Abstract
Purpose
Adaptation to host immune surveillance is now recognized as a hallmark of cancer onset and progression, and represents an early, indispensable event in cancer evolution. This process of evolution is first instigated by an immune selection pressure imposed by natural host surveillance mechanisms and may then be propagated by vaccination or other types of immunotherapy.
Experimental Design
We developed a system to simulate cancer evolution in a live host and to dissect the mechanisms responsible for adaptation to immune selection. Here we show that the oxygen-sensitive α subunit of hypoxia-inducible factor 1 (HIF-1α) plays a central role in cancer immune adaptation under conditions of normal oxygen tension.
Results
We found that tumor cells gain HIF-1α in the course of immune selection under normoxia and that HIF-1α renders tumor cells resistant to lysis by tumor-specific cytotoxic T lymphocytes (CTLs) in culture and in mice. The effects of HIF-1α on immune adaptation were mediated through VEGFA-dependent activation of the AKT and ERK signaling pathways, which induced an anti-apoptotic gene expression network in tumor cells.
Conclusion
Our study therefore establishes a link between immune selection, overexpression of HIF-1α, and cancer immune adaptation under normoxia, providing new opportunities for molecular intervention in cancer patients.
Introduction
Immunotherapy has emerged as a promising approach for the clinical management of cancer. However, in many cases, it has been observed that the generation of a tumor-specific immune response does not translate into tumor regression in cancer patients. A potential explanation for this is the overexpression by tumor cells of proteins that bestow them with enhanced survival, proliferation, and invasion capacity (1). In particular, hypoxia-inducible factor 1 (HIF-1) is a key orchestrator of diverse biochemical pathways, from proliferation and survival to angiogenesis and invasion (2). Overexpression of HIF-1 has been reported in virtually most of carcinomas (3), and HIF-1 has been shown to drive cancer progression as well as resistance to chemotherapy and radiotherapy (2). Moreover, resistance of tumor cells to killing by NK cells or T cells has been reported to occur through HIF-1 under hypoxia (4-9). Although HIF-1 is a gateway to cancer progression, the manner in which it initially arises within tumor cells remains unknown. In fact, HIF-1 is exquisitely sensitive to oxygen tension and is typically only present in stable form under hypoxia (10). Here we found unexpectedly that stable expression of HIF-1 in tumor cells occurs even under normal oxygen tension. We inferred that gain of HIF-1 is a key element of cancer evolution that arises from selection pressure imposed by an antitumor immune response.
To explore this idea, we examined cancer evolution in the context of immune surveillance. Adaptation to immune defenses, in particular those mounted by CD8+ cytotoxic T lymphocytes (CTLs), has emerged as an early, indispensable, and host-intrinsic event in cancer progression (11). Thus, immune surveillance is an ideal selection pressure for the analysis of cancer evolution. We invented a system referred to as VICE, for Vaccination-Induced Cancer Evolution, in which variants of a parental tumor are derived through serial rounds of immune selection either in culture or in mice (12).
We employed VICE to explore the role of HIF-1 in cancer evolution under immune surveillance. Here we show that the α subunit of HIF-1 (HIF-1α) becomes markedly elevated during immune selection even under normoxia, and HIF-1α expression by tumor cells dictates the ability of cognate CTLs to control tumor growth. To our knowledge, gain of HIF-1α in tumor cells under normoxia in response to immune selection has not been previously reported. We found that the effects of HIF-1α on immune adaptation are transmitted through vascular endothelial growth factor A (VEGFA)-mediated activation of the AKT and ERK pathways, which induce the expression of a constellation of anti-apoptotic molecules in tumor cells. Blockade of each of these pathways abrogated resistance of tumor cells to lysis by cognate CTLs, underscoring the importance of the HIF-1α/VEGFA axis in immune adaptation.
Materials and Methods
Cells
HPV-16 E7+ cells (TC-1, TC-1 P3, TC-1 P3 (A17), TC-1/no insert, and TC-1/ HIF-1α) were used as a mouse tumor model. The production and maintenance of TC-1 (13) and TC-1 P3 A17 cells (14, 15) has been described previously. TC-1/HIF-1α cells were generated with the pMSCV/HIF-1α K532R vector (for TC-1/HIF-1α). For the production of human immune-resistant tumor cells, 106 CaSki (CaSki P0) cells were pulsed with 10 μg/ml HLA-A2-restricted MART-1 M27 peptide (AAGIGILTV) for 2 hr and mixed with KKM MART-1-specific human CD8+ T cells (gift from Dr. Cassian Yee) in a 6-well plate at 1:1 ratio for 4 hr. Surviving CaSki cells were expanded and termed CaSki P1 cells. The above procedure was twice repeated to create the CaSki P3 cells. Non-specific antigen NY-ESO-1 (SLLMWITQC) peptide-pulsed CaSki cells pulsed with the irrelevant peptide NY-ESO-1 (SLLMWITQC) were incubated with MART-1-specific human CD8+ T cells to derive control CaSki cells (CaSki N1 and CaSki N3). For hypoxia culture, cells were maintained at 1% O2 in a controlled atmosphere chamber (PLAS-LABS, Lansing, MI) with a gas mixture containing 1% O2, 5% CO2, and 94% N2 at 37°C. Cells were exposed to hypoxia for 14 hr before analysis unless otherwise indicated.
Mice
6- to 8-week-old female C57BL/6 mice were acquired from Daehan Biolink (Chungbuk, Korea). Mice were maintained and handled under protocols approved by the Korea University Institutional Animal Care and Use Committee (KUIACUC-2013-210).
