Abstract
Purpose
Although tyrosine kinase inhibitors (TKI) can be effective therapies for leukemia, they fail to fully eliminate leukemic cells and achieve durable remissions for many patients with advanced BCR-ABL+ leukemias or acute myeloid leukemias (AML). Through a large-scale synthetic lethal RNAi screen, we identified pyruvate dehydrogenase, the limiting enzyme for pyruvate entry into the mitochondrial tricarboxylic acid cycle, as critical for the survival of chronic myeloid leukemia cells upon BCR-ABL inhibition. Here we examined the role of mitochondrial metabolism in the survival of Ph+ leukemia and AML upon TK inhibition.
Experimental Design
Ph+ cancer cell lines, AML cell lines, leukemia xenografts, cord blood, patient samples were examined.
Results
We showed that the mitochondrial ATP-synthase inhibitor oligomycin-A greatly sensitized leukemia cells to TKI in vitro. Surprisingly, oligomycin-A sensitized leukemia cells to BCR-ABL inhibition at concentrations 100–1000-fold below those required for inhibition of respiration. Oligomycin-A treatment rapidly led to mitochondrial membrane depolarization and reduced ATP levels, and promoted superoxide production and leukemia cell apoptosis when combined with TKI. Importantly, oligomycin-A enhanced elimination of BCR-ABL+ leukemia cells by TKI in a mouse model and in primary blast crisis CML samples. Moreover, oligomycin-A also greatly potentiated the elimination of FLT3-dependent AML cells when combined with a FLT3 TKI, both in vitro and in vivo.
Conclusions
TKI therapy in leukemia cells creates a novel metabolic state that is highly sensitive to particular mitochondrial perturbations. Targeting mitochondrial metabolism as an adjuvant therapy could therefore improve therapeutic responses to TKI for patients with BCR-ABL+ and FLT3ITD leukemias.
Keywords: oligomycin-A, pyruvate dehydrogenase, mitochondria, leukemia, imatinib, dasatinib, quizartinib
Introduction
Chronic myeloid leukemia (CML) is a myeloproliferative disorder characterized by a translocation (t(9;22)(q34;q11)) producing the Philadelphia chromosome (Ph) (1). The resultant fusion protein, BCR-ABL, is causative for the disease, and is also present in 20–30% of acute lymphoblastic leukemia (ALL) (2). While the treatment of CML with BCR-ABL tyrosine kinase inhibitors (TKI), like imatinib mesylate and dasatinib, has revolutionized therapy for these leukemias, BCR-ABL TKI typically fail to fully eliminate the leukemia, and thus most patients require lifetime therapy. Moreover, adult Ph+ ALL and advanced stage CML patients exhibit only transient responses to BCR-ABL TKI (3).
Acute myeloid leukemia (AML) is a heterogeneous group of leukemias. AML is the most common adult leukemia and the second most common childhood leukemia (4). About a third of AMLs express mutations in the FMS-like tyrosine kinase 3 receptor (FLT3). The presence of activating FLT3 internal tandem duplications (FLT3ITD) is associated with reduced overall survival (5, 6). Like BCR-ABL, FLT3 signaling provides pro-survival and anti-proliferative signals to AML cells. Although FLT3 TKI as monotherapy have shown promising initial responses in AML, clinical trials with FLT3 inhibitors have so far failed to show durable responses in AML (7–9).
Metabolic adaptations are common in cancer cells, and are thought to enable sustained high rates of proliferation (10, 11). Such adaptations include increased rates of glucose uptake, increased aerobic glycolysis and glutaminolysis, and increased use of the pentose-phosphate pathway, resulting in a decreased dependence on the tricarboxylic acid (TCA) cycle for energy production (11). These changes lead to a “glycolytic phenotype”, in which glycolysis is the main source of energy in the cell and the end product of metabolism is lactate fermentation, also known as the Warburg effect. Bypassing the TCA cycle provides a constant supply of metabolic intermediates for macromolecule biosynthesis without jeopardizing an adequate source of ATP. BCR-ABL confers this phenotype to CML cells, in part via activation of phosphoinositide 3-kinases (PI3K)/protein kinase B (AKT) downstream signaling (12, 13).
Treatment of CML cells with imatinib leads to decreased glucose uptake by suppressing glycolysis, causing the translocation of GLUT-1 transporters away from the surface of CML cells, increasing the flux of residual glucose through the mitochondrial TCA cycle, restricting de novo nucleotide production, and inhibiting fatty acid synthesis (14–16). Furthermore, imatinib-resistance can be mediated in part by HIF1α-dependent upregulation of glycolysis (17, 18).
The role of the TCA cycle and oxidative phosphorylation in the survival of Ph+ and FLT3ITD leukemia is not fully characterized. Oxythiamine, a thiamine analog that inhibits thiamine-dependent TCA cycle and pentose-phosphate pathway enzymes, enhances the efficacy of imatinib towards imatinib-resistant CML cells in vitro and reduces tumor burden in a mouse model of BCR-ABL+ leukemia (17). Recent studies also indicate that AML cells have altered mitochondrial dependencies, including uncoupling of oxygen consumption from ATP production and enhanced dependence on mitochondrial translation (19, 20). These studies suggest that TK-dependent leukemia cells may exhibit specific metabolic dependencies, and that the characterization of these dependencies could reveal processes that can be exploited therapeutically.
We previously performed a large-scale loss of function RNAi screen to identify genes whose inhibition synergizes with imatinib to kill Ph+ leukemia cells (21). This screen identified multiple enzymes involved in glucose metabolism as synthetic lethal. Here, we demonstrate that Ph+ and FLT3ITD leukemia cells become exquisitely sensitive to perturbations in mitochondrial function, specifically upon treatment with TKI.
Materials and Methods
Cell Culture and Generation of Knockdown Cell Lines
KBM7 and KBM5 CML cells were obtained from M. Beran at MD Anderson, Ba/F3 from B. Deininger at Oregon Health & Science University, MV-4–11, MOLM-13 and Kasumi-1 cells from R. Arceci at Phoenix Children’s Hospital, NOMO-1 and OCI-AML-3, K562 and SUP-B15 cells were purchased from the DSMZ and ATCC respectively. Cells were grown in standard culture conditions. MV-4–11 cells express the homozygous insertion D600_L601>HVDFREYEYD in FLT3, while MOLM13 cells express the heterozygous insertion F601_K602>REYEYDL. Kasumi-1 cells express the ligand-independent N822K c-kit activating mutation. Lentiviruses generated using pLKO.1 vectors (Sigma-Aldrich, Table S1) were used to transduce cells as previously described (21). Cells were selected in 2.5 µg/mL puromycin. Ba/F3 murine pro-B cells expressing BCR-ABL and p210 BCR-ABL+ ARF−/− ALL cells were generated as previously described (21–23). Cell lines were authenticated by short tandem repeat examination and tested negative for mycoplasma using the iNtRON e-Myco plus Mycoplasm PCR detection kit in July 2012.
Cell Viability Experiments
For cell line growth curve experiments, 2×104 cells were seeded in 96-well plates and treated with the indicated drugs. After 1–3 days of treatment, cells were washed in PBS, and replated in fresh medium for an additional 2–5 days. At the indicated time, an aliquot was stained with 10 µg/mL propidium iodide and counted using a Beckman Coulter Quanta SC or Guava 8HT flow cytometer. For K562 clonogenic assays, after 3 days of treatment, cells were washed and plated in 1.2% methylcellulose (R&D Systems) and the number of colonies counted under a microscope 7–14 days later.
Pharmacological agents
Imatinib mesylate and dasatinib were obtained from the University of Colorado Hospital Pharmacy, lestaurtinib was obtained from Cephalon, and oligomycin-A, cytarabine, doxorubicin and 2-deoxy-D-glucose (2-DG) purchased from Sigma-Aldrich. Quizartinib (AC220) was synthesized by the University of Colorado Medicinal Chemistry Core (see supplemental methods).
Mouse Leukemia Models
All mouse experiments were approved by our Institutional Animal Care and Use Committee (protocol# 41411(07)1E). The primary human AML samples were obtained under IRB protocol # 12-0173.
Female C57BL/6 mice (4–6 week old) were obtained from the National Cancer Institute. 5×105Arf−/− BCR-ABL/GFP+ B-ALL cells in 100 µL of PBS were injected via tail vein. After 3 days, mice were started on treatment with vehicle (80 mM citric acid pH 2.1 for oral gavage (21) and PBS for intraperitoneal injections, i.p.), 10 mg/kg dasatinib (oral gavage), 100 µg/kg oligomycin-A (i.p.) or combination dasatinib and oligomycin-A. Dasatinib was prepared as previously described (21), and the 10 mg/kg dose has been shown to replicate clinical experience in humans (24). For monitoring leukemic burden in mice, blood collected into heparin was hemolyzed and stained with B220-PE and Mac1-PE-Cy7 and monitored for GFP expression. See Table S1 for antibody information. Samples were run using a Beckman Coulter Gallios Flow Cytometer and analyzed using FloJo software.
