Abstract
The Coxsackievirus and adenovirus receptor (CAR) is an essential cellular protein that is involved in cell-cell adhesion, protein trafficking, and viral infection. The major isoform of CAR is selectively sorted to the basolateral membrane of polarized epithelial cells where it co-localizes with the cellular scaffolding protein membrane-associated guanylate kinase with inverted domain structure-1 (MAGI-1). Previously, we demonstrated CAR interacts with MAGI-1 through a PDZ–domain dependent interaction. Here, we show that the PDZ3 domain of MAGI-1 is exclusively responsible for the high affinity interaction between the seven exon isoform of CAR and MAGI-1 using yeast-two-hybrid analysis and confirming this interaction biochemically and in cellular lysates by in vitro pull down assay and co-immunoprecipitation. The high affinity interaction between the PDZ3 domain and CAR C-terminus was measured by fluorescence resonance energy transfer. Further, we investigated the biological relevance of this high affinity interaction between CAR and the PDZ3 domain of MAGI-1 and found that it does not alter CAR-mediated adenovirus infection. By contrast, interruption of this high affinity interaction altered the localization of MAGI-1 indicating that CAR is able to traffic MAGI-1 to cell junctions. These data deepen the molecular understanding of the interaction between CAR and MAGI-1 and indicate that although CAR plays a role in trafficking PDZ-based scaffolding proteins to cellular junctions, association with a high affinity intracellular binding partner does not significantly alter adenovirus binding and entry via CAR.
Keywords: adenovirus, epithelia, cell junction, PDZ domain
1. Introduction
The Coxsackievirus and adenovirus receptor (CAR) is a primary receptor for two distinct groups of viruses, coxsackie B viruses and most adenoviruses (AdV) (Bergelson, Cunningham et al. 1997, Carson, Chapman et al. 1997, Tomko, Xu et al. 1997). Studies on its endogenous cellular function have shown that CAR is a developmentally essential protein with important cardiac functions (Asher, Cerny et al. 2005, Dorner, Wegmann et al. 2005, Freiberg, Sauter et al. 2014). In polarized epithelia, CAR is primarily a basolateral adhesion protein involved in the maintenance of cell-cell contacts (Walters, Freimuth et al. 2002, Excoffon, Hruska-Hageman et al. 2004). CAR is a 46-kDa glycoprotein with two Ig-like extracellular domains, a transmembrane domain, and a cytoplasmic domain. Two CAR molecules from neighboring cells can interact through homodimerization of the distal immunoglobulin (Ig)-like extracellular domain (D1 domain) with a Kd of ~16 μM (van Raaij, Chouin et al. 2000). Interestingly, the same domain, D1, is necessary and sufficient for adenovirus binding. Thus, adenovirus fiber-knob binding domain overlaps with the CAR-CAR homodimerization interface with a much greater affinity, Kd of ~8 nm (Bewley, Springer et al. 1999, Kirby, Davison et al. 2000, Kirby, Lord et al. 2001). It is generally accepted that extracellular adenovirus fiber-knob will outcompete intercellular CAR-CAR interactions for cell binding. However, it is unknown if this extracellular competition and viral entry is affected by intracellular molecules binding to the cytoplasmic domain of CAR within the cell.
The most abundant, seven exon isoform of CAR contains a 4 aa long class 1 PSD-95/Drosophila discs-large protein/zonula occludens protein-1 (PDZ)-binding domain (PBD) at its C-terminus that interacts with several proteins that contain PDZ domains, which are approximately 80-90 aa long (Excoffon, Gansemer et al. 2010, Ivarsson 2012). Membrane-associated guanylate kinase with inverted domain structure-1 (MAGI-1) is a cellular scaffolding protein that contains up to six alternatively spliced PDZ domains (PDZ0-5), an inactive guanylate kinase domain, and two WW domains. It has three primary alternatively spliced isoforms, MAGI-1a, b and c, which exhibit differential tissue expression. CAR and MAGI-1b have been shown to interact via a PDZ-based interaction and share several interacting partners, such as β-catenin and ASIC3 (Dobrosotskaya and James 2000, Walters, Freimuth et al. 2002, Excoffon, Hruska-Hageman et al. 2004, Hruska-Hageman, Benson et al. 2004). It is known that in order to coordinate signaling, PDZ domain-containing proteins act as molecular scaffolds to co-localize their binding partners to create a signalosome (Kim and Sheng 2004). MAGI-1 facilitates a large number cellular functions as a scaffolding protein including the coordination of large protein complexes for cell signaling, maintenance of cell-cell junction stability, and apoptosis (Dobrosotskaya and James 2000, Ivanova, Repnik et al. 2007, Kranjec, Massimi et al. 2014). MAGI-1 is thought to play pivotal roles in several disease conditions including focal segmental glomerulosclerosis, and the severity of viral-mediated disease and cancer (Kaufman, Potla et al. 2010, Kumar, Liu et al. 2012, Kranjec, Massimi et al. 2014).
While we have previously demonstrated a PDZ-dependent interaction between MAGI-1 and CAR (Excoffon, Hruska-Hageman et al. 2004), the molecular underpinnings of the CAR-MAGI-1 specific interaction remain unclear. Further, the physiological consequence of this interaction on viral infection and MAGI-1 junction localization are unknown. In this study, we show that the CAR PDZ-binding domain (-GSIV) directly interacts with the PDZ3 domain of MAGI-1 with high affinity. While the interaction between MAGI-1 PDZ3 and CAR is critical for MAGI-1 junctional localization, MAGI-1 PDZ3 does not affect CAR-mediated viral infection. As novel experimental therapeutics that target PDZ-based interactions are under development, our study provides critical information that will aid in understanding the molecular mechansims and avoiding off-target effects of molecules that may alter protein interactions with MAGI-1.
