the controls in the same way. Aligned reads were quantitated using cufflinks (Trapnell et al., 2010). Raw FPKM numbers were normalized to external controls, then fitted to single exponential decay curves to calculate the half-lives using the least absolute deviation method to minimize outlier effects. Data was then filtered to exclude dubious ORFs and transcripts with poor fit to the model. Bootstrapped confidence intervals were generated by using un-normalized residuals from the original data to generate simulated data sets.
Statistical Methods
The Codon occurrence to mRNA Stability Correlation coefficient (CSC) was determined by calculating a Pearson correlation coefficient between the frequency of occurrence of individual codons and the half-lives of the messages containing them. To determine the statistical significance, we categorized the CSC as either positive or negative and used a chi-squared test of association. For association between the categories of percent optimal codons and mRNA half-life, an ANOVA f-test with mRNA half-life on the log scale was used.
To mitigate effects of base pair content of the genes, we randomly permuted the sequence and recalculated the test statistic for each of 10,000 permutations. The permutation p-value was calculated as the number of permuted data sets with a test of association stronger than the chi-squared test in the original data. Statistical calculations were done using the R environment. Optimality percentages were calculated by generating a list of optimal and non-optimal codons as previously described (Pechmann and Frydman, 2013).
Supplementary Material
A) Frequency of occurrence (in # per 1000 codons) for each codon is plotted vs the tAI of the codons. Non-optimal codons are colored in red, optimal in green. B) Frequency of occurrence (in # per 1000 codons) for non-optimal codons (in red, on the left) and optimal codons (in green, on the right). Codons are placed in order of decreasing frequency within each group. C) Matrix of Pearson correlation values for previously published RNA-seq-based mRNA decay datasets in yeast. Color coding is based on magnitude of the correlation with darker shading indicating higher correlation. D) Codon occurrence to mRNA Stability Correlation coefficient (as defined in Figure 2A and text) for each codon plotted for the data set presented in this paper (in blue) and a moderately correlated data set from the literature (Miller et al., 2011, in orange). E) Example of CSC value calculation for individual codons. Codon occurrence of selected codons plotted against mRNA half-lives. The R-values of the shown lines are graphed in those figures in the corresponding bars. These R-values are used on the CSC scale in Fig 2A and C and earlier parts of Fig S1.
All mRNA with reliable half-lives are evaluated using a Spearman distance metric and clustered using k-means clustering. An input matrix was generated by assigning each gene a set of rank scores based on its frequency of usage of each codon. The matrix was then clustered using Cluster3 and visualized using Java Treeview. Each column represents the usage of the codon indicated at the top, and columns are ordered by descending optimality. Usage ranks above average are indicated in yellow on the heatmap, scored below average are indicated in blue. Clustered used in further analysis in Figure 3 are indicated with red boxes.
A) Sequence and structure of the synthetic mRNA are shown. It consists of the flanking regions taken from MFA2 or PGK1 with an intervening ORF encoding the peptide shown. The construct is harbored on a GAL-driven expression plasmid. B) Full optimality diagram and sequence of the synthetic optimal mRNA (SYN opt). C) Full optimality diagram and sequence of the synthetic non-optimal mRNA (SYN nonopt). D) Sequence and structure of the synthetic mRNA are shown as in S2a, in this case utilizing PGK1 flanking regions. E) High-resolution acrylamide northern blot showing the results of a transcriptional shut-off experiment demonstrating decay patterns of the SYN reporters. F) Quantitation of (E), with length of poly(A) tail plotted on the left and the percent of RNA remaining plotted on the right. Note that the y-axis of the latter plot is logarithmic. G) Shutoff experiments of the SYN Opt reporter in cell strains indicated to the right of the northern blot results. Deletion of DOM34 or UPF1 has no effect on the half-life of the mRNA, while deletion of CCR4 and DCP2 strongly stabilize the message. H) Shutoff experiments of the SYN Non-opt reporter showing similar results for this mRNA.
Complete list of genes with half-lives used for analyses within the manuscript. Gene list was selected to comply with the parameters described in the extended procedures. Confidence intervals are shown for the half-lives as calculated from the rRNA-depleted libraries (total half-life).
Complete list of yeast strains used in this study as well as the sources of the strains.
Complete list of plasmids used in this study as well as the sources of the plasmids.
Complete list of oligonucleotides used in this study as well as the sources of the oligos.
Acknowledgments
The authors express their appreciation to Dr. Tim Nilsen for his suggestions and critical evaluation of the manuscript, and Dr. Beth Grayhack (University of Rochester) for helpful discussion. We thank the members of the Coller and Baker labs for their input into this work. This work was funded by NIH grant GM080465 to J.C.