Chemicals and reagents
Pharmacologic agents used to inhibit signaling axes were LY294002 (Calbiochem Corp, San Diego, CA) for the PI3K pathway, SB203580 (Calbiochem Corp, San Diego, CA) for p38 MAPK pathway, and PD98059 (Stressgen, MI, USA) for the ERK pathway.
DNA and siRNA constructs
To produce the pMSCV/human HIF-1α vector, DNA fragments encoding the human HIF-1α K532R mutant were PCR-amplified from pCMV/HIF-1α K532R (gift from Dr. Lee Mi Ok) with primers (5’-ATGGAGGGCGCCGGCGGCG-3’ and 5’-CAGAATTCTCAGTTAACTTGATCCAAA-3’). The amplified fragments were cloned into the BglII/EcoRI sites of the pMSCV retroviral vector (Clontech, Mountain View, CA). Synthetic siRNA specific for Hif-1α was synthesized with a 2’-O-ACE-RNA phosphoramides backbone (Invitrogen, Lafayette, CO). The sequences of mouse Hif-1α siRNA are 5’-CCAGATCTCGGCGAAGTAA-3` (sense) and 5’-UUACUUCGCCGAGAUCUGG-3’ (antisense), human Hif-1α siRNA are 5’-CCUAUAUCCCAAUGGAUGAUG-3` (sense) and 5’-CAUCAUCCAUUGGGAUAUAGG-3’ (antisense). 105 tumor cells were transfected in 6-well plates with 100 pmol of synthesized siRNAs with Lipofectamine 2000 (Invitrogen, Carlsbad, CA).
Quantitative RT-PCR
Total RNA was extracted with the RNeasy Mini Kit (Qiagen, Gaithersburg, MD) and treated with DNase (Ambion, Austin, TX). cDNA synthesis was performed with RT&Go Mastermix (MP Biomedicals, Aurora, OH), and real-time PCR was performed with Lightcycler FastStart DNA SYBR Green Master Mix (Roche, Mannheim, Germany) using mouse and human Vegfa-specific primers with the following sequences (Bioneer, Daejeon, Korea): mouse Vegfa, 5’-TGCACCCACGACAGAAGGA-3’ (forward) and 5’-GGCAGTAGCTTCGCTGGTAGAC-3’ (reverse); human Vegfa, 5’-CTGCTGTCTTGGGTGCATTGG-3’ (forward) and 5’-CACCGCCTCGGCTTGTCACAT-3’ (reverse). All data was normalized to expression level of β-Actin mRNA.
Real-time quantitative RT-PCR
Total RNA from TC-1 and CaSki cells was purified using TRIzol reagent (Invitrogen). Real-time PCR was conducted to detect Vegfa with TaqMan Universal SYBR Green Master Mix (Roche, IN) using the mouse and human primer sets for Vegfa.
Western blot
Samples probed with primary antibodies against phospho-AKT (Ser473), AKT, phospho-ERK (T202/Y204), ERK, p38 MAPK, BCL-w, BID, BIM, BAD, phospho-BAD (Ser 136), XIAP, Dual phospho-p38 MAPK (Cell Signaling Technology, MA), HIF-1α, HIF-2α, Bcl-2, Bcl-xL, Bax, (Santa Cruz Biotechnology, Dallas, TX), BAK (BD Biosciences, San Jose, CA), or E7 (gift from Dr. Ju-Hong Jun). Antibodies were diluted 1:1000, and bands were visualized by enhanced chemiluminescence (Elpis Biotech, Daejeon, Korea).
ELISA
VEGFA concentration in supernatant of TC-1 P0, P3 (A17) and CaSki P0, P3, N3 cells collected 48 hr after incubation was measured with mouse and human VEGFA Quantikine ELISA Kit (R&D Systems, Minneapolis, MN). Supernatant of Hif-1α siRNA-treated cells was collected after 72 hr. Supernatant of cells treated with VEGFA neutralizing Ab (100 ng/ml) was collected after 24 hr. VEGFA concentration was normalized to the number of cells in each sample and depicted as pg of VEGFA per 106 cells.
Immunofluorescence microscopy
TC-1 P0, P1, P2, P3, N1, N2 and N3 cells were fixed in 4% paraformaldehyde for 10 min. Cells were washed with PBS, treated with 0.2% Triton X-100, and blocked for 1 hr in 1% BSA. Cells were incubated overnight in the presence of primary mAb against HIF-1α in a humidified chamber at 4°C. Cells were stained with Alexa Flour 555–labeled goat anti-rabbit IgG. Nuclei were stained with SYTOX Green (Invitrogen). Expression of HIF-1α was examined by confocal laser scanning microscopy (Carl Zeiss, Jena, Germany).
CTL-mediated apoptosis assay
TC-1 cells or CaSki cells were labeled with CFSE (10 μM) in DMEM supplemented with 0.1% FBS. Cells were incubated for 10 min at 37°C and then pulsed with E7, NY-ESO-1, or MART1 peptide (10 μg/ml) for 1 hr respectively. Tumor cells were then mixed with E7 or MART-1-specific CD8+ CTLs at a 1:1 ratio and incubated for 4 hr at 37°C. Cells were stained for active caspase-3 as an index of apoptosis and examined by flow cytometry. For CTL-mediated assay upon VEGFA blockade, VEGFA-neutralizing mAb (100ng/ml) was added to cells 24 hr prior to the assay.
Tumor treatment experiments
C57BL/6 mice were inoculated subcutaneously with 105 TC-1 P0/no insert or TC-1 P0/HIF-1α cells. For immunization experiments as previously described (14), mice were administered with Vac-SEL days after tumor challenge . For adoptive transfer experiments, mice were administered with 2×106 E7-specific CTLs or isotonic saline control by tail vein injection 7 days after tumor challenge. Tumor dimensions (length and width) were measured with a calipers every 3-4 days, and tumor volume was calculated as length (mm) × width2 (mm2) × 0.52.
