Abstract
Reactive oxygen species inflict oxidative modifications on various biological molecules, including DNA. One of the most abundant DNA base lesions 8-oxo-7,8-dihydroguanine (8-oxoG) is repaired by 8-oxoguanine DNA glycosylase-1 (OGG1) during DNA base excision repair (OGG1-BER). 8-OxoG accumulation in DNA has been associated with various pathological and aging processes, while its role is unclear. The lack of OGG1-BER in Ogg1-/- mice resulted in decreased inflammatory responses, increased susceptibility to infections and metabolic disorders. Therefore, we proposed that OGG1 and/or 8-oxoG base may have a role in immune and homeostatic processes. To test our hypothesis, we challenged mouse lungs with OGG1-BER product 8-oxoG base and changes in gene expression were determined by RNA sequencing and data were analyzed by gene ontology and statistical tools. RNA-Seq analysis identified 1592 differentially expressed (≥ 3-fold change) transcripts. The upregulated mRNAs were related to biological processes, including homeostatic, immune-system, macrophage activation, regulation of liquid-surface tension, and response to stimulus. These processes were mediated by chemokines, cytokines, gonadotropin-releasing hormone receptor, integrin and interleukin signaling pathways. Taken together, these findings points to a new paradigm showing that OGG1-BER plays a role in various biological processes that may benefit host, but when is in excess could be implicated in disease and/or aging processes.
Keywords: OGG1-BER, 8-oxoguanine, gene expression, biological processes
Introduction
Reactive oxygen species (ROS) are produced as a by-product of cellular metabolism and/or as a consequence of environmental insults. Due to their reactivity, ROS modify biological molecules, including DNA. The accumulation of ROS-induced base and strand modifications, as well as their defective or decreased repair, has been associated with various diseases and aging processes [1, 2]. Due to guanine's highest susceptibility to reactive species among DNA bases [3, 4], 8-oxoG is one of the most abundant. 8-OxoG(syn) mispairs with adenine during DNA replication, resulting in a G:C to T:A transversion mutation. The mutagenic burden imposed by 8-oxoG is prevented primarily by OGG1-initiated DNA base excision repair pathway (OGG1-BER), which is a multi-step process involving recognition and excision of the damaged base, followed by cleavage of the DNA backbone. The resulting apurinic/apyrimidinic site is then processed by the apurinic/apyrimidinic endonuclease 1, and the DNA is repaired in sequential steps of BER [1, 5, 6]. Due to OGG1-BER, 8-oxoG levels in body fluids such as urine are one of the best biomarkers of environmental exposures and inflammatory processes [7, 8].
To study the consequences of 8-oxoG accumulation in DNA and role of OGG1 in patho-physiological processes, OGG1 knock out (Ogg1-/-) mice were developed [9, 10]. Intriguingly, the resulting supraphysiological levels of genomic 8-oxoG did not affect embryonic development or life span. Moreover, increased levels of 8-oxoG in the mitochondrial DNA did not affect mitochondrial respiratory parameters or ROS production [11]. Under conditions of chronic oxidative stress, genomic 8-oxoG levels can be increased up to 250-fold in Ogg1-/- mice, without apparent consequences, including precancerous lesions or tumors in various organs [12]. More surprisingly, Ogg1-/- mice have shown a decreased inflammatory response to various stimuli, including lipopolysaccharide (LPS) [13], consequently increased susceptibility to bacterial infection [14]. A lack of 8-oxoG repair by OGG1 in airway epithelium decreased allergic airway inflammation in mouse models of asthma [15, 16]. Intriguing studies also documented that, following administration of a high-fat diet, Ogg1-/- mice have increased plasma insulin levels, impaired glucose tolerance, enhanced adiposity and hepatic steatosis when compared to wild-type animals [17]. These data together suggest that OGG1 and/or its repair product 8-oxoG base may play an important role in signaling various cellular physiological and pathophysiological processes.
Recent studies have shown that OGG1 binds its repair product, 8-oxoG base, at sites other than its catalytic center, and forms an OGG1•8-oxoG complex. This complex interacts with and activates canonical small GTPases and induces guanine nucleotide exchange (GDP→GTP) in vitro and in cellulo [18-21]. Moreover, studies showed that activation of OGG1-BER was associated with an increase in RAS-GTP levels as well as phosphorylation of downstream RAS targets, including v-raf-1 murine leukemia viral oncogene homolog 1 (RAF1), mitogen-activated protein kinase (MAPK) kinase (MEK1/2), extracellular signal-regulated kinase (ERK1/2), phosphoinositide 3-kinase [18, 19], and an innate immune response [22].
The goal of the present study was to define the role of OGG1-BER in overall gene expression and identify its involvement in biological processes. To mimic OGG1-BER, mouse lungs were challenged with 8-oxoG base via the intranasal route, and changes in whole transcriptome were assessed by RNA-Seq analysis. Results showed that out of 23,337 identified transcripts, 8-oxoG challenge induced upregulation of 983 and downregulation of 1,398. Gene ontology analysis of RNA-Seq data by using the PANTHER classification system showed that the upregulated transcripts were related to various biological processes, among which homeostatic and immune system processes were the most represented. Thus, we document for the first time that, in addition to maintaining genome fidelity, OGG1-BER plays a role in a range of cellular and biological processes that are essential and could either benefit a host or be implicated, when in excess, in diseases or aging processes.
Materials and Methods
Animals and treatment
Animal experiments were performed according to the NIH Guide for Care and Use of Experimental Animals and approved by the University of Texas Medical Branch (UTMB) Animal Care and Use Committee (approval no. 0807044A). Eight-week-old female BALB/c mice (The Jackson Laboratory, Bar Harbor, ME, USA) were used for these studies. Mice (n = 5 per group) were challenged intranasally (i.n.) with 60 μL of pH-balanced 8-oxoG solution (pH: 7.4; 0.005 μg per gram), or saline under mild anesthesia [18]. LPS was below detectable levels in all reagents. Animals were sacrificed at various time points to isolate lung protein/extracts, RNA, and bronchoalveolar lavage fluid (BALF).
RNA isolation
After intranasal challenges, mouse lungs were excised and homogenized with a TissueMiser® (Fisher). RNA was extracted using an RNeasy kit (Qiagen, Valencia, CA) per the manufacturer's instructions. Briefly, lung tissue homogenate was loaded onto an RNeasy column and subjected to washes with RW1 and RPE buffers. RNA was eluted by using the RNase-free water included in the kit. Eluted RNA was digested with RNase-free DNase as previously described [23]. The RNA concentration was determined spectrophotometrically on an Epoch Take-3™ system (Biotek, Winooski, VT) and by using the software Gen5 v2.01. Equal amounts of RNA from each mouse lung within an experimental group (n = 5) were pooled and analyzed in triplicate. Quality of the total RNA was confirmed spectrophotometrically by using the 260/280 nm ratio, which varied from 1.9 to 2.0. RNA integrity was also evaluated by agarose gel electrophoresis.
Library construction and next-generation RNA sequencing
Library construction and deep sequencing analysis were performed in UTMB's Next-Generation Sequencing (NGS) Core Facility (Director: Dr. Thomas G. Wood). RNA sequencing analysis was carried out on an Illumina HiSeq 1000 sequencing system (Illumina Inc., San Diego, CA, USA). Poly-A+ RNA was selected from total RNA (1 μg) using poly-T oligo-attached magnetic beads. Bound RNA was fragmented by incubation at 94°C for 8 minutes in 19.5 μl of fragmentation buffer (Illumina, Part # 15016648). First- and second-strand synthesis, adapter ligation, and amplification of the library were performed using the Illumina TruSeq RNA Sample Preparation kit per the manufacturer's instructions (Illumina). Samples were tracked through the “index tags” incorporated into the adapters. Library quality was evaluated utilizing an Agilent DNA-1000 chip on an Agilent 2100 Bioanalyzer. Library DNA templates were quantitated by qPCR and a known-size reference standard.
Cluster formation of the library DNA templates was performed using the TruSeq PE Cluster Kit v3 (Illumina) and the Illumina cBot workstation under the conditions recommended by the manufacturer. The template input was adjusted to obtain a cluster density of 700-1000 K/mm2. Paired-end, 50-base sequencing by synthesis was performed with a TruSeq SBS kit v3 (Illumina) on an Illumina HiSeq 1000 by protocols defined by the manufacturer. Base call conversion to sequence reads was performed using CASAVA-1.8.2. Sequence data were analyzed with the Bowtie2, Tophat and Cufflinks programs using NCBI's mouse (Mus musculus) genome build reference mm10. RNA-Seq data have been deposited in the NCBI's Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number (GSE61095). Reads per kilobase transcript per million (RPKM) were normalized for each experimental group to its corresponding control [24].
Gene expression analysis
Heat maps and hierarchical clusters from whole transcriptomes were constructed with GENE-E software from Broad Institute (http://www.broadinstitute.org/cancer/software/GENE-E/). Venn diagrams were constructed by using online Venny software (http://bioinfogp.cnb.csic.es/tools/venny/). Gene ontologies (GO) and signaling pathways were analyzed by the PANTHER (Protein ANalysis THrough Evolutionary Relationships) Classification System (http://www.pantherdb.org/) version 9.0. The lists of differentially expressed genes (fold change ≥3) were processed via GO annotations in this database (22160 genes, for Mus musculus). PANTHER's overrepresentation test uses the binomial method with Bonferroni correction for multiple comparisons to annotate classification categories for a list of genes [25, 26]. Significance was considered at P values < 0.05. The overrepresentation test was used to identify functional classes from the submitted gene lists according to PANTHER's reference lists. This test assumes that under the null hypothesis, genes in the uploaded list are sampled from the same general population as are genes from the reference set, i.e., the probability p(C) of observing a gene from a particular category C in the uploaded list is the same as in the database reference list [25].
