Abstract
RAGE is a multi-functional receptor implicated in diverse processes including inflammation and cancer. In this study, we report that RAGE expression is upregulated widely in aggressive triple-negative breast cancer cells, both in primary tumors and lymph node metastases. In evaluating the functional contributions of RAGE in breast cancer, we found RAGE-deficient mice displayed a reduced propensity for breast tumor growth. In an established model of lung metastasis, systemic blockade by injection of a RAGE neutralizing antibody inhibited metastasis development. Mechanistic investigations revealed that RAGE bound to the pro-inflammatory ligand S100A7 and mediated its ability to activate ERK, NF-κB and cell migration. In an S100A7 transgenic mouse model of breast cancer (mS100a7a15 mice), administration of either RAGE neutralizing antibody or soluble RAGE was sufficient to inhibit tumor progression and metastasis. In this model, we found that RAGE/S100A7 conditioned the tumor microenvironment by driving the recruitment of MMP9-positive tumor-associated macrophages. Overall, our results highlight RAGE as a candidate biomarker for triple-negative breast cancers and they reveal a functional role for RAGE/S100A7 signaling in linking inflammation to aggressive breast cancer development.
Keywords: RAGE, S100A7, breast cancer, TNBC, tumor associated macrophages, metastasis
Introduction
RAGE is the signal transduction receptor which senses a variety of signaling molecules (1). The variety of ligands allows RAGE to be implicated in a wide spectrum of pathological conditions such as inflammation and cancer (1-2). Epidemiological and molecular studies, including mouse models, have shown that if inflammation is prolonged it promotes cancer development (3-6). It is now believed that most solid tumors, including those in the breast, have an inflammatory microenvironment (4-5). RAGE expression and activation have been shown to be associated with chronic inflammation, which in turn enhances the malignant transformation of various cancers (1-2,7-11). However, its role in breast cancer, especially in the modulation of breast cancer microenvironment, is unknown.
RAGE was first described as a receptor for advanced glycation end products (AGE), but it has since been shown to be the receptor for several other molecules involved in innate immunity, including high mobility group box 1 peptide (HMGB-1), amyloid-β peptide and the S100 family of proteins (1-2). Phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced pro-inflammatory mediators were shown to be decreased in RAGE-deficient mice (7) which suggests that RAGE expression is involved in sustaining inflammation and skin and pancreatic cancer (1-2,7,9). It has also been well documented that RAGE ligands bind to RAGE and activate its downstream signaling mechanisms that fuel chronic inflammatory conditions leading to neoplastic stage (1,12-13). It is interesting to note that there is very low or no RAGE expression in normal tissues, but enhanced expression in chronic inflammation and cancer (2,10). Although, these features of RAGE make it an ideal candidate for therapeutic strategies against chronic inflammation, not much is known about its role in breast cancer.
RAGE has been shown to bind to its ligand S100A7 in keratinocytes and leukocytes (14-15). S100A7 has been shown to be highly expressed in ERα− breast cancer (16-17). It is believed that S100A7 mediates breast cancer growth and metastasis by recruiting pro-inflammatory cell infiltrates (18-19). Also, pro-inflammatory cytokines enhance TNBC growth and metastasis (20). However, very little is known about mechanisms through which RAGE/S100A7 axis modulates tumor microenvironment and enhances breast cancer growth and metastasis.
Macrophages can be divided into subtypes M1 and M2, where M1 macrophages are associated with an anti-cancer phenotype, and M2 macrophages express a pro-cancer phenotype (21-22). Cytokines/chemokines and growth factors modulate the tumor microenvironment, which could directly/indirectly polarize macrophages toward the M2 tumor-associated macrophages (M2-TAMs) phenotype (22-23).
In this investigation, we for the first time show that RAGE is expressed in a set of aggressive breast cancer cell lines, TNBC and metastatic lymph node deposits. We also demonstrate that blocking of RAGE reduces tumor metastasis, and that RAGE ablation inhibits breast cancer growth. In addition, we show that RAGE mediates its tumor promoting effects in breast cancer through binding to S100A7. Our findings also uncovered that the RAGE/S100A7 pathway enhanced breast cancer growth and metastasis. These studies further demonstrate that RAGE-neutralizing antibodies/soluble RAGE could be used to inhibit breast cancer growth and metastasis, especially in S100A7 expressing invasive cancers. Furthermore, these studies suggest that RAGE could be used as novel biomarker and therapeutic strategy against TNBC.
