Abstract
We propose that the quantitative cancer biology community make a concerted effort to apply lessons from weather forecasting to develop an analogous methodology for predicting and evaluating tumor growth and treatment response. Currently, the time course of tumor response is not predicted; instead, response is- only assessed post hoc by physical exam or imaging methods. This fundamental practice within clinical oncology limits optimization of atreatment regimen for an individual patient, as well as to determine in real time whether the choice was in fact appropriate. This is especially frustrating at a time when a panoply of molecularly targeted therapies is available, and precision genetic or proteomic analyses of tumors are an established reality. By learning from the methods of weather and climate modeling, we submit that the forecasting power of biophysical and biomathematical modeling can be harnessed to hasten the arrival of a field of predictive oncology. With a successful methodology towards tumor forecasting, it should be possible to integrate large tumor specific datasets of varied types, and effectively defeat cancer one patient at a time.
1. Introduction
The past decade has witnessed a dramatic increase in our knowledge on cancer on multiple scales leading to a host of potential drug targets and subsequent clinical trials. Yet the outcome for many cancers has not improved (1). A fundamental reason for this sobering reality is that we do not have a validated theoretical framework to understand how tumors within the individual patient respond to treatment; that is, there is no accepted mathematical description that enables us to generate testable, patient-specific hypotheses. More specifically, we do not have a theory that, given patient-specific data, can we reliably and reproducibly predict the spatiotemporal changes of that patient’s tumor in response to an intervention. Currently, providing optimal therapies for a specific tumor phenotype, particularly with combinations of therapies, is extraordinarily difficult, as the number of potentially important adjustable parameters, such as the order and dosages of therapy, is too large to span in clinical trials and patient heterogeneity in response is large. Clinical trials too frequently lead to inconclusive and confusing results such that approximately half are never even published in the peer reviewed literature (2). As our knowledge of cancer grows there is a desperate need to make real connections between those designing clinical trials and those studying mathematical models of tumor growth and treatment response so that the field of theoretical oncology can provide systematic, testable predictions of the response of individual patients to individual therapeutic regimens. We envision a diagnostic/prognostic toolkit containing experimentally validated, mathematical tumor models coupled with a battery of patient specific measurements to initialize and constrain a patient specific model. Oncologists could then select the most promising approach by systematically and exhaustively exploring, in silico, all relevant combinations of therapies. Unfortunately, there is not yet a theoretical approach that can harness the spectrum of cancer knowledge towards usable predictions.
Cancer is a multiscale problem that extends from the micro (DNA/RNA), to the meso (protein expression, cell behaviors), to the macro (organ function) scales, and critical to understanding cancer will be to model impacts at each level. Multiscale models (and the associated experiments), an active area of cancer research, will need time to develop. However, it may be possible to establish theories of clinical tumor progression that captures the behavior of an individual patient’s tumor well before the influence of underlying events on tumor scale behaviors is elucidated. That is, one does not require an exhaustive understanding, at all scales, of a phenomenon in order to form a useful theory. The history of science teems with examples of extraordinarily useful theories, whose development was driven by a need to quantitatively describe complex systems or complex behaviors, when the underlying principles were not known. One instructive example is weather modeling, which is guided in large part by the fluid equations with hydrostatic balance, also known as the primitive equations (3). We posit that the methods and paradigms of weather forecasting over the last century provide the most pertinent perspective on how to proceed when attempting to build a quantitative theory of cancer. While analogies between weather and tumor forecast are somewhat common in both the scientific and popular press (see. e.g., references 4 and 5), detailed steps for adapting the methods of meteorology to oncology are often overlooked. We establish some relevant parallels in the next section.
2. Guidance from the meteorologist
A “weather forecast” is a scientific estimate of weather conditions at some time in the future. There are essentially two steps in developing a weather forecast: 1) estimate the current physical state of the atmosphere (the “diagnostic phase”), and 2) use techniques to predict the future state of the atmosphere (“prognostic phase”). Once the diagnostic phase is complete, there are two main ways to develop a prognosis: one is based on historical data (both near and long term), while the other employs mathematical models. We provide brief descriptions of both before discussing possible application to cancer.