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) were obtained from The Jackson Laboratory and bred in house. The patient-xenograft sample came from a 54 year-old female with AML expressing FLT3ITD and NPM1 mutations. The vials of primary leukemia were thawed in a water bath, washed in PBS, counted by flow cytometry and resuspended in normal saline for injection. 4- to 6- wk old NSG mice were pre-treated with 25 mg/kg of busulfan (i.p.) 24 h prior to transplantation. After expansion in vivo, the secondary leukemia was harvested from the spleen and bone marrow, and subsequently transplanted into cohorts of mice for drug treatment. 3×106 cells were injected i.v. and treatment started when peripheral blast count was at least 5% (mean 7.85%). Leukemic burden was monitored by flow cytometry staining for human HLA-ABC and CD45.
Statistical Analyses
Statistical significance was determined using one-way ANOVA followed by Tukey post-test using GraphPad Prism. Kaplan-Meier survival curves were analyzed using GraphPad Prism (log-rank test). Combination indices were calculated using the median-effect principle and the Combination Index-Isobologram Theorem (CompuSyn software); and summarized in Table S2. Data shown reflect multiple independent biological replicates, not technical replicates.
See Supplementary Information for additional methods.
Results
Our large-scale RNAi screen in K562 CML blast crisis cells identified multiple metabolic enzymes (Figure S1A), including dihydrolipoamide S-acetyltransferase (DLAT), a component of the pyruvate dehydrogenase (PDH) complex, as synthetic lethal with imatinib (21). DLAT is located in the mitochondria and is a limiting component of PDH, which is essential for pyruvate entry from glycolysis into the TCA cycle.
DLAT knockdown sensitizes CML cells to BCR-ABL TKI
In order to validate DLAT as synthetic lethal with imatinib, K562 cells were transduced with shRNA constructs designed against DLAT or a non-silencing shRNA control (see Table S1 for sequences). Effective DLAT knockdown was demonstrated by Western blot (>90%; Figures 1A and S1B) and biochemically using acidification of the culture medium as a readout for increase lactic acid production, a by-product of blocking pyruvate entry into the TCA cycle (Figure S1C). In addition, DLAT knockdown cells exhibit reduced superoxide levels reflecting decreased oxidative phosphorylation (Figure S1D).
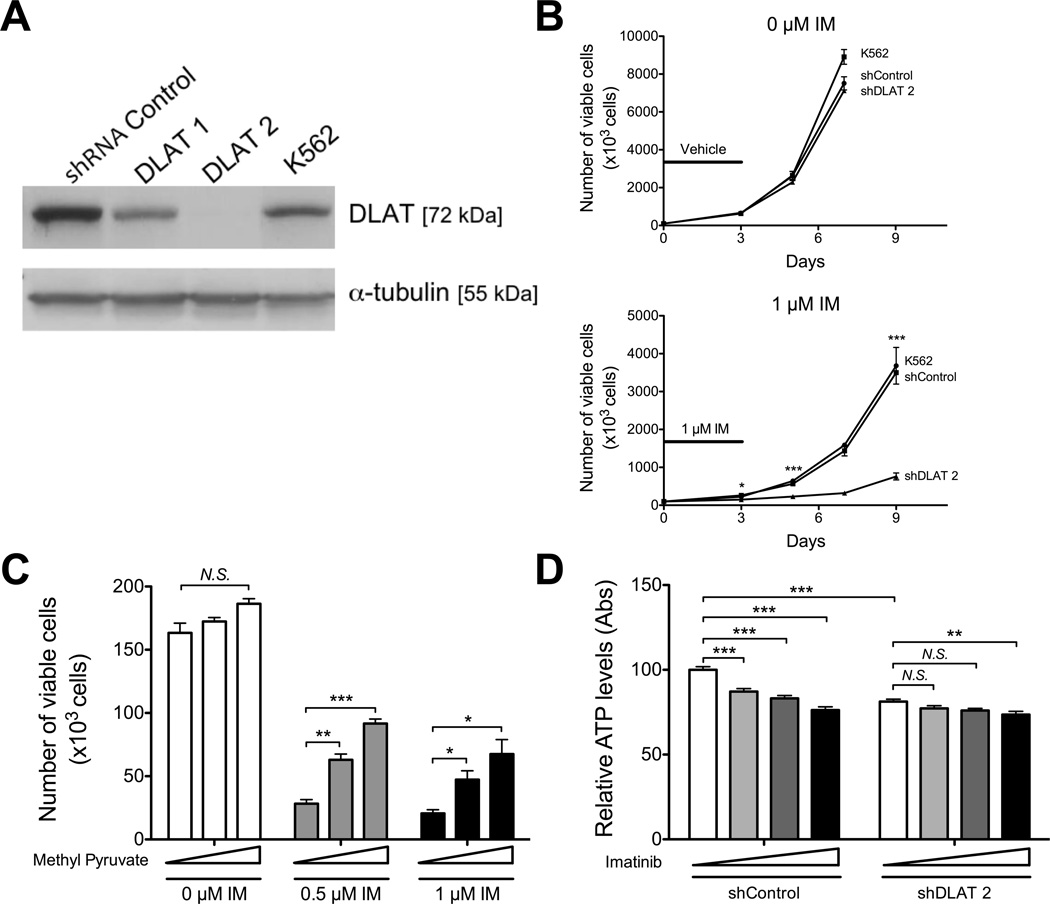
Figure 1. Mitochondrial metabolism becomes essential in TKI-treated Ph+ leukemia cells.
K562 cells were transduced with an shRNA targeting DLAT or non-targeting control and selected in puromycin. A. Expression of DLAT was assessed by Western blot probing for DLAT and α-tubulin. B. K562 parental and knockdown cell lines were treated with vehicle (top) or 1 µM imatinib (bottom) for 3 days, followed by replating the cells without drug to assess remaining proliferative potential. The black line indicates the duration of treatment. At the indicated time points, an aliquot was stained with PI and the number of viable cells determined by flow cytometry. Statistical comparison of shControl to shDLAT2 is shown. C. K562 cells were grown in medium supplemented with increasing concentrations of methyl-pyruvate (0, 5, 10 mM, indicated by triangle) for 3 days and the number of viable cells determined by flow cytometry. D. K562 parental and knockdown cell lines were treated with imatinib for 24 h (0, 1, 2, 5 µM, indicated by triangle) and assayed for ATP levels using the CellTiter-Glo assay and data were normalized to viable cell number. * indicates p<0.05, **p<0.01, and *** p<0.001.
While knocking down DLAT had no detectable impact on the proliferation of vehicle-treated K562 cells (Figures 1B top and S1E top), knockdown greatly sensitized K562 cells to imatinib treatment (Figures 1B bottom and S1E bottom). These data suggest that upon imatinib treatment, CML cells must meet their energetic and anabolic demands via increased reliance on the TCA cycle. Indeed, treatment with methyl-pyruvate, an exogenous substrate for PDH, enhances survival of imatinib-treated CML cells (Figure 1C), supporting the hypothesis that inhibition of glucose utilization is at least partially responsible for the anti-proliferative effects of imatinib but that mitochondrial pyruvate oxidation can provide some protection from BCR-ABL inhibition. Consistent with reduced entry of pyruvate into the TCA cycle for ATP production, knocking down DLAT significantly reduced basal levels of ATP in leukemia cells (Figure 1D). Treatment with imatinib also reduces the levels of ATP in these cells (Figure 1D). While treatment with imatinib at 5 µM has a modest additional effect on ATP levels in combination with DLAT knockdown, synergistic effects of imatinib and DLAT knockdown on leukemia cell killing likely derive from the multiple combined catabolic and anabolic effects of inhibiting glucose metabolism.
Oligomycin-A sensitizes leukemia cells to BCR-ABL TKIs
Given that PDH is not targeted by available therapeutics, we asked how perturbing mitochondrial metabolism using oligomycin-A might sensitize leukemia cells to imatinib. Oligomycin-A binds to mitochondrial ATP-synthase C-subunits which effectively block ATP generation (25, 26). K562 cells were treated with imatinib (0.5 or 1 µM), in the range of doses required to inhibit most BCR-ABL kinase activity (21), in combination with increasing concentrations of oligomycin-A. Notably, while 0.5–2 µM oligomycin-A synergistically eliminated K562 cells when combined with imatinib (Figure S2A–C), we found that treatment with concentrations of oligomycin-A as low as 2–6 nM was equally effective at sensitizing CML cells to imatinib-mediated killing (Figure 2A and Figure S2D). Moreover, low dose oligomycin-A prevented the recovery of imatinib-treated K562 cells after drug removal (Figure 2A bottom), and combined treatment with imatinib and oligomycin-A synergistically reduced colony formation (Figure S2E, see Table S2 for combination indices). Similar results on viability were observed in other Ph+ lines including SUP-B15 B-ALL, the CML blast crisis lines KBM7 and KBM5 (Figure S2F–H); and in two primary blast crisis CML patient samples in colony forming assays (Figure 2B and S2I). While oligomycin-A enhanced imatinib-mediated apoptosis (Figure S3A–B), it did not affect phosphorylation of targets downstream of BCR-ABL, including phospho-ERK1/2 and phospho-STAT5 (Figure S3C–E), indicating that the two drugs have independent mechanisms of action.
