Abstract
Intraflagellar transport (IFT) of ciliary precursors such as tubulin from the cytoplasm to the ciliary tip is involved in the construction of the cilium, a hairlike organelle found on most eukaryotic cells. However, the molecular mechanisms of IFT are poorly understood. Here, we found that the two core IFT proteins IFT74 and IFT81 form a tubulin-binding module and mapped the interaction to a calponin homology domain of IFT81 and a highly basic domain in IFT74. Knockdown of IFT81 and rescue experiments with point mutants showed that tubulin binding by IFT81 was required for ciliogenesis in human cells.
Cilia are microtubule-based organelles that function in motility, sensory reception, and signaling (1). Ciliary dysfunction results in numerous diseases and disorders commonly known as ciliopathies. Intraflagellar transport (IFT) is involved in cilium formation (2, 3) but also functions in other cellular processes, such as the recycling of Tcell receptors at the immune synapse (4). IFT relies on kinesin-2 and IFT-dynein molecular motors moving along the microtubule-based axoneme of cilia (5–7) and on the IFT complex, which contains at least 20 different protein subunits. Although ~600 proteins are known to reside in the cilium (8), we know very little about how they are recognized as ciliary cargo by the IFT machinery (9–11).
To identify potential cargo-binding sites on the IFT complex, we carried out bioinformatical and biochemical screening and identified conserved domains that were not required for IFT complex formation. We reasoned that such domains could protrude from the IFT particle-core structure and would thus be in a prime position for cargo recognition. The two IFT core proteins IFT74 and IFT81 were found to possess N-terminal domains (IFT74N and IFT81N) that were not required for IFT complex formation or stability (fig. S1). Whereas IFT81N is highly conserved in sequence and predicted to be a folded domain, IFT74N was likely to be disordered and was highly basic with an isoelectric point (pI) > 12 (fig. S2). To characterize the properties of IFT74N and IFT81N, we purified recombinant Homo sapiens (Hs) IFT81N, Chlamydomonas reinhardtii (Cr) IFT81N, and a truncated HsIFT74/81 heterodimeric complex (fig. S3) (IFT74N alone degraded rapidly and could not be purified) and determined the crystal structure of CrIFT81N (Fig. 1, A to C; fig. S4, A to D; and table S1). The crystal structure revealed that IFT81N adopts the fold of a calponin homology (CH) domain with unexpected structural similarity to the kinetochore complex component NDC80 with microtubule (MT)–binding properties (12). Given that the cilium consists of a MT-based axoneme, IFT of large quantities of tubulin is required for cilium formation (13). We thus tested the tubulin-binding properties of HsIFT81N using affinity pull-downs (Fig. 1D and fig. S4E) and microscale thermophoresis (MST) with unpolymerized bovine αβ-tubulin (Fig. 1, E and F). HsIFT81N bound tubulin with a dissociation constant (Kd) of 16 μM via a highly conserved, positively charged surface patch, which was enhanced 18-fold by IFT74N (Fig. 1G and fig. S3). Because this result was unexpected, we also carried out MT sedimentation assays and electron microscopy (EM) to visualize IFT81 or IFT74/81 bound to MT (fig. S5). The IFT74/81 complex, but not IFT81N, at low μM concentration cosedimented with MT during ultracentrifugation (fig. S5, D and E) and decorated MT (fig. S5F). Thus, the tubulin-binding module is formed by the IFT74/81 complex rather than by IFT81N alone.
Fig. 1. IFT81 and IFT74 form a tubulin-binding module.
(A) Cartoon representation of the crystal structure of CrIFT81N domain, with conserved lysines and arginines implicated in tubulin binding shown as sticks. (B) Electrostatic surface potential of IFT81N displaying the positively charged patch with the residues labeled according to the HsIFT81 sequence. (C) Surface conservation of IFT81N demonstrates that the basic patch is well conserved among different species (also see fig. S2). (D) Tubulin binding evaluated by glutathione (GSH) affinity pull-down of bovine αβ-tubulin using glutathione S-transferase (GST)–HsIFT81N. Whereas tubulin does not bind the GSH beads and is not pulled down by GST alone, a substantial portion is pulled down by GST-HsIFT81N, demonstrating binding. Whereas the single-point mutation R87E does not strongly impair binding, the K73K75/EE double mutant (mut1) results in reduced amounts of pulled-down tubulin, indicating reduced binding. (E) Quantification of tubulin binding to untagged HsIFT81N by microscale thermophoresis reveals a Kd of 16 μM. (F) The HsIFT81N mut1 has drastically reduced binding with a Kd of 187 μM, showing that the basic patch is required for tubulin binding. (G) Microscale thermophoresis titration of tubulin with truncated HsIFT7481 complex reveals a Kd of 0.9 μM. The curves in (E), (F), and (G) are calculated for three independent experiments, and the error bars represent the mean ± SD. (H) The experiments shown in (D) to (G), along with the data in fig. S5, suggest a model in which IFT81N recognizes the globular domain of tubulin, providing specificity, and IFT74N binds the acidic tail of β-tubulin, providing increased affinity.