Down-regulation of Ogg1
Mouse airways were depleted of OGG1 by RNAi as previously described [15]. We introduced Stealth™ RNAi (Invitrogen Life Technologies, Carlsbad, CA, USA. Cat # MSS237431) i.n. in transfection reagent (Polyplus-transfection SA, New York, NY, Cat# 201-10G) to target Ogg1. Control mice were treated with negative control RNAi (Invitrogen, Cat# 4404020). Oligos were of in vivo purity as described by the manufacturer. Mouse lungs were pre-treated with RNAi at 0 and 25 h and challenged with 8-oxoG or saline 48 h later.
Cytokine Bio-Plex assay
BALF samples were collected after 2h of exposure to 8-oxoG or saline and centrifuged (800 × g for 5 min at 4°C), and the resulting supernatants were stored at -80°C for further analysis of chemokines and cytokines [23, 27]. We used the Bio-Plex Pro™ Mouse Cytokine Assay (Bio-Rad, Hercules, CA), a bead-based, multiplex protein immunoassay (Bio-Rad, Cat # M60-009RDPD) [28], and processed BALF samples (n=3) and cytokine standards (in triplicate) per the manufacturer's instructions. Readings were performed on a Bioplex® 200™ system (Bio-Rad). Data analysis was performed using Bio-Plex Manager™ Software Version 6.0 Built 617 (Bio-Rad).
Assessment of RAS-GTP levels
RAS-GTP levels were quantified with the Active RAS pull-down assay kits (Pierce, Thermo Scientific, Inc) per the manufacturer's instructions, with slight modifications. Freshly isolated mouse lungs were homogenized in 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 60 mM MgCl2, 1% Nonidet P-40, and 5% glycerol, and 500 μg of lung protein homogenate was incubated with the RAS-binding domain of Raf1 immobilized to glutathione resin, as we previously described [18]. After washing with binding buffer, GTP-bound RAS was eluted with Laemmli buffer (0.125 m Tris-HCl, 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, pH 6.8) and subjected to Western blotting. Total RAS was evaluated by Western blotting by using 20 μg of protein from the same lung lysates provided for pull-down assays.
qRT-PCR
Total RNA (1 μg) was reverse-transcribed using the SuperScript® III First-Strand Synthesis System (Invitrogen) per the manufacturer's instructions. To evaluate transcript levels for selected genes, we used the ΔΔCt method, as previously described [23]. The commercial (IDT) validated primers were: Cxcl1, Cat # Mm.PT.58.42076891; Cxcl2, Cat # Mm.PT.530.16380094; Ccl3, Cat # Mm.PT.58.29283216; Tnf, Cat # Mm.PT.S6a.12S7S861; Il1a, Cat # Mm.PT.58.32778767 and Il1b, Cat # Mm.PT.58.41616450. Housekeeping gene primers were: Hprt, Cat # Mm.PT.S30.32092191. qRT-PCR was performed on an ABI7000 thermal cycler. The amplification thermal profile was: 2 min 50 °C, 10 min 95 °C, and 15 sec 95 °C followed by 1 min 60 °C (40 cycles). To confirm the presence of a single product after amplification, a dissociation stage was carried out: 15 sec, 95 °C ; 20 sec, 60 °C ; and 15 sec, 95 °C.
Statistical analysis
Statistical analysis was performed using Student's t-test or ANOVA, followed by post hoc tests: Bonferroni's and Dunnett's T3 with SPSS 14.0 software. The data are presented as the means ± the standard error of the mean. Differences were considered statistically significant at p < 0.05.
Results
The OGG1-BER product 8-oxoG induced changes in global transcriptome
Mouse lungs were challenged via the i.n. route with the 8-oxoG base product of OGG1-BER, and then BALFs, total RNA, and protein extracts were isolated. Preliminary characterizations by qRT-PCR showed that mRNA levels of tumor necrosis factor (Tnf), interleukin 1 alpha (Il1a) and chemokine (C-X-C motif) ligand 1 (Cxcl1) were increased from 30 min on, reaching a maximum at 60 min. Multiplex protein analysis identified de novo-synthesized soluble mediators (e.g., CXCL1, TNF-α) from 120 min on, which was similar to results in our previous studies [22, 29]. Therefore to define the primary effects of OGG1-BER (8-oxoG challenge) on global gene expression by RNA-Seq, we limited the exposure time to 0, 30, 60, and 120 min.
RNAs were pooled from challenged lungs (n = 5), RNA sequencing was carried out twice, and the average of the two consequent analyses is presented. A total of 23,337 transcripts were identified (GEO Series accession number GSE61095). To identify groups of transcripts with similar expression patterns, we performed an unsupervised clustering and generated a heat map from whole-transcriptome data using GENE-E online software (Broad Institute). Hierarchical clustering of whole-genome data from 30, 60 and 120 min rendered a dendogram including three major clusters of transcripts (Fig. 1A). In general, cluster 1 contained 5678 mostly upregulated transcripts at all time points. Transcripts (10106) in cluster 2 show either an immediate increase or decrease, and those that were not altered. Cluster 3 included transcripts (3090) without changes in their expression or that increased only at 120 min post exposure. Red represents expression values greater than 1-fold, and green shows expression less than 1-fold.
Fig. 1.
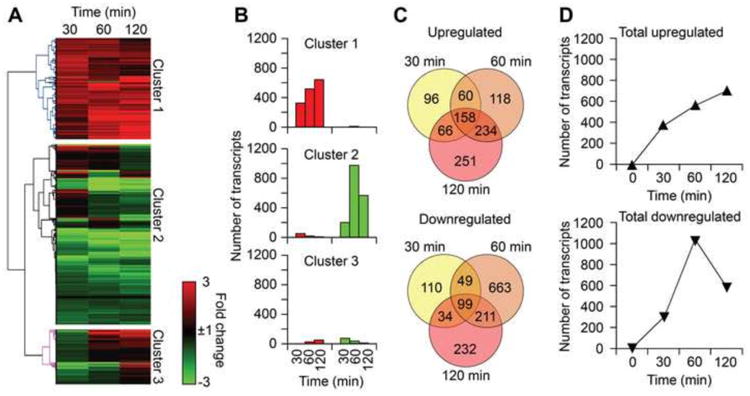
Whole-transcriptome profile induced by 8-oxoG challenge. (A) Graphical depiction of results from hierarchical clustering showing whole-transcriptome profile. Total RNA isolated from challenged lungs at 30, 60 and 120 min were subjected to RNA-Seq, heat maps were generated using GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E/), and transcripts were clustered according to their expression patterns. Transcript levels (RPKM) were normalized to control (time 0 min). Rows represent identified transcripts and columns at time points after 8-oxoG challenge. The intensity of the red or green colors shows the degree of upregulation or downregulation, respectively. (B) The total number of upregulated (≥3-fold) transcripts (red bars) and downregulated (≤3-fold) transcripts (green bars) in each cluster. (C) Number of unique and shared transcripts altered by 8-oxoG challenge. Venn diagrams were constructed by using online software Venny (Materials and Methods). (D) Kinetic changes in transcript levels induced by 8-oxoG challenge.
The number of upregulated and downregulated transcripts (≥ 3-fold, mRNAs, non-coding transcripts and miRNAs) in each cluster at every time point is depicted in Fig. 1B. Venn diagrams show the unique and overlapping differentially expressed transcripts at each time point after challenge with 8-oxoG (Fig. 1C) corresponding to transcript described in Fig. 1B. Whole transcriptome data showed upregulation (≥ 3-fold) of 158 common transcripts and 96, 118 and 251 unique transcripts at 30, 60 and 120 min, respectively. Besides the unique and common transcripts to all time points, only 360 upregulated transcripts were shared between any two time points (Fig. 1C, upper panel). The total of upregulated transcripts (≥ 3-fold) at any time point was 983, of which 570 corresponded to 60 min. Among the 1398 downregulated transcripts, 99 were common, while 110, 663, and 232 were unique at 30, 60, and 120 min, respectively. A total of 294 transcripts were shared between any two time points only (Fig. 1C, lower panel). Together, cumulatively 2,381 transcripts were differentially expressed (≥ 3-fold change) after 8-oxoG challenge.
Results show a continuous increase in the number of transcripts at all time points (Fig. 1D, upper panel). On the other hand, the numbers of downregulated transcripts were decreased from 60 min (Fig. 1D, lower panel). These results could be interpreted as the effect of de novo synthesized, soluble mediators in line with preliminary studies [22, 29]. Therefore, to characterize the impact of 8-oxoG challenge, in our analysis the 60-min time point was examined.
OGG1-BER induces gene expression involved in various biological processes
Next, the list containing 570 upregulated transcripts from RNA-Seq at 60 min post 8-oxoG exposure (Fig. 1C) was searched against the PANTHER classification system [25] which identified 443 mRNAs distributed in various GO categories. The highest overrepresented biological processes were: homeostatic process (P = 9.30E-04), immune system process (P = 5.68E-03), macrophage activation (P = 6.40E-03), regulation of liquid surface tension (P = 7.07E-03), response to stimulus (P = 1.12E-02), and metabolic process (P = 7.30E-01) (Fig. 2A). Overrepresented protein classes were: cytokine (P = 2.01E-07), chemokine (P = 6.37E-05), interleukin superfamily (P = 1.23E-03), surfactant (P = 7.28E-03), microtubule family cytoskeletal (P = 1.11E-02) and signaling molecule (P =1.41E-02) (Fig. 2B). The most overrepresented cellular component was the microtubule (P = 3.33E-02) (Fig. 2C). Among molecular functions, cytokine (P = 4.77E-07), chemokine (P = 5.59E-05) and receptor binding (P = 1.53E-02) were significantly overrepresented (Fig. 2D).