The simplest approach based on historical data is “persistence forecasting”, which assumes that the tendency of the weather is to remain unchanged in the short term. “Climatological forecasting”, employs average weather statistics accumulated over decades to offer a short-term forecast: If a prediction of temperature and precipitation is desired for a particular day, one averages the temperature and precipitation measured on that day for as far back as one’s dataset allows, and from these average values offers a forecast. A third method from historical data is the “analog method”, based on the assumption that weather repeats itself, at least in a general way. The idea is to match current conditions with similar conditions previously observed and construct a forecast based on the observations most frequently seen to follow those conditions in the database. The fourth approach is termed “trend forecasting”, which is based on determining the velocity of certain features (e.g., storm fronts) and extrapolating the future position of those features.
These historical data approaches are useful as they can provide reasonably accurate forecasts for short time periods into the future (generally, less than 24 hours). However, they cannot account for changes in intensity and direction and/or predict the formation or dissipation of major events over extended time (days or weeks). We argue that current clinical methods of predicting treatment response in cancer patients are akin to these “historical trend” methods and, consequently, suffer from the same severe limitations in predictive ability. The clear strategic and economic benefits of knowing what the weather will be tomorrow or next week encouraged the development of numerical weather prediction methods in the last century. The cost of health care and the aging population coupled with recent scientific and technological advancements provide both the incentive and the capacity to develop numerical tumor prediction methods now.
Numerical weather prediction (NWP) methods stem from a set of physical principles that govern the behavior of the atmosphere and that can be expressed by a system of equations. If these equations can be solved, one can provide a description of the future state of the atmosphere (i.e., a forecast) based on its current state. The diagnostic phase of a numerical weather model relies on a vast, global network of surface (land and sea) and air measurements to obtain reliable measures of temperature, cloud cover, humidity, precipitation, and wind velocity. It is difficult to overstress the importance of these measurements. For weather modeling, measurements in one particular region are insufficient to characterize the whole atmosphere because the system is both 1) heterogeneous, and 2) connected. Indeed, the forecaster must have a complete—and maximally accurate—picture of current conditions before the weather at future times can be predicted.
NWP is an “initial value problem”; i.e., in order to integrate the equations of atmospheric motion and project its state at future time points, one must specify the values of the independent variables at an initial time. Mathematically, the atmosphere is described in terms of a state vector, X(r, t0) = {X1(r, t0), X2(r, t0),…, Xn(r, t0)}, of the n model variables at grid points r and initial time t0 (i.e., the diagnostic phase). For meteorology, the Xi vary depending on the form of the equations but include some form of conservation of momentum (horizontal velocity and hydrostatic balance), energy (temperature), air density, and specific humidity. Once obtained, X(r,t0), along with additional diagnostic quantities to close the system, are then substituted into the model equations and evaluated at future time points to generate the forecast (i.e., the prognostic phase).
Two key techniques commonly used in NWP that are concerned with providing the best possible forecast are appropriate for tumor modeling. The first technique is “dynamic model weighting”, by which multiple predictions (from multiple models) are weighed to provide a forecast that combines the most realistic aspects of various models given the particulars of the system under investigation. As it is rarely known, a priori, which model is most appropriate for describing a particular weather system, dynamic model weighing provides a method by which such uncertainty can be minimized by statistically determining how to combine the best predictions from multiple models (6). The second concerns “data assimilation,” which aims to improve upon a short term model forecast by incorporating all available observed data from a diverse suite of in situ and remotely gathered sources that is not regularly available at each temporal or spatial grid point, and using advanced numerical techniques to create an optimized, evenly-gridded representation of the state of the atmosphere in time and space with minimized error (7,8). One application of this technique to determine the relevance for tumor growth showed promise with a moderate degree of model error and measurement noise (9). Related to data assimilation is the process of “reanalysis,” which takes the most advanced data assimilation process to create a long-term data product for comparison to weather and climate models. This provides a consistent data source for analysis and model validation. One challenge for reanalysis products, which is the proper inclusion of changing data quality and coverage over the Earth’s past and present (10), would be minimal for tumor prediction because the methods and data sources could be applied evenly over the lifetime of the tumor prediction window.
3. Numerical Tumor Prediction
An effective field of predictive oncology would develop a comprehensive and systematic method for predicting the future status of an individual tumor given an optimal representation of its initial conditions and an appropriate biophysical model. It is now well known that tumors are a mosaic of different cells with tremendous heterogeneity across scales and between patients even in the same disease sub-type (11,12). The currently employed “climatological” and “analog” approach for tumor forecasting based on an average of prior patients response has limited applicability to forecast an individual tumor development. The persistence and trend forecasting approaches also have limited applicable in oncology as tumors are temporally and spatially dynamic; thus, forecasting that a tumor will maintain its current status (in the case of persistence forecasting) or continue to evolve as it has in the past (in the case of trend forecasting) will be accurate only for a short period of time. We note that persistence forecasting is used for patients for whom “watchful waiting” is appropriate. Of course, persistence forecasting does not provide insights into if and when relapse occurs.)