Footnotes
Author Contributions V.P., K.E.B., B.R.G. and J.C. wrote the manuscript. V.P., Y.H.C, and N.A., S.M. and S.O. performed the experiments. D.W. provided technical expertise, N.K. provided bioinformatic support, and N.M. provided statistical support. All of the authors contributed to discussion, analysis, and the design of the research. All authors commented on the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Akashi H. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G, Muhlrad D, Parker R. A small segment of the MAT alpha 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol Cell Biol. 1993;13:5141–5148. doi: 10.1128/mcb.13.9.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charneski CA, Hurst LD. Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol. 2013;11(3):e1001508. doi: 10.1371/journal.pbio.1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Moqtaderi Z, Fan X, Ozsolak F, Struhl K. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell. 2014;156:812–824. doi: 10.1016/j.cell.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Jacobson A. RNA decay modulates gene expression and controls its fidelity. Wiley Interdiscip Rev RNA. 2010;1:351–361. doi: 10.1002/wrna.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158(6):1281–1292. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Seay DJ, Hook BA, Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J Biol Chem. 2007;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekema A, Kastelein RA, Vasser M, de Boer HA. Codon replacement in the PGK1 gene of Saccharomyces cerevisiae: experimental approach to study the role of biased codon usage in gene expression. Mol Cell Biol. 1987;7:2914–2924. doi: 10.1128/mcb.7.8.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kri Ko A, Copi T, Gabaldón T, Lehner B, Supek F. Inferring gene function from evolutionary change in signatures of translation efficiency. Genome Biol. 2014;15:R44. doi: 10.1186/gb-2014-15-3-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGrandeur T, Parker R. The cis acting sequences responsible for the differential decay of the unstable MFA2 and stable PGK1 transcripts in yeast include the context of the translational start codon. 1999;5:420–433. doi: 10.1017/s1355838299981748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Lykke-Andersen J. Emerging roles for ribonucleoprotein modification and remodeling in controlling RNA fate. Trends Cell Biol. 2013;23:504–510. doi: 10.1016/j.tcb.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C, Schwalb B, Maier K, Schulz D, Dümcke S, Zacher B, Mayer A, Sydow J, Marcinowski L, Dölken L, et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol. 2011;7:458–458. doi: 10.1038/msb.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M, Scafe C, Sexton J, Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Molecular and Cellular Biology. 1987;7(5):1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa EM, Ribas de Pouplana L. Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet. 2012;28:574–581. doi: 10.1016/j.tig.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Olivas W, Parker R. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. Embo J. 2000;19:6602–6611. doi: 10.1093/emboj/19.23.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Frydman J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat Struct Mol Biol. 2013;20:237–243. doi: 10.1038/nsmb.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, Coller J. The DHH1/RCKp54 family of helicases: An ancient family of proteins that promote translational silencing. Biochemica et Biophysica Acta. 2013;1829:817–823. doi: 10.1016/j.bbagrm.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Yang JR, Pearson NM, Maclean C, Zhang J. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012;8(3) doi: 10.1371/journal.pgen.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet T, Kovalak C, Coller J. The DEAD-Box Protein Dhh1 Promotes Decapping by Slowing Ribosome Movement. PLoS Biol. 2012;10:e1001342. doi: 10.1371/journal.pbio.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proc Natl Acad Sci USA. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Weems M, Wilke CO. Translationally optimal codons associate with structurally sensitive sites in proteins. Mol Biol Evol. 2009 Jul;26(7):1571–80. doi: 10.1093/molbev/msp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Frequency of occurrence (in # per 1000 codons) for each codon is plotted vs the tAI of the codons. Non-optimal codons are colored in red, optimal in green. B) Frequency of occurrence (in # per 1000 codons) for non-optimal codons (in red, on the left) and optimal codons (in green, on the right). Codons are placed in order of decreasing frequency within each group. C) Matrix of Pearson correlation values for previously published RNA-seq-based mRNA decay datasets in yeast. Color coding is based on magnitude of the correlation with darker shading indicating higher correlation. D) Codon occurrence to mRNA Stability Correlation coefficient (as defined in Figure 2A and text) for each codon plotted for the data set presented in this paper (in blue) and a moderately correlated data set from the literature (Miller et al., 2011, in orange). E) Example of CSC value calculation for individual codons. Codon occurrence of selected codons plotted against mRNA half-lives. The R-values of the shown lines are graphed in those figures in the corresponding bars. These R-values are used on the CSC scale in Fig 2A and C and earlier parts of Fig S1.
All mRNA with reliable half-lives are evaluated using a Spearman distance metric and clustered using k-means clustering. An input matrix was generated by assigning each gene a set of rank scores based on its frequency of usage of each codon. The matrix was then clustered using Cluster3 and visualized using Java Treeview. Each column represents the usage of the codon indicated at the top, and columns are ordered by descending optimality. Usage ranks above average are indicated in yellow on the heatmap, scored below average are indicated in blue. Clustered used in further analysis in Figure 3 are indicated with red boxes.
A) Sequence and structure of the synthetic mRNA are shown. It consists of the flanking regions taken from MFA2 or PGK1 with an intervening ORF encoding the peptide shown. The construct is harbored on a GAL-driven expression plasmid. B) Full optimality diagram and sequence of the synthetic optimal mRNA (SYN opt). C) Full optimality diagram and sequence of the synthetic non-optimal mRNA (SYN nonopt). D) Sequence and structure of the synthetic mRNA are shown as in S2a, in this case utilizing PGK1 flanking regions. E) High-resolution acrylamide northern blot showing the results of a transcriptional shut-off experiment demonstrating decay patterns of the SYN reporters. F) Quantitation of (E), with length of poly(A) tail plotted on the left and the percent of RNA remaining plotted on the right. Note that the y-axis of the latter plot is logarithmic. G) Shutoff experiments of the SYN Opt reporter in cell strains indicated to the right of the northern blot results. Deletion of DOM34 or UPF1 has no effect on the half-life of the mRNA, while deletion of CCR4 and DCP2 strongly stabilize the message. H) Shutoff experiments of the SYN Non-opt reporter showing similar results for this mRNA.
Complete list of genes with half-lives used for analyses within the manuscript. Gene list was selected to comply with the parameters described in the extended procedures. Confidence intervals are shown for the half-lives as calculated from the rRNA-depleted libraries (total half-life).
Complete list of yeast strains used in this study as well as the sources of the strains.
Complete list of plasmids used in this study as well as the sources of the plasmids.
Complete list of oligonucleotides used in this study as well as the sources of the oligos.