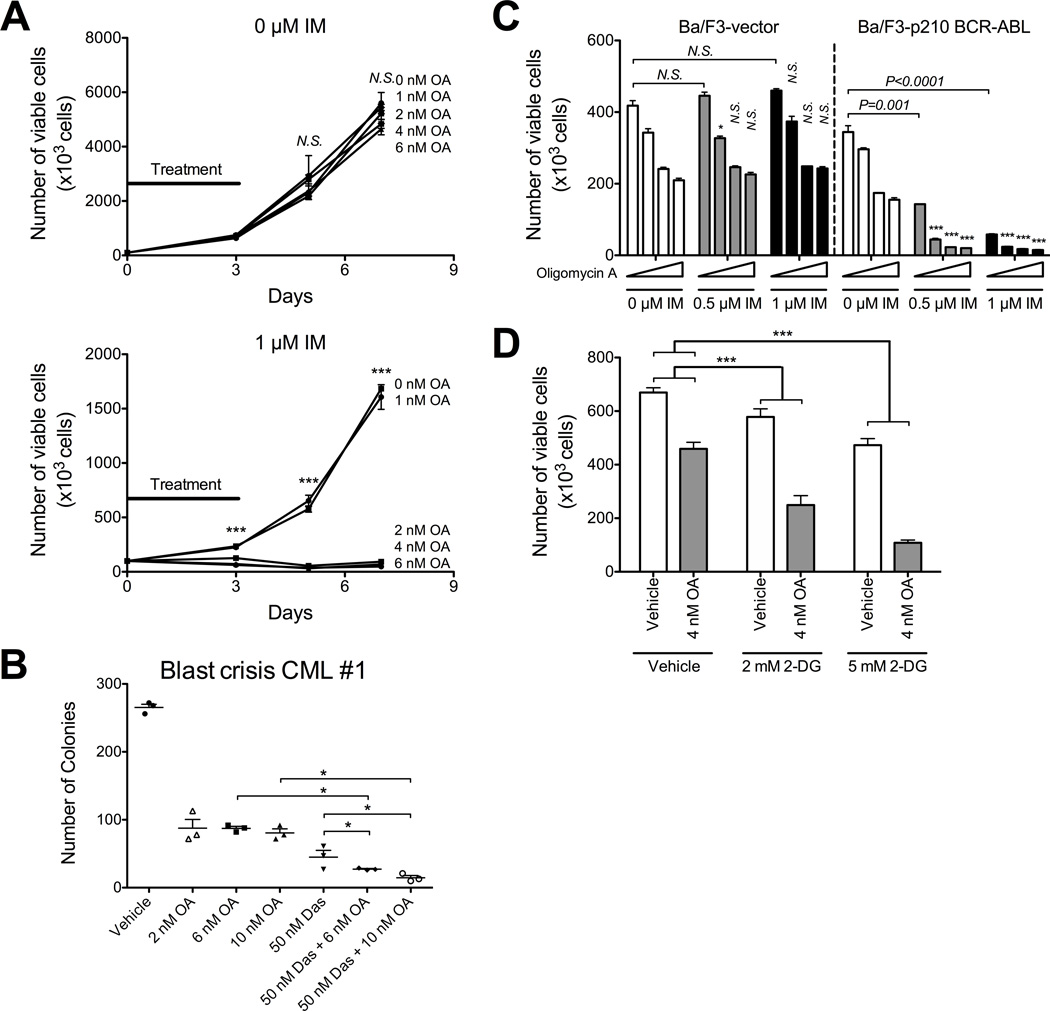
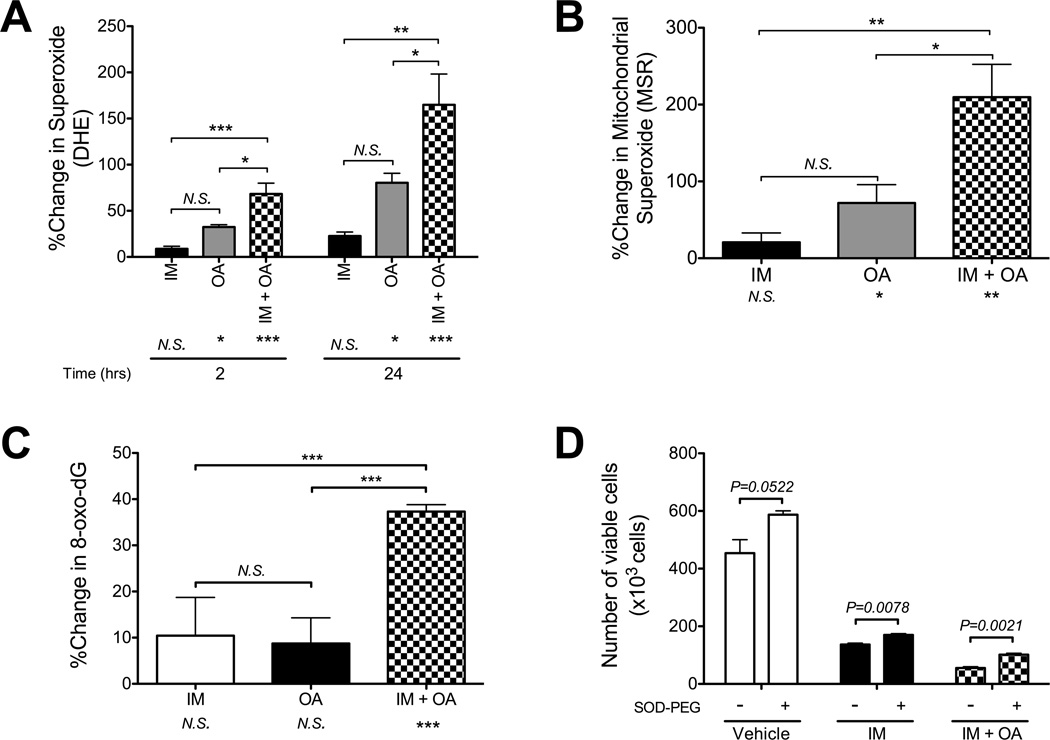
Figure 2. Oligomycin-A sensitizes BCR-ABL+ leukemia cells to BCR-ABL inhibition.
A. K562 cells were treated with the indicated concentrations of oligomycin-A (OA; 0, 1, 2, 4, 6 nM) in combination with vehicle (top) or 1 µM imatinib (bottom) for 3 days, followed by replating without drug. At the indicated time points, an aliquot was stained with PI and the number of viable cells determined by flow cytometry. The black lines indicate the duration of treatment. Statistical comparison of 0 nM to 2, 4 and 6 nM oligomycin-A at each imatinib concentration is shown over each bar and combination indices are shown in Table S2. B. A primary blast crisis CML sample was treated with oligomycin-A (2, 6 or 10 nM) alone or in combination with 50 nM dasatinib for 24 h, washed and seeded into 1.2% methylcellulose. Numbers of colonies were assessed after 14 days. C. Ba/F3 murine pro-B-cells expressing vector or BCR-ABL were treated with vehicle, 0.5 or 1 µM imatinib (IM), in combination with increasing concentrations of oligomycin-A (OA; 0, 1, 2, 4, 6 nM, indicated by triangle). After 3 days, the number of viable cells was determined by flow cytometry. Statistical pairwise comparisons of oligomycin-A-mediated changes are noted above each bar. D. K562 cells were treated with vehicle or 4 nM oligomycin-A and increasing concentration of 2-deoxyglucose (2-DG; 0, 2, 5 mM) for 3 days and viable cells counted by flow cytometry. *indicates p<0.05, **p<0.01, and ***p<0.001.
In order to determine whether efficacy of the combination therapy of imatinib and oligomycin-A is specific to BCR-ABL+ cells, we treated the pro-B-cell line Ba/F3, expressing either vector or p210 BCR-ABL, with increasing concentrations of imatinib and oligomycin-A. Expression of BCR-ABL in Ba/F3 cells confers IL-3 independence (27). As expected, imatinib had no effect on Ba/F3-vector cells, but effectively eliminated Ba/F3-BCR-ABL cells (Figure 2C). We also observed a modest effect (<2 fold inhibition) of oligomycin-A alone in both cell lines. However, oligomycin-A synergized with imatinib only in Ba/F3 cells expressing BCR-ABL, indicating that TK dependency is required for sensitization to the combined treatment. Additionally, inhibition of glycolysis using 2-deoxyglucose, a competitive glucose analog which cannot be metabolized through glycolysis, also sensitized CML cells to oligomycin-A treatment (Figure 2D). These data suggest that the effectiveness of the combination therapy of oligomycin-A and imatinib is in part related to the ability of imatinib to inhibit glycolysis.
FLT3 Inhibition Synergizes with Oligomycin-A to Eliminate FLT3ITD AML Cells
Given that the driving TK in a subset of AML, FLT3 expressing an internal tandem duplication (FLT3ITD), shares similar downstream effectors with BCR-ABL (including AKT) (28), we asked whether the combination of the FLT3 inhibitor quizartinib and oligomycin-A could synergize to eliminate AML cells driven by aberrant FLT3ITD. We treated both FLT3WT (NOMO-1 and OCI-AML-3) and FLT3ITD (MV-4–11 and MOLM-13) cell lines with 0–4 nM quizartinib (AC220) in combination with increasing doses of oligomycin-A (0.5 – 4 nM).