What would clinical oncology look like if a predictive NWP-equivalent were available? Based on numerical tumor prediction (NTP) algorithms, a patient would be given a personalized quantitative output charting the course of the disease. This would include levels of confidence in the prediction itself, as well as a time intervals for future interventions, controls, and updates. While the goal is ambitious (as was the conceit in the early 20th century that we could accurately predict the weather), the success of NWP provides a roadmap and optimism that NTP can be a reality in the near future.
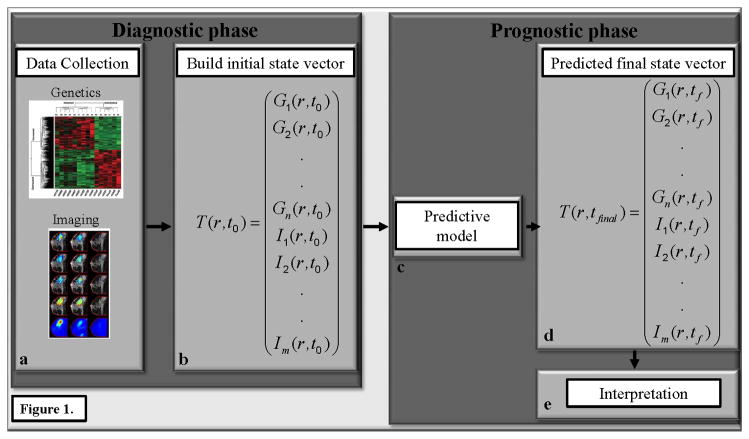
In analogy to NWP, the first step towards developing NTP is to characterize the state vector of the tumor; that is, we need to form a complete diagnosis of the state of the tumor. In order to achieve this we must determine what (independent) tumor variables are the most critical for quantitatively summarizing tumor status. This is no small challenge, but a candidate list has been developed by Hannahan and Weinberg (13). We then form a spatio-temporal equation for each tumor variable and link these to each other so that model variables can influence each other; in the parlance of mathematics, we would build a coupled system of partial differential equations describing the evolution of the tumor mass (rather than also including energy and momentum as is done in NWP). Figure 1 illustrates this approach.
Figure 1.
The figure presents an overview of how numerical tumor prediction would work in practice. The diagnostic phase consists of assembling the appropriate patient specific data (panel a) and building the initial state vector, T(r,t0) (panel b), which provides an initial snapshot of the key biological characteristics of the tumor at position r and initial time t0. In this example, T(r,t0) consists of a series of genomic, Gi(r,t0), and imaging, Ii(r,t0), measurements. The initial state vector then initializes the predictive model (panel c) which is projected forward in time to yield a final state vector (panel d) which is then interpreted (panel e) to give a snapshot of the key biological characteristics of the tumor at some future time point, tf.
We note that there is no shortage of mathematical models of tumor growth in the literature (see, e.g., 14–17), but they are not typically of the kind that can readily be populated by data that are measured clinically at multiple time points with any reasonable spatial resolution. Models must be recast to deal with data that can be measured on an individual basis. Furthermore, dynamic model weighting should be used to eliminate bias that may exist in particular models by combining the predictions of multiple, independent models. Data assimilation can be used to provide updates to the tumor forecasts by including new data points as they become available. It should also be noted that much biological and clinical progress can be made without a comprehensive initial state vector; indeed, the first NWP model used only seven independent variables (pressure, temperature, density, humidity, and three velocity vectors; 18). Regardless of what state variables are chosen for a particular model, the data simply must be acquired from the system (i.e., the patient) for which the prediction is to be made. Initializing predictive models using data culled from population statistics or the literature will not guide the way any more than the historical trend forecasts do for weather prediction.