Materials and Methods
Patients
Institutional review board (IRB) of the Ohio State University (OSU) has approved protocol for the constructed TNBC tissue microarrays (TMAs) (n=173). The clinical and pathological characteristics of TNBC TMA have been recently described (24). TMA for lymph node metastasis (BR1008) was obtained from US Biomax, Inc. (Rockville, MD).
IHC, Immunofluorescence, and ELISA
Samples from mammary glands (MG) and tumors were formalin fixed and paraffin embedded (18). Standard IHC techniques were used according to the manufacturer's recommendations (Vector Laboratories) using antibodies against RAGE (Abcam, 1:400) Ki67 (Neomarkers, 1:100), CD31 (Santa Cruz 1:100), F4/80 (AbD Serotec, 1:50), arginase1 (Santa Cruz, 1:200), and iNOS (Abcam, 1:200) for 60 min at room temperature. Vectastain Elite ABC reagents (Vector Laboratories), using avidin DH:biotinylated horseradish peroxidase H complex with 3,3′-diaminobenzidine (Polysciences) and Mayer's hematoxylin (Fisher Scientific), were used for detection of bound antibodies. Staining of TMAs was graded as previously described (25). Immunofluorescence was performed on paraffin embedded tissues. Briefly, sections were stained with F4/80 (1:75), mS100a7a15 (Santa Cruz, 1:100) and MMP9 (R&D systems, 1:150). Alexa-Fluor conjugated secondary antibodies (Life Technologies) were used to detect primary antibody. Sections were mounted by VECTASHIELD mounting media containing DAPI (Vector Laboratories, Inc.). Images were analyzed by confocal microscopy. Binding of RAGE with human recombinant S100A7 was performed as described (26).
Cancer patient data analysis
High RAGE and S100A7 expressions were defined as over-expression of ager mRNA being greater than 0.5 fold and over-expression of s100a7 is greater than 1.0 fold of standard deviation above the mean, respectively. Association of gene expression alterations was performed based on the TCGA database by Fisher's Exact Test. Analysis of RAGE expression between basal and non-basal breast cancer samples was based on a subtype specific breast cancer study (GEO accession GDS2250) (27). For Kaplan–Meier survival analysis, patient samples with RAGE expression values greater than its median were grouped as high RAGE and the other half as low RAGE.
Cell culture
Murine macrophage cell line RAW264.7 and human breast carcinoma cell lines MDA-MB-231, MDA-MB-453, MCF7, T47D, BT-474 were obtained from ATCC. SCP2 cells were kindly provided by Dr. Massague (28). MVT-1 cells (derived from MMTV-c-Myc; MMTV-VEGF bi-transgenic mice) were obtained from Dr. Johnson and PyMT cells derived from MMTV-PyMT C57BL/6 mice were obtained from Dr. Hai (OSU) (29). MVT-1 highly metastatic clone, PyMT, Met1 and 4T1 cells were cultured as described (18,29).
Chemotaxis
Chemotactic assays were performed using transwell chambers (Costar 8 μm pore size) as described (18,30).
Mice
Nude mice were obtained from Charles River. C57B/6 background RAGE−/− mice were kindly provided by Dr. Schmidt (NYU), and TetO-mS100a7a15 mice were kindly provided by Dr. Yuspa (NIH). TetO-mS100a7a15 mice (15) were cross-bred with MMTV-rtTA mice to generate bi-transgenic MMTV-mS100a7a15 mice. Knockout and transgenic littermates were genotyped by PCR. Female MMTV-mS100a7a15 mice were fed with Dox-chow 1 g/kg (Bio-Serv), and mice with normal diet served as controls. All mice were kept in The OSU's animal facility in compliance with the guidelines and protocols approved by the OSU-IACUC.
Orthotopic injection assay
MVT-1 or PyMT cells were injected into the mammary glands of transgenic or Knockout mice. Transgenic mice injected with MVT-1 cells were either fed with Dox-chow 1 g/kg or normal diet (control). Tumors were measured weekly with external calipers and volume was calculated according to the formula V = 0.52 × a2 × b, where a is the smallest superficial diameter and b is the largest superficial diameter. Orthotopically injected animals were sacrificed and tumors were excised (18). RAGE neutralizing antibody and soluble RAGE were purchased from R&D Systems.