Statistical analysis
All data are representative of at least 3 independent experiments. Individual data points were compared using Student’s t-test. Statistical analysis was performed with SPSS version 17.0 software (SPSS Inc., Chicago, IL). Values of p<0.05 were considered statistically significant.
Results
Tumor cells gain HIF-1α expression in the course of immune selection
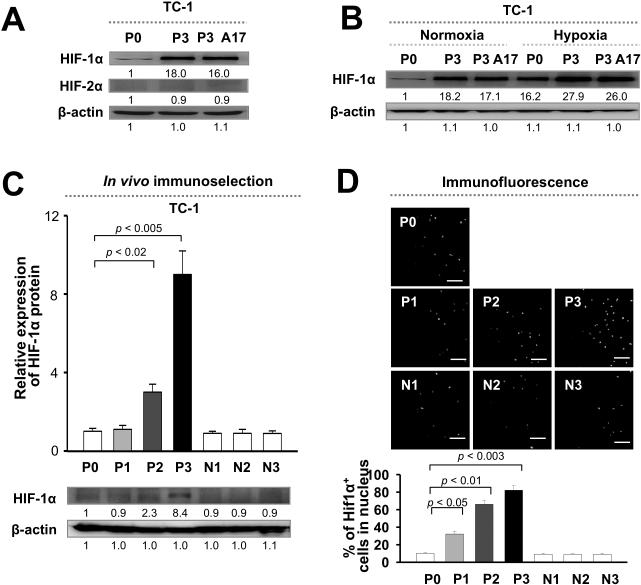
To investigate the molecular evolution of cancer under selection pressure from an antitumor immune response, we created a system termed VICE (Vaccination-Induced Cancer Evolution), in which the tumor is subjected to serial rounds of immune selection by antigen-specific CD8+ cytotoxic T lymphocytes (CTLs) (12). We administered C57BL/6 mice with vaccinia virus encoding the E7 antigen of human papillomavirus type-16 linked to the sorting signal of Lamp-1 (Vac-SEL) to elicit CD8+ CTLs against E7 (16). We then inoculated the mice with parental TC-1 tumor cells (P0), which harbor E7 antigen. Outgrowth tumor was excised and inoculated into naïve mice that previously received Vac-SEL. This process was repeated for 3 cycles to derive the P3 line, which was impervious to lysis by CTLs in vitro and in vivo. As a control, we performed this procedure in mice implanted with TC-1 cells without vaccination to generate N1, N2, and N3 cells without immune selection. Notably, expression of E7 as well as MHC class I was nearly identical in P3 and P0 cells, indicating that the immune resistance of P3 was not due to antigen loss (14). Although the HIF family of proteins has been implicated in chemotherapy resistance, its role in resistance to CTLs has not been previously examined under normal oxygen tension (2). We therefore probed for expression of HIF-1α and HIF-2α protein in P0 and P3 cells. Whereas there was no difference in HIF-2α expression between P0 and P3 cells, HIF-1α was up-regulated nearly 20-fold in P3 relative to P0 cells under normoxia; this was confirmed in a clone from the P3 line (A17) (Figure 1A). Under hypoxia, HIF-1α expression was also elevated in P3 relative to P0 cells (Figure 1B). To verify evolution towards HIF-1α expression under immune surveillance, we measured HIF-1α levels in tumor cells at different stages of selection (P0 to P3). We observed a consistent and gradual increase in HIF-1α from P0 to P3; by contrast, HIF-1α levels remained constant in tumor cells at parallel stages without immune selection (N0 to N3) (Figure 1C). We thus conclude that HIF-1α expression is induced in tumor cells by immune selection.
Figure 1. Immune selection induces HIF-1α in tumor cells.
(A) Western blot analysis of HIF-1α and HIF-2α expression in tumor cells before (P0) or after (P3) immune selection by cytotoxic T lymphocytes. (B) Western blot analysis of HIF-1α expression in P0 or P3 tumor cells in normoxia or hypoxia. (C) Top, Quantification of HIF-1α expression in tumor cells at different stages of immune selection (P0 to P3). Parallel stages without selection are labeled as N1 to N3. Bottom, Representative Western blot images. (D) Top, Representative fluorescence microscopy images of nuclear HIF-1α in tumor cells. Bottom, Quantification of the frequency of tumor cells with nuclear localization of HIF-1α.
The overexpression of HIF-1α could be either due to elevated synthesis of basal HIF-1α by all tumor cells or to an increase the number of HIF-1α+ cells. To address this issue, we labeled P0 to P3 cells with fluorescence-labeled mAb against HIF-1α and determined the frequency of tumor cells with nuclear localization of HIF-1α by fluorescence microscopy. We observed a progressive increase in the frequency of nuclear HIF-1α+ tumor cells as immune selection progressed from the P0 to P3 stages (from 10% to 80% of all tumor cells) (Figure 1D). Our results therefore demonstrate the molecular evolution of tumor cells towards HIF-1α under the immune selection pressure imposed by vaccination.