To dissect the binding mode in the IFT74/81:αβ-tubulin complex, samples were prepared from MT and unpolymerized αβ-tubulin lacking the highly acidic C-terminal tails, often referred to as E-hooks (12) (fig. S5A). αβ-tubulin lacking E-hooks had similar affinity for IFT81N as intact tubulin (fig. S5, B and C), which suggested that IFT81N recognizes the globular domain of αβ-tubulin with no substantial interaction with the E-hooks. IFT74/81 displayed robust MT binding in sedimentation assays, which was, however, reduced to background levels in the absence of the β-tubulin E-hook (fig. S5E). Thus, IFT81N appears to bind the globular domain of tubulin to provide specificity, and IFT74N recognizes the β-tubulin tail to increase affinity (Fig. 1H).
To examine the role of tubulin binding by IFT74/81 in a cellular system, we transiently expressed Flag-HsIFT81 or Flag-HsIFT81ΔN in human RPE-1 cells and induced formation of primary cilia either by treatment with 0.5 μM cytochalasin D (Fig. 2) (14) or serum starvation (fig. S6, A and D). In these experiments, centrioles were visualized by staining for CAP350 and cilia by staining for the small guanosine triphosphatase Arl13b or acetylated tubulin. We detected both Flag-HsIFT81 and Flag-HsIFT81ΔN at the tip of the primary cilium and, to a minor extent, also along the axoneme (fig. S6A), suggesting that IFT81N is not required for the transport of IFT81 within the primary cilium. Remarkably, however, the expression of Flag-HsIFT81ΔN had a strong negative impact on the extent of ciliogenesis (fig. S6, B to D), suggesting that excess IFT81ΔN caused a dominant-negative effect, presumably through formation of IFTcomplexes unable to bind tubulin.
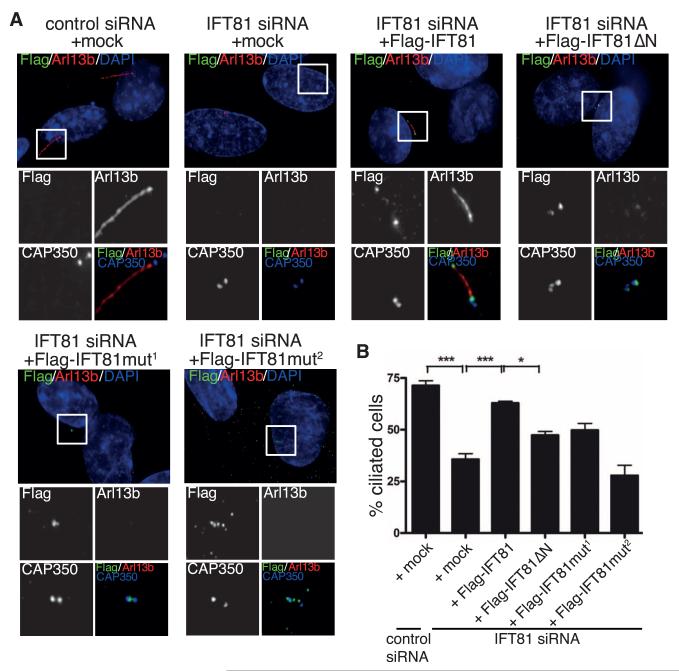
Fig. 2. Tubulin binding by IFT81 is required for ciliogenesis in human cells.