2. Materials and Methods
2.1 Materials
COS-7 and CHO-K1 cells (ATCC, Rockville, MD) were cultured under standard conditions (DMEM with 10% FBS, and 1% penicillin/streptomycin, 2 mM L-glutamine). Individual MAGI-1b PDZ domains (PDZ1, 2, and 3) were cloned into pcDNA/V5/GW/D-TOPO (Invitrogen) for in vitro pull down assays. Plasmids for the myc-tagged full length MAGI-1c and all PDZ domains (PDZ0-5) were kindly provided by Dr. Zhigang Xu (Shandong University, Shandong, China) and Dr. Heller (Stanford University School of Medicine, Stanford, CA) (Xu, Peng et al. 2008) and HA-tagged MAGI-1 domain-deleted mutants were kindly provided by Dr. Ronald Javier (Baylor College of Medicine, Houston, TX) (Glaunsinger, Lee et al. 2000). Xfect transfection reagent (Clontech) was used for plasmid transfection. RmcB and HA Abs were from Millipore, FLAG and actin Abs were from Abcam, myc Ab was from Cell Signaling, and anti-CAR 1605p has previously been described (Sharma, Kolawole et al. 2012). Type 5 AdV-β-Galactosidase (AdV-β-Gal) was from the University of Iowa Vector Core, Iowa City, IA.
2.2 Yeast two-hybrid assay (Y2H)
Y2H screening was performed using the Matchmaker kit (Clontech) according to manufacturer's protocols and as previously described (Kolawole, Sharma et al. 2012). The whole intracellular C-terminus of CAR (CAREx7) was fused with the GAL4 DNA-binding domain and subcloned into the bait vector pGBKT7. The known prey MAGI-1 PDZ domains (0-5) were individually cloned into the GAL4 activation prey vector PGADT7, pre-transformed into yeast cells, and screened by separate matings by plating onto high-stringency selective synthetic dropout media.
2.3 In vitro pull down assay
The TNT T7 quick coupled transcription/translation system (Promega Corporation, Madison, WI) was used to synthesize L-[35S]methionine (PerkinElmer, Waltham, MA) labeled MAGI-1 PDZ domains (PDZ1, 2, 3, and 5) according to the manufacturer's protocol and as previously described (Kolawole, Sharma et al. 2012). COS-7 cells were transfected with the FLAG-tagged CAR isoform plasmid and subjected to immunopreciptitation (IP). In vitro translated MAGI-1 PDZ domain proteins were mixed with CAR-IP lysates at 4°C for 2 hr, followed by washing, centrifugation and elution with 2X denaturing buffer. Labeled proteins were separated by SDS-PAGE and visualized either by autoradiography of dried gels or transferred to PVDF for autoradiography and WB.
2.4 MAGI-1 PDZ and CAR c-terminus protein purification
For FRET and binding assays, MAGI-1 PDZ1 and PDZ3 domains and CAR (CAREx7) C-termini (GAIPVMIPAQSKDGSIV) were cloned (In-Fusion, Clontech, Mountainview, CA) into a pGEX-6p vector (GE Healthcare, Piscataway, NJ) modified to contain a His-tag upstream of the GST-tag (pHH2) (Kolawole, Sharma et al. 2012). pHH2-PDZ and -CAR plasmids transformed into Rosetta (BL21) E. coli cells (EMD Millipore, Gibbstown, NJ) were grown, induced, and purified as previously described (Kolawole, Sharma et al. 2012). Purified GST-tagged and tag-free proteins were quantified using the Bio-Rad protein assay kit and the quality verified by SDS-PAGE.
2.5 Protein labeling and FRET
Tag-free proteins were dialyzed with 10 mM HEPES (pH 8.0), 0.1 mM EDTA, 0.4 mM DTT, 400 mM KCl and 5% glycerol at 4°C overnight using a Slide-a-lyzer dialysis cassette (Thermo Scientific). Purified MAGI-1 PDZ domain proteins were labeled with FluoroLink™ Ab Cy3 labeling kit (GE Healthcare, Piscataway, NJ) while purified CAR C-terminus protein was labeled with FluoroLink™ Ab Cy5 labeling kit according to the manufacturer's recommendations. Absorbance was used to determine protein concentrations and dye to protein ratios. Labeling ratios were approximately 1 dye/peptide. FRET from each Cy3-labeled MAGI-1 PDZ domain to Cy5-labeled CAR was conducted as described (Hostetler, Lupas et al. 2011, Kolawole, Sharma et al. 2012) with a PC1 photon counting spectrofluorometer (ISS Inc., Champaign, IL). Briefly, emission spectra (560–700 nm) were obtained from 30 nM donor (Cy3-labeled PDZ peptides) in PBS at 25°C upon excitation at 550 nm with increasing concentration of acceptor (Cy5-labeled CAR C-terminus peptides). The spectra were corrected for background (buffer only and acceptor only) and the maximal intensities were measured using Vinci 1.5 software (ISS Inc., Champaign, IL). The protein binding affinity, energy transfer efficiency, and intermolecular distance were calculated according to the Förster equation and as described previously (Hostetler, Lupas et al. 2011).