FACS Analysis
Freshly prepared single cell suspensions of tumor-infiltrating cells were incubated with anti-F4/80 PE or anti-CD11b APC (18). RAGE expression was analyzed by staining with RAGE antibody (Abcam) followed by Alexa Fluor 488 antibody. After staining, cells were analyzed by FACS Caliber using CellQuest software (BD Biosciences).
Western Blot and Co-immunoprecipitation
Western blot (WB) analysis of cell or tumor lysates was done as described (30). Co-immunoprecipitation was carried out using protein G plus A-agarose beads as described (31), with S100A7 rabbit (Novus Biologicals) and RAGE mouse (Santa Cruz Biotechnology) antibodies.
Luciferase reporter assay
NF-kB activity was determined using NF-kB luciferase reporter assay (Promega) per manufacture's protocol.
Statistical Analysis
To test the association between two categorical variables, Chi-square tests or Fisher's exact tests were used. For continuous variables, two sample t tests were used if two groups were compared, and ANOVAs were used if more than 2 groups were compared. * indicates P < 0.05; ** indicates P < 0.01.
Results
RAGE is expressed in highly metastatic breast cancer cells and its expression correlates with worse clinical prognosis
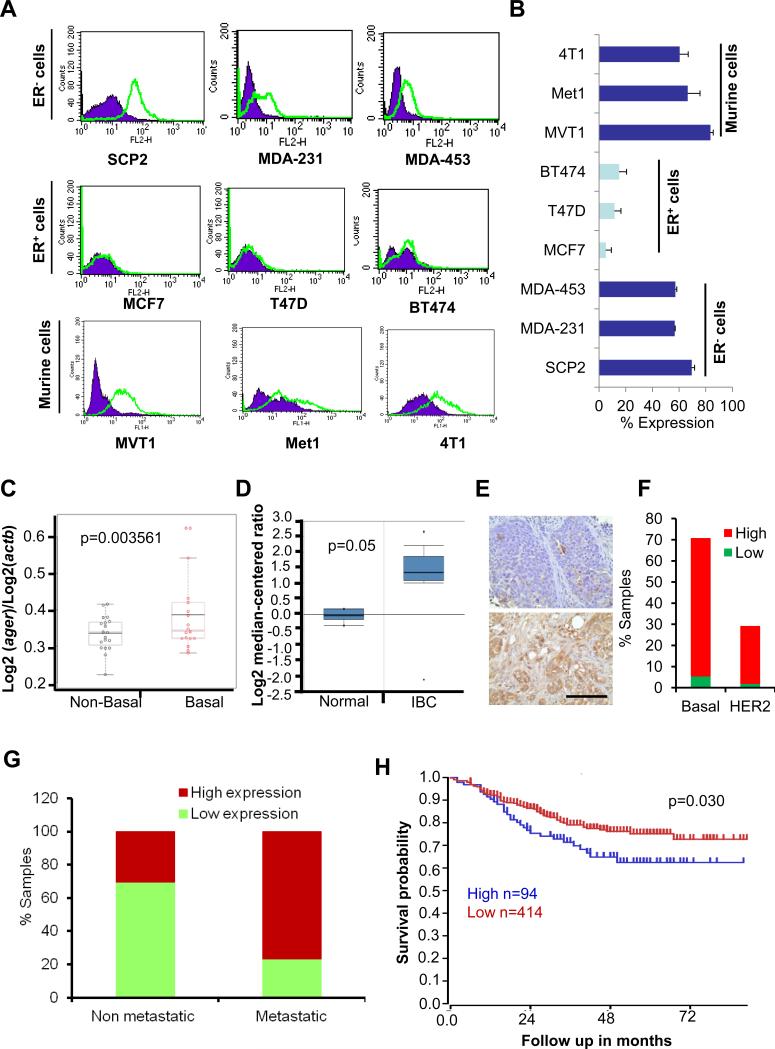
We analyzed RAGE expression in a panel of breast cancer cell lines. RAGE expression was higher in metastatic TNBC cell lines whereas low or no RAGE expression was observed in ERα+ breast cancer cell lines (MCF7, T47D and BT474) (Figure 1A-B) which are weakly metastatic (32-34). This data suggests that RAGE is predominantly expressed in ERα− and highly metastatic breast cancer cell lines. To test the correlation of RAGE with ERα− status, we analyzed open-access Gene Expression Omnibus (GEO) datasets for the expression of RAGE. In a subtype specific breast cancer study (GEO accession GDS2250), RAGE expression is significantly enhanced in basal type (majorly TNBC) and invasive breast cancer patient tumor samples compared to non-basal type tumors (majorly ERα+ cancer) and normal, respectively (Figure 1C). Next, we analyzed open-access dataset for RAGE expression. We found high RAGE expression was observed in invasive breast cancer (IBC) compared with normal control (Figure 1D). Further, we analyzed the expression of RAGE in breast cancer TMAs with accompanying outcome data and other clinical information by immunohistochemistry. We found that 92% of the samples showed high RAGE expression in TNBC tissues (Figure 1E). However, RAGE was expressed only in 29% HER2-positive TMAs (Figure 1E). Using another TMA that contained metastatic and malignant patient samples, we found that metastatic tissue tends to have higher RAGE expression than malignant tissue (p<0.0001) (Figure 1F). Next, we analyzed open-access dataset for clinical outcome of RAGE expression. We found high RAGE expression was associated with poor prognosis (Figure 1G). Taken together, these results show that RAGE expression is associated with highly aggressive and metastatic breast cancers, including TNBC.