4. Clinical Oncology Settings Likely to Benefit
Selecting an optimal treatment regimen for a tumor phenotype, and predicting how an individual patient will respond to a treatment regimen are two areas most likely to benefit quickly from the approach described in Figure 1. Currently, in the best cases, treatment regimens are selected based on molecular phenotype and/or genotype (e.g., breast cancer) while, in the worst cases, untargeted chemotherapy is employed (e.g., pancreatic cancer). If we were able to initialize a reasonable model of tumor growth dynamics with a characterization of an individual tumor’s initial state vector, then it would be possible to systematically run a series of in silico simulations to determine how this particular tumor will respond to an array of treatment regimens. That is, we could run a myriad of patient specific virtual clinical trials to determine the optimal regimen and timing for that particular patient. This is an especially attractive features in the combination therapy setting where one drug is designed to target tumor associated vasculature while another is designed to target the tumor cells themselves (Figure 2); indeed, such trials are common and frequently have unclear results (see. e.g., 19–21). Another promising avenue for this modeling approach is in situations where one drug has the potential to sensitize the tumor to a second therapy. Such is the case in, for example, triple negative breast cancers that are sensitive to PI3K inhibitors which, in turn, may increase their susceptibility to DNA damaging agents (22). An important feature of this theoretical approach is that it generates predictions that experimentally testable in pre-clinical animal models of cancer.) An early, and successful example of this has already been achieved (23) using very limited patient specific data and this speaks to the power of the paradigm. Once a therapeutic approach is selected, we are then faced with the difficulty of using early treatment changes to predict long term response.
Figure 2.
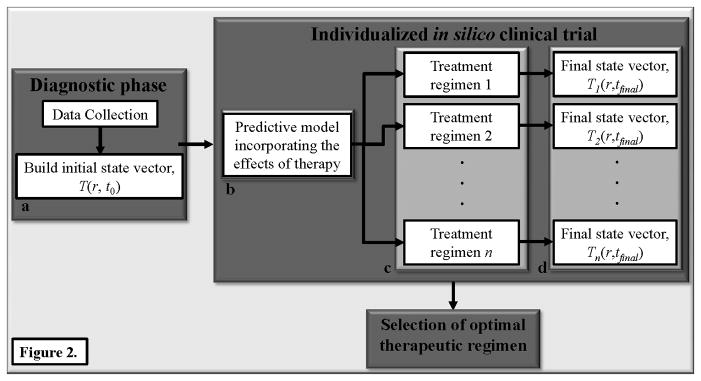
The scheme in figure 1 is easily extended to allow for patient specific clinical trials. Namely, after collecting the data to build the initial state vector, T(r,t0) (panel a), the data is used to initialize a predictive model (panel b) that includes the effects of therapy. Here, “therapy” can be any of n different regimens that systematically vary (for example) dose, timing, and order of multiple treatments (panel c) to yield multiple final state vectors, Ti(r,t0) (panel d). The ith therapy yields the ith final state vector which is then compared to select the optimal therapeutic regimen for the particular patient under investigation.
Currently, responses are not predicted, they are assessed post hoc by physical exam, or structural ultrasound, magnetic resonance imaging, or computed tomography. Many patients are forced to undergo invasive biopsies during their therapy, and others are found to have received ineffective therapy only after months of treatment. The ability to identify—early in the course of therapy—patients that are not responding to a given regimen is highly significant. In addition to limiting patients’ exposure to the toxicities associated with unsuccessful therapies, it would allow patients the opportunity to switch to a potentially more efficacious treatment. As there are typically many therapeutic options available, and many more being developed, switching treatment early in the course of therapy is a very real option—but only if a reliable method to determine early response were available. Unfortunately, existing methods of determining early response are inadequate. A particularly exciting area to apply this approach is in the neoadjuvant setting, where patients who achieve a complete pathological response at the time of surgery is predictive (for many cancers) of improved survival (24–27). Thus, if it were possible to predict before therapy, test early in therapy, and then change (if needed) therapy, long term survival could be significantly improved.
5. Modeling Successes in Clinical Oncology
As was recently, and elegantly, described by Larry Norton (28), mathematical modeling actually guided the development of medical oncology in the 1960s when Skipper et al developed the concept of the “log-kill hypothesis” (29). This hypothesis states that when tumor growth increases by a constant fraction per unit time (i.e., the growth is exponential), then treatment with a cytoxic therapy results in tumor shrinkage by a constant fraction over time. This paradigm led to many ideas (eventually accepted as dogma) on how to properly test and use anticancer drugs (28). Of course, it was subsequently established that tumors do not exhibit unrestrained exponential growth in vivo, and tumor growth is better characterized by logistic or Gompertzian growth (30). This development, in turn, led to important repercussions for dosing schedules for cancer patients. Thus, historically, mathematical modeling has played a key role in drug development and clinical trial design. More recently, there has been promising evidence of the utility of patient specific modeling using clinically available data.