HIF-1α drives immune adaptation
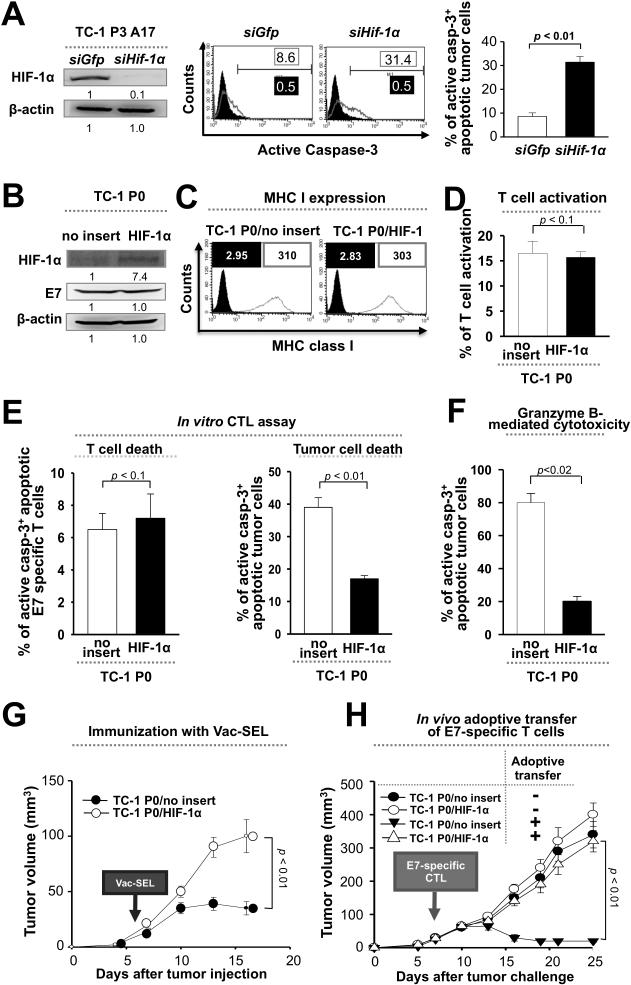
Since HIF-1α is up-regulated in tumor cells during immune surveillance, we reasoned that this expression might be responsible for immune adaptation. To test this, we transfected P3 cells with siRNA against either HIF-1α (siHIF-1α) or GFP control (siGFP), mixed the cells with E7-specific CTLs, and measured the frequency of apoptotic tumor cells. We observed a higher frequency of apoptotic tumor cells in the siHIF-1α- compared to the siGFP-transfected group (Figure 2A), suggesting a role for HIF-1α in immune resistance. To confirm this, we transduced P0 cells with a human HIF-1α K532R mutant (P0/HIF-1α) that is stable in normoxia and characterized their phenotype (Figure 2B). There was no significant difference in surface MHC class I expression between P0/HIF-1α cells or their empty vector-transduced counterparts (P0/no insert) (Figure 2C). Furthermore, the activation and viability of E7-specific CTLs was similar when mixed with HIF-1α or vector-transduced cells (Figure 2, D and E). However, the frequency of apoptotic tumor cells was lower among P0/HIF-1α compared to P0/no insert cells when mixed with either E7-specific CTLs or with granzyme B (Figure 2, E and F).
Figure 2. HIF-1α facilitates tumor immune escape.
(A) E7+ tumor cells after immune selection (P3) were transfected with siRNA against HIF-1α or GFP control, mixed with E7-specific CD8+ cytotoxic T lymphocytes (CTLs), and the frequency of apoptotic tumor cells was determined by flow cytometry analysis of caspase-3 activation. Left, Western blot analysis of HIF-1α expression in P3 cells transfected with siRNA against HIF-1α or GFP. Middle, Representative flow cytometry histograms. Right, Bar graph quantification of the flow cytometry data. Bottom right. (B) Western blot analysis of HIF-1α and E7 expression in E7+ tumor cells before immune selection (P0) transduced with empty vector (P0/no insert) or HIF-1α (P0/HIF-1α). (C) Flow cytometry histograms of MHC class I (H2-Db) expression in P0/no insert or P0/HIF-1α cells (grey outline). Black outline is isotype control. (D) P0/no insert or P0/HIF-1α cells were mixed with E7-specific CTLs. Activation of CTLs was determined by flow cytometry analysis of IFN-γ secretion. (E) The frequency of apoptotic CTLs (left) or tumor cells (right) from the reaction in part (D) was determined by flow cytometry analysis of caspase-3 activation. (F) Granzyme B was delivered into P0/no insert or P0/HIF-1α cells, and the frequency of apoptotic cells was determined by flow cytometry analysis of caspase-3 activation. (G) C57BL/6 mice were inoculated subcutaneously with 105 P0/no insert or P0/HIF-1α cells. 7 days later, mice received vaccination with vaccinia virus encoding E7 fused to the sorting signal of Lamp-1 (Vac-SEL). Tumor size was measured for 17 days. (H) C57BL/6 mice were inoculated subcutaneously with 105 P0/no insert or P0/HIF-1α cells. 7 days later, mice received adoptive transfer through the tail vein of 2×106 E7-specific CTLs or isotonic saline control. Tumor size was measured.
To assess the role of HIF-1α in tumor immune adaptation in the setting of a host immune response, we inoculated C57BL/6 mice with P0/no insert or P0/HIF-1α, and then administered the mice with Vac-SEL 7 days later. Tumor growth was sharply accelerated in the vaccinated mice inoculated with P0/HIF-1α relative to P0/no insert cells (Figure 2G). The presence of HIF-1α also rendered the tumor impervious to adoptive therapy with E7-specific CTLs (Figure 2H). Notably, there was no significant difference in tumor growth between mice inoculated with P0/HIF-1α and P0/no insert cells without adoptive therapy, suggesting that HIF-1α facilitates cancer progression primarily by promoting immune adaptation.