Fig. 2.
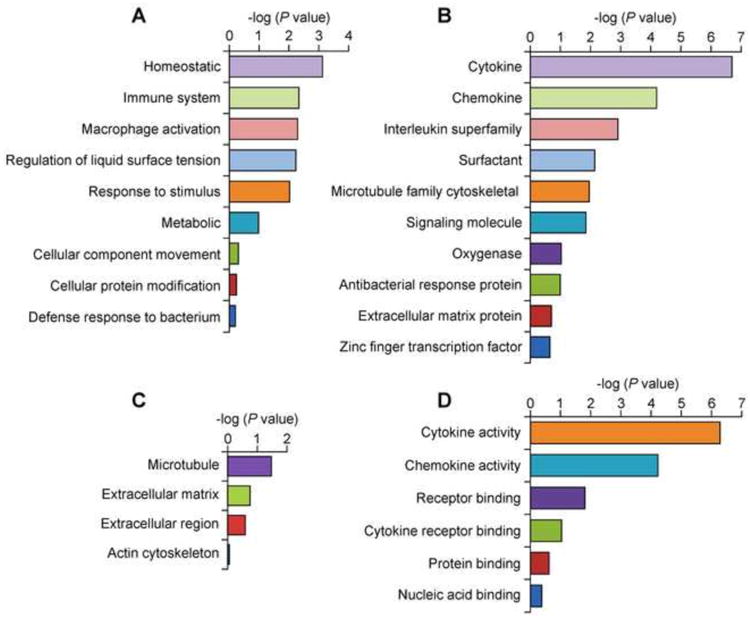
Gene ontology categories represented by the upregulated genes induced after introduction of the OGG1-BER product 8-oxoG into mouse lungs. Overrepresentation test of the genes upregulated (≥ 3 fold) at 60 min was performed by using PANTHER (http://www.pantherdb.org). (A) Biological process. (B) Protein class. (C) Cellular component. (D) Molecular function. X axis represent −log(P value).
The homeostatic process is defined as any biological process involved in the maintenance of an internal steady state according to the Gene Ontology Consortium (http://geneontology.org/). The upregulated (≥ 3-fold) genes involved in homeostatic processes (Fig. 2A, Table 1) were: anterior gradient 2 (Agr2); anterior gradient 3 (Agr3); ATPase, H+/K+ exchanging, gastric, and alpha polypeptide (Atp4a); ATPase, class I, type 8B, member 5 (Atp8b5); chemokine CC motif ligand 9 (Ccl9); collagen type II alpha 1 (Col2a1); collagen type IX alpha 1 (Col9a1); alpha 3 (Col9a3); collagen type X alpha 1 (Col10a1); collagen type XI alpha 1 (Col11a1); calcitonin/calcitonin-related polypeptide alpha (Calca); forkhead box A3 (Foxa3), and G protein-coupled receptor 37 (Gpr37).
Table 1. List of genes up-regulated by OGG1-BER involved in the most represented biological processes and signaling pathways.
| Symbol | RefSeq ID | Name | Biological processes | Signaling pathways | Fold change | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H | I | M | S | R | In | Gn | It | Il | ||||
| Cxcl2 | NM_009140 | Chemokine (C-X-C motif) ligand 2 | X | X | X | 192.95 | ||||||
| Bpifa1 | NM_011126 | BPIa fold containing family A, member 1 | X | X | X | 107.60 | ||||||
| Matn1 | NM_010769 | Matrilin 1 | X | X | X | 83.06 | ||||||
| Ccl20 | NM_001159738 | Chemokine (C-C motif) ligand 20 | X | X | X | 58.87 | ||||||
| Cxcl1 | NM_008176 | Chemokine (C-X-C motif) ligand 1 | X | X | X | 54.45 | ||||||
| Col9a1 | NM_007740 | Collagen, type IX, alpha 1 | X | X | X | X | X | X | 50.12 | |||
| Bpifb1 | NM_001012392 | BPIa fold containing family B, member 1 | X | X | X | 48.76 | ||||||
| Col10a1 | NM_009925 | Collagen, type X, alpha 1 | X | X | X | X | X | X | 45.05 | |||
| Capn13 | NP_001028616 | Calpain 13 | X | 34.00 | ||||||||
| Col2a1 | NM_031163 | Collagen, type II, alpha 1 | X | X | X | X | X | X | 29.07 | |||
| Ccl3 | NM_011337 | Chemokine (C-C motif) ligand 3 | X | X | X | 27.25 | ||||||
| Otx1 | NM_011023 | Orthodenticle homolog 1 | X | 27.16 | ||||||||
| Cxcl3 | NM_008176 | Chemokine (C-X-C motif) ligand 3 | X | X | X | 23.78 | ||||||
| Agr2 | NM_011783 | Anterior gradient 2 | X | X | 22.72 | |||||||
| Matn3 | NM_010770 | Matrilin 3 | X | 20.33 | ||||||||
| Hsf5 | NP_001038992 | Heat shock transcription factor 5 | X | 19.63 | ||||||||
| Tnf | NM_013693 | Tumor necrosis factor | X | X | 16.65 | |||||||
| Klk1b3 | NM_008693 | Kallikrein 1-related peptidase b3 | X | X | 15.42 | |||||||
| Col9a2 | NM_007741 | Collagen, type IX, alpha 2 | X | X | X | X | 13.53 | |||||
| Il17c | NM_145834 | Interleukin 17C | X | 12.62 | ||||||||
| Rgs1 | NM_015811 | Regulator of G-protein signaling 1 | X | 10.54 | ||||||||
| Clec4e | NM_019948 | C-type lectin domain family 4, member e | X | X | X | 10.45 | ||||||
| Il6 | NM_031168 | Interleukin 6 | X | X | 10.44 | |||||||
| Lypd2 | NP_080947 | Ly6/Plaur domain containing 2 | X | 10.23 | ||||||||
| Klk1b21 | NM_010642 | Kallikrein 1-related peptidase b21 | X | X | 10.16 | |||||||
| Pgc | NM_025973 | Progastricsin (pepsinogen C) | X | 9.81 | ||||||||
| Lyzl4 | NP_081191 | Lysozyme-like 4 | X | 9.81 | ||||||||
| Nr4a1 | NM_010444 | Nuclear receptor subfamily 4, group A | X | X | X | 8.36 | ||||||
| Tacr1 | NM_009313 | Tachykinin receptor 1 | X | X | 7.84 | |||||||
| Vmn1r3 | NM_001167535 | Vomeronasal 1 receptor 3 | X | 7.71 | ||||||||
| Col11a1 | NM_007729 | Collagen, type XI, alpha 1 | X | X | X | X | X | X | 7.56 | |||
| Ctla4 | NM_009843 | Cytotoxic T-lymphocyte-associated protein 4 | X | X | 7.08 | |||||||
| Calca | NM_007587 | Calcitonin/calcitonin-related polypeptide, alpha | X | 6.63 | ||||||||
| Wfikkn1 | NP_001093924 | WAPb, follistatin/kazal immunoglobulin KKN1c | X | X | 6.53 | |||||||
| Col9a3 | NM_009936 | Collagen, type IX, alpha 3 | X | X | X | X | X | X | 6.13 | |||
| Fcrlb | NM_001160215 | Fragment crystallizable (Fc) receptor-like B | X | X | X | 5.89 | ||||||
| Vmn1r2 | NM_001167534 | Vomeronasal 1 receptor 2 | X | 5.84 | ||||||||
| Il10 | NM_010548 | Interleukin 10 | X | X | X | X | 5.84 | |||||
| Sik1 | NM_010831 | Salt inducible kinase 1 | X | 5.82 | ||||||||
| Klk1b24 | NM_010643 | Kallikrein 1-related peptidase b24 | X | X | 5.61 | |||||||
| Ngp | NM_008694 | Neutrophilic granule protein | X | X | 5.51 | |||||||
| Klkb1 | NM_008455 | Kallikrein B, plasma 1 | X | X | 5.37 | |||||||
| Wfdc8 | NM_029325 | WAPb four-disulfide core domain 8 | X | X | 5.26 | |||||||
| Asgr1 | NM_009714 | Asialoglycoprotein receptor 1 | X | X | X | 5.22 | ||||||
| Crb1 | NM_133239 | Crumbs homolog 1 | X | 5.21 | ||||||||
| Ccl4 | NM_13652 | Chemokine (C-C motif) ligand 4 | X | X | X | 5.17 | ||||||
| Atf3 | NM_007498 | Activating transcription factor 3 | X | X | 5.03 | |||||||
| Alox12e | NM_145684 | Arachidonate lipoxygenase, epidermal | X | X | 5.02 | |||||||
| Camp | NM_009921 | Cathelicidin antimicrobial peptide | X | X | 4.98 | |||||||
| Tbr1 | NM_009322 | T-box brain gene 1 | X | 4.91 | ||||||||
| Ccl28 | NM_007930 | Chemokine (C-C motif) ligand 28 | X | X | X | 4.80 | ||||||
| Klk1b27 | NM_020268 | Kallikrein 1-related peptidase b27 | X | X | 4.74 | |||||||
| Nfkbia | NM_010907 | NF-LPd gene enhancer in B cells inhibitor, alpha | X | X | X | 4.73 | ||||||
| Acsm1 | NM_054094 | Acyl-CoA synthetase medium-chain family member 1 | X | 4.69 | ||||||||
| Pigr | NM_011082 | Polymeric immunoglobulin receptor | X | X | 4.68 | |||||||
| Agr3 | NP_997414 | Anterior gradient 3 | X | X | 4.39 | |||||||
| Cd14 | NM_009841 | CD14 antigen | X | 4.38 | ||||||||
| Dusp1 | NM_013642 | Dual specificity phosphatase 1 | X | X | 4.37 | |||||||
| Foxa3 | NM_008260 | Forkhead box A3 | X | 4.36 | ||||||||
| Gpr37 | NM_010338 | G protein-coupled receptor 37 | X | X | X | 4.33 | ||||||
| Col19a1 | NM_007733 | Collagen, type XIX, alpha 1 | X | 4.21 | ||||||||
| Vmn1r79 | NM_001166835 | Vomeronasal 1 receptor 79 | X | 4.21 | ||||||||
| Gnat3 | NM_001081143 | Guanine nucleotide binding protein alpha T3e | X | 4.21 | ||||||||
| Il1a | NM_010554 | Interleukin 1 alpha | X | X | X | X | 4.17 | |||||
| Il1b | NM_008361 | Interleukin 1 beta | X | X | X | 4.15 | ||||||
| Nfkbiz | NM_030612 | NF-LPd gene enhancer biz | X | X | 4.10 | |||||||