Chmielecki et al employed mathematical modeling to develop a theoretically optimum dosing schedule for non-small cell lung cancers with mutations in the epidermal growth factor receptor (EGFR) gene (31). Briefly, the group incorporated knowledge of the different growth kinetics of drug-sensitive and resistant EGFR-mutant cells into an evolutionary model constrained by clinically available data. Their model led to the prediction that high-dose pulses of erlotinib combined with a continuous low dose of the drug can significantly delay the onset of resistance. This hypothesis is now being tested in a prospective clinical trial (32).
Bozic et al also employed evolutionary dynamics to predict the effects of combination therapy for patients receiving the targeted therapy vemurafenib (33). One exciting result generated by their model is that combination therapy with two drugs given simultaneously is more effective than if the therapies are given sequentially thereby providing important guidance for the design of subsequent clinical trials.
There have also been efforts to use clinically available data to predict response, after the treatment regimen has been selected. Neal et al developed and applied a patient specific model of glioblastoma multiforme (GBM) growth to predict, at the first post-radiation treatment time point, progression free and overall survival (34). Their model is based on a realization of the reaction diffusion model that describes GBM growth in terms of cellular proliferation and random diffusion. Those two parameters are then determined on a patient specific basis and used to project tumor development forward in time. An extension of the “personalized” reaction diffusion equation was put forward by Weis et al who coupled the random diffusion term to tissue mechanical properties (35). Applying this model to breast cancer patients receiving neoadjuvant chemotherapy achieved an area under the receiver operator characteristic curve of 0.81 for predicting, after the first cycle of treatment, which patients would go on to achieve a pathological complete response (36).
It is important to stress that each of these (illustrative) studies was based on clinically available data and their results indicate that clinically relevant “forecasts” can be made to both optimize the therapeutic regimen, and then predict which patients will respond to that regimen.
6. Potential Limitations
In general, the factors that dramatically affect the accuracy of initial value problems are: 1) inadequate mathematical representation of the processes being modeled, 2) errors in the initial conditions (i.e., errors in the diagnostic phase), and 3) inadequate model resolution (i.e., physical spacing between the measurements). Unfortunately, all three of these limitations are well-represented in oncology. In addition to lacking clinically relevant mathematical descriptions of tumor growth and treatment response, we have an incomplete description of the state vector described in Figure 1. Our assessment of initial conditions is frequently based on sampling a small portion of tissue via, for example a biopsy, and this is almost certainly not adequate to characterize the entire tumor (12). One would desire the state vector used to initialize the NTP model to be populated at every grid point associated with the scale at which the tumor forecast is to be made. Furthermore, just as in NWP, if too small an area is chosen to make a prediction, then the forecast will be quite poor as information from outside the forecast area spreads into the forecast area. Similarly, in cancer, it is critical to not only characterize the tumor itself, but also the surrounding healthy-appearing tissue.
We have previously stressed the importance of using quantitative noninvasive imaging methods to help address this issue (37). Of course, the spatial resolution available from (even advanced) imaging methods are limited to a several hundred microns and this coarseness will obscure more subtle features whose importance will almost certainly grow with time. However, it could be that such subtleties are not required to answer many clinically relevant questions; that is, perhaps a mesoscale model making predictions on bulk tumor properties is appropriate for predicting (say) progression free survival in a particular patient. Indeed, such “mesoscale” predictions that lack local area specifics and tend to be less precise (e.g., long range forecasts are not specific—make claims like warmer/cooler, wetter/drier than historical mean) are still of critical use in NWP for the transportation, agricultural, and energy industries. Thus, it may be that similar predictive power in oncology is “good enough” to indicate whether or not an individual patient will beat the average progression free or overall survival?
7. Conclusion
In this paper we have proposed that the cancer community can learn much from the phenomenological models that enable numerical weather prediction. Following such an approach potentially enables optimal pairing of treatment regimen with tumor phenotype, and predicting how an individual patient will respond to a particular regimen. All of this is predicated on initializing models with patient specific data obtained early after diagnosis to predict future patient status. Just as the problem of calculating future events based on present events has seen enormous advances in the physical sciences over the last century, it must be tacked in all earnest for oncology. Indeed, this is the whole goal of science: the calculation of future events based on present events. Oncology should be no different.