HIF-1α turns on an anti-apoptotic network in tumor cells through AKT and ERK signaling
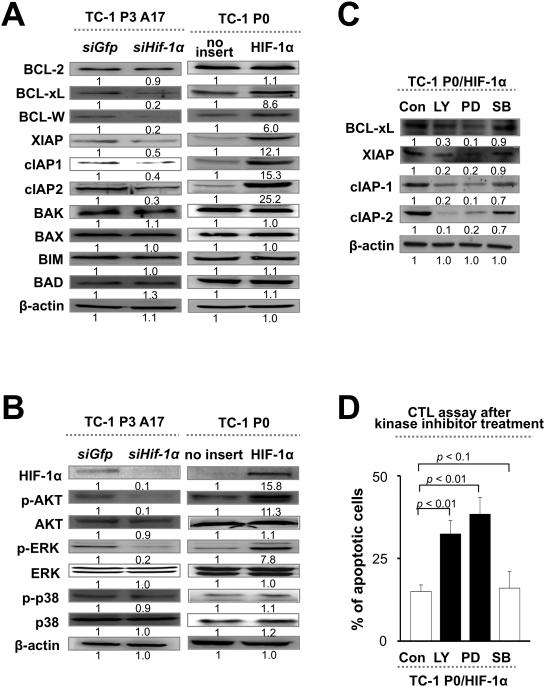
We hypothesized that HIF-1α may cause immune adaptation by establishing an anti-apoptotic state in tumor cells. To test this, we probed for the expression of a panel of pro- or anti-apoptotic molecules in P3 cells transduced with siHIF-1α (relative to siGFP control) or in P0 cells transduced with HIF-1α (relative to empty vector control). We found that an array of anti-apoptotic molecules, including Bcl-2, Bcl-xL, Bcl-W, XIAP, cIAP1, and cIAP2, were induced by HIF-1α (Figure 3A). To elucidate the pathways by which HIF-1α controls expression of these molecules, we measured phosphorylation of AKT, ERK, and P38. We found that HIF-1α drives phosphorylation of AKT and ERK (Figure 3B), and that pharmacologic inhibition of these pathways strongly reduces expression of anti-apoptotic molecules (Figure 3C) and renders tumor cells sensitive to lysis by CTLs (Figure 3D). Therefore, HIF-1α-mediated immune adaptation occurs through the AKT and ERK signaling pathways, which trigger the activation of an anti-apoptotic network within tumor cells.
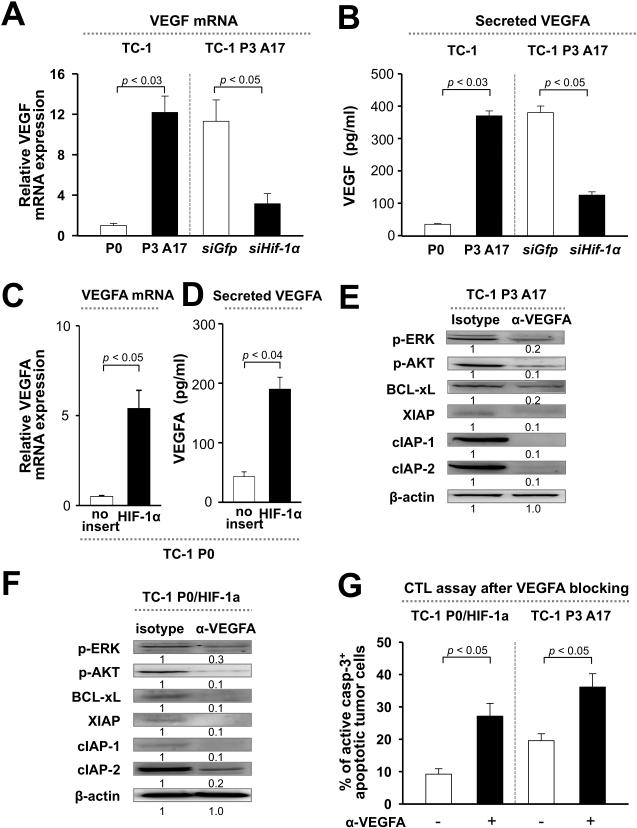
Figure 3. VEGFA facilitates HIF-1α-mediated immune adaptation through activation of the AKT and ERK pathways.
(A and B) qRT-PCR analysis of VEGFA mRNA levels (A) or ELISA for VEGFA secretion (B) in tumor cells before (P0) or after (P3) immune selection (left), or in P3 cells transfected with siRNA against GFP or HIF-1α (right). (C and D) qRT-PCR analysis (C) or ELISA (D) of VEGFA in P0 cells transduced with empty vector or HIF-1α (P0/HIF-1α). (E and F) Western blot analysis of AKT and ERK phosphorylation, as well as expression of anti-apoptotic factors in P3 cells (E) or in P0/HIF-1α cells (F) pre-treated with VEGFA blocking mAb or isotype control. (G) P0/HIF-1α or P3 cells treated with or without α-VEGFA mAb were mixed with E7-specific CTLs. The frequency of apoptotic cells was determined by flow cytometry analysis of caspase-3 activation. All experiments were repeated independently at least 3 times.
VEGFA ties HIF-1α expression to AKT and ERK signaling
We next sought to uncover the connection between HIF-1α expression and activation of AKT and ERK signaling. It has previously been shown that HIF-1α drives expression of its key target VEGFA, which is reported to induce the expression of anti-apoptotic molecules in tumor cells (17, 18). We thus measured VEGFA expression and secretion in P0 versus P3 cells, and found both to be over 10-fold higher in P3 relative to P0 cells (Figure 4, A and B). We confirmed that HIF-1α controls expression of VEGFA by knocking down this transcription factor in P3 cells with siRNA (Figure 4, A and B), or by introducing it into P0 cells (Figure 4, C and D). To explore whether VEGFA triggers AKT and ERK signaling and the downstream anti-apoptotic network, we incubated P3 or P0/HIF-1α cells with VEGFA blocking mAb and then measured AKT and ERK phosphorylation, as well as expression of anti-apoptotic molecules. We observed reduced AKT and ERK phosphorylation among P3 or P0/HIF-1α cells incubated with α-VEGFA mAb relative to isotype control, together with lower expression of anti-apoptotic molecules (Figure 4, E and F). Consistent with this data, P3 and P0/HIF-1α cells incubated with α-VEGFA mAb were more sensitive to killing by E7-specific CTLs compared to cells incubated with isotype control (Figure 4G). We conclude that a core signaling axis linking HIF-1α, VEGFA, AKT/ERK, and an array of anti-apoptotic molecules drives cancer immune adaptation.