2.6 Immunoprecipitation and Western blot (WB)
Cells were lysed, subjected to IP, and blotted as previously described (Excoffon, Gansemer et al. 2010, Sharma, Kolawole et al. 2012). Briefly, for IP, primary Ab was added to equal amounts of protein for 2 hr to overnight at 4°C. Labeled proteins were isolated with protein G sepharose (GE Healthcare), eluted with SDS-PAGE sample buffer, followed by SDS-PAGE and WB. Protein bands were detected with ECL reagents (Pierce, Rockford, IL) and imaged on a Fuji LAS 4000 and/or developed in an X-ray Medical film processor (Konica SRX 101).
2.7 MAGI-1 PDZ mutants
MAGI-1b PDZ-domain mutants were generated by mutating the “GFGF” carboxylate binding motif, required for binding PBD, to “GFRE” by the Quikchange Lightning Site Directed mutagenesis kit according to the manufacturer's instructions. Mutations were confirmed by sequencing.
2.8 AdV-β-Gal infection and β-galactosidase assay
Transfected CHO-K1 cells were infected with type 5 AdV-β-Gal at a multiplicity of infection of 100 for 1 hr at 37°C. β-galactosidase expression was determined with Galacto-Light Plus System (Applied Biosystems, Carlsbad, CA), as previously described (Excoffon, Gansemer et al. 2010, Kolawole, Sharma et al. 2012).
2.9 Immunostaining
COS-7 cells grown on collagen coated chamber slides were washed once with PBS, fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with 2% BSA in SuperBlock (Pierce, Rockford, IL). Cells were incubated with primary Ab, washed extensively and incubated with Alexa-labeled secondary Ab. After washing, slides were coverslipped with Vectashield mounting media (Vector Laboratories, Burlingame, CA). Images were acquired with an FV1000 Laser Scanning Confocal Microscope (Olympus, CenterValley, PA) using a 60× oil immersion lens.
2.9 Statistical analysis
All experiments were performed independently at least three times. Data were expressed as either a representative experiment or mean ± standard error of mean of three separate experiments. Data were analyzed using ANOVA and/or Student's t test with a two-tailed distribution, and were considered statistically significant if p ≤ 0.05.
3. Results
3.1. CAR Interacts with the MAGI-1 PDZ3 Domain
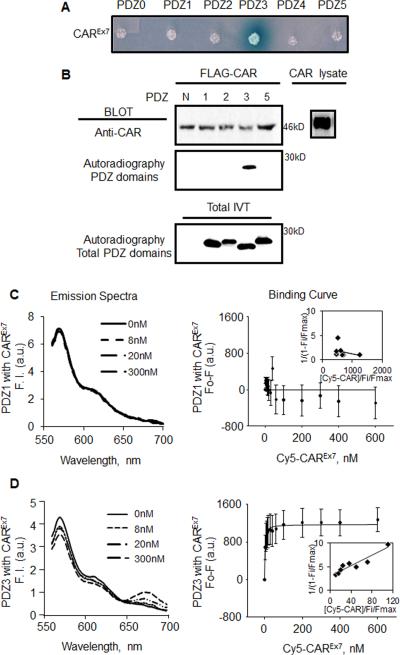
Previous work indicated that the last four amino acids of the C-terminus of CAR (-GSIV) comprise a PBD that is required for the interaction between CAR and MAGI-1 (Excoffon, Hruska-Hageman et al. 2004). To understand the molecular basis of this interaction, we first asked which MAGI-1 PDZ domain(s) CAR interacts with. Yeast-two-Hybrid (Y2H) analysis was initially used to screen interactions between the last 17 aa of the intracellular C-terminus of CAR and all of the individual MAGI-1 PDZ domains (PDZ0-5; Fig. 1A). Only the MAGI-1 PDZ3 domain produced a blue colony when incubated with the CAR C-terminus bait, suggesting a direct interaction between the CAR C-terminus and PDZ3. We then performed in vitro pull down assays to confirm Y2H results. As expected, CAR only pulled down 35S-labeled MAGI-1 PDZ3 indicating a specific interaction in vitro (Fig. 1B).
Fig. 1. CAR has a high affinity interaction with MAGI-1 PDZ3.
A) Yeast Y187 cells were transformed with pairs of pGBKT7-derived bait (CAR C-terminus) and pGADT7-derived prey vectors (PDZ domains), and plated on selective media containing X-α-gal. Blue colony indicates an interaction between CAR and PDZ3. B) In vitro pull down assay. Individual 35S-methionine-labeled MAGI-1 PDZ domains generated by in vitro translation were added to immunoprecipitated FLAG-tagged CAR from transfected COS-7 cells (CAR lysate). Western blot for immunoprecipitated CAR (FLAG-CAR), and autoradiography for PDZ domain binding (N, no PDZ domain). In vitro translation of all PDZ domains was confirmed by autoradiography. Emission spectra, linear plot of binding curve and average change in maximal fluorescence intensity at 570 nm (F0-F) of Cy3-PDZ domains and Cy5 CAR C-termini upon excitation of Cy3 at 550nm with increasing concentration of Cy5 CAR C-termini (0-500nM). C) Cy3-PDZ1 and Cy5-CAR (no interaction). D) Cy3-PDZ3 and Cy5-CAR. All emission spectrum were corrected for background signals. NA = no interaction. Representative data shown from 3 replicate experiments.