Acknowledgments
This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
The authors thank the National Cancer Institute for support through 1U01CA142565, 1U01CA174706, R01 CA138599, and P30 CA68485. Parts of this material is based upon work supported by the U.S. Department of Energy, Office of Science under the Scientific Discovery Through Advanced Computing (SciDAC) project on Multiscale Methods for Accurate, Efficient, and Scale-Aware Models of the Earth System.
References
- 1.Marshall E. Cancer and the $90 billion metaphor. Science. 2011;331:1540–1. doi: 10.1126/science.331.6024.1540-a. [DOI] [PubMed] [Google Scholar]
- 2.Riveros C, Dechartres A, Perrodeau E, Haneef R, Boutron I, Ravaud P. Timing and completeness of trial results posted at ClinicalTrials.gov and published in journals. PLoS Med. 2013;10:e1001566. doi: 10.1371/journal.pmed.1001566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lauritzen PH, Jablonowski C, Taylor MA, Nair RD, editors. Lecture Notes in Computational Science and Engineering. New York: Springer; 2011. Numerical Techniques for Global Atmospheric Models; p. 564. [Google Scholar]
- 4.Chauviere AH, Hatzikirou H, Lowengrub JS, Frieboes HB, Thompson AM, Cristini V. Mathematical Oncology: How Are the Mathematical and Physical Sciences Contributing to the War on Breast Cancer? Curr Breast Cancer Rep. 2010;2:121–9. doi: 10.1007/s12609-010-0020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nazaryan A. Getting Cancer Wrong. 2014 Mar 20; Newsweek. Published on http://www.newsweek.com/2014/03/28/getting-cancer-wrong-248029.html.
- 6.Räisänen J, Ylhäisi JS. Can model weighting improve probabilistic projections of climate change? Climate Dynamics. 2012;39:1981–98. [Google Scholar]
- 7.Kalnay E. Atmospheric Modeling, Data Assimilation, and Predictability. Cambridge: Cambridge University Press; 2002. [Google Scholar]
- 8.Lorenc AC. Analysis methods for numerical weather prediction. Quart J Roy Meteor Soc. 1986;112:1177–94. [Google Scholar]
- 9.Kostelich EJ, Kuang Y, McDaniel JM, Moore NZ, Martirosyan NL, Preul MC. Accurate state estimation from uncertain data and models: an application of data assimilation to mathematical models of human brain tumors. Biol Direct. 2011;6:64. doi: 10.1186/1745-6150-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Screen JA, Simmons I. Erroneous Arctic Temperature Trends in the ERA-40 Reanalysis: A Closer Look. J Climate. 2011;24:2620–7. [Google Scholar]
- 11.Kaiser J. The Downside of Diversity. Science. 20013;339:1543–5. doi: 10.1126/science.339.6127.1543. [DOI] [PubMed] [Google Scholar]
- 12.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hannahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Deisboeck TS. Mathematical modeling in cancer drug discovery. Drug Discov Today. 2014;19:145–50. doi: 10.1016/j.drudis.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 15.Kim M, Gillies RJ, Rejniak KA. Current advances in mathematical modeling of anti-cancer drug penetration into tumor tissues. Front Oncol. 2013;3:278. doi: 10.3389/fonc.2013.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baldock AL, Rockne RC, Boone AD, Neal ML, Hawkins-Daarud A, Corwin DM, et al. From patient-specific mathematical neuro-oncology to precision medicine. Front Oncol. 2013;3:62. doi: 10.3389/fonc.2013.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li F, Tan H, Singh J, Yang J, Xia X, Bao J, et al. A 3D multiscale model of cancer stem cell in tumor development. BMC Syst Biol. 2013;7(Suppl 2):S12. doi: 10.1186/1752-0509-7-S2-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bjerknes V. Das Problem der Wettervorhersage, betrachtet vom Standpunkte der Mechanik und der Physik. Metero Zeit. 1904;21:1–7. [Google Scholar]
- 19.Brandes AA, Mason W, Pichler J, Nowak AK, Gil M, Saran F, et al. Can bevacizumab prolong survival for glioblastoma patients through multiple lines of therapy? Future Oncol. 2014;10:1137–45. doi: 10.2217/fon.14.75. [DOI] [PubMed] [Google Scholar]