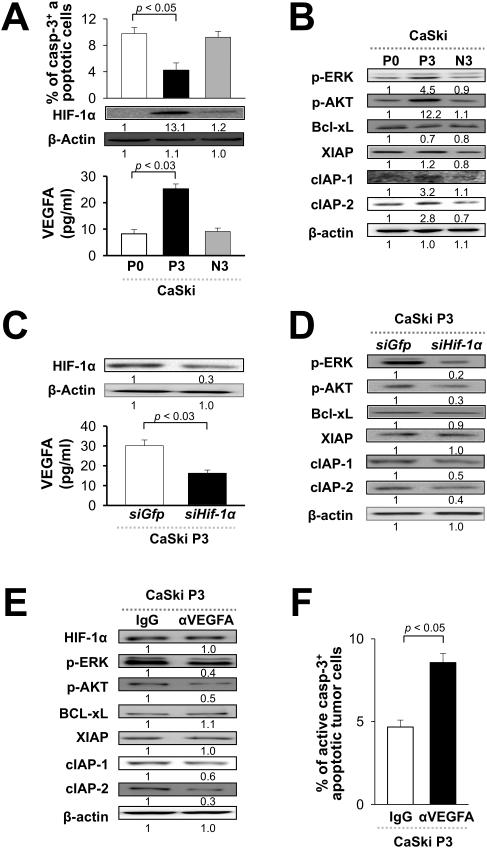
Figure 4. HIF-1α-triggered VEGFA-AKT/ERK axis promotes immune adaptation in human cancer.
Parental (P0) CaSki human cancer cells were pulsed with MART-1 peptide or irrelevant E7 peptide and subjected to selection with MART-1-specific human CTLs for 3 rounds to produce immune-resistant (P3) or control (N3) cells. (A) Top, P0, P3, or N3 CaSki cells were incubated with MART-1-specific human CTLs and the frequency of apoptotic tumor cells was determined by flow cytometry analysis of caspase-3 activation. Middle, Western blot analysis of HIF-1α expression in P0, P3 and N3 cells. Bottom, VEGFA concentration (pg/ml) in the supernatant of P0, P3 and N3 cells in culture. (B) Western blot analysis of AKT and ERK phosphorylation, as well as expression of anti-apoptotic proteins in P0, P3, and N3 cells. (C) Top, Western blot analysis of HIF-1α expression in P3 cells transfected with siRNA against HIF-1α or GFP. Bottom, VEGFA concentration (pg/ml) in the supernatant of HIF-1α or GFP siRNA-treated CaSki P3 cells. (D, E) Western blot analysis of AKT and ERK phosphorylation, as well as expression of anti-apoptotic proteins in HIF-1α or GFP siRNA and α-VEGFA mAb-treated CaSki P3 cells. (F) Control IgG or α-VEGFA mAb-treated CaSki P3 cells were pulsed with MART-1 peptide and mixed with MART-1-specific human CTLs. The frequency of apoptotic cells was determined by flow cytometry analysis of caspase-3 activation.
The HIF-1α-triggered VEGFA-AKT/ERK axis mediates immune adaptation of human cancer cells
Having established that the VEGFA-AKT/ERK axis induces HIF-1α-driven immune adaptation in a mouse model, we next sought to determine if this is also true in human cancer. To do so, we pulsed parental CaSki human cervical cancer cells (P0) with a model peptide antigen, MART-1, (or an irrelevant antigen, NY-ESO-1 peptide) and then subjected these cells to immune selection with MART-1-specific human CTLs. After 3 rounds of selection, CaSki cells pulsed with MART-1 were termed P3, while those pulsed with irrelevant NY-ESO-1 peptide were termed N3. We mixed P0, P3, or N3 cells with MART-1-specific CTLs and determined the frequency of apoptotic tumor cells by flow cytometry analysis of caspase-3 activation. We found that there were over 2 times less apoptotic P3 cells compared to either N3 or parental P0 cells, confirming, as expected, that immune selection of human cancer cells promotes immune adaptation (Figure 5A). We also probed for HIF-1α expression in P0, P3, or N3 cells by Western blot and observed over 10 times higher HIF-1α expression in P3 cells compared to either N3 or P0 cells (Figure 5A). We also created 2 additional immune-resistant human tumor model systems by in vitro selection of parental MDA-MB-231 breast cancer cells or 526mel melanoma cells; we observed HIF-1α overexpression by immune-resistant cells compared to their parental counterparts in both of these systems (data not shown). Consistently, VEGFA mRNA level and protein secretion, AKT and ERK phosphorylation, and expression of the anti-apoptotic factors cIAP-1 and cIAP-2, were up-regulated in P3 cells relative to N3 and P0 cells as well (Figure 5A, 5B, Supplementary Figure 1 and 2). These results demonstrate that immune selection induces HIF-1α expression, VEGFA secretion, and activation of the AKT/ERK axis in human cancer cells, which prompts the expression of anti-apoptotic factors. We found that silencing HIF-1α with siRNA reduced VEGFA secretion, AKT/ERK phosphorylation, and cIAP expression in CaSki P3 cells, showing that this sequence of signaling events is dependent on the presence of HIF-1α in immune-resistant human cancer cells (Figure 5, C and D). HIF-1α knockdown also rendered immune-resistant, MART-1-pulsed CaSki cells sensitive to lysis by human MART-1-specific CTLs (Supplementary Figure 3). Likewise, HIF-1α knockdown in immune-resistant MDA-MB-231 or 526mel cells rendered them sensitive to these CTLs (data not shown). Furthermore, blockade of the VEGFA pathway with α-VEGFA mAb diminished AKT/ERK phosphorylation and cIAP expression, and restored sensitivity of P3 cells to MART-1-specific CTLs, illustrating that HIF-1α triggers the AKT/ERK axis, anti-apoptotic signaling, and immune adaptation directly through VEGFA (Figure 5, E and F) in human cancer.