| Osm | NP_001013383 | Oncostatin M | X | 3.90 | ||||||||
| Il23r | NM_144548 | Interleukin 23 receptor | X | X | X | 3.86 | ||||||
| Il17b | NM_019508 | Interleukin 17B | X | 3.86 | ||||||||
| Trim30b | NM_175648 | Tripartite motif-containing 30B | X | X | 3.84 | |||||||
| Tlr2 | NM_011905 | Toll-like receptor 2 | X | 3.81 | ||||||||
| Vtcn1 | NM_178594 | V-set domain containing T cell activation inhibitor 1 | X | 3.75 | ||||||||
| Fos | NM_010234 | Finkel–Biskis–Jinkins oncogene | X | X | X | 3.71 | ||||||
| Cd69 | NP_001028294 | CD69 antigen | X | X | 3.69 | |||||||
| Bdkrb1 | NM_007539 | Bradykinin receptor, beta 1 | X | X | 3.67 | |||||||
| Lcn4 | NM_010695 | Lipocalin 4 | X | 3.61 | ||||||||
| Nr4a2 | NM_013613 | Nuclear receptor subfamily 4, group A, member 2 | X | X | 3.61 | |||||||
| Sntn | NM_177624 | Sentan, cilia apical structure protein | X | X | 3.60 | |||||||
| Dnajb13 | NM_153527 | DnaJ related, subfamily B 13 (Hsp40f) | X | X | 3.58 | |||||||
| Per1 | NM_011065 | Period circadian clock 1 | X | 3.53 | ||||||||
| Vmn1r238 | NM_001167539 | Vomeronasal 1 receptor, 238 | X | 3.50 | ||||||||
| Ms4a15 | NM_001034898 | Membrane-spanning 4a15 | X | 3.50 | ||||||||
| Klk1b9 | NM_010116 | Kallikrein 1-related peptidase b9 | X | X | 3.50 | |||||||
| Atp4a | NM_018731 | ATPase, H+/K+ exchanging, gastric alpha ppg | X | 3.50 | ||||||||
| Nlrp3 | NM_145827 | NOD-like receptor family, pyrin domain containing 3h | X | X | 3.48 | |||||||
| Proc | NM_008934 | Protein C | X | 3.40 | ||||||||
| Lrrc26 | NM_146117 | Leucine rich repeat containing 26 | X | 3.39 | ||||||||
| Atp8b5 | NM_177195 | ATPase, class I, type 8B, member 5 | X | 3.30 | ||||||||
| Mapk15 | NM_177922 | Mitogen-activated protein kinase 15 | X | X | X | 3.28 | ||||||
| Junb | NM_008416 | Jun B proto-oncogene | X | X | 3.28 | |||||||
| Klra5 | NM_008463 | Killer cell lectin-like receptor A, member 5 | X | X | 3.27 | |||||||
| Mog | NM_010814 | Myelin oligodendrocyte glycoprotein | X | 3.15 | ||||||||
| Pkd1l2 | NM_029686 | Polycystic kidney disease 1 like 2 | X | 3.15 | ||||||||
| Zfp352 | NM_153102 | Zinc finger protein 352 | X | X | 3.15 | |||||||
| Gadd45g | NM_011817 | Growth arrest and DNA-damage-inducible 45 | X | X | 3.13 | |||||||
| Ccl9 | NM_011338 | Chemokine (C-C motif) ligand 9 | X | X | X | 3.09 | ||||||
Biological processes: H = Homeostatic process (GO:0042592); I = Immune system process (GO:0002376);M = Macrophage activation (GO:0042116); S = Regulation of liquid surface tension (GO:0050828); R = Response to stimulus (GO:0050896). “X” denotes presence of the genes in each biological process category. Signaling pathways: In = Inflammation mediated by chemokine and cytokine signaling (P00031), Gn = Gonadotropin releasing hormone receptor (P06664), It = Integrin signaling, Il = Interleukin signaling (P00036).
Bactericidal/permeability-increasing fold containing family a1;
Whey acidic protein;
Knitz and netrin domain containing 1;
Nuclear factor of kappa light polypeptide;
Heat shock protein 40 kD;
Guanine nucleotide binding protein alpha transducing 3,
ATPase, H+/K+ exchanging, gastric alpha polypeptide,
Nucleotide-binding oligomerization domain-containing protein 1.
The immune system process was represented by C-X-C chemokines (e.g., Cxcl1, Cxcl2, Cxcl3) C-C motif chemokines (e.g., Ccl3, Ccl4, Ccl9, Ccl20 and Ccl28); interleukins Il1a, Il1b, Il6, Il10, Il17b, Il17c. Tnf and the cytokine receptor Il23r were also upregulated by 8-oxoG challenge. The Gene Ontology Consortium (http://geneontology.org/) considers collagen genes (Col2a1, Col9a1, Col9a2, Col9a3, Col10a1, Col11a1 and Col19a1) part of the immune system and homeostasic process. Moreover, the 8-oxoG challenge (OGG1-BER mimic) upregulated the kallikrein family of serine proteases kallikrein B, plasma 1 (Klkb1), kallikrein 1-related peptidases Klk1b3, Klk1b9, Klk1b21, Klk1b24, and Klk1b27. All fold-increase values are shown in Table 1.
The macrophage activation process is defined by the Gene Ontology Consortium as a change in morphology and behavior of macrophages resulting from exposure to a cytokine, chemokine, cellular ligand, or soluble factor. This GO term was represented by 22 genes, which are overlapping with those listed in immune system processes (above and listed in Table 1).
Another, significantly upregulated biological process was regulation of liquid surface tension, which included genes encoding primarily for collagens (Col2a1, Col9a1, Col9a2, Col9a3, Col10a1, Col11a1 and Col19a1). Genes included in response to stimulus were chemokines, cytokines, interleukins, and collagens (listed above and Table 1). Interestingly, a large number of genes were significantly upregulated in lungs and involved in metabolic processes (lipid, fatty acid, protein, nucleobase metabolic, and primary metabolic process). However, due to the large number of genes included in the PANTHER' metabolic categories, representation of 8-oxoG challenge–induced genes was below significance (P = 7.30E-01).
Signaling pathways induced by OGG1-BER
PANTHER analysis identified and ranked 443 upregulated (≥ 3 fold) genes (induced at 60 min after 8-oxoG challenge) contributing to signaling pathways. Fig. 3 shows the distribution by percentages of genes for various signaling pathways. Among signaling pathways, the most represented one was inflammation mediated by chemokines and cytokines, followed by gonadotropin-releasing hormone receptor (GnRHR), integrin signaling, and interleukin signaling.
Fig. 3.
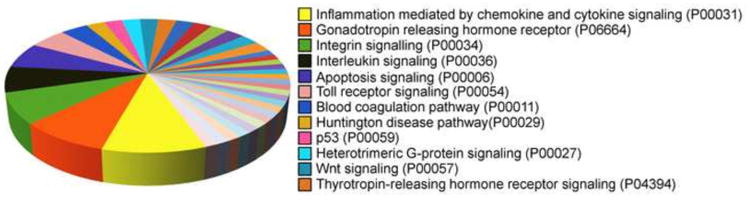
Signaling pathways induced by OGG1-BER product, 8-oxoG in mouse lungs. Pie chart was generated by using PANTHER (Materials and Methods). Pathway analysis included only those genes with expression levels ≥ 3-fold at 60 min.
The inflammatory signaling pathway included various significantly upregulated genes (≥ 3 fold, Table 1). For example, among C-X-C chemokines, Cxcl1 (54.45-fold) and Cxcl2 (192.95-old) are involved in multiple inflammatory processes, such as chemo-attraction of neutrophils [30]. Cxcl3 (23.78-fold) of which protein product CXCL3 (also known as macrophage inflammatory protein-2-beta, or MIP2b) controls migration and adhesion of monocytes has chemotactic activity for neutrophils [31]. Members of the C-C chemokine family represented in this signaling pathway included: Ccl3 (27.25-fold), which encodes for protein CCL3, a chemoattractant for polymorphonuclear leukocytes, and regulates macrophage function [32], and Ccl20 (58.87-fold), implicated in the mucosal immunity via chemoattraction of lymphocytes (T-cells and B-cells) and DCs [33].