Figure 1. RAGE expression in breast cancer cell lines and patient samples.
(A) FACS analysis of RAGE in human TNBC, ERα+ and highly metastatic murine mammary tumor cell lines. (B) Quantification of RAGE expression obtained by FACS. (C) RAGE expression in basal and non-basal type breast cancer. (D) RAGE expression in normal and invasive breast cancer. (E left panel) Representative photographs of RAGE expression in TNBC TMA. (E right panel) Bar graph showing RAGE expression in TNBC (n=80) and HER2 (n=33) TMA samples. (F) Bar graph showing RAGE expression in metastatic breast cancer. We used TMA (n=100) that contained n=40 lymph node metastasis, n=50 malignant and n=10 normal tissues. (G) Expression level of RAGE predicts survival differences by Kaplan-Meier (KM) analysis using R2 microarray dataset. Scale bar: 100μm. *, p<0.05
Blockade of RAGE inhibits tumor growth and metastasis in vivo
To investigate the role of RAGE in metastasis, we used the IVIS imaging system to analyze the metastatic potential of RAGE expressing SCP2 cells. We injected highly metastatic single cell progeny clone 2 (SCP2) of MDA-MB-231 cell lines intracardially into nude mice and blocked RAGE activity with RAGE neutralizing antibody (naRAGE). As shown in Figures 2A-B, naRAGE treatment significantly reduced the metastatic potential of SCP2 cells as compared to IgG control treated mice. Next, we elucidated the role of host RAGE on mammary cancer progression and development using the RAGE−/− model. First, we analyzed surface expression of RAGE on PyMT cells by FACS analysis. As shown in Figure 2C, PyMT cells expressed high RAGE expression. Next, mice were injected with PyMT cells and observed for tumor growth for 35 days (Figure 2D). Interestingly, PyMT-derived tumor growth was significantly inhibited in RAGE−/− mice as compared to RAGE+/+ mice (p=0.0125) (Figure 2E-F). To identify the signaling molecules that are associated with breast cancer tumor growth, we examined the expression of ERK, cyclinD1, and MMP2 in PyMT-derived tumor tissues obtained from RAGE−/− mice. We observed reduced phospho-ERK, MMP2, cyclinD1, CD31 and Ki67 expression in RAGE−/− mice compared to RAGE+/+ mice (Figure 2G-H). CyclinD1 and Ki67 are markers for active proliferation (35-36). This data demonstrates that RAGE blockade significantly inhibits mammary tumor development and progression.
Figure 2. Inhibition of RAGE reduces breast cancer growth and distant metastasis.