As expected, only the two FLT3ITD cell lines, MV-4–11 and MOLM13, were sensitive to quizartinib (Figures 3A left and S4A). These cell lines also showed modest reductions in cell numbers with oligomycin-A alone. Importantly, the combination of quizartinib and oligomycin-A synergized to eliminate FLT3ITD cells. In contrast, the two FLT3WT cell lines showed no sensitivity to either quizartinib and/or oligomycin-A (Figures 3A right and S4B). In addition, we observe similar combinatorial efficacy for the primary patient FLT3ITD and FLT3D835 samples (Figure 3B and Figure S4H) when treated with quizartinib and oligomycin-A. Moreover, synergism in FLT3ITD cells can be observed with a different FLT3 inhibitor lestaurtinib (Figure S4C) and with treatment duration as short as 24 hours with quizartinib (Figure S4D–E; note log10 scale). Similar results were obtained by combining oligomycin-A with imatinib-mediated inhibition of KIT in Kasumi-1 AML cells, which express the ligand-independent N822K activating KIT mutation (Figure S4F).
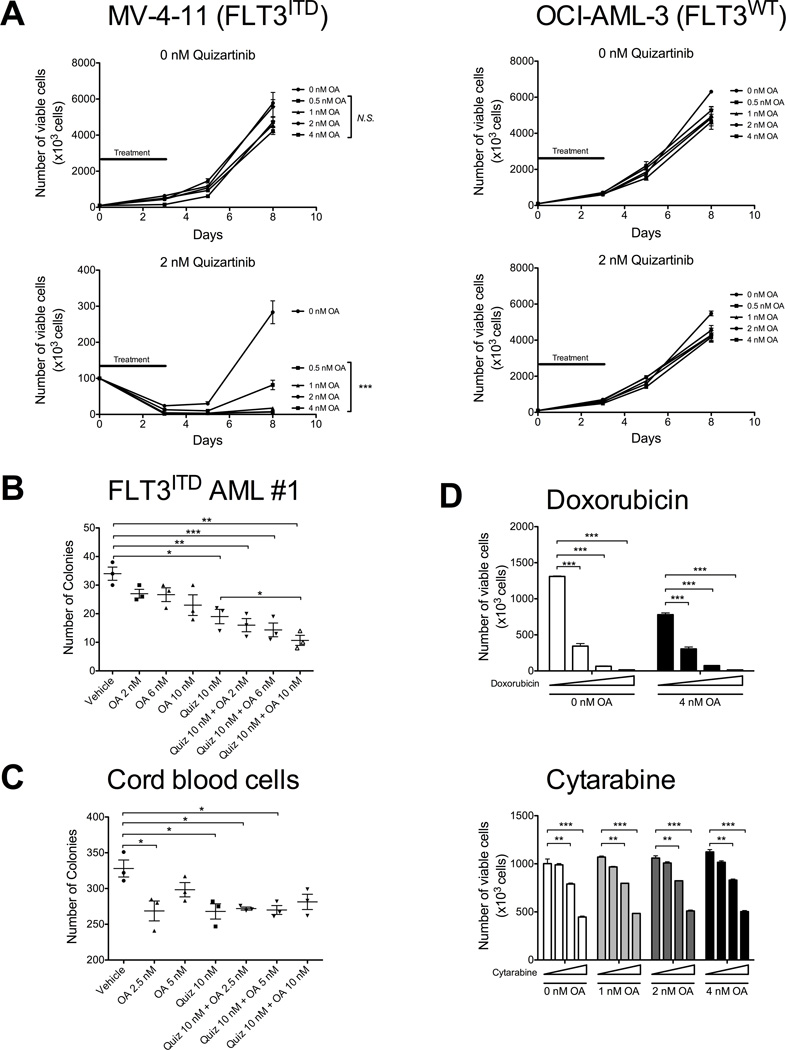
Figure 3. Oligomycin-A sensitizes acute myeloid leukemia cells to FLT3 inhibition.
A. MV-4-11 cells (FLT3ITD, left) and OCI-AML-3 cells (FLT3WT, right) were treated with vehicle (top) or 2 nM quizartinib (bottom), in combination with increasing concentrations of oligomycin-A (OA; 0, 0.5, 1, 2, 4 nM) for 3 days, followed by replating the cells without drug. The black line indicates the duration of treatment. At the indicated time points, an aliquot was stained with PI and the number of viable cells determined by flow cytometry. Statistical comparison of 0 nM oligomycin-A to 0.5, 1, 2 and 4 nM oligomycin-A at each quizartinib concentration is shown next to the legend and combination indices are shown in Table S2. B–C. Cells from a patient with FLT3ITD AML (panel B) and human cord blood cells from a healthy newborn (panel C) were treated with vehicle or 10 nM quizartinib, in combination with increasing concentration of oligomycin-A (OA; 0, 2, 6, 10 nM) for 24 h, washed and seeded in methylcellulose. After 2 weeks, the number of colonies was counted using light microscopy. D. MOLM13 (FLT3ITD) were treated with increasing concentrations of doxorubicin (top; 0, 2.5, 5, 10 nM), or cytarabine (bottom; 0, 2.5, 5, 10 nM) in combination with oligomycin-A (0, 1, 2, 4 nM) for 3 days and the number of viable cells determined by flow cytometry. *indicates p<0.05, **p<0.01, and ***p<0.001.
To assess whether normal human hematopoietic progenitors would be affected by the combination therapy, we treated whole cord blood or CD34+ selected cord blood cells from healthy newborns with oligomycin-A and/or quizartinib for 24 hours and plated cells in methylcellulose for clonogenic assays. Treatment with quizartinib or oligomycin-A alone showed a modest reduction in colony numbers, but the effects were not further reduced by the combination therapy (Figure 3C and S4G) suggesting that oligomycin-A has minimal effects on healthy hematopoietic progenitors.
To assess whether the addition of oligomycin-A to a TKI is specific to TK inhibition, we treated MOLM13 (FLT3ITD cells) with doxorubicin and cytarabine, two standard chemotherapies in AML treatment regimens, in combination with oligomycin-A (Figure 3D). Doxorubicin and cytarabine impaired AML cell survival as expected, but these chemotherapies failed to synergize with oligomycin-A. These data further indicate that oligomycin-A becomes effective only upon inhibition of a driving tyrosine kinase.
Synergistic concentrations of oligomycin-A do not impair mitochondrial TCA cycle or respiration
Oligomycin-A inhibits respiration with an IC50 of 500 nM to 1 µM (25, 29). To confirm this, we measured the oxygen consumption rate (OCR) in K562 cells treated with oligomycin-A or two additional electron transport chain (ETC) inhibitors, antimycin (complex III inhibitor) and rotenone (inhibitor of electron transfer from complex I to ubiquinone; Figure 4A, top). All three ETC drugs inhibited the oxygen consumption rate at the expected concentrations. For oligomycin-A, doses below 500 nM of oligomycin-A have no effect on the OCR (Figure 4A, bottom). Thus, we observed no appreciable effect of low nM oligomycin-A treatment on respiration, despite potent synergistic killing of leukemia cells when combined with TKI at these concentrations. In contrast, antimycin treatment of CML cells at concentrations that inhibit respiration showed only additive effects when combined with imatinib (Figure S5A), further substantiating that the effects of oligomycin-A on leukemia cell killing when combined with TKI are separable from its ability to inhibit respiration.
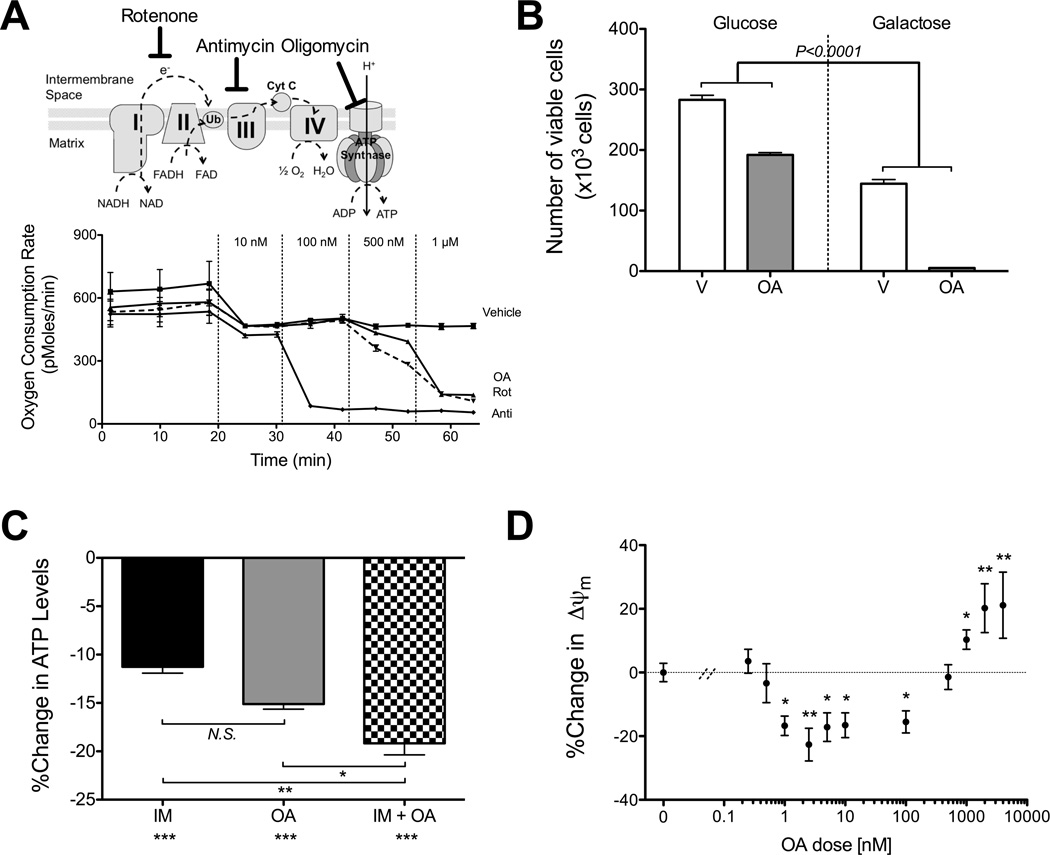
Figure 4. Synergistic concentrations of oligomycin-A do not impair mitochondrial TCA cycle or respiration.