- 20.Kerbel RS. Reappraising antiangiogenic therapy for breast cancer. Breast. 2011;20(Suppl 3):S56–60. doi: 10.1016/S0960-9776(11)70295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aghajanian C, Goff B, Nycum LR, Wang Y, Husain A, Blank S. Independent radiologic review: bevacizumab in combination with gemcitabine and carboplatin in recurrent ovarian cancer. Gynecol Oncol. 2014;133:105–10. doi: 10.1016/j.ygyno.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Mayer IA, Abramson VG, Lehmann BD, Pietenpol JA. New strategies for triple-negative breast cancer--deciphering the heterogeneity. Clin Cancer Res. 2014;20:782–90. doi: 10.1158/1078-0432.CCR-13-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chmielecki J, Foo J, Oxnard GR, Hutchinson K, Ohashi K, Somwar R, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci Transl Med. 2011;3:90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;12:1275–81. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 25.Hou JY, Kelly MG, Yu H, McAlpine JN, Azodi M, Rutherford TJ, et al. Neoadjuvant chemotherapy lessens surgical morbidity in advanced ovarian cancer and leads to improved survival in stage IV disease. Gynecol Oncol. 2007;105:211–7. doi: 10.1016/j.ygyno.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 26.Mahmoud O, Dosch A, Kwon D, Pitcher JD, Conway S, Benedetto P, et al. The Impact of Perioperative Chemotherapy Timing in Conjunction With Postoperative External-Beam Radiation Therapy on Extremity Soft-Tissue Sarcomas Outcome. Am J Clin Oncol. 2014 May 29; doi: 10.1097/COC.0000000000000087. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 27.Porten S, Siefker-Radtke AO, Xiao L, Margulis V, Kamat AM, Wood CG, et al. Neoadjuvant chemotherapy improves survival of patients with upper tract urothelial carcinoma. Cancer. 2014;120:1794–9. doi: 10.1002/cncr.28655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norton L. Cancer log-kill revisited. Am Soc Clin Oncol Educ Book. 2014:3–7. doi: 10.14694/EdBook_AM.2014.34.3. [DOI] [PubMed] [Google Scholar]
- 29.Skipper HE, Schabel FR, Jr, Wilcox WS. Experimental evaluation of potential anticancer agents. XII. On the criteria and kinetics associated with “curability” of experimental leukemia. Cancer Chemother Rep. 1964;35:1–111. [PubMed] [Google Scholar]
- 30.Norton L, Simon R, Brereton HD, Bogden AE. Predicting the course of Gompertzian growth. Nature. 1976;264:542–5. doi: 10.1038/264542a0. [DOI] [PubMed] [Google Scholar]
- 31.Chmielecki J, Foo J, Oxnard GR, Hutchinson K, Ohashi K, Somwar R, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci Transl Med. 2011;3:90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dolgin E. The mathematician versus the malignancy. Nature Medicine. 2014;20:460–3. doi: 10.1038/nm0514-460. [DOI] [PubMed] [Google Scholar]
- 33.Bozic I, Reiter JG, Allen B, Antal T, Chatterjee K, Shah P, et al. Evolutionary dynamics of cancer in response to targeted combination therapy. eLife. 2013;2:e00747. doi: 10.7554/eLife.00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neal ML, Trister AD, Cloke T, Sodt R, Ahn S, Baldock AL, et al. Discriminating survival outcomes in patients with glioblastoma using a simulation-based, patient-specific response metric. PLoS One. 2013;8:e51951. doi: 10.1371/journal.pone.0051951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weis JA, Miga MI, Arlinghaus LR, Li X, Chakravarthy AB, Abramson V, et al. A mechanically coupled reaction-diffusion model for predicting the response of breast tumors to neoadjuvant chemotherapy. Phys Med Biol. 2013;58:5851–66. doi: 10.1088/0031-9155/58/17/5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weis JA, Miga MI, Arlinghaus LR, Li X, Chakravarthy AB, Abramson V, et al. Prediction of Response to Neoadjuvant Chemotherapy Using a Mechanical Coupled Reaction-Diffusion Model. AAPM 2014 Annual Meeting Med Phys. 2014;41:509. [Google Scholar]
- 37.Yankeelov TE, Atuegwu N, Hormuth D, Weis JA, Barnes SL, Miga MI, et al. Clinically relevant modeling of tumor growth and treatment response. Sci Transl Med. 2013;5:187ps9. doi: 10.1126/scitranslmed.3005686. [DOI] [PMC free article] [PubMed] [Google Scholar]