(A) Transient expression of Flag-IFT81, but not the tubulin-binding–deficient IFT81 mutants (in green), rescues the ciliogenesis defect after IFT81 siRNA knockdown. Primary cilia formation was induced by 0.5 μM cytochalasin D and detected by antibody to Arl13b (in red). CAP350 (in blue, inset images only) was used to visualize centrosomes. Mut1 and Mut2 are K73K75/EE and K73K75K113K114R115/EEEEE tubulin-binding mutants, respectively. Scale bar, 5 μm. (B) Quantification of the rescue experiment shown in (A). n = 3 independent experiments; statistical analyses by one-way analysis of variance.
To further investigate the function of the tubulin-binding domain of IFT81 in ciliogenesis, we carried out small interfering RNA (siRNA)–rescue experiments. siRNA-mediated depletion of IFT81 (fig. S6E) strongly reduced the percentage of ciliated cells (Fig. 2), which could be rescued by coexpression of an siRNA-resistant full-length IFT81, as expected. In contrast, none of the IFT81 mutants deficient in tubulin binding in vitro (HsIFT81mut1 and HsIFT81mut2) compensated fully for the depletion of endogenous IFT81. Whereas expression of the deletion mutant (HsIFT81ΔN) or the mutant with reduced tubulin-binding ability (HsIFT81mut1) resulted in partial rescue, expression of HsIFT81mut2, in which the entire tubulin-binding patch was mutated, completely failed to rescue the siRNA-mediated knockdown of IFT81 (Fig. 2B). Thus, the entire negative effect on cilium formation by IFT81 depletion was recapitulated with a specific tubulin-binding–deficient mutant.
The fact that IFT81ΔN formed stable IFT core complexes (fig. S1) suggested that the ciliogenesis phenotype was because of reduced tubulin binding and not a general failure of IFT. To rule out whether mutation of the IFT81N CH domain resulted in general IFT deficiency, we turned to the unicellular protozoan parasite Trypanosoma brucei, where IFT has been well studied (15), and tested the effect of IFT81N CH-domain disruption on IFT. Yellow fluorescent protein (YFP)–tagged but otherwise normal IFT81 or mutant IFT81 (IFT81I46D,L47D, Dm) where the CH domain was unfolded (fig. S7A) were expressed at wild-type levels. One of the two IFT81 alleles was replaced with either YFP::IFT81 or YFP::IFT81Dm, leaving one WT IFT81 allele unaltered (fig. S7B). Both the localization and the IFT speed of IFT81 and IFT81Dm were similar to that observed for other IFT proteins as judged by live-cell imaging and kymographic analysis (Fig. 3 and movies S1 and 2) (15). Thus, IFT81N is not required for IFT complex assembly or normal IFTin vivo, which corroborates that the ciliogenesis phenotype observed upon IFT81N CH-domain mutation (Fig. 2) was indeed because of tubulin-binding deficiency.
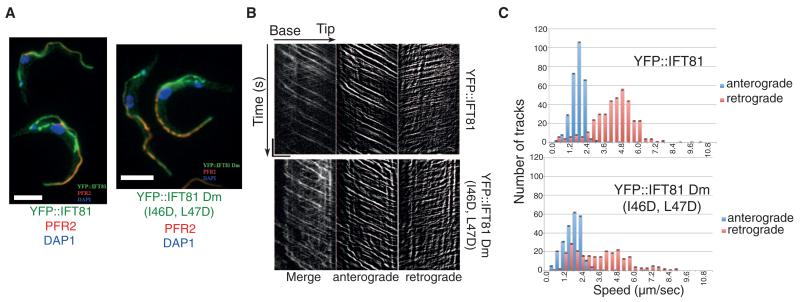
Fig. 3. IFT81N is not required for normal IFT.
(A) Immunofluorescence analysis of methanol-fixed trypanosomes expressing the indicated YFP fusion proteins from the endogenous locus stained with an antibody to green fluorescent protein (GFP) (green) and with the antibody to PFR2 L8C4 to visualize the flagellum (red). The left panel corresponds to a control strain expressing YFP::IFT81 and the right panel to the mutant YFP::IFT81Dm, where the IFT81N CH domain is unfolded. Scale bar, 5 μm. (B) Kymograph generation and separation of anterograde and retrograde traces. Kymographs were extracted from videos of cells expressing YFP::IFT81 (movie S1) or YFP::IFT81Dm (movie S2). Panels show the complete kymograph, anterograde events, and retrograde events (from left to right). The x axis corresponds to the length of the flagellum (horizontal scale bar, 5 μm) and the y axis to the elapsed time (vertical scale bar, 5 s). (C) Quantitation of the kymograph analysis shown in (B). Anterograde (blue) and retrograde velocity (red) distribution of IFT particles are calculated from cells expressing YFP::IFT81 and YFP::IFT81Dm. The kymographic analysis reveals robust anterograde trafficking with a speed of 1.75 ± 0.55 μm/s for YFP::IFT81 (n = 294 tracks from 15 cells) and 1.68 ± 0.72 μm s−1 for YFP::IFT81Dm (n = 244 tracks from 15 cells). These values are in line with those reported for anterograde movement of GFP-IFT52 (15). Curiously, retrograde transport was slowed down in the case of YFP::IFT81Dm, where a second population of relatively slow trains was detected.
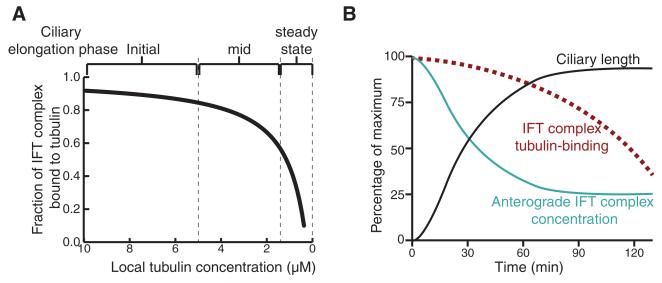
Axonemal precursors such as tubulin are added to the tip of the cilium in a length-dependent manner (16, 17). The removal of tubulin from the axonemal tip, on the other hand, appears to be constant, with no dependence on cilium length (17, 18). These observations inspired the balance-point model, in which the length of a mature cilium is the result of equal delivery and removal rates for axonemal precursors (17, 19). Furthermore, the concentration of tubulin in the cytoplasm influences ciliogenesis and cilium length in mammalian cells (20). Based on the measured affinity between IFT74/81 and tubulin (Kd = 0.9 μM) (Fig. 1G), we calculated the fraction of IFT complexes bound to αβ-tubulin as a function of tubulin concentration (Fig. 4A). Because the cellular tubulin concentration is estimated to be in the low μM range (21) and tubulin expression is induced at the onset of ciliogenesis (22), the IFT74/81:tubulin affinity is optimal for regulating cilium length via tubulin transport (Fig. 4B). The prediction is thus that most IFT complexes are loaded with tubulin during early stages of ciliogenesis, whereas lower occupancies are found during steady-state cilium length (Fig. 4), which agrees well with previously obtained data demonstrating that tubulin transport in full-length cilia yields only faint traces on kymographs, likely due to low tubulin occupancy on IFTcomplexes (13). During cilium growth, both anterograde IFTcomplex concentration and tubulin binding are negatively correlated with cilia length, resulting in a decreasing assembly rate as the cilium approaches steady-state length.
Fig. 4. Model for tubulin transport and ciliary length control.
(A) Fraction of IFT complex bound to tubulin at varying tubulin concentrations is plotted using the equation OIFT = [Tub]/{Kd + [Tub]}. OIFT is the fraction of IFT bound to tubulin, Kd is the binding constant that is experimentally determined in this study as 0.9 μM, and [Tub] is the local concentration of free tubulin at the base of the cilium. (B) From the point of initiation of flagellar regeneration, the relationship between the ciliary length, the concentration of anterograde IFT particles, and OIFT is plotted.
Here, we have shown that the two core IFT proteins IFT74 and IFT81 form a tubulin-binding module required for ciliogenesis, which suggests a role of IFT74/81 in the transport of tubulin within cilia. The fact that the high-affinity binding of tubulin occurs only for the IFT74/81 complex and not for IFT81 alone could help ensure that tubulin cargo only binds in the context of properly assembled IFT complexes. Because tubulin constitutes the backbone of all cilia, it makes sense that the tubulin has a dedicated cargo-binding site on the IFT core complex (23). We hypothesize that, although abundant ciliary cargo proteins such as tubulin may undergo IFT via dedicated transport modules, less abundant ciliary proteins are likely to compete for generic cargo-binding sites on the IFT complex.