We also observed increased expression levels of Ccl4 (5.15-fold), Ccl9 (3.09-fold) and Ccl28 (4.80-fold) (Table1). The upregulated interleukins included Il17c (12.62-fold); Il1a (4.17-fold); Il1b (4.15-fold); and Il6 (4.15-fold); and these have the following attributes: Il17c has a protein product with a critical role in innate immunity of the epithelium, that stimulates the release of TNF-α and IL1B from monocytic cells, and maintains epithelial homeostasis during inflammation [34]. Il1a has a protein product released by damaged airway epithelial cells acting as an alarmin and triggering inflammatory responses in lung fibroblasts [35]. Il1b has a protein product that is a mediator of the inflammatory response and also involved in various cellular processes, including cell proliferation, differentiation, and apoptosis [36]. Il6 encodes a protein that contributes to acute and chronic inflammation and plays an active role, e.g., in the pathogenesis of asthma and COPD [37]. Tnf (16.65-fold) encodes for TNF-α a pleiotropic cytokine involved in various immune responses, inflammatory processes, hematopoiesis, cell proliferation, differentiation, and apoptosis [38]. The highly expressed Bpifa1 encodes for palate, lung, and nasal epithelium, and its protein is reported to be involved in innate immune responses in the upper airways in addition to other biological processes [39].
The gonadotropin-releasing, hormone receptor-signaling pathway was represented by: nuclear receptor subfamily 4, group A (Nr4a1; 8.36-fold); dual specificity phosphatase 1 (Dusp1; 4.37-fold); orthodenticle homolog 1 (Otx1; 27.16-fold); period circadian clock 1 (Per1; 3.53-fold), activating transcription factor 3 (Atf3; 5.03-fold), Finkel–Biskis–Jinkins oncogene (Fos; 3.71-fold); and Jun B proto-oncogene (Junb; 3.28-fold). Nr4a1's protein product is a nuclear receptor that regulates gene transcription and has an anti-inflammatory effect [40]. Dusp1 encodes for a dual specificity phosphatase 1, which dephosphorylates the oxidative-stress stimulated MAPKs p38, c-Jun N-terminal kinase and ERK [41]. Otx1 plays a role in cell differentiation of the mammalian cortex [42], and Per1 encodes for the protein PER1, which regulates the expression of genes involved in maintaining homeostasis of blood pressure [43]. In addition, a significant increase in immediate early genes, Atf3, Fos, and Junb, was observed.
The integrin signaling pathway was represented by members of the collagen family: Col2a1 (29.07-fold); Col9a2 (13.53-fold); Col11a1 (7.56-fold); Col9a1 (50.12-fold); Col9a3 (6.13-fold); Col10a1 (45.05-fold); and integrin beta 2-like (Itgb2l). The interleukin signaling pathway was represented by: Il1a, Il6, and Il10 (5.84-fold); Il17c (12.62-fold); Il17b (3.9-fold); interleukin-23 receptor (Il23r, 3.9-fold); Il23a (alpha subunit, 3.2-fold); Fos (3.71-fold); and Mapk15 (3.28-fold). One of the highly expressed genes in the Il pathway was Il10, which encodes for a pleiotropic cytokine having multiple functions, including inhibition of pro-inflammatory cytokine production in macrophages and other immune cells regulating mucosal immune homeostasis [44]. The expression of Mapk15, also known as Erk8, and the activity of its protein product are regulated by DNA damage, which does not appear to require an upstream activating kinase [45].
Validation of RNA-Seq by qRT-PCR
To confirm the results of RNA-Seq analysis, we verified the expression levels of selected highly (Ccl20, Ccl3, Cxcl1, Cxcl2) and moderately expressed (Il1a, Il1b, and Tnf) pro-inflammatory chemokines and cytokines by qRT-PCR by using the same RNAs as those utilized for RNA-Seq analysis isolated from 8-oxoG-challenged mouse lungs. Fig. 4 shows the comparison of fold changes in RPKM by RNA-Seq analysis (Fig. 4 A) and mRNA levels as determined by qRT-PCR (Fig. 4B). Results from these studies demonstrated that fold changes in RPKMs from RNA-Seq and in mRNA levels determined by qRT-PCR are consistent. The observed differences in fold changes between transcript and mRNA levels were probably due to differences in sensitivity of the two methods.
Fig. 4.
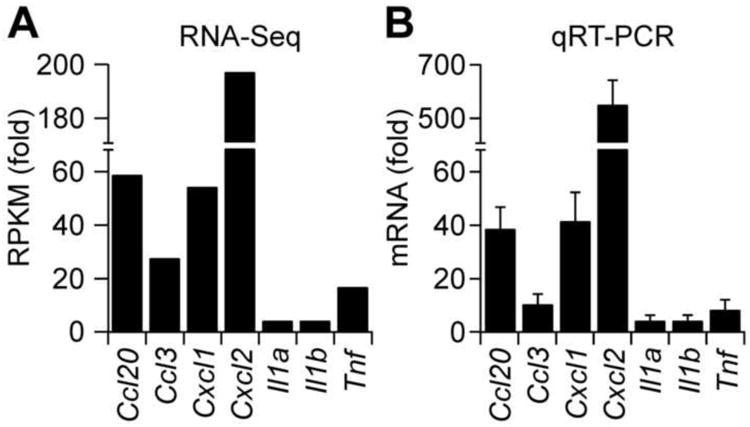
Validation of RNA-Seq data. (A) Fold changes in RPKM of selected genes as determined by RNA-Seq analysis. RNA was isolated from lungs at 60 min post challenge and pooled (n = 5) or kept separately. One μg of pooled RNA was subjected to RNA-Seq as in Materials and Methods. (B) qRT-PCR confirmation of RNA-Seq data. One μg RNA isolated from individual control and challenged mice (n = 5) was reverse transcribed, and cDNAs were amplified by using specific primer pairs as described in Materials and Methods.
RAS signaling-dependent genes induced by 8-oxoG challenge
It has been documented that OGG1-BER, or addition of 8-oxoG to cells, increased the GTP-bound levels of small GTPase (RAS, RHOA, RAC1) family proteins [18-20]. Among small GTPases, RAS is one of the central modulators of signal transduction and gene expression related to cell proliferation and differentiation and inflammation [46-48]. To define whether some of the differentially expressed genes are associated with RAS activation, we first tested whether 8-oxoG challenge altered the expression of genes previously defined to be dependent on RAS signaling. To do so, we utilized a published RAS signature gene list [49], which includes 105 upregulated (Fig. 5A) and 42 downregulated genes (Fig. 5B) and then overlaid the 570 upregulated transcripts from RNA-Seq.
Fig. 5.
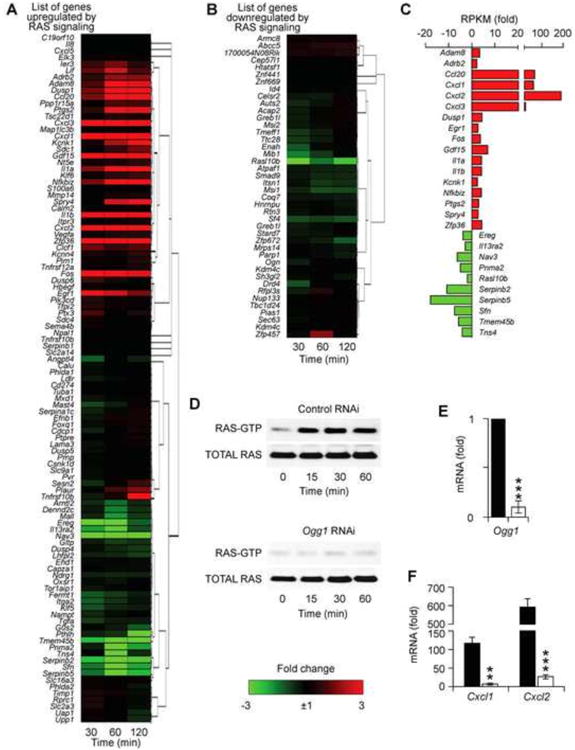
OGG1-BER alters expression of RAS signature genes. Effect of 8-oxoG challenge on the expression of genes reported to be up-regulated (A) or down regulated (B) by RAS signaling. (C) Differentially expressed RAS signature genes with ≥ 3-fold expression levels. Upregulated genes are shown in red bars and down regulated ones in green bars. (D) Activation of RAS-GTPase by 8-oxoG challenge in lungs (upper panel) and dependency on OGG1 expression in 8-oxoG-exposed lungs. (E) Down-regulation of Ogg1 in mouse airway epithelium by RNAi. (F) Decreased expression of Cxcl1 and Cxcl2 in Ogg1-depleted airways. **p < 0.01, ***p < 0.001.
From the 105 genes, 8-oxoG challenge increased the expression levels of 28 genes and downregulated 36 genes (Fig. 5A). The remaining 41 genes were not significantly affected. Among the 42 genes downregulated by RAS, 26 genes were downregulated, while 4 genes showed some increase in expression levels, and 16 remained unaltered (Fig. 5B). Fig. 5C shows a subset of RAS signature genes with significant differential expression (≥3-fold) levels. Notably, the highest upregulated (>20-fold) RAS signaling-dependent genes were Ccl20, Cxcl1, Cxcl2 and Cxcl3 (red bars), all related to immune system and cellular processes and response to stimulus as described above. The most downregulated (>5-fold) genes were the tumor suppressors: serine (or cysteine) peptidase inhibitor, clade B, members 2 (Serpinb2) and 5 (Serpinb5) and the adapter protein implicated in the regulation of various signaling pathways: stratifin (Sfn) as shown in Fig. 5C (green bars).
To validate the role of RAS activation-dependent gene expression, we examined whether there was an increase in RAS-GTP levels in lungs after 8-oxoG challenge. As described (first section of results) in parallel with isolation of RNA for RNA-seq, protein extracts were prepared and subjected to active RAS pull-down assays. Results in Fig. 5D, upper panel, show a time-dependent increase in RAS-GTP levels. These results imply an association between RAS activation and expression of RAS-dependent genes, which are in line with the results of previous studies showing an activation of RAS GTPases by OGG1-BER or 8-oxoG challenge[18, 19, 50].