3.2. CAR Has a High Affinity Interaction with MAGI-1 PDZ3
To establish a direct interaction, determine the binding affinity (Kd), and determine the intermolecular distance between the CAR C-terminus and MAGI-1 PDZ3 domain, in vitro Förster fluorescence resonance energy transfer (FRET) was utilized (Fig. 1C, D). Purified MAGI-1 PDZ1 (used as a negative control; Fig. 1C) or MAGI-1 PDZ3 (Fig. 1D) were labeled with Cy3 while CAR C-terminus was labeled with Cy5. Cy3/Cy5 labels form a FRET donor/acceptor pair, and FRET only occurs if the labeled proteins are in close proximity (less than 100 Å) (Hostetler, Lupas et al. 2011). FRET was observed as a decrease in Cy3 fluorescence intensity (decrease at 570 nm), and a subsequent increase in Cy5 fluorescence intensity (increase at approximately 670 nm). The maximum peak values were recorded at 570 nm (Cy3) and 670 nm (Cy5, data not shown) for each titration and were used to create binding curves (Fig. 1 C, D). In contrast to the lack of interaction between CAR and PDZ1 (Kd > 500 nM), the change in fluorescence intensity as a function of PDZ3 concentration yielded a saturable hyperbolic binding curve indicating high-affinity binding for PDZ3-CAR (Kd = 2.0 ± 0.9 nM) and an intermolecular distance of 46.7 Å, further indicating a direct interaction. Double reciprocal plots of the data revealed a single site binding isotherm (shown as a single straight line, inset) for CAR and PDZ3.
3.3. CAR Interacts with the MAGI-1 PDZ3 Domain in Cells
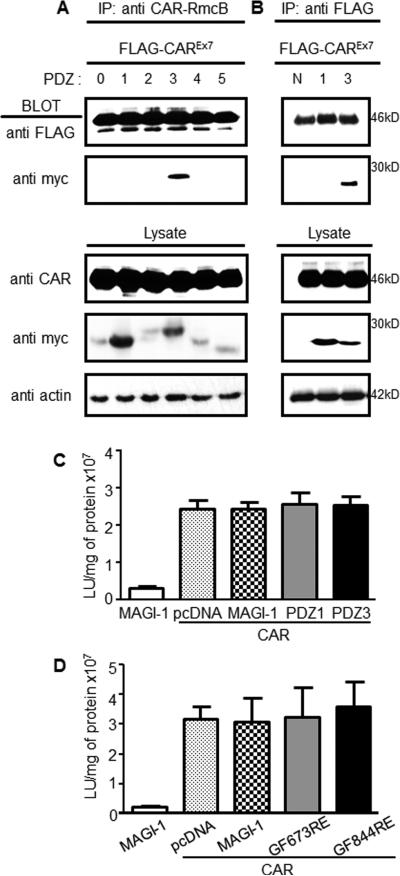
Co-IP from cell lysates was investigated in order to demonstrate that the CAR-MAGI-1 PDZ3 interaction occurs in vivo (Fig. 2A, B). FLAG-tagged CAR plasmid co-transfected with plasmids for each individual myc-tagged MAGI-1 PDZ domain into COS-7 cells was immunoprecipitated with monoclonal anti-CAR-RmcB antibody. WB analysis, using FLAG-tag antibody to recognize CAR and anti-myc-tag antibody to recognize the PDZ domains, was consistent with the above results. CAR only pulled down the MAGI-1 PDZ3 domain (Fig. 2A). Overexpression of CAR and each MAGI-1 PDZ domain was confirmed by blotting total lysate. Immunoprecipitaion with FLAG antibody directed against the overexpressed FLAG-tagged CAR excluded the possibility that IP was a result of endogenous CAR recognized by anti-CAR-RmcB antibody (Fig. 2B).
Fig. 2. CAR interacts with MAGI-1 PDZ3 in cells and does not alter adenovirus infection.
COS-7 cells were co-transfected with each individual myc-tagged MAGI-1 PDZ domain and FLAG-tagged CAR. CAR was immunoprecipitated with A) mouse anti-CAR RmcB or B) anti-FLAG and probed with rabbit anti-FLAG (CAR, ~46 kDa) and rabbit anti-myc (MAGI-1 PDZ domains; 20-28kDa). Western blot of total lysates confirmed expression of all PDZ domains, CAR, and equal loading (actin). C) CAR-deficient CHO-K1 cells were co-transfected with CAR, MAGI-1 (dotted bars), PDZ3 (black bars), or PDZ1 (gray), and balanced with empty pcDNA3.1 plasmid (white bar, control), followed by AdV-β-Gal (MOI 100) transduction. D) CAR-deficient CHO-K1 cells were co-transfected with CAR, MAGI-1 (dotted bars), MAGI-1 PDZ3 mutant (GF844RE; black bars), or MAGI-1 PDZ1 mutant (GF673RE; gray), and balanced with empty pcDNA3.1 plasmid (white bar, control), followed by AdV-β-Gal (MOI 100) transduction. Representative data from 3 replicate experiments.
3.4. MAGI-1 Does Not Alter the Adenovirus Receptor Activity of CAR
MAGI-1 is a scaffolding protein that interacts with several interacting partners simultaneously (Dobrosotskaya and James 2000, Kotelevets, van Hengel et al. 2005). Given the high affinity interaction between CAR and PDZ3, we hypothesized that full length MAGI-1 or the isolated MAGI-1 PDZ3 domain may influence CAR-mediated adenovirus infection. In order to test this, we co-transfected the CAR expression plasmid together with equal amounts of plasmid encoding full length MAGI-1, the PDZ3 domain, PDZ1 domain, or empty plasmid (pcDNA3.1) into CAR-negative CHO-K1 cells. Transfected CHO-K1 cells were infected with adenovirus carrying the β-galactosidase reporter gene (AdV-β-Gal) 48hr later. AdV-β-Gal transduction was measured by the level of β-galactosidase expression 24 hr post-infection. Although transfection with CAR significantly increased adenovirus transduction, there was no significant difference in viral transduction between CAR co-transfection with pcDNA3.1 (control), full length MAGI-1, or either PDZ3 or PDZ1 (control) domains (Fig. 2C).