Figure 5. HIF-1α induces an anti-apoptotic program through the AKT and ERK pathways.
(A) Western blot analysis of a panel of pro- or anti-apoptotic factors in post-selection tumor cells (P3) transfected with siRNA against GFP or HIF-1α, or in pre-selection tumor cells (P0) transduced with empty vector or HIF-1α (P0/HIF-1α). β-actin was included as an internal control. (B) Western blot analysis of AKT, ERK, and p38 phosphorylation in P3 and P0 cells with indicated HIF-1α expression. (C) Western blot analysis of anti-apoptotic molecules in P0/HIF-1α cells treated with LY-294002 (LY; AKT inhibitor), PD-98059 (PD; ERK inhibitor), SB-203580 (SB; p38 inhibitor), or DMSO control (Con). (D) P0/HIF-1α cells treated as indicated were mixed with E7-specific CTLs. The frequency of apoptotic cells was determined by flow cytometry analysis of caspase-3 activation.
Discussion
The immune adaption of tumor cells in response to host immune defenses has emerged as a major driving force for cancer onset and progression (19). This process may occur through host-intrinsic events, such as the induction of tolerance to tumor antigen in tumor-specific CTLs or the recruitment of regulatory T cells or myeloid-derived suppressor cells to the tumor microenvironment (16). Alternatively, it may be controlled by tumor-intrinsic events, such as loss of antigen or gain of resistance to apoptosis (17, 18).
Here we demonstrate cancer immune adaption in response to a selection pressure imposed by tumor-specific CTLs can be triggered via the HIF-1α/VEGFA pathway.
We found that expression of HIF-1α is profoundly up-regulated in tumor cells subjected to immune selection by tumor-specific CTLs compared to those without selection. Expression of HIF-1α in tumor cells mediated resistance to lysis by CTLs both in a culture system and in mice in a tumor-intrinsic manner; the activation, survival, and function of CTLs was not influenced by the HIF-1α expression status of tumor cells. Furthermore, we did not detect any effects of HIF-1α expression on either MHC class I expression or antigen processing and presentation in tumor cells, suggesting that HIF-1α conferred a direct survival capacity to tumor cells. We found that HIF-1α up-regulated expression of a constellation of anti-apoptotic molecules—Bcl-xL, Bcl-W, XIAP, cIAP1, and cIAP2—in an AKT- and ERK-dependent manner. Furthermore, the effects of HIF-1α on AKT/ERK activation and tumor survival were mediated through VEGFA signaling, as VEGFA blockade abolished the anti-apoptotic properties of HIF-1α. It is well-established that HIF-1α stability is exquisitely sensitive to oxygen tension, such that HIF-1 is typically only present in abundant amounts under hypoxia. In the context of the tumor microenvironment, the core of a solid tumor mass is characterized by hypoxia, and thus, HIF-1 expression is frequently high in these areas. However, our data suggest that HIF-1 expression is not only limited to the core of the tumor mass but may also extend into the marginal areas of the tumor (which have normal oxygen tension). What would be the benefit for tumor cells of HIF-1 expression in these areas? We believe that, since tumor cells in the periphery are the first to encounter infiltrating lymphocytes, these cells would be the most susceptible to immune attack. Therefore, gain of HIF-1α by these marginal tumor cells would confer a strong ‘fitness’ advantage, potentially explaining the evolution of tumor cells towards HIF-1 expression even under normoxia in response to an immune selection pressure. Independently, HIF-1 expression under normoxia would induce VEGF secretion and promote angiogenesis, thereby contributing to tumor progression (3, 23). For instance, HIF-1α under normoxia has been implicated in patients with highly vascularized hemangioblastoma, in which tumor cells immediately adjacent to blood vessels were found to stain intensely for HIF-1α (3, 23, 24). Future work should aim to explore these theories.
Finally, it is intriguing to consider our results in light of the “adaptation model” of the immune system that has been recently proposed (25), in which the expression of certain receptors and corresponding ligands by immune cells or peripheral tissue cells tunes the magnitude of the immune response. In this context, HIF-1α may represent an adaptation-inducing factor that turns on the expression of ligands which directly modulate lymphocyte activation. This will be an interesting area for future investigation.
In conclusion, our study unravels a signaling pathway comprised of VEGFA, AKT/ERK, and anti-apoptotic molecules linking HIF-1α to cancer immune adaptation. Inhibitors of each component of this pathway should in principle be effective against cancer, particularly as adjuvants for immune-based therapies.
Supplementary Material
Statement of Translational Relevance
Our study identifies HIF-1α as a key instigator of tumor immune adaptation, even under conditions of normal oxygen tension. We discovered that HIF-1α mediates immune adaptation through VEGFA-dependent activation of the AKT and ERK signaling pathways. Therefore, our study suggests that pharmacologic blockade of HIF-1α or these associated signaling pathways may represent an effective strategy for the clinical management of cancer, particularly in combination with immune-based modalities.