Next, we examined whether an increase in RAS-GTP levels is OGG1-dependent. To do so, we depleted Ogg1 expression from the airways by RNAi [15], the lungs were challenged with saline containing 8-oxoG base, and protein extracts were prepared. The extent of Ogg1 down regulation in airway epithelium is shown in Fig. 5E. Ogg1-RNAi prevented activation of RAS after 8-oxoG challenge (Fig. 5D, lower panel). Next, RNAs isolated (at 60 min) from OGG1-deficient mice (n = 5) and mRNA levels of Cxcl1 and Cxcl2 were compared to those of OGG1-proficient ones using qRT-PCR. Fig. 5F shows that Cxcl1 and Cxcl2 were increased 117-fold and 592-fold, respectively, in OGG1-proficient airways, while OGG1 depletion nearly prevented their expression (***p < 0.001).
Discussion
Oxidative modifications of biomolecules, including DNA, are inevitable consequences of environmental exposures and cellular metabolic processes. DNA base and strand lesions impose a continuous threat to the genomic integrity of all organisms. Among such lesions, one of the most abundant is 8-oxoG, and its accumulation in DNA has been linked to carcinogenesis and to various age-related diseases and aging processes [51]; however, the mechanism is not fully elucidated. Results from the present study show that exposure of airways to the specific product of OGG1-BER, 8-oxoG free base, induced differential gene expression associated with various signaling pathways and biological processes. Although a great amount of work remains to be completed in defining the precise role of OGG1-BER-driven cellular physiological responses, these results suggest that it may not be the genomic accumulation of 8-oxoG that has biological significance, but rather its release from DNA. Elucidation of the significance of each induced pathway identified in this study will be useful for rational design of therapeutics to treat diseases and delay aging processes that previously linked DNA damage/repair.
Upon challenge, Ogg1-/- mice exhibit decreased innate and allergic inflammatory responses and increased susceptibility to infections and metabolic disorders [15, 16, 17, 22]. These disorders have been linked to a lack of signaling by 8-oxoG base in some studies [17, 18, 20, 22]. To extend our understanding, we examined the overall impact of 8-oxoG challenge on lungs at the whole-transcriptome level utilizing RNA-Seq and bioinformatics for analysis. The 8-oxoG base is rapidly up taken by cells [20], and, thus, instead of inducing oxidative stress and consequent DNA damage repair in these studies, we challenged animals with 8-oxoG base, to mimic OGG1-initiated BER, to avoid signaling by ROS and generation of oxidatively modified molecules. RNA sequencing identified a total of 23,337 transcripts, out of which cumulatively (at 30, 60 and 120 min) 2,381 were differentially expressed (≥ 3-fold change) after 8-oxoG challenge. Changes in transcript levels were continuously increased from 30 min and peaked at 60 min. Preliminary characterization of bronchoalveolar fluids derived from challenged lungs showed that from 120 min on de novo synthesized soluble mediators (e.g., CXCL1, TNF-α) were present in high titers. These data were similar to those in our previous studies [22, 29]. Therefore, to define the primary effects of OGG1-BER (8-oxoG challenge) on global gene expression, RNA-Seq data derived from the 60-min-time point were analyzed in detail. At 60 min post-challenge, 570 transcripts were up-regulated, while 1022 were down-regulated. The down-regulated genes did not represent any biological processes based on the PANTHER data base.
The PANTHER classification system [25], identified 443 (out of 570) mRNAs at 60 min post 8-oxoG exposure distributed in various GO categories. In addition, there were 71 microRNAs and 55 non-coding, full-length RNAs identified according to Mouse Genome Informatics. The functions of these transcripts will be examined in future studies. The encoded proteins from mRNAs are significant contributors to various biological processes (e.g., homeostasis, immune system processes, regulation of liquid surface tension, metabolic), and signaling pathways (e.g., driven by chemokines/cytokines, gonadotropin-releasing hormone receptor, integrins and interleukins). The most overrepresented protein classes were cytokines, chemokines, interleukins, integrins, and the microtubule cytoskeletal family. Experimental validation of these biological processes and signaling pathways will be undertaken in future studies. These results points to the role of OGG1-BER in wide-range of cellular responses, which primarily associated with immune and homeostatic processes. One may also suggest that OGG1-BER can trigger immune defense upon environmental challenge.
Biological processes such as liquid surface tension and homeostatic/metabolic processes are in a broad sense, associated with the regulation of cellular homeostasis. It is well-documented that exposure to various environmental chemicals, and physical and biological agents dysregulate cellular/tissue physiological states (redox, osmolarity, energy demand), induces oxidative DNA damage, and activates repair pathways including OGG1-BER. We speculate that OGG1-BER and consequent activation of signaling pathways are links between environmental exposures and cellular responses, including reestablishment of homeostasis. Indeed, genes upregulated by 8-oxoG challenge (e.g., ATPases participating in H+/K+ exchange, GTPase alpha polypeptide, class 1 type 8B ATPases, C-C chemokines, collagen types II, IX, X, XI or G protein-coupled receptors) were previously associated with homeostatic states (PANTHER). Another, significantly overrepresented biological process was that of immune system processes, mediated by chemokines and cytokines. This finding leads to the likely scenario that 8-oxoG challenge may not be inducing homeostasis per se, but rather inducing inflammatory responses. One may argue that this only seems to be contradictory, in that an immune response is an attempt to eliminate environmental insult and thus re-establish homeostatic pre-exposure conditions. By the same token, our data also imply that an 8-oxoG base may be classified as an “alarmin,” an endogenous molecule released by OGG1-BER upon oxidative challenge of DNA. In general, alarmins play a role in the various tissue homeostatic processes and benefit a host. However, there is evidence that excessive release of alarmin(s) contributes to dysregulated processes such as inflammation. Indeed, challenge with the 8-oxoG base (but not with guanine, 8-oxodeoxyguanosine, or 2,6-diamino-4-hydroxy-5-formamidopyrimidine, a ring-opened derivative of 8-oxoG) or when the base was released from DNA by OGG1-BER induced activation of NF-κB pathway, expression of chemokines, cytokines and recruitment of neutrophils to the airways as previously documented [22].
Intriguing studies have documented changes in whole-body energy homeostasis and susceptibility to obesity of OGG1-BER-deficient (Ogg1-/-) mice [17]. Specifically, when Ogg1-/- mice were kept on a high-fat diet, they showed increased plasma insulin levels, impaired glucose tolerance, enhanced adiposity and hepatic steatosis [17]. In addition to other explanations for these observations authors proposed that increased susceptibility to obesity of Ogg1-/- mice could be due to a lack of 8-oxoG base released from DNA by OGG1-BER. Therefore, we examined whether challenge with 8-oxoG base modulated expression of genes participating in metabolic processes in lungs. Indeed, we identified genes involved in lipid, fatty acid, carbohydrate, protein, and nucleobase metabolism; however, due to the large number of genes cataloged in each subcategory in the PANTHER data base, number of upregulated genes in our data sets did not reach statistical significance. Future studies are warranted to sort out the roles of 8-oxoG (OGG1•8-oxoG complex) in regulating gene expression involved in metabolic processes. In our present studies, we did not utilize Ogg1-/- mice, as we have documented both OGG1 and 8-oxoG base are required for activation of small GTPases and down-stream signaling [18, 20, 22].
Studies undertaken previously show that OGG1 binds the repair product, 8-oxoG base (but not guanine or 8-oxodesoxyguanosine), with high affinity (Kd = 0.56 nM) and that the complex physically interacts with and activates small GTPases in vitro and in cultured cells [18, 20, 21]. Therefore, critically addressing the means by which 8-oxoG challenge of OGG1-expressing lungs could induce biological responses, we asked whether it could be linked to small GTPases. Small GTPases play an important role in various biological processes by regulating the expression of multiple genes [47]. To examine the role of the most relevant GTPase in signaling 8-oxoG-induced gene expression, we matched our RNA-Seq datasets to a published list of RAS signaling-dependent genes [49]. As described in the “Results” section, among the 147 genes for which the expression has been reported to be dependent on RAS signaling, only 28 (out of 105) were up-regulated and 26 (out of 42) down-regulated by 8-oxoG challenge. In theory, all RAS-dependent genes should be modulated, as 8-oxoG challenge induces a robust RAS activation; however, this was not the case. Although we cannot fully explain these observations, these data suggest that gene activation was dependent on cell types and organ and whether cells were normal or had an oncogenic phenotype. Indeed, the RAS signature set of genes was developed based on cell lines mostly isolated from tumors [49]. It is also possible that the OGG1•8-oxoG complex induced a subset of RAS-dependent genes that mediate signaling cascades and biological processes primarily serving cellular/host immune homeostasis.
Challenging airways with 8-oxoG increased the levels of GTP-bound RAS in an OGG1-dependent fashion similar to that in previous studies [18, 20, 22]. Activation of RAS was observed as early as 15 min, and mRNA levels were increased from 30 min on, so we propose that these events are not only overlapping, but are also etiologically related. In support, Sparmann and coworkers showed that activated RAS led to upregulation of C-X-C chemokines [53]. Another study documented that introduction of Kras into the bronchiolar epithelium is associated with robust pro-inflammatory gene expression (e.g., MIP-2, KC, MCP-1 and LIX chemokines) [54]. These data are consistent with an increase in RAS-GTP and Cxcl1 and Cxcl2 mRNA levels after 8-oxoG challenge. Moreover, both the activation of RAS and an increase in mRNA levels of Cxcl1 and Cxcl2 were significantly prevented in OGG1-depleted airways [22].