A. The sites of action are depicted on the top panel. At the indicated time points, vehicle or increasing concentrations of oligomycin-A (OA), rotenone (Rot) or antimycin A (Anti) were sequentially added to K562 cells plated on Seahorse microplates and the oxygen consumption rate measured. The initial drop in OCR is due to equilibration of the probe after the addition of fresh media containing vehicle. B. K562 cells were treated with vehicle or 10 nM oligomycin-A for 24 h in media containing 20 mM of glucose or galactose and counted by flow cytometry. Statistical pairwise comparison of oligomycin-A mediated changes in cell numbers are shown.C. The relative ATP levels were measured and normalized to vehicle. K562 cells were treated for 1 h with 1 µM imatinib and/or 10 nM oligomycin-A. Statistical comparisons to vehicle are shown below x-axis labels. D. K562 cells were treated with increasing concentrations of oligomycin-A (0.25 nM to 4 µM) for 1 h and stained with Mitotracker Orange (MTO) to measure ΔΨm. Apoptotic cells were excluded by DAPI stain and mitochondrial mass measured with Mitotracker Green stain. The change relative to vehicle is shown.*indicates p<0.05, **p<0.01, and ***p<0.001.
We also performed NMR spectroscopy on the cellular extracts and medium of K562 cells treated with imatinib and/or oligomycin-A. As expected, imatinib decreased levels of glycolytic and TCA cycle intermediates, but oligomycin-A had no discernable additional effects (Figure S5B–E and Table S3). Taken together, these data indicate that oligomycin-A greatly potentiates imatinib-dependent CML cell killing at concentrations that do not appear to impact either the TCA cycle or respiration.
Oligomycin-A impairs mitochondrial function
We next asked whether the mechanism of action of low dose oligomycin-A is still through impairment of mitochondrial function or integrity. Cells with dysfunctional mitochondria are unable to efficiently utilize galactose as a primary carbon source and rely on glycolysis for energy production (30). In the presence of glucose, treatment with oligomycin-A alone only modestly affected the expansion of K562 cells (Figure 4B). In contrast, in the presence of galactose, treatment with oligomycin-A led to effective killing of K562 cells. Furthermore, both imatinib and oligomycin-A treatments decrease ATP levels in K562 within 30 min, with greater decreases observed in cells treated with both drugs (Figure 4C). This inhibition of ATP levels is not sustained and ATP levels are restored by 6 hours of treatment (Figure S6A). The decrease in ATP levels correlated with an increase in phosphorylated AMPK, indicating increases in the AMP:ATP ratio (Figure S6B).
Notably, treatment with concentrations of oligomycin-A as low as 1 nM, with or without imatinib, rapidly decreased the mitochondrial membrane potential (ΔΨm) in CML cells, as early as 1-hour post-treatment (Figure 4D and S6C). Interestingly, while low nanomolar concentrations of oligomycin-A caused depolarization of ΔΨm, higher concentrations (>500 nM) caused hyperpolarization of ΔΨm (Figure 4D). ΔΨm depolarization was observed with two different dyes, from 1 to 24 h post oligomycin-A treatment (before any appreciable cell death occurs), and was not affected by co-treatment with imatinib (Figure S6D–E). Similar depolarization is observed in MOLM-13 (FLT3ITD) AML cells treated with oligomycin-A (Figure S6F). In all, these results indicate that while low dose oligomycin-A does not appreciably inhibit the TCA cycle or respiration, key mitochondrial functions including ATP production and maintenance of ΔΨm are significantly impaired in oligomycin-A treated leukemia cells.
Oligomycin-A in combination with TKI increases superoxide levels
The process of electron transfer in the ETC, which is coupled to ATP-synthase, is not completely efficient resulting in generation of ROS that can oxidize cellular components. K562 cells were treated with imatinib and/or oligomycin-A for different periods of time, and the levels of total and mitochondrial superoxide levels were measured using flow cytometry (Figure 5A–B). The combination of oligomycin-A with imatinib caused an immediate and sustained increase in superoxide, suggestive of a perturbation in ETC function. The immediate increases in superoxide observed in the combination therapy suggest that the superoxide levels are affected by actions of the drugs themselves and not by the subsequent apoptotic process. We observed similar effects in Ph+ SUP-B15 ALL cells (Figure S6G). In addition, we observed increased levels of the oxidized derivative of deoxyguanosine, 8-oxo-dG (a major product of DNA oxidation (31)), in imatinib and oligomycin-A treated cells (Figures 5C and hydrogen peroxide dose response controls in S6H). Thus, the combination of oligomycin-A with imatinib leads to significant increases in superoxide and subsequent oxidative damage.
Figure 5. Oligomycin-A increases levels of superoxide.
A–B. K562 cells were treated with vehicle, 1 µM imatinib, 10 nM oligomycin-A or the combination of both drugs. After 2 or 24 h of treatment, cells were stained with DHE to measure total superoxide levels (A) or after 18 h with MitoSox-Red (MSR) to measure mitochondrial superoxide levels (B) using flow cytometry. Changes are normalized to vehicle control. Apoptotic cells were excluded by DAPI stain. C. After 48 h of treatment, cells were stained with avidin to measure the levels of 8-oxo-deoxyguanosine (normalized to vehicle control). For A, B and C, statistical comparisons to vehicle are shown below x-axis labels. D. K562 cells were treated with vehicle, 1 µM imatinib and/or 10 nM oligomycin-A for 3 days with vehicle or 200 U/mL of SOD-PEG. After 3 days, viable cell numbers were determined by flow cytometry. *indicates p<0.05, **p<0.01, and ***p<0.001.
To further characterize the role of superoxide, we treated K562 cells with increasing concentrations of the superoxide dismutase (SOD) inhibitor 2-methoxyestradiol (32). SODs catalyze the conversion of superoxide into oxygen and hydrogen peroxide, and are the major enzymes responsible for detoxification of ROS in mammalian cells. Concentrations of 2-methoxyestradiol that increase the levels of superoxide in K562 cells (Figure S6I) to levels similar to those observed in the combination therapy were sufficient to elicit cell death (Figure S6J). Finally, treating cells with methoxy-polyethylene glycol-coupled SOD1 reduced the ability of oligomycin-A to potentiate imatinib-mediated killing (~2X; Figure 5D). These data suggest that the mechanism of action of oligomycin-A in promoting apoptosis in TKI-treated leukemia cells is partially mediated by enhanced superoxide production.
Combined treatment with TKI and oligomycin-A effectively eliminates leukemia cells in vivo
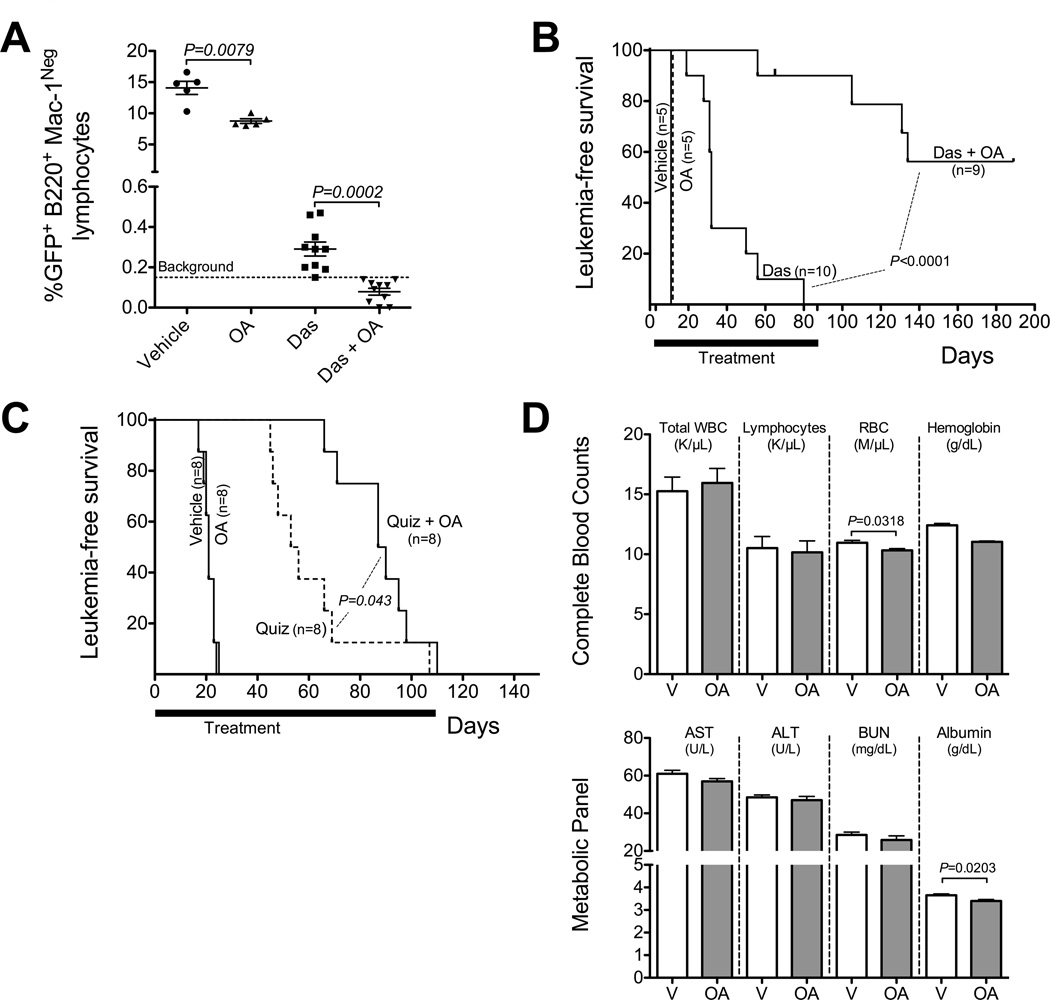
In order to test whether oligomycin-A could enhance the efficacy of TKI to eliminate BCR-ABL+ leukemia cells in vivo, we tested the combination therapy in a mouse model of BCR-ABL+ B-ALL. This model has previously been shown to closely mimic human BCR-ABL+ B-ALL, including responsiveness to the BCR-ABL inhibitor dasatinib (22, 23). Mice were inoculated with ARF−/− p185-BCR-ABL/GFP leukemia, and after 3 days, started on daily treatments with vehicle, dasatinib, oligomycin-A or the combination therapy of dasatinib plus oligomycin-A. After 4 days of therapy, leukemic burden in the combination therapy group was significantly reduced compared to the dasatinib alone group (Figures 6A and S7A). Mice treated with either vehicle or oligomycin-A alone succumbed to leukemia on day 10 with postmortem analysis showing extensive leukemia in the blood, bone marrow and spleen. Dasatinib treated mice had a median survival of 32 days, but all mice succumbed to leukemia by day 80 (Figure 6B). In order to determine if all leukemia cells had indeed been eliminated in the remaining 8/9 mice in the combination therapy, therapy was stopped on day 85. While three of the mice eventually relapsed with leukemia, five of the remaining mice remained healthy up to day 189 when the experiment was terminated. Post-mortem analysis revealed no detectable leukemia cells in peripheral blood, bone marrow or spleens by flow cytometry and in bone marrow samples by RT-PCR analysis for BCR-ABL mRNA (Figure S7B), indicating that the combination therapy of dasatinib and oligomycin-A completely eliminated leukemia cells in these five mice.
Figure 6. Low concentration oligomycin-A synergizes with TKIs to eliminate leukemia cells in vivo.
A–B. 5×105Arf−/− BCR-ABL/GFP leukemia cells were injected i.v. into non-irradiated C57BL/6 mice. Mice were treated with either vehicle, oligomycin-A (OA; 100 µg/kg i.p.), dasatinib (Das; 10 mg/kg o.g.) or both drugs. Percent GFP in peripheral B-cells (B220+Mac1Neg) after 4 days of therapy was determined by flow cytometry (A). Kaplan-Meier survival curve for the indicated drug treatment is shown in B. The tick on the Das+OA curve at day 65 indicates a mouse that died with no signs of leukemia. C. NSG mice treated with busulfan and transplanted with 3×106 cells. Mice were treated with either vehicle, oligomycin-A (OA; 100 µg/kg i.p,), quizartinib (Quiz; 10 mg/kg o.g.) or both drugs. Mice were monitored by peripheral bleed staining for human CD45 and HLA-ABC. Kaplan-Meier survival curve for the indicated drug treatment is shown in C. D. Mice were treated with either vehicle or oligomycin-A (OA; 100 µg/kg i.p.) daily for 15 days. Complete blood counts (top) and metabolic markers of renal and hepatic toxicities in serum (bottom) were analyzed on day 15.
We next asked whether similar efficacy could be observed for FLT3ITD AML. NOD/SCID/γ-chain−/− (NSG) mice were transplanted with a primary human AML sample (FLT3ITD), monitored by flow cytometry and after one month randomized into groups with similar peripheral blood burdens (Figure S7C–D). The mice were treated with quizartinib at a clinically relevant dose (33) and/or oligomycin-A, and sacrificed when the mice lost over 15% of their body weight or showed signs of disease. As shown in Figure 6C, we observed a significant extension of survival in the group treated with the combination of quizartinib and oligomycin-A (median survival of 54.5 vs. 88.5 days). Mice treated with quizartinib alone or in combination with oligomycin-A had a dramatic initial response but they eventually relapse with AML (Figure S7E). In all, these data show that low doses of oligomycin-A can improve therapeutic outcomes for TKI for both Ph+ and FLT3ITD leukemias in vivo. Note also that mice treated with TKI (dasatinib or quizartinib) and oligomycin-A for over two months appeared healthy.
Oligomycin-A treatment exhibits minimal toxicity in vivo
In order to determine potential toxicity of low-dose oligomycin-A, we treated mice with either vehicle or oligomycin-A for 15 days. After 15 days of therapy, the mice receiving oligomycin-A exhibited no evident changes in appearance or behavior, but did show a slight decrease in weight (<1% from initial weight) compared to the vehicle controls (Table S4). Complete blood counts revealed small differences in the number of monocytes and RBCs, and hematocrit and hemoglobin levels in oligomycin-A treated mice, but these parameters remained within the reference normal ranges (Figure 6D top and Table S4). Thus, oligomycin-A does not appear to be myelosuppressive in vivo. In addition, we measured markers of hepatic and renal toxicity. Serum levels of total protein, ALT, AST, creatinine, and blood urea nitrogen remained the same for the two treatment groups. Minor differences in albumin levels (Figure 6D bottom) were observed but values remained within the reference range (Table S4).
Due to concerns of mitochondrial toxicity from treatment with oligomycin-A, we tested mitochondrial integrity in vivo. The mitochondrial permeability transition pore (PTP) and cyclophilin-D regulate platelet activation, and thus mitochondria play a key role in the function of platelets (34). If oligomycin-A disrupts mitochondrial ΔΨm in platelets, we would expect treatment with oligomycin-A to disrupt platelet activation. However, we did not observe changes in the number of circulating platelets, their activation state or their ability to respond to platelet agonists in oligomycin-A treated mice (Figure S7F–G and Table S4). These data suggest that therapy with low dose oligomycin-A has minimal impact, at least in the short-term, on normal mouse physiology at a concentration that exhibits potent anti-leukemic effects.
Discussion
Our results reveal that perturbing mitochondrial function can severely limit the survival of leukemia cells upon TKI treatment (Figure S8). First, we identified that pyruvate entry into the TCA cycle becomes important upon BCR-ABL inhibition, providing these cells an alternative mechanism of glucose metabolism to compensate for the inhibition of glycolysis driven by BCR-ABL. PDH, the gatekeeping enzyme linking glycolysis to the TCA cycle, converts pyruvate to acetyl-CoA which is decarboxylated via the TCA cycle to generate the electron donors used to generate the mitochondrial membrane potential. Still, our results contrast with those observed by Bonnet et al., in which forcing entry of pyruvate into the TCA cycle by dichloroacetate-mediated inhibition of pyruvate dehydrogenase kinase impairs the survival of cancer cells in vitro and tumor growth in mice (35). In fact, cancer cells generally limit pyruvate entry into the TCA, such as by negative regulation of pyruvate kinase, which generates ATP and pyruvate from phosphoenolpyruvate and ADP (11). Moreover, pyruvate kinase activators can inhibit tumor growth (36). We propose that imatinib treatment of BCR-ABL+ leukemia cells reverses the leukemic cell dependency on pyruvate entry into the TCA cycle. It is well established that imatinib treatment in leukemia cells reduces glucose uptake and flux through glycolysis, which in turn limits the amount of pyruvate available in the cell (14). Under these conditions, our data suggest that upon TKI treatment CML cells become dependent on pyruvate entry into the TCA to sustain metabolic demands for continued survival.
Importantly, our studies have identified low nM concentrations of oligomycin-A as an effective adjuvant therapy to TKI in the treatment of BCR-ABL+ and FLT3ITD leukemias. Oligomycin-A binds the C-subunit of the ATP-synthase and inhibits ATP production (25, 26, 29). Although low nM oligomycin-A concentrations do not inhibit oxygen consumption, these concentrations of oligomycin-A lead to transient decreases in ATP levels and ΔΨm depolarization in BCR-ABL+ cells. Moreover, combining oligomycin-A with TKI increases the generation of superoxides, which have been shown to result in oxidation of critical cellular macromolecules and cell death (37, 38). We show that increases in superoxides contribute to, but do not fully account for, the ability of oligomycin-A to synergize with BCR-ABL TKIs to eliminate leukemia cells.
Recent studies have shown that ATP-synthase subunits also function as the mitochondrial PTP (39, 40). Opening of the PTP causes increased ion flux, leading to ΔΨm depolarization across the inner mitochondrial membrane and matrix swelling (41, 42), which can promote outer mitochondrial membrane disruption during apoptosis. Indeed, given that oligomycin-A leads to ΔΨm depolarization at low nM concentrations similar to those that synergize with BCR-ABL and FLT3 TKI to eliminate leukemia cells, ATP-synthase’s function as the PTP may be the key target of oligomycin-A in leukemia cells. ΔΨm depolarization would also be expected to decrease ATP production, which is dependent on the electrochemical gradient across the inner mitochondrial membrane, and increase superoxide production by uncoupling the ETC (41, 42). Thus, investigations presented here have potentially discovered an exciting means to selectively promote partial ΔΨm depolarization opening in leukemia cells, leading to apoptosis when combined with TKI treatment.
Based on the known inhibitory effects of TKI on glucose uptake and glycolysis, and that either 2-DG treatment or glucose substitution with galactose sensitizes leukemia cells to oligomycin-A, we propose that inhibition of glycolysis is key to sensitization of leukemia cells to oligomycin-A. In fact, leukemia cells are quite tolerant of low levels of oligomycin-A (and the consequent modest mitochondrial depolarization) in their usual highly glycolytic states. The inhibition of glycolysis will force cells to depend on mitochondrial carbon metabolism (as evidenced by the dependency on PDH engendered by TKI treatment), and this new state of mitochondrial dependency makes the leukemia cells highly sensitive to oligomycin-A treatment (Figure S8). While our evidence points to mitochondrial action of oligomycin-A (including the ability of galactose to sensitize leukemia cells to oligomycin-A, as well as oligomycin-A-mediated induction of superoxides, transient reductions of ATP levels, and ΔΨm depolarization), we cannot definitively conclude that the PTP is the relevant target of oligomycin-A. That said, given that the PTP has recently been shown to be composed of ATP-synthase subunits (including the oligomycin-sensitive F0 subunit) (39, 40), that ATP-synthase is the conserved target for oligomycin-A from yeast to humans (43), and that ΔΨm depolarization shows a concentration dependence for oligomycin-A that closely mirrors synergism with TKI, the ATP-synthase in the PTP is currently the strongest candidate as the relevant target for low concentrations of oligomycin-A.
Importantly, clonogenic assays with healthy human cord blood cells and studies of treated mice reveal minimal side effects of oligomycin-A on normal cells, which highlight its therapeutic potential for the elimination of TK-addicted leukemia. TK inhibition in leukemia cells makes the cells hypersensitive to oligomycin-A in mouse models, at concentrations that have no substantial negative impact on mouse physiology, perhaps due to reduced glycolysis and/or reduced dependency of non-cancer cells for the targeted TK. Although oligomycin-A has been previously reported to induce cell death in leukemia, the effects were observed at concentrations (~12 µM) at which the ATP-synthase is completely inhibited (44). At this concentration, the toxicity of such therapy will outweigh any clinical benefit. In contrast, the concentrations of oligomycin-A that synergize with TKI are ~1000X less and do not impede oxygen consumption. Interestingly, other studies have shown that inhibition of mitochondrial ATP-synthase, by increasing ROS-dependent NFκB activation, is essential for cancer cell survival (45, 46). Again, we observe the opposite effects of low concentration oligomycin-A in leukemias, but specifically in the context of TK inhibition. It is notable that low nM oligomycin-A causes ΔΨm depolarization, while µM oligomycin-A causes hyperpolarization (Figure 4F). Other groups have also reported hyperpolarization of ΔΨm at µM concentrations of oligomycin-A (47). Thus, we do not believe that our studies are in conflict with these previous studies, but reveal how inhibition of a dominant TK oncogene in a cancer can radically change mitochondrial dependencies, with clear therapeutic implications. Indeed, a recent study showed that inhibition of mitochondrial respiration (including with oligomycin-A) blocked the emergence of a slow-cycling tumor maintaining melanoma cell subpopulation in response to various therapies (48)
Understanding how glucose utilization and energy production pathways are altered in cancers upon treatment with both conventional and targeted therapeutics will be critically important for the development of metabolically-targeted drug therapies. Our studies reveal that inhibition of the dominant oncogene in Ph+ and FLT3ITD leukemias can engender mitochondrial dependencies that can be exploited therapeutically. Notably, the Warburg effect is shared by many tumor types (10, 11, 49, 50), which may suggest that perturbing mitochondrial function could be a ubiquitous target for improved elimination of cancer cells upon therapy-mediated reversal of the glycolytic phenotype.
Supplementary Material
Translational Relevance.
Studies described here reveal aberrant mitochondrial metabolic dependencies in BCR-ABL and FLT3ITD driven leukemias. We propose that the inhibition of glycolysis forces leukemia cells to depend more on mitochondrial carbon metabolism, as evidenced by the TKI-engendered dependency on pyruvate dehydrogenase, and this new state of mitochondrial dependency makes the leukemia cells highly sensitive to oligomycin-A treatment. Of importance, we show in mouse models that this combination therapy is very effective against BCR-ABL+ leukemias and FLT3ITD acute myeloid leukemia. Thus, oligomycin-A could be a safe and effective adjuvant therapy that enhances the efficacy of tyrosine kinase inhibitors in treating leukemias. While cancer cells normally limit pyruvate entry in the TCA cycle, and inhibition of ATP-synthase has been shown to protect cancer cells from apoptosis, our studies reveal how inhibition of a dominant oncogenic tyrosine kinase in a cancer can radically change metabolic dependencies, with clear therapeutic implications.
Acknowledgements
We would like to thank Jorge DiPaola, Chris Rivard, Steve Anderson, Kristen Boyle, Cheng-Jun Hu and Andrea Merz for help with experiments, and Chris Porter, Jennifer Salstrom and the rest of the DeGregori lab for their comments.
Financial Support: These studies were supported by grants from the NIH (K01-CA133182 and K22-CA172757 to M.A.G and F31-CA157166 to F.A.C) and the Leukemia Lymphoma Society (J.D. and D.K.G.). The NMR Core and Medicinal Chemistry Core are supported by NCRR CCTSI/5UL1-RR025780 and Shared Resources by Cancer Center Support Grant P30-CA046934.
Footnotes
Conflict of Interest: All authors declare that they have no conflict of interest relevant to this study.
Authorship
Conception and design: F.A.-C., M.G., C.P.-D. and J.D.
Acquisition of data: F.A.-C., M.G., C.P.-D., L.G., V.Z., D.D., A.H. B.S. and N.S.
Analysis and interpretation of data: F.A.-C., M.G., C.P.-D. and B.S.
Writing, review and/or revision of the manuscript: F.A.-C., M.G., C.P.-D., and J.D.
Material support: D.P., A.K., V.K. and M.W.
Study supervision: C.T.J., D.K.G, N.S., and J.D.
References
- 1.Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96:3343–3356. [PubMed] [Google Scholar]
- 2.Faderl S, O'Brien S, Pui C-H, Stock W, Wetzler M, Hoelzer D, et al. Adult acute lymphoblastic leukemia: concepts and strategies. Cancer. 2010;116:1165–1176. doi: 10.1002/cncr.24862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foa R, Vitale A, Vignetti M, Meloni G, Guarini A, De Propris MS, et al. Dasatinib as first-line treatment for adult patients with Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood. 2011;118:6521–6528. doi: 10.1182/blood-2011-05-351403. [DOI] [PubMed] [Google Scholar]
- 4.Smith A, Howell D, Patmore R, Jack A, Roman E. Incidence of haematological malignancy by sub-type: a report from the Haematological Malignancy Research Network. British journal of cancer. 2011;105:1684–1692. doi: 10.1038/bjc.2011.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santos FP, Jones D, Qiao W, Cortes JE, Ravandi F, Estey EE, et al. Prognostic value of FLT3 mutations among different cytogenetic subgroups in acute myeloid leukemia. Cancer. 2011;117:2145–2155. doi: 10.1002/cncr.25670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic Relevance of Integrated Genetic Profiling in Acute Myeloid Leukemia. New England Journal of Medicine. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortes JE, Kantarjian H, Foran JM, Ghirdaladze D, Zodelava M, Borthakur G, et al. Phase I Study of Quizartinib Administered Daily to Patients With Relapsed or Refractory Acute Myeloid Leukemia Irrespective of FMS-Like Tyrosine Kinase 3-Internal Tandem Duplication Status. Journal of Clinical Oncology. 2013;31:3681–3687. doi: 10.1200/JCO.2013.48.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith BD, Levis M, Beran M, Giles F, Kantarjian H, Berg K, et al. Single-agent CEP-701, a novel FLT3 inhibitor, shows biologic and clinical activity in patients with relapsed or refractory acute myeloid leukemia. Blood. 2004;103:3669–3676. doi: 10.1182/blood-2003-11-3775. [DOI] [PubMed] [Google Scholar]
- 9.Knapper S, Burnett AK, Littlewood T, Kell WJ, Agrawal S, Chopra R, et al. A phase 2 trial of the FLT3 inhibitor lestaurtinib (CEP701) as first-line treatment for older patients with acute myeloid leukemia not considered fit for intensive chemotherapy. Blood. 2006;108:3262–3270. doi: 10.1182/blood-2006-04-015560. [DOI] [PubMed] [Google Scholar]
- 10.Tennant DA, Durán RV, Boulahbel H, Gottlieb E. Metabolic transformation in cancer. Carcinogenesis. 2009;30:1269–1280. doi: 10.1093/carcin/bgp070. [DOI] [PubMed] [Google Scholar]
- 11.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 13.Gottlob K, Majewski N, Kennedy S, Kandel E, Robey RB, Hay N. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes & Development. 2001;15:1406–1418. doi: 10.1101/gad.889901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottschalk S, Anderson N, Hainz C, Eckhardt SG, Serkova NJ. Imatinib (STI571)-mediated changes in glucose metabolism in human leukemia BCR-ABL-positive cells. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:6661–6668. doi: 10.1158/1078-0432.CCR-04-0039. [DOI] [PubMed] [Google Scholar]
- 15.Barnes K, McIntosh E, Whetton AD, Daley GQ, Bentley J, Baldwin SA. Chronic myeloid leukaemia: an investigation into the role of Bcr-Abl-induced abnormalities in glucose transport regulation. Oncogene. 2005;24:3257–3267. doi: 10.1038/sj.onc.1208461. [DOI] [PubMed] [Google Scholar]
- 16.Kominsky DJ, Klawitter J, Brown JL, Boros LG, Melo JV, Eckhardt SG, et al. Abnormalities in glucose uptake and metabolism in imatinib-resistant human BCR-ABL-positive cells. Clin Cancer Res. 2009;15:3442–3450. doi: 10.1158/1078-0432.CCR-08-3291. [DOI] [PubMed] [Google Scholar]
- 17.Zhao F, Mancuso A, Bui TV, Tong X, Gruber JJ, Swider CR, et al. Imatinib resistance associated with BCR-ABL upregulation is dependent on HIF-1alpha-induced metabolic reprograming. Oncogene. 2010 doi: 10.1038/onc.2010.67. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kluza J, Jendoubi M, Ballot C, Dammak A, Jonneaux A, Idziorek T, et al. Exploiting mitochondrial dysfunction for effective elimination of imatinib-resistant leukemic cells. PLoS One. 2011;6:e21924. doi: 10.1371/journal.pone.0021924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samudio I, Fiegl M, Andreeff M. Mitochondrial uncoupling and the Warburg effect: molecular basis for the reprogramming of cancer cell metabolism. Cancer research. 2009;69:2163–2166. doi: 10.1158/0008-5472.CAN-08-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skrtic M, Sriskanthadevan S, Jhas B, Gebbia M, Wang X, Wang Z, et al. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2011;20:674–688. doi: 10.1016/j.ccr.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregory MA, Phang TL, Neviani P, Alvarez-Calderon F, Eide CA, O'Hare T, et al. Wnt/Ca2+/NFAT signaling maintains survival of Ph+ leukemia cells upon inhibition of Bcr-Abl. Cancer cell. 2010;18:74–87. doi: 10.1016/j.ccr.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams RT, den Besten W, Sherr CJ. Cytokine-dependent imatinib resistance in mouse BCR-ABL+, Arf-null lymphoblastic leukemia. Genes & Development. 2007;21:2283–2287. doi: 10.1101/gad.1588607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams RT, Sherr CJ. The INK4-ARF (CDKN2A/B) locus in hematopoiesis and BCR-ABL-induced leukemias. Cold Spring Harb Symp Quant Biol. 2008;73:461–467. doi: 10.1101/sqb.2008.73.039. [DOI] [PubMed] [Google Scholar]
- 24.Williams RT, Sherr CJ. The INK4-ARF (CDKN2A/B) Locus in Hematopoiesis and BCR-ABL-induced Leukemias. Cold Spring Harb Symp Quant Biol. 2008 doi: 10.1101/sqb.2008.73.039. [DOI] [PubMed] [Google Scholar]
- 25.Roberton AM, Holloway CT, Knight IG, Beechey RB. A comparison of the effects of NN'-dicyclohexylcarbodi-imide, oligomycin A and aurovertin on enrgy-linked reactions in mitochondria and submitochondrial particles. Biochem J. 1968;108:445–456. doi: 10.1042/bj1080445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Symersky J, Pagadala V, Osowski D, Krah A, Meier T, Faraldo-Gomez JD, et al. Structure of the c(10) ring of the yeast mitochondrial ATP synthase in the open conformation. Nat Struct Mol Biol. 2012;19(S1):485–491. doi: 10.1038/nsmb.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daley GQ, Baltimore D. Transformation of an interleukin 3-dependent hematopoietic cell line by the chronic myelogenous leukemia-specific P210bcr/abl protein. Proc Natl Acad Sci U S A. 1988;85:9312–9316. doi: 10.1073/pnas.85.23.9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brandts CH, Sargin B, Rode M, Biermann C, Lindtner B, Schwable J, et al. Constitutive activation of Akt by Flt3 internal tandem duplications is necessary for increased survival, proliferation, and myeloid transformation. Cancer research. 2005;65:9643–9650. doi: 10.1158/0008-5472.CAN-05-0422. [DOI] [PubMed] [Google Scholar]
- 29.Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292:C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 30.Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer research. 2004;64:985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 31.de Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, et al. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer research. 2001;61:5378–5381. [PubMed] [Google Scholar]
- 32.Gao N, Rahmani M, Dent P, Grant S. 2-Methoxyestradiol-induced apoptosis in human leukemia cells proceeds through a reactive oxygen species and Akt-dependent process. Oncogene. 2005;24:3797–3809. doi: 10.1038/sj.onc.1208530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chao Q, Sprankle KG, Grotzfeld RM, Lai AG, Carter TA, Velasco AM, et al. Identification of N-(5-tert-butyl-isoxazol-3-yl)-N'-{4-[7-(2-morpholin-4-yl-ethoxy)imidazo[2,1-b][1 ,3]benzothiazol-2-yl]phenyl}urea dihydrochloride (AC220), a uniquely potent, selective, and efficacious FMS-like tyrosine kinase-3 (FLT3) inhibitor. Journal of medicinal chemistry. 2009;52:7808–7816. doi: 10.1021/jm9007533. [DOI] [PubMed] [Google Scholar]
- 34.Jobe SM, Wilson KM, Leo L, Raimondi A, Molkentin JD, Lentz SR, et al. Critical role for the mitochondrial permeability transition pore and cyclophilin D in platelet activation and thrombosis. Blood. 2008;111:1257–1265. doi: 10.1182/blood-2007-05-092684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonnet S, Archer SL, Allalunis-Turner J, Haromy A, Beaulieu C, Thompson R, et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer cell. 2007;11:37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 36.Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012 doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martindale JL, Holbrook NJ. Cellular response to oxidative stress: signaling for suicide and survival. J Cell Physiol. 2002;192:1–15. doi: 10.1002/jcp.10119. [DOI] [PubMed] [Google Scholar]
- 38.Barzilai A, Yamamoto K-I. DNA damage responses to oxidative stress. DNA Repair. 2004;3:1109–1115. doi: 10.1016/j.dnarep.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 39.Giorgio V, von Stockum S, Antoniel M, Fabbro A, Fogolari F, Forte M, et al. Dimers of Mitochondrial ATP Synthase form the Permeability Transition Pore. Proc Natl Acad Sci USA. In Press doi: 10.1073/pnas.1217823110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonora M, Bononi A, De Marchi E, Giorgi C, Lebiedzinska M, Marchi S, et al. Role of the c subunit of the FO ATP synthase in mitochondrial permeability transition. Cell cycle (Georgetown, Tex) 2013;12:674–683. doi: 10.4161/cc.23599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinnally KW, Antonsson B. A tale of two mitochondrial channels, MAC and PTP, in apoptosis. Apoptosis. 2007;12:857–868. doi: 10.1007/s10495-007-0722-z. [DOI] [PubMed] [Google Scholar]
- 42.Rasola A, Bernardi P. The mitochondrial permeability transition pore and its involvement in cell death and in disease pathogenesis. Apoptosis. 2007;12:815–833. doi: 10.1007/s10495-007-0723-y. [DOI] [PubMed] [Google Scholar]
- 43.Symersky J, Osowski D, Walters DE, Mueller DM. Oligomycin frames a common drug-binding site in the ATP synthase. Proc Natl Acad Sci U S A. 2012;109:13961–13965. doi: 10.1073/pnas.1207912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolvetang EJ, Johnson KL, Krauer K, Ralph SJ, Linnane AW. Mitochondrial respiratory chain inhibitors induce apoptosis. FEBS Letters. 1994;339:40–44. doi: 10.1016/0014-5793(94)80380-3. [DOI] [PubMed] [Google Scholar]
- 45.Santamaria G, Martinez-Diez M, Fabregat I, Cuezva JM. Efficient execution of cell death in non-glycolytic cells requires the generation of ROS controlled by the activity of mitochondrial H+-ATP synthase. Carcinogenesis. 2006;27:925–935. doi: 10.1093/carcin/bgi315. [DOI] [PubMed] [Google Scholar]
- 46.Formentini L, Sanchez-Arago M, Sanchez-Cenizo L, Cuezva JM. The mitochondrial ATPase inhibitory factor 1 triggers a ROS-mediated retrograde prosurvival and proliferative response. Mol Cell. 2012;45:731–742. doi: 10.1016/j.molcel.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 47.Johnson KM, Chen X, Boitano A, Swenson L, Opipari AW, Jr, Glick GD. Identification and validation of the mitochondrial F1F0-ATPase as the molecular target of the immunomodulatory benzodiazepine Bz-423. Chem Biol. 2005;12:485–496. doi: 10.1016/j.chembiol.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 48.Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann Katharina M, Speicher D, et al. Overcoming Intrinsic Multidrug Resistance in Melanoma by Blocking the Mitochondrial Respiratory Chain of Slow-Cycling JARID1Bhigh Cells. Cancer Cell. 23:811–825. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tennant DA, Durán RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 50.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.