(A) Luciferase positive SCP2 cells (1×105/100μL) were injected intr-acardially into nude mice (n=6) pre-treated with RAGE neutralizing (naRAGE) or IgG antibody. Representative BLI images show comparative metastases of naRAGE or IgG treated mice. (B) Normalized photon flux of mice treated with naRAGE or IgG. (C) RAGE expression was analyzed in PyMT cells by FACS. (D) PyMT (1×106) cells were injected into the mammary gland of RAGE+/+ and RAGE−/− mice (n=5) and tumor volume was measured every week. (E) After five weeks, tumors were excised from mice and weighed. (F) Representative photograph of mice showing tumors dissected from different experimental groups. (G) PyMT cell line derived tumors from RAGE+/+ and RAGE−/− mice were subjected to IHC staining for Ki67 and CD31 expression (H) Tumor lysates (50 μg) from RAGE+/+ and RAGE−/− mice were subjected to Western blot using phospho-ERK (P-ERK), total ERK (T-ERK), cyclinD1 and MMP-2 antibodies. GAPDH served as loading control. Data represent mean ± SD of three independent experiments. Scale bar: 100μm. *, p<0.05 and **, p<0.01.
RAGE mediates its effect in breast cancer cells through S100A7
RAGE has been shown to bind to S100A7 in various immune cells (14). When analyzing the RAGE and S100A7 expression in breast cancer (TCGA), we discovered that RAGE and S100A7 are often simultaneously upregulated in breast tumors among patients of the IDC cohort (p=0.0055, Fisher's Exact Test); whereas an irrelevant gene ERK (mapk1) showed no correlation with RAGE or S100A7 (Figure 3A). This co-currency of gene upregulation implies a functional link between RAGE and S100A7 in breast cancer. Furthermore, we observed enhanced expression of RAGE in S100A7- overexpressing MDA-MB-231 cells by Western blot, FACS and immunofluorescence analysis (Figure 3B-C-D). This suggests that RAGE could be a receptor for S100A7 as ligand in breast cancer cells. We further verified the direct interaction between human RAGE and human S100A7 protein using an ELISA-based binding assay, in which EGFR, an irrelevant receptor, was used as a negative control. As shown in Figure 3E, the total binding was dependent on RAGE-Fc concentration, whereas no binding was observed between EGFR-Fc and S100A7 protein. To further confirm the association of RAGE with S100A7, we performed a co-immunoprecipitation (IP) assay. S100A7 co-immunoprecipitated with RAGE in S100A7-overexpressing MDA-MB-231 cells (Figure 1F). Next, we analyzed the effect of human S100A7 on MDA-MB-231 and SCP2 cell migration and that of its murine paralog mS100A7a15 on MVT-1 cell migration. S100A7 enhanced the migration of MDA-MB-231 and SCP2 cells, respectively and these effects were significantly abrogated by blocking RAGE using neutralizing antibodies (Figure 3G-H). We also showed that its murine ortholog mS100a7a15 enhanced migration of MVT1 cells (Figure S1A). Next, we analyzed the S100A7-induced wound healing capacity of MDA-MB-231 and MDA-MB-453 cell lines and observed that RAGE neutralizing antibody significantly abrogated this effect (Figure S1B-C). In addition, we observed that S100A7 or mS100a7a15 significantly enhanced the invasion of SCP2 and MVT-1 cell lines (Figure S1D-E). To examine whether activation of RAGE/S100A7 enhanced downstream signaling in breast cancer cells, we analyzed ERK activation. We showed that S100A7-induced ERK activation was inhibited by blocking S100A7 with soluble RAGE (sRAGE) in MDA-MB-231 and SCP2 cells (Figure 3I-J). To further confirm these effects, we used RAGE neutralizing antibody to block S100A7-induced ERK activation in MDA-MB-231 cells (Figure S1F). In addition, we observed that sRAGE inhibits S100A7-induced MMP9 activation in SCP2 cells (Figure S1F). NF-κB has also been shown to be the downstream target of RAGE (37). Using NF-κB reporter assay, we showed that S100A7 significantly enhanced NF-κB activity and this effect was inhibited by naRAGE (Figure 3K). Taken together, these results imply that the RAGE/S100A7/mS100a7a15 signaling axis is necessary for breast cancer cell migration and invasion.
Figure 3. RAGE receptor binds to S100A7 and enhances the migration of breast cancer cell lines.
(A) Co-currency of ager (rage) and s100a7 gene upregulation (>1.0 SD) was analyzed as described (TCGA, Nature, 2012) (50). An irrelevant gene mapk1 (ERK gene) is used as control, which showed no association with ager and s100a7. (B) RAGE expression was analyzed in S100A7-overexpressing MDA-MB-231 (SA7) cells compared to vec control (Vec) as determined by (D) Western blot (C) