ABSTRACT
The high mobility group box protein SOX9 and the GLI1 transcription factor play protumorigenic roles in pancreatic ductal adenocarcinoma (PDA). In Kras transgenic mice, each of these factors are crucial for the development of PDA precursor lesions. SOX9 transcription is directly regulated by GLI1, but how SOX9 functions downstream of GLI1 is unclear. We observed positive feedback, such that SOX9-deficient PDA cells have severely repressed levels of endogenous GLI1, attributed to loss of GLI1 protein stability. SOX9 associated with the F-box domain of the SKP1/CUL1/F-box (SCF) E3 ubiquitin ligase component, β-TrCP (also known as F-box/WD repeat-containing protein 1A), and suppressed its association with SKP1 and GLI1, a substrate of SCF-β-TrCP. SOX9 also tethered β-TrCP within the nucleus and promoted its degradation. SOX9 bound to β-TrCP through the SOX9 C-terminal PQA/S domain that mediates transcriptional activation. Suppression of β-TrCP in SOX9-deficient PDA cells restored GLI1 levels and promoted SOX9-dependent cancer stem cell properties. These studies identify SOX9–GLI1 positive feedback as a major determinant of GLI1 protein stability and implicate β-TrCP as a latent SOX9-bound tumor suppressor with the potential to degrade oncogenic proteins in tumor cells.
KEY WORDS: Pancreatic ductal adenocarcinoma, SOX9, GLI1, β-TrCP
INTRODUCTION
The evolution of metazoans required new strategies for proper regulation of cell fate, including intercellular signaling, signal reception and signal transduction, processes in which transcription factors play crucial roles (Nusse, 2003; Weinberg, 2007). Central to cancer etiology is the dysregulation of cell fate, often through genetic changes that impinge on transcription factor signaling (Taipale and Beachy, 2001; Kalderon, 2002). GLI1, a member of the GLI family of zinc finger transcription factors, is a central regulator of cell fate that is deregulated in diverse tumor types (Lauth and Toftgård, 2007; Ruiz i Altaba et al., 2007; Stecca and Ruiz i Altaba, 2010; Morris et al., 2010; Hui and Angers, 2011). Increased levels of GLI1 mRNA and protein can result from genetic inactivation of tumor suppressors, such as the Hedgehog pathway receptor Patched1 (PTCH1), or mutational activation of factors such as Smoothened (SMO). GLI1 signaling impacts on multiple cancer-relevant cellular processes, promoting dedifferentiation, the generation of cancer stem cells (CSCs), tumor progression and metastasis. In addition, GLI1 can directly induce the transcription of its own mRNA through a well-characterized autoregulatory feedback, and therefore GLI1 mRNA levels often reflect the overall GLI transactivation capacity (Dai et al., 1999; Vokes et al., 2007).
Pancreatic ductal adenocarcinoma (PDA) is an aggressively metastatic tumor type that is often diagnosed at a later clinical stage (Koorstra et al., 2008; Feig et al., 2012). Although GLI1 is expressed in both epithelial PDA cells and stromal cells, a cell autonomous role within carcinoma cells appears central to the pathogenesis of this disease (Feldmann et al., 2007; Nolan-Stevaux et al., 2009; Tian et al., 2009; Lauth et al., 2010). Indeed, suppression of GLI1 in human PDA cells leads to loss of malignant properties (Ji et al., 2007; Feldmann et al., 2007; Nolan-Stevaux et al., 2009). In a Kras-dependent mouse model of PDA, either Cre-mediated excision of Gli1 or expression of a dominant-negative GLI factor suppresses tumorigenesis, including the outgrowth of precursor lesions termed pancreatic intraepithelial neoplasia (PanIN) (Rajurkar et al., 2012; Mills et al., 2013). Conversely, enforced expression of an active GLI factor in pancreatic epithelial cells promotes tumorigenesis in mice (Pasca di Magliano et al., 2006). In the canonical Hedgehog–GLI pathway, GLI activity is dependent upon signaling by Hedgehog through PTCH1 and SMO, whereas in PDA cells GLI1 is instead maintained by activated KRAS (Hingorani et al., 2005; Pasca di Magliano et al., 2006; Ji et al., 2007; Nolan-Stevaux et al., 2009; Tian et al., 2009; Lauth et al., 2010).
The protein stability of GLI1 is regulated by two E3 ubiquitin ligases, the Skp/Cul/F-box complex SCFβ-TrCP and the E3 ligase ITCH in conjunction with the adaptor protein NUMB (Huntzicker et al., 2006; Di Marcotullio et al., 2006). Similar to slmb regulation of the Drosophila GLI homolog cubitus interruptus, the mammalian SCFβ-TrCP is a major regulator of the protein stability and/or proteolytic cleavage of mammalian GLI1 and its paralogs GLI2 and GLI3 (Jiang, 2006; Huntzicker and Oro, 2008). SCFβ-TrCP is comprised of the bridging protein SKP1, the scaffolding protein CUL1, the substrate-recognizing F-box protein β-TrCP (also known as F-box/WD repeat-containing protein 1A) and the RING finger protein RBX1. This complex catalyzes the transfer of ubiquitin from E2 ligase to the substrate, leading to degradation by the ubiquitin proteasome system (UPS) (Skaar et al., 2013). In cultured human keratinocytes, GLI1 stability is dependent upon epidermal growth factor (EGF) signaling through the MEK1/2–ERK1/2 pathway (Kasper et al., 2006). Similarly, in cultured human PDA cells, activated KRAS can stabilize the GLI1 protein through ERK1/2 (also known as MAPK3/1) signaling (Ji et al., 2007). These results suggest a broader role of RAS, MEK1/2 and ERK1/2 in stabilization of GLI1.
GLI1 directly induces the transcription of SOX9, an Sry-like high mobility group (HMG) box transcription factor that plays key roles in sex determination, chondrogenesis and cell differentiation (de Crombrugghe et al., 2001; Kashimada and Koopman, 2010; Barrionuevo and Scherer, 2010). SOX9 responds to Hedgehog–Gli signaling in multiple contexts, including chondrocytes, retinal progenitor cells and developing hair follicles (Tavella et al., 2004; Vidal et al., 2005; McNeill et al., 2012; Eberl et al., 2012). Consistent with these results, the SOX9 promoter and upstream flanking region contains consensus GLI-binding sites that, when linked to a transcriptional reporter, can be regulated by GLI1 in cultured cells (Bien-Willner et al., 2007; Eberl et al., 2012).
In the developing pancreas, SOX9 is expressed in stem- or progenitor-like cells and is required for normal organogenesis (Seymour et al., 2007; Lynn et al., 2007). In the adult pancreas, SOX9 is expressed in ductal and centroacinar cells, but is normally expressed at low levels in or absent from acinar cells. Two types of studies have documented a protumorigenic role for SOX9 in PDA. First, xenograft experiments utilizing human PDA cells such as Panc-1 cells indicate that SOX9 promotes the maintenance of tumor-initiating cells (Eberl et al., 2012; Sun et al., 2013). Second, the induction of PanIN lesions in the conditional KrasG12D mouse model of PDA involves the early induction of SOX9 in acinar cells, followed by acinar-ductal metaplasia and tumor progression (Kopp et al., 2012). Conditional gene knockout or enforced expression reveals that SOX9 is critical for the occurrence of PanIN lesions. Similarly SOX9 is protumorigenic in other contexts, including colorectal cancer and mammary cancer, promoting the induction of cancer stem cell (CSC) factors such as BMI1 and/or cooperating with mesenchyme-inducing factors such as Snail or Slug (Guo et al., 2012; Matheu et al., 2012).
In the current study, we found that SOX9 is important for efficient in vitro transformation by GLI1, a feature attributed to its stabilization of GLI1. Like other SCFβ-TrCP substrates, GLI1 interacted with the C-terminal WD domain of β-TrCP. SOX9 instead interacted with the N-terminal F-box domain, and yet inhibited the association of GLI1 and β-TrCP. Consistent with a crucial role of the SOX9–β-TrCP interaction for stabilization of GLI1, suppression of β-TrCP in SOX9-deficient PDA cells led to restoration of GLI1 and promoted malignant properties. Because SCFβ-TrCP can promote the ubiquitylation of functionally diverse proteins, its ultimate role as pro- versus anti-tumorigenic might be context dependent (Frescas and Pagano, 2008; Lau et al., 2012; Shaik et al., 2012; Skaar et al., 2013). Our data suggest that β-TrCP-associated proteins such as SOX9 could be a crucial aspect of this context, capable of suppressing SCF activity against multiple pro-tumorigenic substrates such as GLI1, β-catenin and the anti-apoptotic factor MCL1. These results identify reactivation of latent β-TrCP as a strategy for the disruption of KRAS-mediated protumorigenic signaling.
RESULTS
A role for SOX9 in GLI1-mediated epithelial transformation in vitro
RK3E cells, derived from rat kidney cells by immortalization with adenovirus E1A, undergo malignant transformation in response to GLI1 and provide an epithelial context for functional studies (Foster et al., 1999; Li et al., 2006). Utilizing this model, we identified increased levels of Sox9 mRNA and the corresponding protein as early responses to exogenous human GLI1 (HsGli1, Fig. 1A; supplementary material Fig. S1A). In RK3E cells engineered to induce GLI1 when exposed to tetracycline (RK3E-TO GLI1 cells), the Sox9 mRNA was induced between 1 and 3 h after drug treatment. Kinetics were similar to those of other well-established GLI1-regulated genes, including Ptch1 and Bcl2. Consistent with a direct interaction, ChIP analyses and luciferase reporter studies identified a candidate enhancer element containing a GLI1 consensus site, located downstream of the rat Sox9 coding region and ∼1.0 kb downstream of exon 3 (supplementary material Fig. S1B–D).
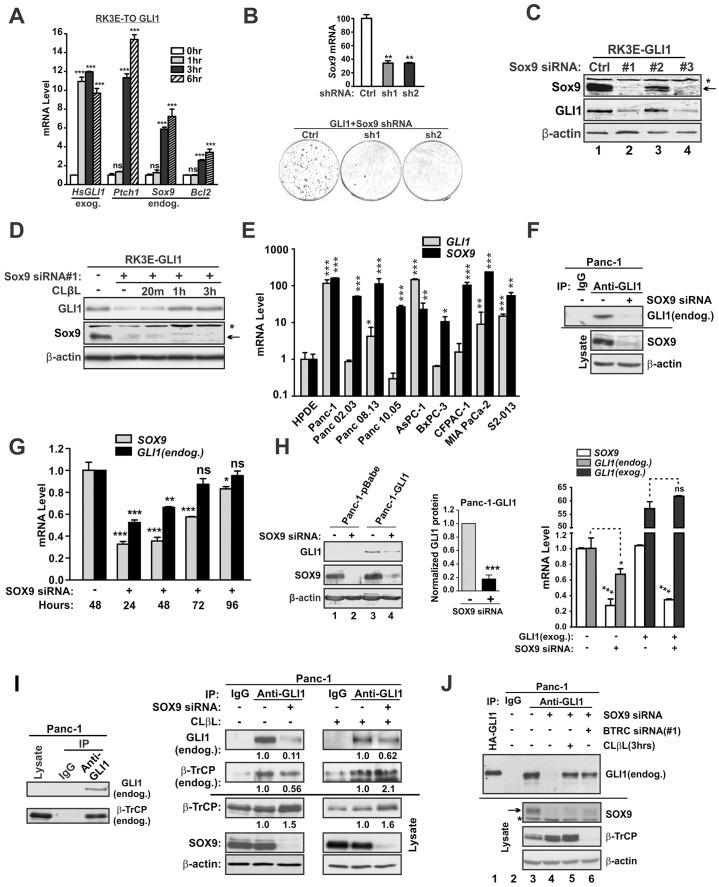
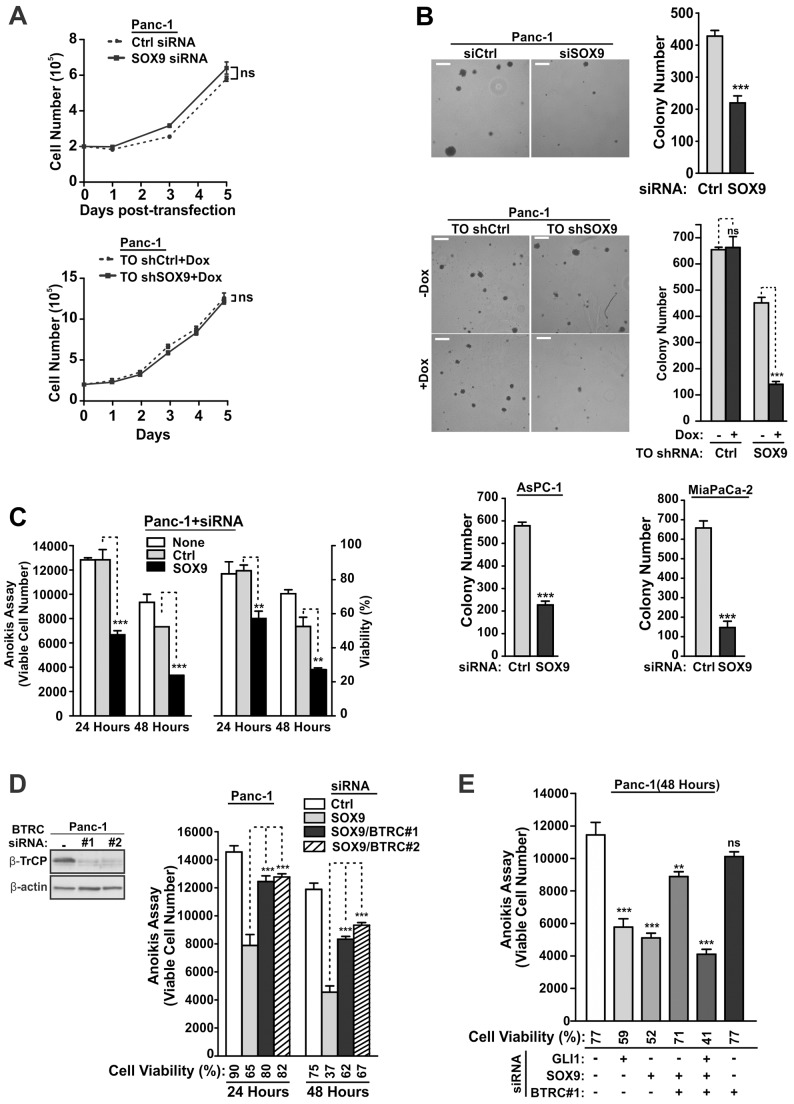
Fig. 1.
SOX9 stabilizes GLI1 in the rat RK3E epithelial model and in human PDA cells. (A) Rapid induction of SOX9 by GLI1. RK3E-derived, tet-on (TO) GLI1 cells were induced with tet or with vehicle control, and gene expression was analyzed by qRT-PCR (n = 4). exog., exogenous; endog., endogenous. (B) Role of SOX9 for GLI1-mediated in vitro transformation. Upper panel, Sox9 shRNA constructs were transfected into RK3E cells. Sox9 mRNA was analyzed by qRT-PCR following drug selection. Lower panel, GLI1-mediated in vitro transformation was assayed in RK3E cells by counting morphologically transformed foci. Dishes are representative of three independent experiments. Background was determined using empty vector and was <1 focus per dish (not shown). (C) Dependence of GLI1 protein expression on SOX9. GLI1-transformed RK3E cells were transfected with the indicated siRNA. Protein expression was analyzed at 48 h post-transfection. The arrow shows the position of SOX9. The asterisk shows a nonspecific band. (D) Proteasome inhibition restores GLI1 in SOX9-deficient cells. GLI1-transformed RK3E cells were transfected with Sox9 siRNA. Cells were treated with vehicle (DMSO, 1 h) or with proteasome inhibitor (CLβL) for the indicated interval. Cell extracts for protein analysis were prepared at 48 h post-transfection. (E) SOX9 and GLI1 were analyzed in human PDA cell lines. mRNA levels were normalized to those of immortalized pancreatic ductal epithelial cells (HPDE). (F) Dependence of GLI1 protein expression on SOX9. Panc-1 cells were treated with control or SOX9 siRNA, and endogenous GLI1 protein levels were determined by immunoprecipitation (IP)-immunoblot analysis. (G) Dependence of GLI1 mRNA expression on SOX9. Panc-1 cells were treated with siRNA and SOX9 and GLI1 mRNA levels were assessed at the indicated post-transfection interval. (H) SOX9 can regulate GLI1 protein levels independently of the mRNA. To circumvent the GLI1 autoregulation of its own transcription, a GLI1 retroviral vector was transduced into Panc-1 cells such that the preponderance of GLI1 mRNA was transcribed under control of the LTR promoter (Panc-1-GLI1) versus the endogenous GLI1 promoter (Panc-1-pBabe). The impact of SOX9 deficiency on the GLI1 protein level in Panc-1-GLI1 cells was then analyzed by immunoblotting (left panel). Following normalization to β-actin, the GLI1 protein level in control and SOX9-deficient cells was quantified in three independent assays (middle panel). Similarly, mRNA levels were analyzed by qRT-PCR (right panel). All quantitative data show the mean±s.d. *P<0.05; **P<0.01; ***P<0.001; ns, not significant. Data were analyzed using the unpaired Student's t-test (two-tailed) or one-way analysis of variance (ANOVA) followed by Tukey's multiple comparison ad hoc post-test. (I) SOX9 regulates the association of GLI1 and β-TrCP. Left panel, endogenous GLI1–β-TrCP interaction in Panc-1 cells was determined by co-IP-immunoblot analysis. Right panel, Panc-1 cells were treated with control siRNA, SOX9 siRNA and/or CLβL as indicated; the endogenous GLI1–β-TrCP interaction was evaluated by co-IP-immunoblot analysis. (J) The role of β-TrCP and the proteasome in the regulation of GLI1 by SOX9. Panc-1 cells were treated as indicated and endogenous protein levels were determined by immunoprecipitation-immunoblot analysis.
To analyze SOX9, we generated shRNA expression vectors (shSox9-1 and -2) that stably suppressed the Sox9 mRNA in RK3E cells (Fig. 1B, upper panel). As compared to a non-targeting control (Ctrl), plasmid co-transfection of GLI1 vector with either shRNA construct efficiently inhibited the outgrowth of transformed foci (Fig. 1B, lower panel). By contrast, SOX9 appeared largely dispensable when transformed foci were instead generated using an ERBB2 vector (supplementary material Fig. S1E).
Following small interfering (si)RNA-mediated suppression of SOX9 in GLI1-transformed RK3E cells, we found that GLI1 protein levels were reduced, suggesting a role for SOX9 in the maintenance of GLI1 (Fig. 1C, lanes 2 and 4). Treatment with the proteasome inhibitor clasto-lactacystin β-lactone (CLβL) rapidly restored GLI1 protein levels (Fig. 1D). These results identified SOX9 as a transcription factor with a potential role in GLI1 protein stability.
Endogenous SOX9 stabilizes GLI1 in pancreatic cancer cells through regulation of β-TrCP-mediated degradation
Analysis of conditionally deficient mice has shown that SOX9 and GLI1 play crucial roles in PDA development. We analyzed nine PDA tumor cell lines and found that, relative to non-malignant human pancreatic ductal epithelial (HPDE) cells, SOX9 levels were markedly upregulated (10- to 240-fold), and most lines likewise expressed increased levels of GLI1 (Fig. 1E).
To examine a role for endogenous SOX9, we utilized transient or inducible siRNAs that target two distinct sequences (supplementary material Fig. S2A). To detect endogenous GLI1, a low abundance factor, we utilized immunoprecipitation and immunoblot analysis. Using this assay, either of two different GLI1 antibodies identified a protein of 150 kDa (supplementary material Fig. S2B and data not shown). Repression of SOX9 in PDA cells resulted in the reduction of endogenous GLI1 protein levels (Fig. 1F), and mRNA analysis revealed synchronous loss of both SOX9 and GLI1 signals after siRNA transfection (Fig. 1G).
To analyze how SOX9 regulates GLI1, we engineered PDA cells that independently regulate the GLI1 mRNA and protein, through introduction of a GLI1 transgene under the control of a retroviral promoter (Panc-1-GLI1). In these cells, the exogenous GLI1 protein was reduced following SOX9 knockdown (Fig. 1H, lanes 3, 4). Three independent experiments indicated loss of 82% of the GLI1 (Fig. 1H, middle panel). By contrast, the GLI1 transgene-derived mRNA levels were not dependent upon SOX9 (Fig. 1H, right panel). In vector control cells, the suppression of SOX9 repressed the endogenous GLI1 mRNA. These results indicate that loss of GLI1 mRNA following SOX9 suppression is due to destabilization of the GLI1 protein and its autoregulation of GLI1 transcription.
Interaction of endogenous GLI1 and β-TrCP proteins was readily detected in human PDA cells (Fig. 1I, left panel). In the absence or presence of proteasome inhibitor the suppression of endogenous SOX9 increased the ratio of β-TrCP∶GLI1 in immunoprecipitates (Fig. 1I, right panel). Similar to results observed following treatment with proteasome inhibitor, GLI1 expression was rescued by suppression of β-TrCP (Fig. 1J, lanes 5, 6). Similar results were obtained using transient SOX9 siRNA or the TO conditional approach in either Panc-1 or AsPC-1 cells (supplementary material Fig. S2C).
SOX9 disrupts the interaction of GLI1 with β-TrCP
SOX9 was previously shown by co-immunoprecipitation (co-IP) and colocalization studies to associate with β-TrCP, likely through the SOX9 C-terminus, as deletion of this region abrogated binding (Topol et al., 2009). To investigate whether the association of SOX9 with β-TrCP can modulate the targeting of GLI1 for ubiquitylation, we utilized HEK293 cells that contain very low levels of endogenous SOX9. When expressed by transient transfection, GLI1 was found to associate with both endogenous and exogenous β-TrCP (Fig. 2A). Similar to previous reports (Topol et al., 2009), the SOX9–β-TrCP interaction was readily detected (Fig. 2B). However, in multiple experiments we were unable to observe any association of GLI1 and SOX9 (Fig. 2C). Negative results were obtained for co-IP of endogenous proteins in Panc-1 cells as well as overexpressed proteins in HEK293, using both forward and reverse co-IP strategies. These results suggested that SOX9 might not regulate GLI1 through a direct interaction but rather could regulate its association with the E3 ligase component β-TrCP.
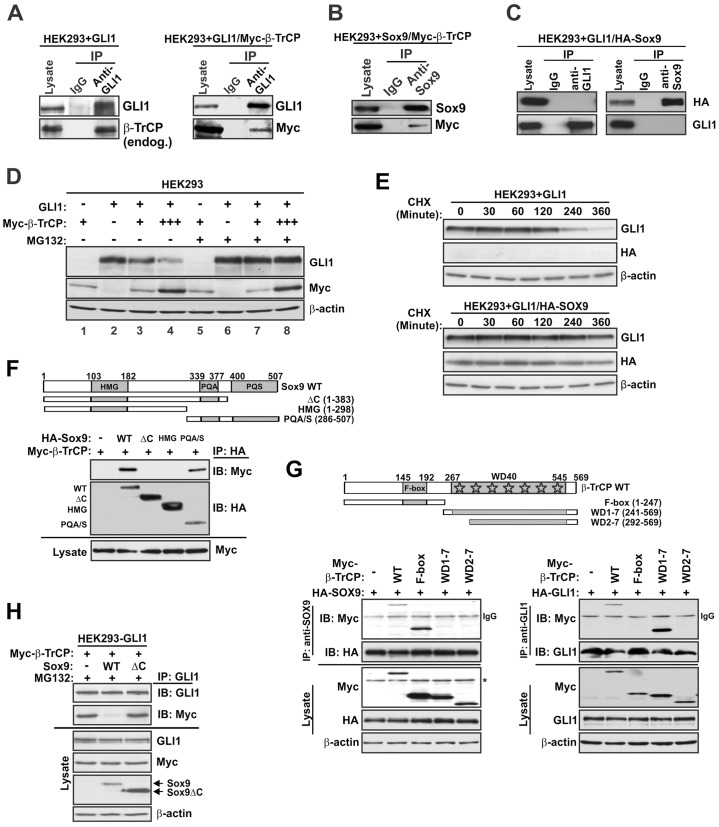
Fig. 2.
SOX9 and GLI1 interact with distinct regions of β-TrCP in a mutually incompatible fashion. Enforced expression studies were performed in HEK293 cells (A–H). (A) Co-IP analysis of GLI1 and β-TrCP. Immunoprecipitated (IP) proteins were analyzed by immunoblot analysis. Lysate lanes represent 5% of the input extract. endog., endogenous. (B) Co-IP analysis of SOX9 and β-TrCP. (C) Co-IP analysis of GLI1 and SOX9. (D) Role of the proteasome in the regulation of GLI1 by β-TrCP. Cells were transfected with the indicated vectors and treated with proteasome inhibitor (MG132) or vehicle (DMSO) for 6 h prior to preparation of cell extracts for immunoblot analysis. (E) Stabilization of GLI1 by SOX9. Cells transfected with the indicated vectors were treated with cycloheximide (CHX) prior to preparation of cell extracts for immunoblot analysis. (F) Co-IP analysis of Myc–β-TrCP with wild-type (WT) or truncated HA–SOX9 constructs. Precipitated proteins were detected by immunoblotting (IB). (G) SOX9 and GLI1 interact with distinct regions of β-TrCP. Co-IP analysis of GLI1 or SOX9 with wild-type or truncated Myc–β-TrCP constructs. Precipitated proteins were detected by immunoblot analysis. The asterisk indicates a nonspecific species. (H) The SOX9 C-terminal region is required for disruption of the GLI1 interaction with β-TrCP. Cells stably transduced with a GLI1 retroviral vector were transfected with the indicated expression vectors. The Myc–β-TrCP plasmid amount was doubled when co-transfected with wild-type SOX9 (middle lane) to achieve similar Myc–β-TrCP protein levels. Cells were treated with MG132 for 3 h before they were harvested for Co-IP analysis. Similar results were obtained in an independent experiment.
Consistent with the established role of β-TrCP in GLI1 regulation, GLI1 levels were suppressed by transient delivery of exogenous β-TrCP, and restored by addition of proteasome inhibitor (Fig. 2D). In contrast to GLI1, there was no suppression of SOX9 levels in response to β-TrCP (supplementary material Fig. S2D). These results suggest that exogenous SOX9 might prevent the binding of GLI1 to its E3 ligase and the subsequent ubiquitylation (Huntzicker et al., 2006). Indeed, an assay of GLI1 stability using the protein synthesis inhibitor cycloheximide (CHX) showed that SOX9 could extend the half-life of this protein (Fig. 2E).
SOX9 contains three conserved domains – HMG, PQA and PQS (Fig. 2F). The HMG box mediates DNA binding, whereas PQA and PQS promote transactivation. Deletion analysis indicated that the transactivation region (fragment PQA/S) is sufficient to interact with β-TrCP, and with comparable efficiency to that of wild-type SOX9 (Fig. 2F). As reported previously (Topol et al., 2009), deletions within the transactivation domain (ΔC or HMG) abrogated the interaction.
The WD40 repeats of β-TrCP mediate substrate binding, and the first WD40 repeat plays a crucial role in substrate recognition (Skaar et al., 2013). By contrast, the F-box region mediates interaction with SKP1. Consistent with lack of a suppressive effect of β-TrCP on SOX9 (supplementary material Fig. S2D), and unlike SCFβ-TrCP substrates or pseudosubstrates (Davis et al., 2002), deletion analysis indicated that SOX9 associated with the F-box region of β-TrCP (Fig. 2G, left panel). As expected for a substrate, GLI1 interacted with the WD40 repeats of β-TrCP (fragment WD1-7), and deletion of the first WD40 repeat (fragment WD2-7) abrogated this interaction (Fig. 2G, right panel). Although they interacted with distinct β-TrCP domains, co-IP of GLI1 and β-TrCP was nevertheless markedly suppressed in the presence of SOX9 (Fig. 2H, lane 2). By contrast, the co-expression of SOX9ΔC, deficient for β-TrCP binding, had little effect (Fig. 2H, lane 3). Unlike for β-TrCP, using reciprocal co-IPs, we observed no interaction of SOX9 with other F-box proteins, including FBXW7 and SKP2 (supplementary material Fig. S2E; data not shown).
SOX9 interferes with SKP1 binding and facilitates β-TrCP turnover in pancreatic cancer cells
As SOX9 interacted with F-box region of β-TrCP, we asked whether this association would interfere with β-TrCP–SKP1 binding. 293T cells contain much lower levels of endogenous SOX9 compared with Panc-1 cells (Fig. 3A). In 293T cells, the binding of Myc–β-TrCP to endogenous SKP1 was suppressed by a mean of 67% in the presence of exogenous SOX9 (Fig. 3B; three independent experiments). Conversely, the binding of Myc–β-TrCP to endogenous SKP1 was increased by a mean of 76% following SOX9 knockdown in Panc-1 cells (Fig. 3C; three independent experiments). These results identify interference with the β-TrCP–SKP1 association as a mechanism by which SOX9 can stabilize GLI1.
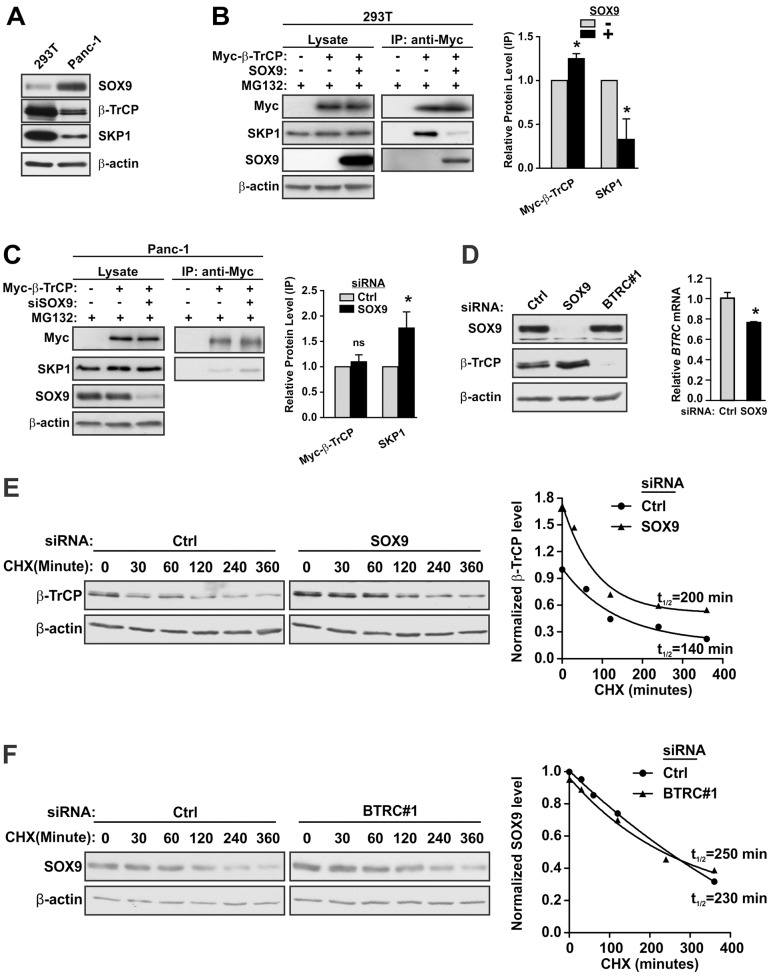
Fig. 3.
SOX9 disrupts the β-TrCP–SKP1 association and destabilizes β-TrCP in PDA cells. (A) Relative levels of SOX9, β-TrCP and SKP1 protein in HEK293T and Panc-1 cells. (B) The effect of exogenous SOX9 on the association of SKP1 and β-TrCP. 293T cells were transfected with the indicated expression vectors. Myc–β-TrCP plasmid amounts were doubled when co-transfected with wild-type SOX9 to achieve similar levels of Myc–β-TrCP protein expression. Cells were treated with MG132 for 3 h before they were harvested for co-IP analysis. The results of three independent experiments are indicated. IP, immunoprecipitation. (C) Effect of endogenous SOX9 on the association of SKP1 and β-TrCP. Panc-1 cells were transfected with the indicated expression vectors and siRNAs. Cells were treated with MG132 for 3 h before being harvested for co-IP analysis. The results of three independent experiments are indicated. (D) The effect of SOX9 on expression of β-TrCP. Panc-1 cells were treated with the indicated siRNA and then analyzed by immunoblot analysis (left panel) or by qRT-PCR (right panel). Quantitative data in B–D show the mean±s.d. *P<0.05; ns, not significant. Data were analyzed using the unpaired Student's t-test (two-tailed). (E,F) Analyses of protein stability. Panc-1 cells were transfected with the indicated siRNA and then treated with cycloheximide (CHX) for the indicated interval prior to the preparation of extracts for immunoblot analysis at 48 h post-transfection. The scanned images were quantified using NIH ImageJ and normalized to β-actin. Rate constants and protein half-lives were determined using the first order rate law.
In PDA cells, SOX9 suppression resulted in consistent increases in β-TrCP protein levels relative to the mRNA (Fig. 3D; see also the lysate panels in Fig. 1I, right panel; Fig. 1J; supplementary material Fig. S2C). Coexpression of SOX9 with β-TrCP in HEK293 or Panc-1 cells promoted β-TrCP degradation (supplementary material Fig. S2D; data not shown). We therefore determined the half-life of SOX9 and β-TrCP proteins in control PDA cells and in cells deficient in SOX9 or β-TrCP. Knockdown of SOX9 increased the steady-state abundance of β-TrCP and extended its half-life from 140 to 200 minutes (Fig. 3E). By contrast, β-TrCP suppression did not significantly alter the expression or stability of SOX9, which had a half-life of ∼230 minutes in control cells (Fig. 3F). These results identify distinct inhibitory effects of SOX9 on SCFβ-TrCP.
SOX9 tethers β-TrCP within the nucleus and selectively protects nuclear GLI1 from degradation
Both SOX9 and β-TrCP are primarily localized in the cell nucleus (Davis et al., 2002; Vidal et al., 2005; Wang et al., 2007; Topol et al., 2009). We analyzed the effect of endogenous SOX9 on the localization of transiently expressed Myc–β-TrCP in PDA cells. As previously shown for primary chondrocytes (Topol et al., 2009), SOX9 appeared to efficiently tether β-TrCP in this compartment (Fig. 4A). In SOX9-deficient PDA cells, the β-TrCP was instead dispersed throughout the cell such that the nuclear staining was reduced and cytoplasmic staining was increased.
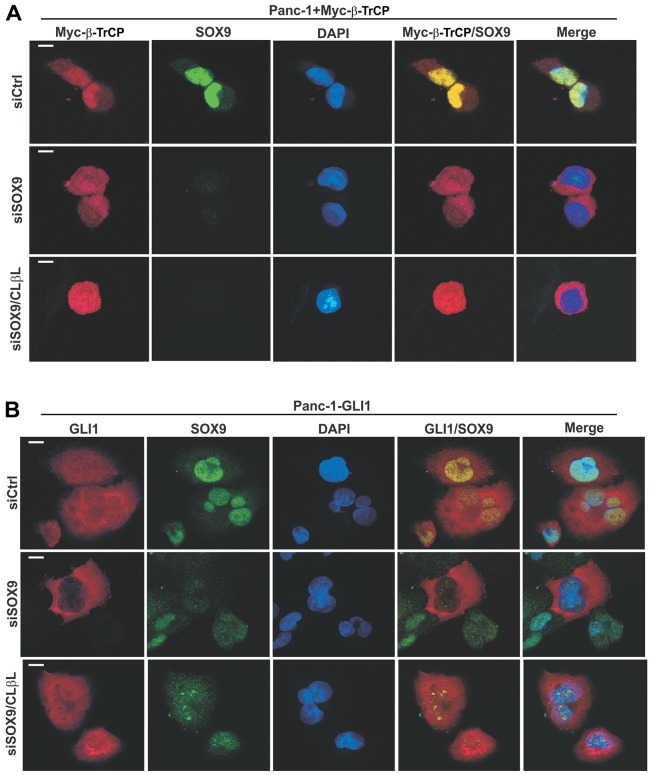
Fig. 4.
SOX9 tethers β-TrCP within the nuclei of PDA cells and protects nuclear GLI1 from degradation. (A) Panc-1 cells were transfected with exogenous Myc-tagged β-TrCP together with the indicated siRNA. (B) Panc-1 cells were infected with the lentiviral GLI1 vector and then with the indicated siRNA. Cells were treated with CLβL or vehicle (DMSO) for the final 3 h prior to fixation. Cells were fixed and processed for indirect immunofluorescence analysis at 48 h post-transfection. Scale bars: 10 µm.
Exogenous GLI1 was detected in both the nucleus and the cytoplasm (Fig. 4B, upper row). In SOX9-deficient cells, GLI1 was preferentially lost from the cell nucleus, but cytoplasmic staining was preserved (Fig. 4B, middle row). Consistent with a UPS role in the suppression of nuclear GLI1, treatment of cells with CLβL restored nuclear staining (Fig. 4B, lower row). This immunostaining data appeared to be consistent with the partial suppression of exogenous GLI1 observed by immunoblot analysis (Fig. 1H). The more complete suppression observed for endogenous GLI1 (Fig. 1F,I,J; supplementary material Fig. S2C) remains unexplained, but could reflect its preferential nuclear localization relative to that of the overexpressed protein. These results support a role for SOX9 in tethering β-TrCP within the nucleus of PDA cells. Selective loss of nuclear GLI1 upon suppression of SOX9 indicates that nuclear SCFβ-TrCP activity is increased despite the dispersal of β-TrCP throughout the cell. These results suggest that the apparent β-TrCP concentration can be discordant with its activity when SOX9 is present.
SOX9 promotes the malignant properties of PDA cells in a fashion dependent upon β-TrCP and GLI1
Both SOX9 and GLI1 appear to be crucial for tumorigenesis in PDA (Rajurkar et al., 2012; Kopp et al., 2012; Mills et al., 2013). Although SOX9-deficient Panc-1 cells fail to form tumors in a mouse xenograft assay, potential links to GLI1 function remain unexplored (Eberl et al., 2012). For SOX9-deficient and control PDA cells we analyzed several in vitro correlates of the malignant phenotype, including proliferation, anchorage independence and survival. SOX9-deficient Panc-1 cells proliferated in two-dimensional (2D) culture similarly to control cells (Fig. 5A), but were unable to efficiently form colonies in soft agar over a period of 2 weeks (Fig. 5B). Other PDA cell lines, AsPC-1 and MiaPaCa-2, responded similarly. When cells were suspended in low attachment plates for 24 or 48 h (anoikis assay), deficiency in SOX9 resulted in a marked reduction in the overall viable cell number and the proportion of viable cells (Fig. 5C). These results confirm that SOX9 deficiency can abrogate the malignant properties of PDA cells.
Fig. 5.
In a GLI1-dependent fashion, β-TrCP suppression rescues malignant properties in SOX9-deficient PDA cells. (A) Panc-1 cell growth in 2D culture was analyzed following transfection of SOX9 or control siRNAs (upper panel). Alternatively, Panc-1-TO shRNA cells were treated with Dox (lower panel). (B) Analysis of anchorage-independent growth regulation by SOX9. A soft agar assay was performed using Panc-1 cells (n = 3). Similar results are shown for AsPC-1 and MiaPaCa-2 cell lines (lower panels). Scale bars: 400 µm. (C) Analysis of anoikis regulation by SOX9. The anoikis assay was performed using Panc-1 cells. The number of surviving cells (left y-axis) and the percentages of viable cells (right y-axis) are indicated for each timepoint (n = 3). (D,E) Role of β-TrCP in the phenotype of SOX9-deficient cells. Following siRNA transfection, anoikis assays were performed in Panc-1 cells. All quantitative data show the mean±s.d. **P<0.01; ***P<0.001; ns, not significant. Growth curves were analyzed using non-linear regression curve fitting. Other quantitative data were analyzed using the unpaired Student's t-test (two-tailed) or one-way ANOVA followed by Tukey's multiple comparison ad hoc post-test.
We used the anoikis assay to determine whether deficiency of SCFβ-TrCP substrates such as GLI1 might be responsible for the loss of malignant properties. We restored GLI1 expression in SOX9-deficient cells using β-TrCP siRNAs (Fig. 5D; supplementary material Fig. S3A). The cell death phenotype of SOX9-deficient PDA cells was attenuated by co-suppression of β-TrCP in both Panc-1 and AsPC-1 cells. Consistent with a functional role for GLI1 as a mediator of the increased cell survival following β-TrCP suppression, co-suppression of GLI1 promoted cell death (Fig. 5E).
SOX9 promotes the CSC-like properties of PDA cells in a β-TrCP-dependent fashion
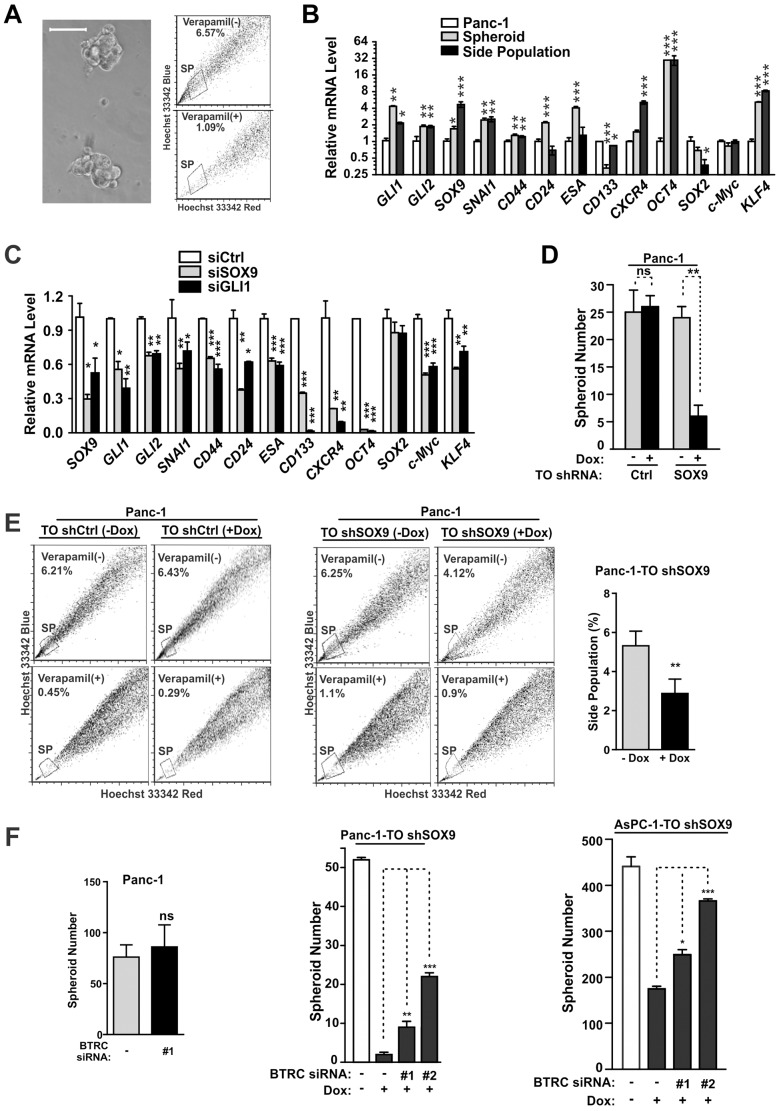
CSCs are an aggressively malignant subset of tumor cells (Clevers, 2011). In PDA, both SOX9 and GLI1 are important for the maintenance of this subpopulation (Eberl et al., 2012; Tang et al., 2012; Sun et al., 2013; Li et al., 2013). To determine whether there is phenotypic overlap following modulation of SOX9 or GLI1, we enriched for CSCs using spheroid formation or dye efflux assays (i.e. side population cells) (Bhagwandin and Shay, 2009) (Fig. 6A). In addition to GLI or GLI-regulated factors (GLI1, GLI2, SOX9, SNAI1), these subpopulations had increased levels of pancreatic CSC markers (CD24, CD44, ESA, CD133 and CXCR4) as well as factors important for the generation of induced pluripotent stem cells (OCT4 and KLF4) (Takahashi and Yamanaka, 2006; Li et al., 2007; Hermann et al., 2007) (Fig. 6B). Consistent with a role for GLI1 in the phenotype of SOX9-deficient cells, suppression of either gene product in 2D cultures of PDA cells reduced the expression of CSC markers, resulting in very similar profiles (Fig. 6C).
Fig. 6.
β-TrCP suppression rescues CSC properties in SOX9-deficient PDA cells. (A) Panc-1 cells form spheroids when suspended in stem cell growth medium (left panel). Scale bar: 200 µm. Side population (SP) Panc-1 cells were identified by verapamil-sensitive dye efflux (right panel). (B) Expression profiling of spheroid cells and side population cells. (C) Roles of SOX9 and GLI1 in the regulation of CSC markers. The expression of putative CSC markers was determined in Panc-1 cells treated with the indicated siRNA. (D) Spheroid formation assay of Panc-1-TO shRNA cells (n = 3). (E) Side population analysis of PDA cells. Cells were treated with Dox or vehicle for 5 days and then stained with Hoechst 33342. The results of three independent experiments are presented (right panel). (F) Role of β-TrCP in the growth of tumor spheroids. siβ-TrCP had little effect on its own in parental Panc-1 cells (left panel). The effect of siβ-TrCP was then analyzed in the context of SOX9-deficient PDA cells (middle and right panels). All quantitative data show the mean±s.d. *P<0.05; ** P<0.01; *** P<0.001; ns, not significant.
We next examined whether β-TrCP suppression could rescue the CSC phenotype in SOX9-deficient cells. Stable suppression of SOX9 reduced spheroid formation by 75% (Fig. 6D). Consistent with this result, SOX9 suppression in 2D-cultured PDA cells also reduced the abundance of side population cells (Fig. 6E; supplementary material Fig. S3B). Without SOX9 suppression, transient suppression of β-TrCP had little effect on spheroid growth (Fig. 6F, left panel). However, in SOX9-deficient cells, siβ-TrCP partially rescued spheroid formation (Fig. 6F; supplementary material Fig. S3C). The remarkably similar expression profile in SOX9- and GLI1-deficient tumor cells, and restoration of spheroid formation by siβ-TrCP support a role for the suppression of SCFβ-TrCP activity as a mechanism by which SOX9 can promote GLI1 expression and the CSC-like phenotype.
SOX9 is increased whereas β-TrCP is decreased in primary human PDA
The above data indicate that SOX9 functions in cultured PDA cells to antagonize β-TrCP expression and its activity towards GLI1. We utilized Oncomine microarray data (Compendia Bioscience) to analyze SOX9 and β-TrCP expression in primary human PDA specimens (Rhodes et al., 2004). Four studies each detected increased levels of PDA signature genes, typified by SFN (also known as 14-3-3σ), in PDA tumor tissue relative to morphologically normal tissue (Logsdon et al., 2003; Iacobuzio-Donahue et al., 2003; Segara et al., 2005; Badea et al., 2008; Pei et al., 2009) (supplementary material Table S1 and Fig. S4). In these studies, SOX9 mRNA expression was upregulated (1.00- to 1.87-fold), whereas transcripts encoding β-TrCP (BTRC) were consistently repressed (1.12- to 1.25-fold). This increase in SOX9 and decrease in BTRC expression might contribute to the upregulation of GLI1 in PDA (Thayer et al., 2003; Feldmann et al., 2007; Nolan-Stevaux et al., 2009).
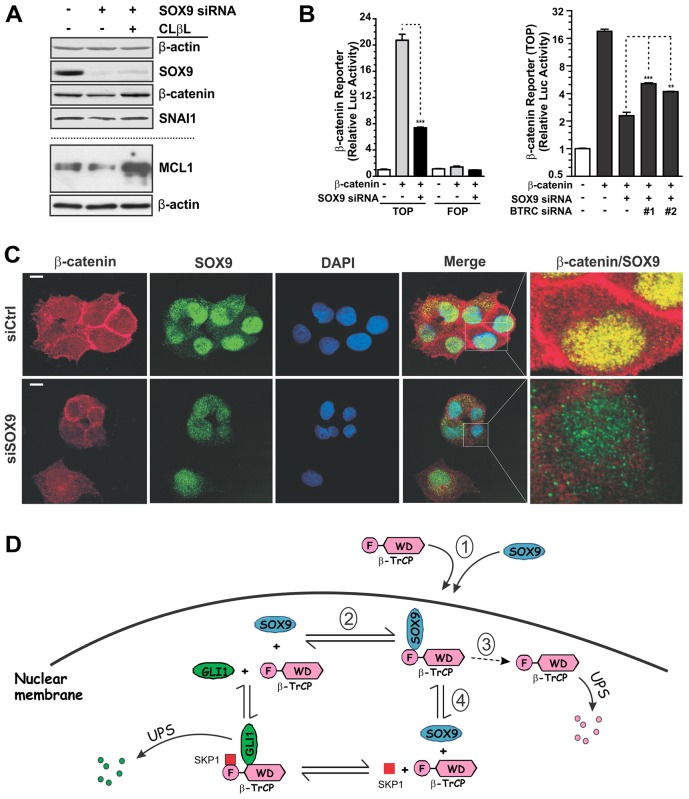
SOX9 stabilizes other β-TrCP-regulated proteins
If SOX9 selectively stabilizes nuclear GLI1 by antagonizing β-TrCP activity in this compartment, then other substrates might be similarly affected, potentially with more subtle effects for cytoplasmic substrates. Consistent with their known regulation by SCFβ-TrCP, β-catenin, Snail and MCL1 protein levels were each reduced by similar amounts to 50% of control levels following SOX9 knockdown in PDA cells, and CLβL treatment restored the levels (Fig. 7A) (Polakis, 1999; Zhou et al., 2004; Ding et al., 2007). For β-catenin, consistent results were obtained with a transcriptional reporter assay (Fig. 7B) and immunofluorescence analysis (Fig. 7C). Cells transfected with SOX9 siRNA had similar to 50% reduction in the expression of both nuclear and cytoplasmic β-catenin (Fig. 7C; supplementary material Fig. S3D). These data are consistent with a predominant role of SOX9 in the nuclear tethering and suppression of SCFβ-TrCP activity through protein–protein interaction, with more subtle effects in the cytoplasm attributed to alterations in β-TrCP concentration (Fig. 4A). Fig. 7D presents a model summarizing the observed effects of SOX9 on β-TrCP and nuclear GLI1.
Fig. 7.
SOX9 stabilizes β-catenin in Panc-1 cells. (A) Regulation of diverse SCFβ-TrCP substrates by SOX9. Panc-1 cells were treated with the indicated siRNA and protein expression was analyzed by immunoblotting. (B) Regulation of β-catenin activity by SOX9 and β-TrCP. Panc-1 cells were transfected with the indicated siRNA and TCF4-dependent transcriptional activity was determined by using a luciferase reporter assay. Data show the mean±s.d. **P<0.01; ***P<0.001. (C) Regulation of β-catenin protein expression by SOX9. Panc-1 cells were treated with the indicated siRNA and then analyzed by indirect immunofluorescence analysis. Scale bars: 10 µm. Quantification of β-catenin is shown in supplementary material Fig. S3D. (D) A model depicting the effects of SOX9 on β-TrCP in PDA cells, including nuclear tethering (1), disruption of its association with GLI1 (2), promotion of UPS-mediated turnover (3) and disruption of its association with SKP1 (4).
DISCUSSION
SOX9 is crucial for diverse developmental processes including sex determination, chondrogenesis and pancreatogenesis (de Crombrugghe et al., 2001; Kashimada and Koopman, 2010; Barrionuevo and Scherer, 2010). For cancers of the prostate, colorectum, pancreas and breast, SOX9 plays important roles in carcinoma cell malignant properties (Vidal et al., 2005; Wang et al., 2008; Eberl et al., 2012; Kopp et al., 2012; Guo et al., 2012; Matheu et al., 2012; Sun et al., 2013). In chondrocytes, SOX9 induces the transcription of genes such as COL2A1, COL11A2 and aggrecan (de Crombrugghe et al., 2001; Oh et al., 2010; Pritchett et al., 2011). Although SOX9-responsive genes were analyzed in several contexts, the mechanisms by which SOX9 promotes malignant properties remain poorly understood, and the relative importance of transcriptional signaling versus other biochemical activities is unclear (Lynn et al., 2007; Bhandari et al., 2012; Guo et al., 2012).
In rat RK3E epithelial cells, we identified Sox9 as an early transcriptional response to GLI1, and linked this regulation to a downstream Sox9 enhancer element using ChIP and transcriptional reporter studies. These data are consistent with previous reports indicating regulation of mouse or human SOX9 by Hedgehog and/or GLI1 (Tavella et al., 2004; Vidal et al., 2005; McNeill et al., 2012; Eberl et al., 2012). In analyzing GLI1-transformed cells we were surprised to observe a pronounced loss of GLI1 protein stability upon suppression of SOX9, attributed to UPS-mediated degradation. This effect was conserved in human PDA cells and indicated a consistent role for SOX9 in stabilization of GLI1.
In a previous study focused upon chondrocytes, SOX9 was found to associate with β-TrCP and to promote β-TrCP nuclear localization and the degradation of nuclear β-catenin (Topol et al., 2009). In the current study focused upon PDA, we found that SOX9 likewise functions as a nuclear tether for β-TrCP, but few other aspects of the chondrocyte study appeared to extend to the PDA context. We instead obtained extensive data supporting a potent inhibitory role for SOX9 on β-TrCP activity in the nucleus. Suppression of β-TrCP in SOX9-deficient cells restored GLI1 levels and promoted phenotypes attributed to SOX9 or GLI1. Supporting the identification of GLI1 as a major factor in the altered phenotype of SOX9-deficient cells, the CSC marker profile of SOX9- or GLI1-deficient PDA cells was remarkably similar.
Consistent with an inhibitory effect on SCFβ-TrCP, SOX9 interacted through its PQA/S region with the F-box domain of β-TrCP and suppressed the association of β-TrCP with either endogenous or exogenous SKP1. Typical of other substrates, GLI1 associated with the WD domain. Co-IP assays indicated that GLI1 and SOX9 are mutually incompatible for β-TrCP association. Indeed, in SOX9-deficient PDA cells, the association of GLI1 with β-TrCP was increased by several fold. It is unclear why SOX9 association with the F-box region would prevent GLI1 from binding to the WD domain. Possibilities include steric hindrance, a conformational shift or that SOX9 could facilitate the binding of other nuclear proteins such as hnRNP-U, a WD domain-binding pseudosubstrate, and thereby prevent the association of β-TrCP with GLI1 (Davis et al., 2002).
An additional effect that strongly supported the inhibition of SCFβ-TrCP by SOX9 was the destabilization of β-TrCP. Whether this instability results from the reduced association with SKP1, from tethering within the nuclear compartment and/or from some other effect of SOX9 or yet another associated factor is currently unclear.
In SOX9-deficient PDA cells, β-TrCP was more widely distributed throughout the cell, with reduced nuclear staining and approximately a twofold increase in cytoplasmic staining compared with that of control cells. This redistribution appeared to impact on not only nuclear GLI1 but also cytoplasmic proteins such as the SCFβ-TrCP substrate β-catenin. While nuclear GLI1 was markedly destabilized, the cytoplasmic substrate β-catenin was suppressed by ∼50%. Similar effects were observed for two other well-established substrates of SCFβ-TrCP, Snail and MCL1. These results indicate a broad role for SOX9 in the regulation of protein stability.
In SOX9-deficient PDA cells, the effects of untethered β-TrCP were quite distinct for nuclear and cytoplasmic GLI1. These results appear to indicate that nuclear GLI1 is selectively targeted by β-TrCP, although the mechanism(s) responsible for this selectivity is unclear. The results are overall consistent with identification of the Drosophila homolog as a short-lived nuclear transcriptional activator in response to Hedgehog signaling (Ohlmeyer and Kalderon, 1998). We conclude that when SOX9 is present, the β-TrCP nuclear staining intensity can be discordant with nuclear SCFβ-TrCP activity.
Mouse model data have established clear roles for GLI1 and SOX9 in the malignant progression of PDA (Pasca di Magliano et al., 2006; Ji et al., 2007; Feldmann et al., 2007; Nolan-Stevaux et al., 2009; Rajurkar et al., 2012; Eberl et al., 2012; Kopp et al., 2012; Mills et al., 2013). Interestingly, both SOX9 transcript levels and GLI1 protein stability appear to be crucially dependent upon upstream signaling by RAS–MEK–ERK, but it remains unclear how either factor is regulated by this signaling (Murakami et al., 2000; Kasper et al., 2006; Ji et al., 2007; Eberl et al., 2012). Whereas a Sox9 transgene directed to the pancreatic acinar cells is sufficient on its own to initiate early PanIn lesions in mice, a GLI transgene does not induce these lesions, suggesting that GLI is the more dependent factor (Pasca di Magliano et al., 2006; Kopp et al., 2012). These considerations suggest that KRAS might stabilize GLI1 by signaling through SOX9. Alternatively, it is possible that activated KRAS could more directly regulate GLI1 or both transcription factors in parallel. Blockade of this KRAS signaling in tumor cells, a current focus of research, has the potential to suppress SOX9 and to activate β-TrCP-mediated degradation of multiple oncoproteins.
MATERIALS AND METHODS
Expression vectors and plasmid transfection
pcDNA4/TO-HA-GLI1 was described previously (Li et al., 2006). Retroviral vectors were made by inserting the HAGLI1 cassette of pcDNA4/TO-HA-GLI1 into pB-puro and pLUT7. pLUT7 was provided by Alexey Ivanov (West Virginia University School of Medicine). The human SOX9 expression vector, HA3 SOX9 pcDNA3.1, was a gift from Benoit de Crombrugghe (MD Anderson Cancer Center).
Mouse β-TrCP1 vector containing a 3×Myc epitope tag at the N-terminus (Wan et al., 2004), a gift from Xu Cao (Johns Hopkins University School of Medicine), was used to generate truncated constructs by PCR. All cloned PCR products were verified by sequencing. A human ubiquitin vector, Myc-Ub, was provided by Alexey Ivanov. pcDNA3-β-catenin (Addgene 16828) was a gift from Eric Fearon (University of Michigan Medical School) (Kolligs et al., 1999). pCMV-Myc CDC4 WT* was a gift from Bert Vogelstein (Johns Hopkins University School of Medicine; FBXW7, Addgene plasmid 16652). pcDNA3-myc-Skp2 was a gift from Yue Xiong (University of North Carolina at Chapel Hill; Addgene plasmid 19947).
pTRIPZ-TOshSOX9 lentivector was generated by transfer of the shRNAmir cassette from pGIPZ (V3LHS_396212, Thermo Scientific). Non-targeting TRIPZ lentiviral shRNAmir RHS4743 served as a control. shRNA vectors targeting rat Sox9 (RnSox9) were constructed in pSilencer 2.1-U6 neo as described previously (Li et al., 2006). shRNA target sequences are listed in supplementary material Table S2.
Retroviral transduction was performed as described previously (Foster et al., 1999). Transient transfections were performed as described previously (Lin et al., 2011) using Lipofectamine 2000 Transfection Reagent (Life Technologies) for Panc-1 cells and TransIT-LT1 Reagent (Mirus) for other cells.
Isolation of rat Sox9 cDNA and generation of Sox9 expression vectors
PCR was used to synthesize cDNA fragments containing the RnSox9 5′UTR and protein coding regions. The sequence has been deposited with the NCBI (accession number KP732536). pBpuro-RnSox9 was used for stable expression studies. RnSox9 vectors tagged with hemagglutinin (HA), including wild-type HA–Sox9 and truncation mutants, were generated by PCR followed by insertion into pcDNA3.1. All cloned PCR products were verified by sequencing.
Cell culture and small molecules
The immortalized human pancreatic ductal epithelial cell line HPDE (H6c7), provided by Ming-Sound Tsao (University of Toronto, Canada), was cultured in keratinocyte serum-free (KSF) medium supplemented with bovine pituitary extract and epidermal growth factor (Life Technologies). S2-013 PDA cells were a gift of Martin Johnson (University of Alabama at Birmingham). S2-013, HEK293, 293T and RK3E cells were cultured in high-glucose DMEM supplemented with L-glutamine, penicillin, streptomycin and 10% (v/v) fetal bovine serum. Other PDA cell lines were from American Type Culture Collection (ATCC) and were cultured as recommended by ATCC. Doxycycline (Dox, 0.5 µg/ml), tetracycline (tet, 1.0 µg/ml), cycloheximide (CHX, 50 µg/ml), clasto-lactacystin β-lactone (CLβL, 10 µM) and MG132 (20 µM) were from Sigma-Aldrich.
siRNA transfection, RNA isolation and real-time quantitative PCR
siRNA target sequences are listed in supplementary material Table S2. siRNA transfection was performed using Lipofectamine RNAiMax (Life Technologies) as reported previously (Lin et al., 2011). For some experiments, the Panc-1 cells were re-transfected after 24 h. Cell extracts were prepared 48 h after the start of the initial transfection. Co-transfection of plasmids with siRNAs was as reported previously (Lin et al., 2011).
Total RNA was purified using the RNeasy Plus Mini kit (Qiagen). Reverse transcription was performed using SuperScript II (Life Technologies). Real-time quantitative (qRT)-PCR reactions utilized Brilliant II SyBr green QPCR master mix, GAPDH served as the internal control, and reactions were analyzed on an Mx3005P system (Agilent). Oligonucleotides are shown in supplementary material Table S3. qRT-PCR results represent three or more independent experiments (error bars show the s.d.).
Luciferase reporter assay
Topflash (wild-type TCF4-binding sites) or Fopflash (mutated sites) reporter constructs were gifts from Bert Vogelstein. The Renilla luciferase vector pRL-TK served as an internal control. For siRNA co-transfection, siRNA Ctrl2 served as the control (supplementary material Table S2). Reporter assays were performed on 12-well plates at 48 h post-transfection using Dual Luciferase Reporter Assays (Promega) as described previously (Lin et al., 2011). Data represent three or more independent experiments (error bars show the s.d.).
Protein expression studies
For immunoblotting, mouse anti-GLI1 and rabbit anti-β-TrCP, anti-SKP1 and anti-SNAI1/Snail were from Cell Signaling Technology. Rabbit anti-SOX9 was from EMD Millipore. Mouse anti-Myc (9E10) and anti-β-catenin were from BD Biosciences. Rat anti-HA (3F10) was from Roche Applied Science. Anti-MCL1 was from Santa Cruz Biotechnology (S-19).
Whole-cell lysates for immunoblotting were obtained by extraction in ice-cold RIPA buffer without SDS as described previously (Lin et al., 2011). For immunoprecipitation and co-IP, cells were resuspended in NE-A buffer and then lysed by addition of NaCl and glycerol as described previously (Chen and Bieker, 2001). GLI1 protein was precipitated with rabbit anti-GLI1 (Li et al., 2006), whereas rabbit anti-SOX9 (H-90, Santa Cruz Biotechnology) or a monoclonal anti-HA (12CA5, Roche) was used for SOX9. Normal rabbit or mouse IgG (Sigma-Aldrich) was used as a control. Protein complexes were recovered using Protein-A–Sepharose (Sigma-Aldrich).
For immunoblotting, proteins were transferred to nitrocellulose membrane and detected using chemiluminescence (Pierce ECL, Thermo-Scientific). Quantitative analysis of autoradiographic images was performed using ImageJ v1.42 (National Institutes of Health), and the normalized results were transferred to GraphPad Prism (version 5) for statistical analysis.
In vitro transformation assay
RK3E transformation assays were performed as described previously (Li et al., 2006). Cells were fixed and stained using Modified Wright Stain (Sigma-Aldrich) at 2–3 weeks post-transfection, and foci with a diameter >1.0 mm were counted.
Soft agar assay
Anchorage-independent cell growth assays were performed on six-well plates. A 2.0-ml underlay composed of 0.63% (w/v) agar in complete DMEM was placed in each well. A volume of 1.0 ml of cells (3.0×103) in DMEM containing 0.33% agar was plated on top. Cells were fed with 1.0 ml of 0.63% agar in DMEM each week. For induction of shRNA expression, 0.5 µg/ml of doxycycline was included. Cells were fixed after 2–3 weeks in 10% methanol/10% acetic acid and stained with 0.1% Crystal Violet (Sigma-Aldrich) in 50% methanol. Colonies >50 µm in diameter were counted under phase contrast microscopy.
Anoikis assay
Matrix deprivation assays were performed as described previously (Kumar et al., 2011). At 24 h post siRNA transfection, 2.0×104 cells were resuspended in 1.0 ml of complete DMEM containing 1% methylcellulose (Sigma-Aldrich) and plated in six-well low-attachment plates. Assays were performed in triplicate (error bars show the s.d.). At 24 and 48 h later, cells were collected, resuspended in Accumax (Innovative Cell Technologies) to generate single cell suspensions, and mixed with 0.4% Trypan Blue (Cellgro). Trypan Blue staining was scored using a hemocytometer.
Spheroid formation assay
PDA cells were trypsinized, counted and suspended in six-well low-attachment plates using 1∶1 DMEM/F-12 supplemented with 1× B27, 4.0 µg/ml heparin, 20 ng/ml EGF, 20 ng/ml FGF and 1% methylcellulose. For each cell line, 2000–5000 cells were plated in triplicate in a final volume of 2.0 ml. The spheroids in each well were quantified by phase contrast microscopy. For RNA isolation, spheroids were harvested after 7–10 days by centrifugation at 200 g for 1 min.
Flow cytometry
For side population analysis, cells were trypsinized, washed with PBS and resuspended in staining buffer [DMEM, 2% (v/v) FBS, 1.0 mM HEPES-KOH pH 7.0]. Samples were incubated for 30 min at 37°C with or without 50 µg/ml verapamil (Sigma-Aldrich), mixed with Hoechst 33342 (Sigma-Aldrich) to a final concentration of 5.0 µg/ml and incubated for 90 min at 37°C with intermittent mixing. The cells were washed twice with PBS, resuspended in sorting buffer [1×PBS, 5.0 mM EDTA, 1% (w/v) BSA, 25 mM HEPES-KOH pH 7.0], and maintained at 4°C in the dark. Analysis was performed using a FACSAria III cell sorter (BD Biosciences), and both Hoechst Red and Hoechst Blue were measured.
Immunofluorescent staining and quantification
Cells were treated with CLβL or DMSO for 3 h before fixation. Cells were fixed and stained as described previously (Pandya et al., 2004) at 48 h post-transfection using rabbit anti-SOX9 (1∶1000, EMD Millipore), mouse anti-GLI1 (1∶100, Cell Signaling Technologies), anti-Myc (9E10, 2.5 µg/ml, BD Biosciences) and/or anti-β-catenin (0.17 µg/ml, BD Biosciences). Secondary antibodies were cross-absorbed Alexa-Fluor-647-conjugated goat anti-rabbit-IgG and Alexa-Fluor-555-conjugated goat anti-mouse-IgG (Life Technologies). Nuclei were counterstained in DAPI prior to mounting of coverslips using ProLong Antifade (Life Technologies).
Digital images were captured and pseudocolored using a Zeiss LSM 510 laser scanning confocal on an upright Zeiss AxioImager and Zeiss LSM software (Carl Zeiss). Images were exported as tiff files, and Corel software was used to make minor identical adjustments to the experimental and control panels in parallel. β-catenin was quantified using Image J.
Supplementary Material
Acknowledgments
We thank Benoit de Crombrugghe (MD Anderson Cancer Center), Bert Vogelstein (Johns Hopkins University School of Medicine), Xu Cao (Johns Hopkins University School of Medicine), Eric Fearon (University of Michigan), Yue Xiong (University of North Carolina at Chapel Hill) and Alexey Ivanov (West Virginia University) for providing expression vectors. We thank Ming-Sound Tsao (University of Toronto) and Martin Johnson (University of Alabama at Birmingham) for providing cell lines.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
W.D. and D.B.V. performed assays of gene expression, protein stability, protein-protein interaction and cancer stem cell properties. C.-C.L. performed some of the flow cytometry studies. K.H.M. assisted with immunofluorescence microscopy and K.M.B. assisted with flow cytometry studies. J.M.R. assisted with experimental design and the interpretation of data.
Funding
This work was supported by the National Cancer Institute [grant number RO1 CA127405 to J.M.R.]; the Jo and Ben Statler Chair in Breast Cancer Research; and a Pilot grant from the West Virginia University Research Foundation. Flow cytometry experiments were performed in the West Virginia University Flow Cytometry Core Facility, supported by a National Institutes of Health (NIH) equipment grant [grant number RR020866]; and grants from the Institutional Development Award (IDeA) from the National Institute of General Medical Sciences [grant numbers P30GM103488 (CoBRE) and P20GM103434 (INBRE)]. Imaging experiments and image analysis were performed in the West Virginia University Microscope Imaging Facility, supported by the Mary Babb Randolph Cancer Center; and the NIH [grant numbers P20 RR016440, P30 RR032138/GM103488 and P20 RR016477]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.162164/-/DC1
References
- Badea L., Herlea V., Dima S. O., Dumitrascu T., Popescu I. (2008). Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology 55, 2016–2027. [PubMed] [Google Scholar]
- Barrionuevo F., Scherer G. (2010). SOX E genes: SOX9 and SOX8 in mammalian testis development. Int. J. Biochem. Cell Biol. 42, 433–436 10.1016/j.biocel.2009.07.015 [DOI] [PubMed] [Google Scholar]
- Bhagwandin V. J., Shay J. W. (2009). Pancreatic cancer stem cells: fact or fiction? Biochim. Biophys. Acta 1792, 248–259 10.1016/j.bbadis.2009.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari R. K., Haque M. M., Skinner M. K. (2012). Global genome analysis of the downstream binding targets of testis determining factor SRY and SOX9. PLoS ONE 7, e43380 10.1371/journal.pone.0043380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien-Willner G. A., Stankiewicz P., Lupski J. R. (2007). SOX9cre1, a cis-acting regulatory element located 1.1 Mb upstream of SOX9, mediates its enhancement through the SHH pathway. Hum. Mol. Genet. 16, 1143–1156 10.1093/hmg/ddm061 [DOI] [PubMed] [Google Scholar]
- Chen X., Bieker J. J. (2001). Unanticipated repression function linked to erythroid Krüppel-like factor. Mol. Cell. Biol. 21, 3118–3125 10.1128/MCB.21.9.3118-3125.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. (2011). The cancer stem cell: premises, promises and challenges. Nat. Med. 17, 313–319 10.1038/nm.2304 [DOI] [PubMed] [Google Scholar]
- Dai P., Akimaru H., Tanaka Y., Maekawa T., Nakafuku M., Ishii S. (1999). Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J. Biol. Chem. 274, 8143–8152 10.1074/jbc.274.12.8143 [DOI] [PubMed] [Google Scholar]
- Davis M., Hatzubai A., Andersen J. S., Ben-Shushan E., Fisher G. Z., Yaron A., Bauskin A., Mercurio F., Mann M., Ben-Neriah Y. (2002). Pseudosubstrate regulation of the SCFβ-TrCP ubiquitin ligase by hnRNP-U Genes Dev. 16, 439–451 10.1101/gad.218702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crombrugghe B., Lefebvre V., Nakashima K. (2001). Regulatory mechanisms in the pathways of cartilage and bone formation. Curr. Opin. Cell Biol. 13, 721–728 10.1016/S0955-0674(00)00276-3 [DOI] [PubMed] [Google Scholar]
- Di Marcotullio L., Ferretti E., Greco A., De Smaele E., Po A., Sico M. A., Alimandi M., Giannini G., Maroder M., Screpanti I. et al. (2006). Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat. Cell Biol. 8, 1415–1423 10.1038/ncb1510 [DOI] [PubMed] [Google Scholar]
- Ding Q., He X., Hsu J. M., Xia W., Chen C. T., Li L. Y., Lee D. F., Liu J. C., Zhong Q., Wang X. et al. (2007). Degradation of Mcl-1 by β-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol. Cell. Biol. 27, 4006–4017 10.1128/MCB.00620-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl M., Klingler S., Mangelberger D., Loipetzberger A., Damhofer H., Zoidl K., Schnidar H., Hache H., Bauer H. C., Solca F. et al. (2012). Hedgehog-EGFR cooperation response genes determine the oncogenic phenotype of basal cell carcinoma and tumour-initiating pancreatic cancer cells. EMBO Mol. Med. 4, 218–233 10.1002/emmm.201100201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig C., Gopinathan A., Neesse A., Chan D. S., Cook N., Tuveson D. A. (2012). The pancreas cancer microenvironment. Clin. Cancer Res. 18, 4266–4276 10.1158/1078-0432.CCR-11-3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann G., Dhara S., Fendrich V., Bedja D., Beaty R., Mullendore M., Karikari C., Alvarez H., Iacobuzio-Donahue C., Jimeno A. et al. (2007). Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 67, 2187–2196 10.1158/0008-5472.CAN-06-3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster K. W., Ren S., Louro I. D., Lobo-Ruppert S. M., McKie-Bell P., Grizzle W., Hayes M. R., Broker T. R., Chow L. T., Ruppert J. M. (1999). Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ. 10, 423–434. [PubMed] [Google Scholar]
- Frescas D., Pagano M. (2008). Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 8, 438–449 10.1038/nrc2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Keckesova Z., Donaher J. L., Shibue T., Tischler V., Reinhardt F., Itzkovitz S., Noske A., Zürrer-Härdi U., Bell G. et al. (2012). Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148, 1015–1028 10.1016/j.cell.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann P. C., Huber S. L., Herrler T., Aicher A., Ellwart J. W., Guba M., Bruns C. J., Heeschen C. (2007). Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 1, 313–323 10.1016/j.stem.2007.06.002 [DOI] [PubMed] [Google Scholar]
- Hingorani S. R., Wang L., Multani A. S., Combs C., Deramaudt T. B., Hruban R. H., Rustgi A. K., Chang S., Tuveson D. A. (2005). Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7, 469–483 10.1016/j.ccr.2005.04.023 [DOI] [PubMed] [Google Scholar]
- Hui C. C., Angers S. (2011). Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 27, 513–537 10.1146/annurev-cellbio-092910-154048 [DOI] [PubMed] [Google Scholar]
- Huntzicker E. G., Oro A. E. (2008). Controlling hair follicle signaling pathways through polyubiquitination. J. Invest. Dermatol. 128, 1081–1087 10.1038/sj.jid.5700957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzicker E. G., Estay I. S., Zhen H., Lokteva L. A., Jackson P. K., Oro A. E. (2006). Dual degradation signals control Gli protein stability and tumor formation. Genes Dev. 20, 276–281 10.1101/gad.1380906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobuzio-Donahue C. A., Maitra A., Olsen M., Lowe A. W., van Heek N. T., Rosty C., Walter K., Sato N., Parker A., Ashfaq R. et al. (2003). Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am. J. Pathol. 162, 1151–1162 10.1016/S0002-9440(10)63911-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z., Mei F. C., Xie J., Cheng X. (2007). Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J. Biol. Chem. 282, 14048–14055 10.1074/jbc.M611089200 [DOI] [PubMed] [Google Scholar]
- Jiang J. (2006). Regulation of Hh/Gli signaling by dual ubiquitin pathways. Cell Cycle 5, 2457–2463 10.4161/cc.5.21.3406 [DOI] [PubMed] [Google Scholar]
- Kalderon D. (2002). Similarities between the Hedgehog and Wnt signaling pathways. Trends Cell Biol. 12, 523–531 10.1016/S0962-8924(02)02388-7 [DOI] [PubMed] [Google Scholar]
- Kashimada K., Koopman P. (2010). Sry: the master switch in mammalian sex determination. Development 137, 3921–3930 10.1242/dev.048983 [DOI] [PubMed] [Google Scholar]
- Kasper M., Schnidar H., Neill G. W., Hanneder M., Klingler S., Blaas L., Schmid C., Hauser-Kronberger C., Regl G., Philpott M. P. et al. (2006). Selective modulation of Hedgehog/GLI target gene expression by epidermal growth factor signaling in human keratinocytes. Mol. Cell. Biol. 26, 6283–6298 10.1128/MCB.02317-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolligs F. T., Hu G., Dang C. V., Fearon E. R. (1999). Neoplastic transformation of RK3E by mutant β-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol. Cell. Biol. 19, 5696–5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koorstra J. B., Hustinx S. R., Offerhaus G. J., Maitra A. (2008). Pancreatic carcinogenesis. Pancreatology 8, 110–125 10.1159/000123838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp J. L., von Figura G., Mayes E., Liu F. F., Dubois C. L., Morris J. P., IV, Pan F. C., Akiyama H., Wright C. V., Jensen K. et al. (2012). Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 22, 737–750 10.1016/j.ccr.2012.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Park S. H., Cieply B., Schupp J., Killiam E., Zhang F., Rimm D. L., Frisch S. M. (2011). A pathway for the control of anoikis sensitivity by E-cadherin and epithelial-to-mesenchymal transition. Mol. Cell. Biol. 31, 4036–4051 10.1128/MCB.01342-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A. W., Fukushima H., Wei W. (2012). The Fbw7 and betaTRCP E3 ubiquitin ligases and their roles in tumorigenesis. Front. Biosci. (Landmark Ed) 17, 2197–2212 10.2741/4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauth M., Toftgård R. (2007). Non-canonical activation of GLI transcription factors: implications for targeted anti-cancer therapy. Cell Cycle 6, 2458–2463 10.4161/cc.6.20.4808 [DOI] [PubMed] [Google Scholar]
- Lauth M., Bergström A., Shimokawa T., Tostar U., Jin Q., Fendrich V., Guerra C., Barbacid M., Toftgård R. (2010). DYRK1B-dependent autocrine-to-paracrine shift of Hedgehog signaling by mutant RAS. Nat. Struct. Mol. Biol. 17, 718–725 10.1038/nsmb.1833 [DOI] [PubMed] [Google Scholar]
- Li X., Deng W., Nail C. D., Bailey S. K., Kraus M. H., Ruppert J. M., Lobo-Ruppert S. M. (2006). Snail induction is an early response to Gli1 that determines the efficiency of epithelial transformation. Oncogene 25, 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Heidt D. G., Dalerba P., Burant C. F., Zhang L., Adsay V., Wicha M., Clarke M. F., Simeone D. M. (2007). Identification of pancreatic cancer stem cells. Cancer Res. 67, 1030–1037 10.1158/0008-5472.CAN-06-2030 [DOI] [PubMed] [Google Scholar]
- Li S. H., Fu J., Watkins D. N., Srivastava R. K., Shankar S. (2013). Sulforaphane regulates self-renewal of pancreatic cancer stem cells through the modulation of Sonic hedgehog-GLI pathway. Mol. Cell. Biochem. 373, 217–227 10.1007/s11010-012-1493-6 [DOI] [PubMed] [Google Scholar]
- Lin C. C., Liu L. Z., Addison J. B., Wonderlin W. F., Ivanov A. V., Ruppert J. M. (2011). A KLF4-miRNA-206 autoregulatory feedback loop can promote or inhibit protein translation depending upon cell context. Mol. Cell. Biol. 31, 2513–2527 10.1128/MCB.01189-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon C. D., Simeone D. M., Binkley C., Arumugam T., Greenson J. K., Giordano T. J., Misek D. E., Kuick R., Hanash S. (2003). Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 63, 2649–2657. [PubMed] [Google Scholar]
- Lynn F. C., Smith S. B., Wilson M. E., Yang K. Y., Nekrep N., German M. S. (2007). Sox9 coordinates a transcriptional network in pancreatic progenitor cells. Proc. Natl. Acad. Sci. USA 104, 10500–10505 10.1073/pnas.0704054104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu A., Collado M., Wise C., Manterola L., Cekaite L., Tye A. J., Canamero M., Bujanda L., Schedl A., Cheah K. S. et al. (2012). Oncogenicity of the developmental transcription factor Sox9. Cancer Res. 72, 1301–1315 10.1158/0008-5472.CAN-11-3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill B., Perez-Iratxeta C., Mazerolle C., Furimsky M., Mishina Y., Andrade-Navarro M. A., Wallace V. A. (2012). Comparative genomics identification of a novel set of temporally regulated hedgehog target genes in the retina. Mol. Cell. Neurosci. 49, 333–340 10.1016/j.mcn.2011.12.008 [DOI] [PubMed] [Google Scholar]
- Mills L. D., Zhang Y., Marler R. J., Herreros-Villanueva M., Zhang L., Almada L. L., Couch F., Wetmore C., Pasca di Magliano M., Fernandez-Zapico M. E. (2013). Loss of the transcription factor GLI1 identifies a signaling network in the tumor microenvironment mediating KRAS oncogene-induced transformation. J. Biol. Chem. 288, 11786–11794 10.1074/jbc.M112.438846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. P., IV W. a. n. g., S. C. and Hebrok M. (2010). KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat. Rev. Cancer 10, 683–695 10.1038/nrc2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S., Kan M., McKeehan W. L., de Crombrugghe B. (2000). Up-regulation of the chondrogenic Sox9 gene by fibroblast growth factors is mediated by the mitogen-activated protein kinase pathway. Proc. Natl. Acad. Sci. USA 97, 1113–1118 10.1073/pnas.97.3.1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan-Stevaux O., Lau J., Truitt M. L., Chu G. C., Hebrok M., Fernández-Zapico M. E., Hanahan D. (2009). GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev. 23, 24–36 10.1101/gad.1753809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R. (2003). Wnts and Hedgehogs: lipid-modified proteins and similarities in signaling mechanisms at the cell surface. Development 130, 5297–5305 10.1242/dev.00821 [DOI] [PubMed] [Google Scholar]
- Oh C. D., Maity S. N., Lu J. F., Zhang J., Liang S., Coustry F., de Crombrugghe B., Yasuda H. (2010). Identification of SOX9 interaction sites in the genome of chondrocytes. PLoS ONE 5, e10113 10.1371/journal.pone.0010113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmeyer J. T., Kalderon D. (1998). Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature 396, 749–753 10.1038/25533 [DOI] [PubMed] [Google Scholar]
- Pandya A. Y., Talley L. I., Frost A. R., Fitzgerald T. J., Trivedi V., Chakravarthy M., Chhieng D. C., Grizzle W. E., Engler J. A., Krontiras H. et al. (2004). Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin. Cancer Res. 10, 2709–2719 10.1158/1078-0432.CCR-03-0484 [DOI] [PubMed] [Google Scholar]
- Pasca di Magliano M., Sekine S., Ermilov A., Ferris J., Dlugosz A. A., Hebrok M. (2006). Hedgehog/Ras interactions regulate early stages of pancreatic cancer. Genes Dev. 20, 3161–3173 10.1101/gad.1470806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H., Li L., Fridley B. L., Jenkins G. D., Kalari K. R., Lingle W., Petersen G., Lou Z., Wang L. (2009). FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell 16, 259–266 10.1016/j.ccr.2009.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis P. (1999). The oncogenic activation of β-catenin. Curr. Opin. Genet. Dev. 9, 15–21 10.1016/S0959-437X(99)80003-3 [DOI] [PubMed] [Google Scholar]
- Pritchett J., Athwal V., Roberts N., Hanley N. A., Hanley K. P. (2011). Understanding the role of SOX9 in acquired diseases: lessons from development. Trends Mol. Med. 17, 166–174 10.1016/j.molmed.2010.12.001 [DOI] [PubMed] [Google Scholar]
- Rajurkar M., De Jesus-Monge W. E., Driscoll D. R., Appleman V. A., Huang H., Cotton J. L., Klimstra D. S., Zhu L. J., Simin K., Xu L. et al. (2012). The activity of Gli transcription factors is essential for Kras-induced pancreatic tumorigenesis. Proc. Natl. Acad. Sci. USA 109, E1038–E1047 10.1073/pnas.1114168109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D. R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A. M. (2004). ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6 10.1016/S1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz i Altaba A., Mas C., Stecca B. (2007). The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol. 17, 438–447 10.1016/j.tcb.2007.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segara D., Biankin A. V., Kench J. G., Langusch C. C., Dawson A. C., Skalicky D. A., Gotley D. C., Coleman M. J., Sutherland R. L., Henshall S. M. (2005). Expression of HOXB2, a retinoic acid signaling target in pancreatic cancer and pancreatic intraepithelial neoplasia. Clin. Cancer Res. 11, 3587–3596 10.1158/1078-0432.CCR-04-1813 [DOI] [PubMed] [Google Scholar]
- Seymour P. A., Freude K. K., Tran M. N., Mayes E. E., Jensen J., Kist R., Scherer G., Sander M. (2007). SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc. Natl. Acad. Sci. USA 104, 1865–1870 10.1073/pnas.0609217104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaik S., Nucera C., Inuzuka H., Gao D., Garnaas M., Frechette G., Harris L., Wan L., Fukushima H., Husain A. et al. (2012). SCF(β-TRCP) suppresses angiogenesis and thyroid cancer cell migration by promoting ubiquitination and destruction of VEGF receptor 2. J. Exp. Med. 209, 1289–1307 10.1084/jem.20112446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar J. R., Pagan J. K., Pagano M. (2013). Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14, 369–381 10.1038/nrm3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B., Ruiz I Altaba A. (2010). Context-dependent regulation of the GLI code in cancer by HEDGEHOG and non-HEDGEHOG signals. J. Mol. Cell Biol. 2, 84–95 10.1093/jmcb/mjp052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Mathews L. A., Cabarcas S. M., Zhang X., Yang A., Zhang Y., Young M. R., Klarmann K. D., Keller J. R., Farrar W. L. (2013). Epigenetic regulation of SOX9 by the NF-κB signaling pathway in pancreatic cancer stem cells. Stem Cells 31, 1454–1466 10.1002/stem.1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J., Beachy P. A. (2001). The Hedgehog and Wnt signalling pathways in cancer. Nature 411, 349–354 10.1038/35077219 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Tang S. N., Fu J., Nall D., Rodova M., Shankar S., Srivastava R. K. (2012). Inhibition of sonic hedgehog pathway and pluripotency maintaining factors regulate human pancreatic cancer stem cell characteristics. Int. J. Cancer 131, 30–40 10.1002/ijc.26323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavella S., Biticchi R., Schito A., Minina E., Di Martino D., Pagano A., Vortkamp A., Horton W. A., Cancedda R., Garofalo S. (2004). Targeted expression of SHH affects chondrocyte differentiation, growth plate organization, and Sox9 expression. J. Bone Miner. Res. 19, 1678–1688 10.1359/JBMR.040706 [DOI] [PubMed] [Google Scholar]
- Thayer S. P., di Magliano M. P., Heiser P. W., Nielsen C. M., Roberts D. J., Lauwers G. Y., Qi Y. P., Gysin S., Fernández-del Castillo C., Yajnik V. et al. (2003). Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature 425, 851–856 10.1038/nature02009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., Callahan C. A., DuPree K. J., Darbonne W. C., Ahn C. P., Scales S. J., de Sauvage F. J. (2009). Hedgehog signaling is restricted to the stromal compartment during pancreatic carcinogenesis. Proc. Natl. Acad. Sci. USA 106, 4254–4259 10.1073/pnas.0813203106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topol L., Chen W., Song H., Day T. F., Yang Y. (2009). Sox9 inhibits Wnt signaling by promoting β-catenin phosphorylation in the nucleus. J. Biol. Chem. 284, 3323–3333 10.1074/jbc.M808048200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal V. P., Chaboissier M. C., Lützkendorf S., Cotsarelis G., Mill P., Hui C. C., Ortonne N., Ortonne J. P., Schedl A. (2005). Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr. Biol. 15, 1340–1351 10.1016/j.cub.2005.06.064 [DOI] [PubMed] [Google Scholar]
- Vokes S. A., Ji H., McCuine S., Tenzen T., Giles S., Zhong S., Longabaugh W. J., Davidson E. H., Wong W. H., McMahon A. P. (2007). Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development 134, 1977–1989 10.1242/dev.001966 [DOI] [PubMed] [Google Scholar]
- Wan M., Tang Y., Tytler E. M., Lu C., Jin B., Vickers S. M., Yang L., Shi X., Cao X. (2004). Smad4 protein stability is regulated by ubiquitin ligase SCFβ-TrCP1. J. Biol. Chem. 279, 14484–14487 10.1074/jbc.C400005200 [DOI] [PubMed] [Google Scholar]
- Wang H., McKnight N. C., Zhang T., Lu M. L., Balk S. P., Yuan X. (2007). SOX9 is expressed in normal prostate basal cells and regulates androgen receptor expression in prostate cancer cells. Cancer Res. 67, 528–536 10.1158/0008-5472.CAN-06-1672 [DOI] [PubMed] [Google Scholar]
- Wang H., Leav I., Ibaragi S., Wegner M., Hu G. F., Lu M. L., Balk S. P., Yuan X. (2008). SOX9 is expressed in human fetal prostate epithelium and enhances prostate cancer invasion. Cancer Res. 68, 1625–1630 10.1158/0008-5472.CAN-07-5915 [DOI] [PubMed] [Google Scholar]
- Weinberg R. A. (2007). Growth factors, receptors, and Cancer. The Biology of Cancer Weinberg R A, ed119–158New York, NY: Garland Science, Taylor and Francis Group, LLC. [Google Scholar]
- Zhou B. P., Deng J., Xia W., Xu J., Li Y. M., Gunduz M., Hung M. C. (2004). Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat. Cell Biol. 6, 931–940 10.1038/ncb1173 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.