These results were further confirmed by mutating the conserved PDZ-binding domain binding site within the MAGI-1 PDZ3 domain (GFGF to GF844RE) (Ivarsson 2012) in order to eliminate the interaction between CAR and full length MAGI-1. The conserved binding site within PDZ1 was mutated (GF673RE) in order to serve as control. CHO-K1 cells co-transfected with CAR and parental MAGI-1 or each of the two PDZ domain mutants were infected with AdV-β-Gal 48 hr post-transfection and β-Gal activity 24 hr post-infection. No significant difference in viral transduction was measured in cells co-expressing CAR and MAGI-1 as compared to cells expressing mutated PDZ3 or PDZ1 domain (Fig. 2D). Taken together, these data indicate that although MAGI-1 is a scaffolding protein binds that CAR, and presumably co- interacts with other cellular proteins, the high affinity intracellular interaction does not significantly alter CAR-mediated adenovirus infection.
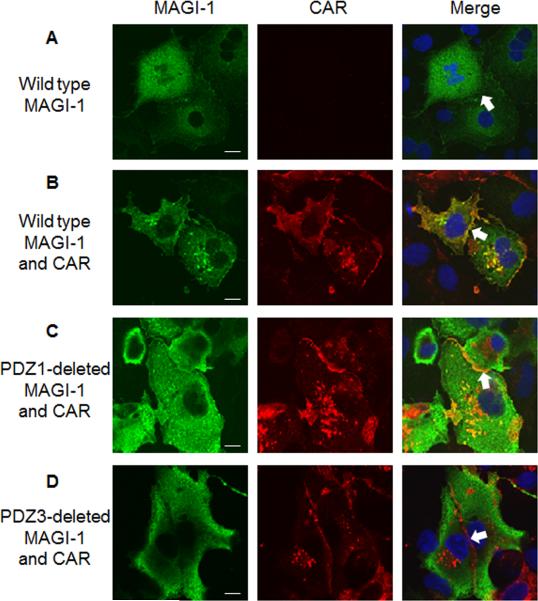
3.5. MAGI-1 PDZ3 is required for CAR-mediated enrichment of MAGI-1 at cell junctions
Previously we have shown that MAGI-1 is largely cytoplasmic when expressed in COS-7 cells (Fig. 3A) and that co-expression of CAR and MAGI-1 together leads to MAGI-1 enrichment at cell-cell junction (Fig. 3B, white arrow, yellow at junctions indicate overlap) (Excoffon, Hruska-Hageman et al. 2004). Based on our findings in this study that CAR directly binds to the PDZ3 domain of MAGI-1 with high affinity, we hypothesized that MAGI-1 junctional localization requires the MAGI-1 PDZ3 domain. As expected, the selective deletion of the MAGI-1 PDZ1 domain did not affect CAR-mediated MAGI-1 junctional localization (Fig. 3C, white arrow, yellow cell-cell junctions). Virtually no junctional localization was observed upon deletion of MAGI-1 PDZ3 domain (Fig. 3D, white arrow, red (CAR) at junctions). These data show that the interaction between CAR and MAGI-1 PDZ3 is critical for efficient junctional localization of MAGI-1.
Fig. 3. MAGI-1 PDZ3 is required for CAR-mediated enrichment of MAGI-1 at cell junctions.
COS-7 cells were transfected with A) MAGI-1, B) CAR and MAGI-1, C) CAR and MAGI-1 with the PDZ1 domain deleted, or D) CAR and MAGI-1 with the PDZ3 domain deleted, and immunostained with an anti-CAR Ab (1605p; red) and anti-HA Ab (directed against MAGI-1; green). Nuclei are stained with DAPI (blue). 60 x oil immersion confocal microscopy. Single X-Y optical sections are shown. Merge of optical sections shown in final panel. White arrow identifies a representative junction between two adjacent cells taken from 3 replicate experiments. White bar = 10 μm.
4. Discussion
Understanding the biology and regulation of CAR and MAGI-1 is important due to their multiple cellular roles. CAR is a virus receptor that mediates adenovirus and coxsackievirus infection. CAR is also a critical cell adhesion protein that plays a role in the junctional localization of important scaffolding proteins and cognate signaling proteins, such as PSD-95 and ASIC3 (Excoffon, Kolawole et al. 2012). Moreover, the essential nature of CAR may be due to its ability to traffic or modulate the junctional stability of important proteins, such as MAGI-1, connexin 45, β-catenin, and ZO-1, particularly at cell junctions and the cardiac intercalated disc (Lim, Xiong et al. 2008, Pazirandeh, Sultana et al. 2011, Freiberg, Sauter et al. 2014). MAGI-1 is an important scaffolding protein that can create a signalosome with over 20 distinct proteins that are involved in diverse cellular pathways, from adhesion to the regulation of signaling. In this study, we demonstrate that CAR interacts directly with the third PDZ domain of MAGI-1 and that this high affinity intracellular interaction is required for MAGI-1 cell junction localization but does not significantly influence the adenovirus receptor activity of CAR.