For RNA-sequencing the entire mouse lung was used to isolate RNA by raising the possibility that fold changes in mRNA and transcript levels could be different (actually be higher) in the airway epithelium which constitutes ∼25% of total lung cells [52]. At this point we cannot exclude the contribution of other cells types (e.g., mast and dendritic cells, which represent <1% of all cells in the lung epithelium). Resident macrophages are affected by 8-oxoG challenge (unpublished data); however, they were removed before RNA extraction by lavage of airways with ice-cold PBS. Therefore, we speculate that the observed changes in mRNA levels primarily represent the response of airway epithelium to 8-oxoG challenge. In support, a) 8-oxoG is rapidly taken up by cells [20], primarily by the epithelium in our model; b) RAS GTPases were activated from 15 min on; and c) from 30 min on (60-min maximum) robust changes in gene expression were observed. This hypothesis is supported by the overrepresentation of immune response processes and is in line with the innate immune defense functions of airway epithelium [52].
In summary, we document for the first time that challenge of lungs with 8-oxoG base induced the expression of large numbers of transcripts, contributing to the activation of various signaling pathways and biological processes, primarily triggering cellular homeostatic and immune responses. Although future studies are required, these data are in accord with decreased innate and allergic immune responses, and the susceptibility of Ogg1 null mice to obesity. We may speculate that 8-oxoG serves as an endogenously generated alarmin that benefits the host, while its excessive release contributes to dysregulated processes. Finally, we raise the possibility that OGG1-BER and consequent activation of signaling pathways could be a link between environmental exposures and cellular responses.
Research Highlights.
8-Oxoguanine DNA glycosylase1 (OGG1) prevents accumulation of genomic 8-oxoguanine
The released 8-oxoG base, in complex with OGG1, induces signaling via small GTPases
OGG1-BER signaling increases levels of 2381 transcripts, out of which 443 are mRNAs
These genes regulate essential biological processes and signaling pathways
OGG1-BER signaling may be a link between DNA damage-repair and cellular responses
Acknowledgments
This work was supported by grants NIEHS RO1 ES018948 (IB), NIAID/AI062885 (ARB, IB), NHLBI Proteomic Center, N01HV00245 (IB, SS, Dr. A. Kurosky director); NIEHS Center Grant P30 ES006676; International Science-Technology Collaboration Foundation (20120728) of Jilin Province in China (XB), the European Union and the European Social Fund, TAMOP 4.2.2.A-11/1/KONV-2012-2023 (AB). L. Aguilera-Aguirre is an Environmental Toxicology Research Training Fellow (NIEHS T32 ES007254-22). We thank Mardelle Susman (Department of Microbiology and Immunology) for critically editing the manuscript and Dr. David Konkel (Institute for Translational Sciences, UTMB) both for his scientific input and for editing the manuscript. We also thank unknown reviewers for constructive comments that helped us to improve the quality of the manuscript.
Abbreviations
- 8-oxoG
7,8-dihydro-8-oxoguanine
- BALF
bronchoalveolar lavage fluid
- BER
base-excision repair
- CASAVA
Consensus Assessment of Sequence and Variation
- COPD
chronic obstructive pulmonary disease
- C-X-C
C-X-C motif chemokines
- C-C
C-C motif chemokines
- i.n.
intranasal
- GENE-E
is a matrix visualization and analysis platform
- GEO
Gene Expression Omnibus
- GO
gene ontology
- Kras
Kirsten rat sarcoma viral oncogene homolog
- LPS
lipopolysaccharide
- miRNA
micro RNA(s)
- OGG1
8-oxoguanine DNA glycosylase-1
- OGG1-BER
OGG1-initiated DNA base excision repair
- PANTHER
Protein ANalysis THrough Evolutionary Relationships
- RAC1
Ras-related C3 botulinum toxin substrate 1
- RHOA
Ras homolog gene family, member A
- RPKM
reads per kb transcript per million
- RNA-Seq
RNA sequencing
- Ras
rat viral sarcoma oncogene homolog
- RNAi
RNA interference
- ROS
reactive oxygen species
- TNF-α
tumor necrosis factor alpha
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacob KD, Noren Hooten N, Trzeciak AR, Evans MK. Markers of oxidant stress that are clinically relevant in aging and age-related disease. Mech Ageing Dev. 2013;134:139–157. doi: 10.1016/j.mad.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanvah S, Joseph J, Schuster GB, Barnett RN, Cleveland CL, Landman U. Oxidation of DNA: damage to nucleobases. Acc Chem Res. 2010;43:280–287. doi: 10.1021/ar900175a. [DOI] [PubMed] [Google Scholar]
- 4.Steenken S, Jovanovic S. How Easily Oxidizable Is DNA? One-Electron Reduction Potentials of Adenosine and Guanosine Radicals in Aqueous Solution. J Am Chem Soc. 1997;119:617–618. [Google Scholar]
- 5.Mitra S, Hazra TK, Roy R, Ikeda S, Biswas T, Lock J, Boldogh I, Izumi T. Complexities of DNA base excision repair in mammalian cells. Mol Cells. 1997;7:305–312. [PubMed] [Google Scholar]
- 6.Dizdaroglu M. Substrate specificities and excision kinetics of DNA glycosylases involved in base-excision repair of oxidative DNA damage. Mutat Res. 2003;531:109–126. doi: 10.1016/j.mrfmmm.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Svoboda P, Maekawa M, Kawai K, Tominaga T, Savela K, Kasai H. Urinary 8-hydroxyguanine may be a better marker of oxidative stress than 8-hydroxydeoxyguanosine in relation to the life spans of various species. Antioxid Redox Signal. 2006;8:985–992. doi: 10.1089/ars.2006.8.985. [DOI] [PubMed] [Google Scholar]
- 8.Dedon PC, DeMott MS, Elmquist CE, Prestwich EG, McFaline JL, Pang B. Challenges in developing DNA and RNA biomarkers of inflammation. Biomark Med. 2007;1:293–312. doi: 10.2217/17520363.1.2.293. [DOI] [PubMed] [Google Scholar]
- 9.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci U S A. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minowa O, Arai T, Hirano M, Monden Y, Nakai S, Fukuda M, Itoh M, Takano H, Hippou Y, Aburatani H, Masumura K, Nohmi T, Nishimura S, Noda T. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc Natl Acad Sci U S A. 2000;97:4156–4161. doi: 10.1073/pnas.050404497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stuart JA, Bourque BM, de Souza-Pinto NC, Bohr VA. No evidence of mitochondrial respiratory dysfunction in OGG1-null mice deficient in removal of 8-oxodeoxyguanine from mitochondrial DNA. Free Radic Biol Med. 2005;38:737–745. doi: 10.1016/j.freeradbiomed.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Arai T, Kelly VP, Minowa O, Noda T, Nishimura S. The study using wild-type and Ogg1 knockout mice exposed to potassium bromate shows no tumor induction despite an extensive accumulation of 8-hydroxyguanine in kidney DNA. Toxicology. 2006;221:179–186. doi: 10.1016/j.tox.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Mabley JG, Pacher P, Deb A, Wallace R, Elder RH, Szabo C. Potential role for 8-oxoguanine DNA glycosylase in regulating inflammation. Faseb J. 2005;19:290–292. doi: 10.1096/fj.04-2278fje. [DOI] [PubMed] [Google Scholar]
- 14.Touati E, Michel V, Thiberge JM, Ave P, Huerre M, Bourgade F, Klungland A, Labigne A. Deficiency in OGG1 protects against inflammation and mutagenic effects associated with H. pylori infection in mouse. Helicobacter. 2006;11:494–505. doi: 10.1111/j.1523-5378.2006.00442.x. [DOI] [PubMed] [Google Scholar]
- 15.Bacsi A, Aguilera-Aguirre L, Szczesny B, Radak Z, Hazra TK, Sur S, Ba X, Boldogh I. Down-regulation of 8-oxoguanine DNA glycosylase 1 expression in the airway epithelium ameliorates allergic lung inflammation. DNA Repair (Amst) 2013;12:18–26. doi: 10.1016/j.dnarep.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li G, Yuan K, Yan C, Fox J, 3rd, Gaid M, Breitwieser W, Bansal AK, Zeng H, Gao H, Wu M. 8-Oxoguanine-DNA glycosylase 1 deficiency modifies allergic airway inflammation by regulating STAT6 and IL-4 in cells and in mice. Free Radic Biol Med. 2012;52:392–401. doi: 10.1016/j.freeradbiomed.2011.10.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sampath H, Vartanian V, Rollins MR, Sakumi K, Nakabeppu Y, Lloyd RS. 8-Oxoguanine DNA glycosylase (OGG1) deficiency increases susceptibility to obesity and metabolic dysfunction. PLoS One. 2012;7:e51697. doi: 10.1371/journal.pone.0051697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boldogh I, Hajas G, Aguilera-Aguirre L, Hegde ML, Radak Z, Bacsi A, Sur S, Hazra TK, Mitra S. Activation of ras signaling pathway by 8-oxoguanine DNA glycosylase bound to its excision product, 8-oxoguanine. J Biol Chem. 2012;287:20769–20773. doi: 10.1074/jbc.C112.364620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.German P, Szaniszlo P, Hajas G, Radak Z, Bacsi A, Hazra TK, Hegde ML, Ba X, Boldogh I. Activation of cellular signaling by 8-oxoguanine DNA glycosylase-1-initiated DNA base excision repair. DNA Repair (Amst) 2013;12:856–863. doi: 10.1016/j.dnarep.