Supplementary Material
Acknowledgments
We thank the staff at SLS for help with x-ray diffraction data collection; the crystallization facility of the Max Planck Institute of Biochemistry (Munich) for access to crystallization screening; the staff at Biozentrum Imaging Core Facility for assistance; C. Basquin for static light-scattering experiments; C. Jung for help with microscale thermophoresis; and S. Chakrabarti, A. Cook, and B. D. Engel for carefully reading and correcting the manuscript. B. D. Engel is additionally acknowledged for assistance with Fig. 4 and for many valuable discussions concerning ciliary length control. We thank the Plateforme d’Imagerie Dynamique for providing access to their equipment and Jean-Yves Tinevez for technical advice. We acknowledge M. Morawetz and M. Stiegler for technical assistance with molecular biology. This work was funded by an Emmy Noether grant (DGF; LO1627/1-1), the European Research Council (ERC grant 310343), and the European Molecular Biology Organization Young Investigator program. Work at the Institut Pasteur was funded by Agence Nationale de la Recherche grant ANR-11-BSV8-016. C.F. is funded by a fellowship from the Ministère de l’Enseignement et de la Recherche (ED387). E.A.N. acknowledges support from the University of Basel and the Swiss National Science Foundation (31003A_132428/1). M.T. is the recipient of an Erwin Schroedinger stipend granted by the Austrian Science Fund J3148-B12, S.B was supported by the International Max Planck Research School for Molecular and Cellular Life Sciences (IMPRS), and L.C. was supported by the FEBS long-term fellowship. The authors declare that they have no conflict of interest. The data presented in this paper are tabulated in the main paper and the supplementary materials. Structural coordinates have been deposited at the Protein DataBank, accession nos. 4LVP and 4LVR.
Footnotes
Supplementary Materials www.sciencemag.org/cgi/content/full/341/6149/1009/DC1 Materials and Methods, Supplementary Text, Figs. S1 to S8, Table S1, References (24–47)
References and Notes
- 1.Ishikawa H, Marshall WF. Nat. Rev. Mol. Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- 2.Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenbaum JL, Witman GB. Nat. Rev. Mol. Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 4.Finetti F, et al. Nat. Cell Biol. 2009;11:1332–1339. doi: 10.1038/ncb1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozminski KG, Beech PL, Rosenbaum JL. J. Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole DG, et al. J. Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pazour GJ, Dickert BL, Witman GB. J. Cell Biol. 1999;144:473–481. doi: 10.1083/jcb.144.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pazour GJ, Agrin N, Leszyk J, Witman GB. J. Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL. J. Cell Biol. 2004;164:255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou Y, et al. J. Cell Biol. 2007;176:653–665. doi: 10.1083/jcb.200608041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piperno G, Mead K. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4457–4462. doi: 10.1073/pnas.94.9.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciferri C, et al. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao L, et al. Nat. Cell Biol. 2011;13:790–798. doi: 10.1038/ncb2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, et al. Nature. 2010;464:1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buisson J, et al. J. Cell Sci. 2013;126:327–338. doi: 10.1242/jcs.117069. [DOI] [PubMed] [Google Scholar]
- 16.Johnson KA, Rosenbaum JL. J. Cell Biol. 1992;119:1605–1611. doi: 10.1083/jcb.119.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall WF, Rosenbaum JL. J. Cell Biol. 2001;155:405–414. doi: 10.1083/jcb.200106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song L, Dentler WL. J. Biol. Chem. 2001;276:29754–29763. doi: 10.1074/jbc.M103184200. [DOI] [PubMed] [Google Scholar]
- 19.Marshall WF, Qin H, Rodrigo Brenni M, Rosenbaum JL. Mol. Biol. Cell. 2005;16:270–278. doi: 10.1091/mbc.E04-07-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma N, Kosan ZA, Stallworth JE, Berbari NF, Yoder BK. Mol. Biol. Cell. 2011;22:806–816. doi: 10.1091/mbc.E10-03-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiller G, Weber K. Cell. 1978;14:795–804. doi: 10.1016/0092-8674(78)90335-5. [DOI] [PubMed] [Google Scholar]
- 22.Stephens RE. Dev. Biol. 1977;61:311–329. doi: 10.1016/0012-1606(77)90301-3. [DOI] [PubMed] [Google Scholar]
- 23.Lucker BF, et al. J. Biol. Chem. 2005;280:27688–27696. doi: 10.1074/jbc.M505062200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.