The CAR isoform investigated in this study is derived from an mRNA that terminates at the end of the seventh exon of the eight-exon containing CXADR gene, and is also known as CAREx7 in humans (used hereafter) or mCAR2 in mice (Andersson, Tomko et al. 2000, Excoffon, Gansemer et al. 2010). The other transmembrane isoform splices from a cryptic splice donor site within the seventh exon of CXADR and terminates at the end of the eighth exon (CAREx8) (Excoffon, Gansemer et al. 2010). Although these two isoforms differ only in the last 26 or 13 amino acids (aa) in the cytoplasmic domain, respectively, localization within a polarized epithelium and interaction with MAGI-1 differ (Excoffon, Hruska-Hageman et al. 2004, Excoffon, Gansemer et al. 2010). CAREx7 localizes at the basolateral surface as an adhesion protein. In contrast, CAREx8 appears at the apical surface where it is able to mediate adenovirus infection from the apical surface of the polarized epithelium (Excoffon, Gansemer et al. 2010, Kolawole, Sharma et al. 2012). Previously, we demonstrated that MAGI-1 is a major negative regulator of cellular CAREx8 protein levels and apical adenovirus infection (Kolawole, Sharma et al. 2012). Further, we demonstrated that the CAREx8 PBD (-ITVV) interacts with two MAGI-1 PDZ domains, PDZ1 and PDZ3, and that these two domains have opposing activities. Expression of the individual PDZ3 domain suppresses cell surface CAREx8 protein levels and adenovirus infection, while, paradoxically, expression of PDZ1 is able to rescue CAREx8 from MAGI-1-mediated degradation and increases adenovirus infection. The goal of the current study was to identify the specific MAGI-1-PDZ domain interaction with the basolateral isoform of CAREx7 and determine whether the interacting domain(s) had any effect on adenovirus infection. Similar to CAREx8, CAREx7 interacts with PDZ3. However, in contrast to the inhibitory effect of PDZ3 on CAREx8 protein levels, MAGI-1 PDZ3 does not affect CAREx7 protein levels or CAREx7-mediated adenovirus infection. Although the affinity between PDZ3 and CAREx8 is greater than CAREx7 (~0.4 nM versus ~2 nM, respectively), the ten-fold higher level of CAREx7 expression in epithelial cells (Excoffon, Gansemer et al. 2010) may sequester MAGI-1 away from nascent CAREx8 allowing increased levels of apical receptor and adenovirus infection. Future work will focus on the potential competition between these two isoforms with cellular MAGI-1. By understanding the dynamics between MAGI-1, CAREx7 and CAREx8, we may be able to manipulate the expression level of apical CAREx8, and hence CAREx8-mediated viral infection. Whereas downregulation of CAREx8 would allow us to potentially decrease wild-type AdV infection, upregulation may allow improved AdV-mediated gene therapy.
MAGI-1 is associated with a diverse interactome mostly through its individual PDZ domains in a PDZ domain – PBD manner. The PDZ3 domain of MAGI-1 is responsible for its interaction with brain-specific angiogenesis inhibitor (BAI-1) (also interacts with MAGI-1 PDZ4 domain) (Shiratsuchi, Futamura et al. 1998), ESAM (Wegmann, Ebnet et al. 2004, Kimura, Ishida et al. 2010), nephrin (also interacts with MAGI-1 PDZ2 domain) (Hirabayashi, Mori et al. 2005), Sidekick1 and 2 (also interacts with MAGI-1 PDZ2 domain) (Yamagata and Sanes 2010), the primary oncoprotein E4 region-encoded open-reading-frame 1 (E4-ORF1) of type 9 human adenovirus (AdV9), and CAREx8 (also interacts with MAGI-1 PDZ1). Here, we add CAREx7 into this PDZ3 domain interaction pool. Among these proteins, CAREx7 and ESAM are capable of properly localizing MAGI-1 to the cell-cell junctions through their highly homologous last 21 aa of the C-termini, including the 4 aa PDZ-binding domain (GSLV in ESAM and GSIV in CAREx7). Despite the similarities between CAREx7 and ESAM, ESAM is restricted to endothelial cells, activated platelets and primitive hematopoietic progenitors, while CAREx7 is expressed in a much wider array of cell types. Nephrin appears to recruit MAGI-1 to this silt diaphragm in neighboring foot processes of podocytes within the kidney, where these proteins play a role in separating the apical and basolateral membranes of this complex tissue. It is possible that the junction localization of MAGI-1 is regulated by a diverse set of distinct proteins in different cell types and cell-cell contacts. Junction localized MAGI-1 plays an important role in maintaining the cell-cell junction integrity and trafficking other protein partners involved in ligand-receptor interactions in multiple cell signaling pathways. Thus, the identification of proteins that regulate MAGI-1 junctional localization, such as CAREx7, will improve our understanding of pathway dysfunction in diseases such as glomerulosclerosis and cancer, and our understanding of viral infections.
5. Conclusions
We have limited this discussion to proteins that interact with MAGI-1 PDZ3. However, there are numerous other known interactions mediated by MAGI-1, many of which do not yet have a defined domain interaction. Interestingly, several proteins interact with more than one MAGI-1 PDZ domain. However, it is not yet clear which residues are responsible for both the promiscuity and over-lapping specificity of PDZ-based interactions. Cellular and molecular analysis of the interaction between CAREx7 and MAGI-1 also reinforce the understanding of the crosstalk among MAGI-1 related scaffolding complex. By gaining a deeper knowledge of the molecular basis of the MAGI-1 interactome, it may be possible target the various PDZ domains of MAGI-1 to regulate the expression of proteins of interest, such as CAREx8, in vivo. As many of the proteins in the MAGI-1 interactome are of high clinical relevance, novel approaches to target the PDZ domains of MAGI-1 could lead to new medical treatments for conditions such as cancer and viral infection.
Acknowledgements
This work was supported by NIH/NIAID Award R15AI090625 (KE), NIH/NIDDK Award R00DK77573 (HH), a Wright State University (WSU) Research Incentive Seed Grant (KE), and WSU Women in Science Giving Circle Award (KE). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or WSU.