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hajas G, Bacsi A, Aguilera-Aguirre L, Hegde ML, Tapas KH, Sur S, Radak Z, Ba X, Boldogh I. 8-Oxoguanine DNA glycosylase-1 links DNA repair to cellular signaling via the activation of the small GTPase Rac1. Free Radic Biol Med. 2013;61C:384–394. doi: 10.1016/j.freeradbiomed.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo J, Hosoki K, Bacsi A, Radak Z, Hegde ML, Sur S, Hazra TK, Brasier AR, Ba X, Boldogh I. 8-Oxoguanine DNA glycosylase-1-mediated DNA repair is associated with Rho GTPase activation and alpha-smooth muscle actin polymerization. Free Radic Biol Med. 2014:430–438. doi: 10.1016/j.freeradbiomed.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aguilera-Aguirre L, Bacsi A, Radak Z, Hazra TK, Mitra S, Sur S, Brasier AR, Ba X, Boldogh I. Innate Inflammation Induced by the 8-Oxoguanine DNA Glycosylase-1-KRAS-NF-kappaB Pathway. J Immunol. 2014:4643–4653. doi: 10.4049/jimmunol.1401625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aguilera-Aguirre L, Bacsi A, Saavedra-Molina A, Kurosky A, Sur S, Boldogh I. Mitochondrial dysfunction increases allergic airway inflammation. J Immunol. 2009;183:5379–5387. doi: 10.4049/jimmunol.0900228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dillies MA, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N, Keime C, Marot G, Castel D, Estelle J, Guernec G, Jagla B, Jouneau L, Laloe D, Le Gall C, Schaeffer B, Le Crom S, Guedj M, Jaffrezic F. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 2013;14:671–683. doi: 10.1093/bib/bbs046. [DOI] [PubMed] [Google Scholar]
- 25.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mi H, Thomas P. PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol Biol. 2009;563:123–140. doi: 10.1007/978-1-60761-175-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yadav UC, Ramana KV, Aguilera-Aguirre L, Boldogh I, Boulares HA, Srivastava SK. Inhibition of aldose reductase prevents experimental allergic airway inflammation in mice. PLoS One. 2009;4:e6535. doi: 10.1371/journal.pone.0006535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yadav UC, Naura AS, Aguilera-Aguirre L, Ramana KV, Boldogh I, Sur S, Boulares HA, Srivastava SK. Aldose reductase inhibition suppresses the expression of Th2 cytokines and airway inflammation in ovalbumin-induced asthma in mice. J Immunol. 2009;183:4723–4732. doi: 10.4049/jimmunol.0901177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ba X, Aguilera-Aguirre L, Rashid QT, Bacsi A, Radak Z, Sur S, Hosoki K, Hegde ML, Boldogh I. The Role of 8-Oxoguanine DNA Glycosylase-1 in Inflammation. Int J Mol Sci. 2014;15:16975–16997. doi: 10.3390/ijms150916975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. 2013;13:159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 31.Smith DF, Galkina E, Ley K, Huo Y. GRO family chemokines are specialized for monocyte arrest from flow. Am J Physiol Heart Circ Physiol. 2005;289:H1976–1984. doi: 10.1152/ajpheart.00153.2005. [DOI] [PubMed] [Google Scholar]
- 32.de Jager SC, Bot I, Kraaijeveld AO, Korporaal SJ, Bot M, van Santbrink PJ, van Berkel TJ, Kuiper J, Biessen EA. Leukocyte-specific CCL3 deficiency inhibits atherosclerotic lesion development by affecting neutrophil accumulation. Arterioscler Thromb Vasc Biol. 2013;33:e75–83. doi: 10.1161/ATVBAHA.112.300857. [DOI] [PubMed] [Google Scholar]
- 33.Schutyser E, Struyf S, Van Damme J. The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor Rev. 2003;14:409–426. doi: 10.1016/s1359-6101(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 34.Wang CQ, Akalu YT, Suarez-Farinas M, Gonzalez J, Mitsui H, Lowes MA, Orlow SJ, Manga P, Krueger JG. IL-17 and TNF synergistically modulate cytokine expression while suppressing melanogenesis: potential relevance to psoriasis. J Invest Dermatol. 2013;133:2741–2752. doi: 10.1038/jid.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suwara MI, Green NJ, Borthwick LA, Mann J, Mayer-Barber KD, Barron L, Corris PA, Farrow SN, Wynn TA, Fisher AJ, Mann DA. IL-1alpha released from damaged epithelial cells is sufficient and essential to trigger inflammatory responses in human lung fibroblasts. Mucosal Immunol. 2014;7:684–693. doi: 10.1038/mi.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dinarello CA. Overview of the interleukin-1 family of ligands and receptors. Semin Immunol. 2013;25:389–393. doi: 10.1016/j.smim.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Marini M, Vittori E, Hollemborg J, Mattoli S. Expression of the potent inflammatory cytokines, granulocyte-macrophage-colony-stimulating factor and interleukin-6 and interleukin-8, in bronchial epithelial cells of patients with asthma. J Allergy Clin Immunol. 1992;89:1001–1009. doi: 10.1016/0091-6749(92)90223-o. [DOI] [PubMed] [Google Scholar]
- 38.Gaur U, Aggarwal BB. Regulation of proliferation, survival and apoptosis by members of the TNF superfamily. Biochem Pharmacol. 2003;66:1403–1408. doi: 10.1016/s0006-2952(03)00490-8. [DOI] [PubMed] [Google Scholar]
- 39.Bingle CD, Bingle L. Characterisation of the human plunc gene, a gene product with an upper airways and nasopharyngeal restricted expression pattern. Biochim Biophys Acta. 2000;1493:363–367. doi: 10.1016/s0167-4781(00)00196-2. [DOI] [PubMed] [Google Scholar]
- 40.Ipseiz N, Uderhardt S, Scholtysek C, Steffen M, Schabbauer G, Bozec A, Schett G, Kronke G. The nuclear receptor Nr4a1 mediates anti-inflammatory effects of apoptotic cells. J Immunol. 2014;192:4852–4858. doi: 10.4049/jimmunol.1303377. [DOI] [PubMed] [Google Scholar]
- 41.Patterson KI, Brummer T, O'Brien PM, Daly RJ. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem J. 2009;418:475–489. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- 42.Panto MR, Zappala A, Tuorto F, Cicirata F. Role of the Otx1 gene in cell differentiation of mammalian cortex. Eur J Neurosci. 2004;19:2893–2902. doi: 10.1111/j.0953-816X.2004.03326.x. [DOI] [PubMed] [Google Scholar]
- 43.Stow LR, Richards J, Cheng KY, Lynch IJ, Jeffers LA, Greenlee MM, Cain BD, Wingo CS, Gumz ML. The circadian protein period 1 contributes to blood pressure control and coordinately regulates renal sodium transport genes. Hypertension. 2012;59:1151–1156. doi: 10.1161/HYPERTENSIONAHA.112.190892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shouval DS, Biswas A, Goettel JA, McCann K, Conaway E, Redhu NS, Mascanfroni ID, Al Adham Z, Lavoie S, Ibourk M, Nguyen DD, Samsom JN, Escher JC, Somech R, Weiss B, Beier R, Conklin LS, Ebens CL, Santos FG, Ferreira AR, Sherlock M, Bhan AK, Muller W, Mora JR, Quintana FJ, Klein C, Muise AM, Horwitz BH, Snapper SB. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity. 2014;40:706–719. doi: 10.1016/j.immuni.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klevernic IV, Martin NM, Cohen P. Regulation of the activity and expression of ERK8 by DNA damage. FEBS Lett. 2009;583:680–684. doi: 10.1016/j.febslet.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 46.Johnson DS, Chen YH. Ras family of small GTPases in immunity and inflammation. Curr Opin Pharmacol. 2012;12:458–463. doi: 10.1016/j.coph.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heo J. Redox control of GTPases: from molecular mechanisms to functional significance in health and disease. Antioxid Redox Signal. 2011;14:689–724. doi: 10.1089/ars.2009.2984. [DOI] [PubMed] [Google Scholar]
- 48.Reedquist KA, Tak PP. Signal transduction pathways in chronic inflammatory autoimmune disease: small GTPases. Open Rheumatol J. 2012;6:259–272. doi: 10.2174/1874312901206010259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loboda A, Nebozhyn M, Klinghoffer R, Frazier J, Chastain M, Arthur W, Roberts B, Zhang T, Chenard M, Haines B, Andersen J, Nagashima K, Paweletz C, Lynch B, Feldman I, Dai H, Huang P, Watters J. A gene expression signature of RAS pathway dependence predicts response to PI3K and RAS pathway inhibitors and expands the population of RAS pathway activated tumors. BMC Med Genomics. 2010;3:26. doi: 10.1186/1755-8794-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al-Afaleg NO, Al-Senaidy A, El-Ansary A. Oxidative stress and antioxidant status in Saudi asthmatic patients. Clin Biochem. 2011;44:612–617. doi: 10.1016/j.clinbiochem.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 51.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. Faseb J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 52.Strengert M, Knaus UG. Analysis of epithelial barrier integrity in polarized lung epithelial cells. Methods Mol Biol. 2011;763:195–206. doi: 10.1007/978-1-61779-191-8_13. [DOI] [PubMed] [Google Scholar]
- 53.Sparmann A, Bar-Sagi D. Ras-induced interleukin-8 expression plays a critical role in tumor growth and angiogenesis. Cancer Cell. 2004;6:447–458. doi: 10.1016/j.ccr.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 54.Ji H, Houghton AM, Mariani TJ, Perera S, Kim CB, Padera R, Tonon G, McNamara K, Marconcini LA, Hezel A, El-Bardeesy N, Bronson RT, Sugarbaker D, Maser RS, Shapiro SD, Wong KK. K-ras activation generates an inflammatory response in lung tumors. Oncogene. 2006;25:2105–2112. doi: 10.1038/sj.onc.1209237. [DOI] [PubMed] [Google Scholar]