Acknowledgements
This work was funded by the National Research Foundation of Korea (2012R1A2A2A01007527, 2013M3A9D3045881, and 2009-0086652), the Korea Healthcare Technology R&D Project (HI11C-0052-030013), the United States National Institutes of Health (NIH) Cervical Cancer Specialized Program of Research Excellence (SPORE) (P50 CA098252), Head and Neck Cancer SPORE (P50 CA96784-06), and R01 grant (CA114425-01, and CA183040). Chih-Ping Mao is a recipient of the NIH F30 Research Fellowship (CA177221).
Footnotes
Conflict of interest: The authors declare that there is no conflict of interest
References
- 1.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2002;481:306. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 3.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, et al. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830. [PubMed] [Google Scholar]
- 4.Barsoum IB, Hamilton TK, Li X, Cotechini T, Miles EA, Siemens DR, et al. Hypoxia induces escape from innate immunity in cancer cells via increased expression of ADAM10: role of nitric oxide. Cancer Res. 2011;71:7433. doi: 10.1158/0008-5472.CAN-11-2104. [DOI] [PubMed] [Google Scholar]
- 5.Barsoum IB, Smallwood CA, Siemens DR, Graham CH. A mechanism of hypoxia-mediated escape from adaptive immunity in cancer cells. Cancer Res. 2014;74:665. doi: 10.1158/0008-5472.CAN-13-0992. [DOI] [PubMed] [Google Scholar]
- 6.Noman MZ, Buart S, Van Pelt J, Richon C, Hasmim M, Leleu N, Chouaib S. The cooperative induction of hypoxia-inducible factor-1 alpha and STAT3 during hypoxia induced an impairment of tumor susceptibility to CTL-mediated cell lysis. J Immunol. 2009;182:3510. doi: 10.4049/jimmunol.0800854. [DOI] [PubMed] [Google Scholar]
- 7.Noman MZ, Janji B, Kaminska B, Van Moer K, Pierson S, Przanowski P, Chouaib S. Blocking hypoxia-induced autophagy in tumors restores cytotoxic T-cell activity and promotes regression. Cancer Res. 2011;71:5976. doi: 10.1158/0008-5472.CAN-11-1094. [DOI] [PubMed] [Google Scholar]
- 8.Hasmim M, Noman MZ, Lauriol J, Benlalam H, Mallavialle A, Rosselli F, Chouaib S. Hypoxia-dependent inhibition of tumor cell susceptibility to CTL-mediated lysis involves NANOG induction in target cells. J Immunol. 2011;187:4031. doi: 10.4049/jimmunol.1101011. [DOI] [PubMed] [Google Scholar]
- 9.Noman MZ, Buart S, Romero P, Ketari S, Janji B, Chouaib S. Hypoxia-inducible miR-210 regulates the susceptibility of tumor cells to lysis by cytotoxic T cells. Cancer Res. 2012;72:4629. doi: 10.1158/0008-5472.CAN-12-1383. [DOI] [PubMed] [Google Scholar]
- 10.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 11.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 331:1565. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 12.Lin KY, Lu D, Hung CF, Peng S, Huang L, Jie C, et al. Ectopic expression of vascular cell adhesion molecule-1 as a new mechanism for tumor immune evasion. Cancer res. 2007;67:1832. doi: 10.1158/0008-5472.CAN-06-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin KY, Guarnieri FG, Staveley-O'Carroll KF, Levitsky HI, August JT, Pardoll DM, et al. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer res. 1996;56:21. [PubMed] [Google Scholar]
- 14.Noh KH, Kang TH, Kim JH, Pai SI, Lin KY, Hung CF, et al. Activation of Akt as a mechanism for tumor immune evasion. Mol Ther. 2009;17:439. doi: 10.1038/mt.2008.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noh KH, Lee YH, Jeon JH, Kang TH, Mao CP, Wu TC, et al. Cancer vaccination drives Nanog-dependent evolution of tumor cells toward an immune-resistant and stem-like phenotype. Cancer res. 2012;72:1717. doi: 10.1158/0008-5472.CAN-11-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu TC, Guarnieri FG, Staveley-O'Carroll KF, Viscidi RP, Levitsky HI, Hedrick L, et al. Engineering an intracellular pathway for major histocompatibility complex class II presentation of antigens. Proc Natl Acad Sci U S A. 1995;92:11671. doi: 10.1073/pnas.92.25.11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, et al. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pidgeon GP, Barr MP, Harmey JH, Foley DA, Bouchier-Hayes DJ. Vascular endothelial growth factor (VEGF) upregulates BCL-2 and inhibits apoptosis in human and murine mammary adenocarcinoma cells. Br J Cancer. 2001;20:273. doi: 10.1054/bjoc.2001.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 20.Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural innate and adaptive immunity to cancer. Annu Rev Immunol. 2001;29:235. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- 21.DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature. 2012;482:405. doi: 10.1038/nature10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Igney FH, Krammer PH. Immune escape of tumors: apoptosis resistance and tumor counterattack. J Leukoc Biol. 2002;71:907–920. [PubMed] [Google Scholar]
- 23.Birner P, Preusser M, Gelpi E, Berger J, Gatterbauer B, Hainfellner JA. Expression of hypoxia-related tissue factors correlates with diminished survival of adjuvantly treated patients with chromosome 1p aberrant oligodendroglial neoplasms and therapeutic implications. Clin Cancer Res. 2004;10:6567. doi: 10.1158/1078-0432.CCR-04-0617. [DOI] [PubMed] [Google Scholar]
- 24.Interstitial pH and pO2 gradients in solid tumors in vivo: high-resolution measurements reveal a lack of correlation. Nat. Med. 1997;3:177. doi: 10.1038/nm0297-177. [DOI] [PubMed] [Google Scholar]
- 25.Manjili MH. The adaptation model of immunity. Immunotherapy. 2014;6:59. doi: 10.2217/imt.13.157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.