Abbreviations
- Ab
antibody
- AdV
adenovirus
- CAR
coxsackievirus and adenovirus receptor
- CAREx7
the seven exon-containing isoform of CAR
- CAREx8
the eight exon-containing isoform of CAR
- FRET
Förster resonance energy transfer
- PBD
PDZ-binding domain
- Y2H
yeast-2-hybird assay
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersson B, Tomko Richard P., Edwards K, Mirza M, Darban H, Öncü D, Sonnhammer E, Sollerbrant K, Philipson L. Putative regulatory domains in the human and mouse CVADR genes. Gene Function & Disease. 2000;1(2):82–86. [Google Scholar]
- Asher DR, Cerny AM, Weiler SR, Horner JW, Keeler ML, Neptune MA, Jones SN, Bronson RT, Depinho RA, Finberg RW. Coxsackievirus and adenovirus receptor is essential for cardiomyocyte development. Genesis. 2005;42(2):77–85. doi: 10.1002/gene.20127. [DOI] [PubMed] [Google Scholar]
- Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, Horwitz MS, Crowell RL, Finberg RW. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275(5304):1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- Bewley MC, Springer K, Zhang YB, Freimuth P, Flanagan JM. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science. 1999;286(5444):1579–1583. doi: 10.1126/science.286.5444.1579. [DOI] [PubMed] [Google Scholar]
- Carson SD, Chapman NN, Tracy SM. Purification of the putative coxsackievirus B receptor from HeLa cells. Biochem Biophys Res Commun. 1997;233(2):325–328. doi: 10.1006/bbrc.1997.6449. [DOI] [PubMed] [Google Scholar]
- Dobrosotskaya IY, James GL. MAGI-1 interacts with beta-catenin and is associated with cell-cell adhesion structures. Biochem Biophys Res Commun. 2000;270(3):903–909. doi: 10.1006/bbrc.2000.2471. [DOI] [PubMed] [Google Scholar]
- Dorner AA, Wegmann F, Butz S, Wolburg-Buchholz K, Wolburg H, Mack A, Nasdala I, August B, Westermann J, Rathjen FG, Vestweber D. Coxsackievirus-adenovirus receptor (CAR) is essential for early embryonic cardiac development. J Cell Sci. 2005;118(Pt 15):3509–3521. doi: 10.1242/jcs.02476. [DOI] [PubMed] [Google Scholar]
- Excoffon KJ, Gansemer ND, Mobily ME, Karp PH, Parekh KR, Zabner J. Isoform-specific regulation and localization of the coxsackie and adenovirus receptor in human airway epithelia. PLoS One. 2010;5(3):e9909. doi: 10.1371/journal.pone.0009909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffon KJ, Hruska-Hageman A, Klotz M, Traver GL, Zabner J. A role for the PDZ-binding domain of the coxsackie B virus and adenovirus receptor (CAR) in cell adhesion and growth. J Cell Sci. 2004;117(Pt 19):4401–4409. doi: 10.1242/jcs.01300. [DOI] [PubMed] [Google Scholar]
- Excoffon KJ, Kolawole AO, Kusama N, Gansemer ND, Sharma P, Hruska-Hageman AM, Petroff E, Benson CJ. Coxsackievirus and adenovirus receptor (CAR) mediates trafficking of acid sensing ion channel 3 (ASIC3) via PSD-95. Biochem Biophys Res Commun. 2012;425(1):13–18. doi: 10.1016/j.bbrc.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiberg F, Sauter M, Pinkert S, Govindarajan T, Kaldrack J, Thakkar M, Fechner H, Klingel K, Gotthardt M. Interspecies differences in virus uptake versus cardiac function of the coxsackievirus and adenovirus receptor (CAR). J Virol. 2014 doi: 10.1128/JVI.00104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaunsinger BA, Lee SS, Thomas M, Banks L, Javier R. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene. 2000;19(46):5270–5280. doi: 10.1038/sj.onc.1203906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi S, Mori H, Kansaku A, Kurihara H, Sakai T, Shimizu F, Kawachi H, Hata Y. MAGI-1 is a component of the glomerular slit diaphragm that is tightly associated with nephrin. Lab Invest. 2005;85(12):1528–1543. doi: 10.1038/labinvest.3700347. [DOI] [PubMed] [Google Scholar]
- Hostetler HA, Lupas D, Tan Y, Dai J, Kelzer MS, Martin GG, Woldegiorgis G, Kier AB, Schroeder F. Acyl-CoA binding proteins interact with the acyl-CoA binding domain of mitochondrial carnitine palmitoyl transferase I. Mol Cell Biochem. 2011;355(1-2):135–148. doi: 10.1007/s11010-011-0847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruska-Hageman AM, Benson CJ, Leonard AS, Price MP, Welsh MJ. PSD-95 and Lin-7b interact with acid-sensing ion channel-3 and have opposite effects on H+-gated current. J Biol Chem. 2004;279(45):46962–46968. doi: 10.1074/jbc.M405874200. [DOI] [PubMed] [Google Scholar]
- Ivanova S, Repnik U, Banks L, Turk V, Turk B. Cellular localization of MAGI-1 caspase cleavage products and their role in apoptosis. Biol Chem. 2007;388(11):1195–1198. doi: 10.1515/BC.2007.141. [DOI] [PubMed] [Google Scholar]
- Ivarsson Y. Plasticity of PDZ domains in ligand recognition and signaling. FEBS Lett. 2012;586(17):2638–2647. doi: 10.1016/j.febslet.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman L, Potla U, Coleman S, Dikiy S, Hata Y, Kurihara H, He JC, D'Agati VD, Klotman PE. Up-regulation of the homophilic adhesion molecule sidekick-1 in podocytes contributes to glomerulosclerosis. J Biol Chem. 2010;285(33):25677–25685. doi: 10.1074/jbc.M110.133959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5(10):771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Kimura R, Ishida T, Kuriyama M, Hirata K, Hayashi Y. Interaction of endothelial cell-selective adhesion molecule and MAGI-1 promotes mature cell-cell adhesion via activation of RhoA. Genes Cells. 2010;15(4):385–396. doi: 10.1111/j.1365-2443.2010.01387.x. [DOI] [PubMed] [Google Scholar]
- Kirby I, Davison E, Beavil AJ, Soh CP, Wickham TJ, Roelvink PW, Kovesdi I, Sutton BJ, Santis G. Identification of contact residues and definition of the CAR-binding site of adenovirus type 5 fiber protein. J Virol. 2000;74(6):2804–2813. doi: 10.1128/jvi.74.6.2804-2813.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby I, Lord R, Davison E, Wickham TJ, Roelvink PW, Kovesdi I, Sutton BJ, Santis G. Adenovirus type 9 fiber knob binds to the coxsackie B virus-adenovirus receptor (CAR) with lower affinity than fiber knobs of other CAR-binding adenovirus serotypes. J Virol. 2001;75(15):7210–7214. doi: 10.1128/JVI.75.15.7210-7214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolawole AO, Sharma P, Yan R, Lewis KJ, Xu Z, Hostetler HA, Ashbourne Excoffon KJ. The PDZ1 and PDZ3 domains of MAGI-1 regulate the eight-exon isoform of the coxsackievirus and adenovirus receptor. J Virol. 2012;86(17):9244–9254. doi: 10.1128/JVI.01138-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotelevets L, van Hengel J, Bruyneel E, Mareel M, van Roy F, Chastre E. Implication of the MAGI-1b/PTEN signalosome in stabilization of adherens junctions and suppression of invasiveness. FASEB J. 2005;19(1):115–117. doi: 10.1096/fj.04-1942fje. [DOI] [PubMed] [Google Scholar]
- Kranjec C, Massimi P, Banks L. Restoration of MAGI-1 Expression in HPV Positive Tumour Cells Induces Cell Growth Arrest and Apoptosis. J Virol. 2014 doi: 10.1128/JVI.03247-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Liu H, Rice AP. Regulation of interferon-beta by MAGI-1 and its interaction with influenza A virus NS1 protein with ESEV PBM. PLoS One. 2012;7(7):e41251. doi: 10.1371/journal.pone.0041251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim BK, Xiong D, Dorner A, Youn TJ, Yung A, Liu TI, Gu Y, Dalton ND, Wright AT, Evans SM, Chen J, Peterson KL, McCulloch AD, Yajima T, Knowlton KU. Coxsackievirus and adenovirus receptor (CAR) mediates atrioventricular-node function and connexin 45 localization in the murine heart. J Clin Invest. 2008;118(8):2758–2770. doi: 10.1172/JCI34777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazirandeh A, Sultana T, Mirza M, Rozell B, Hultenby K, Wallis K, Vennstrom B, Davis B, Arner A, Heuchel R, Lohr M, Philipson L, Sollerbrant K. Multiple phenotypes in adult mice following inactivation of the Coxsackievirus and Adenovirus Receptor (Car) gene. PLoS One. 2011;6(6):e20203. doi: 10.1371/journal.pone.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Kolawole AO, Wiltshire SM, Frondorf K, Excoffon KJ. Accessibility of the coxsackievirus and adenovirus receptor and its importance in adenovirus gene transduction efficiency. J Gen Virol. 2012;93(Pt 1):155–158. doi: 10.1099/vir.0.036269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratsuchi T, Futamura M, Oda K, Nishimori H, Nakamura Y, Tokino T. Cloning and characterization of BAI-associated protein 1: a PDZ domain-containing protein that interacts with BAI1. Biochem Biophys Res Commun. 1998;247(3):597–604. doi: 10.1006/bbrc.1998.8603. [DOI] [PubMed] [Google Scholar]
- Tomko RP, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci U S A. 1997;94(7):3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Raaij MJ, Chouin E, van der Zandt H, Bergelson JM, Cusack S. Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 A resolution. Structure. 2000;8(11):1147–1155. doi: 10.1016/s0969-2126(00)00528-1. [DOI] [PubMed] [Google Scholar]
- Walters RW, Freimuth P, Moninger TO, Ganske I, Zabner J, Welsh MJ. Adenovirus fiber disrupts CAR-mediated intercellular adhesion allowing virus escape. Cell. 2002;110(6):789–799. doi: 10.1016/s0092-8674(02)00912-1. [DOI] [PubMed] [Google Scholar]
- Wegmann F, Ebnet K, Du Pasquier L, Vestweber D, Butz S. Endothelial adhesion molecule ESAM binds directly to the multidomain adaptor MAGI-1 and recruits it to cell contacts. Exp Cell Res. 2004;300(1):121–133. doi: 10.1016/j.yexcr.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Xu Z, Peng AW, Oshima K, Heller S. MAGI-1, a candidate stereociliary scaffolding protein, associates with the tip-link component cadherin 23. J Neurosci. 2008;28(44):11269–11276. doi: 10.1523/JNEUROSCI.3833-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. Synaptic localization and function of Sidekick recognition molecules require MAGI scaffolding proteins. J Neurosci. 2010;30(10):3579–3588. doi: 10.1523/JNEUROSCI.6319-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]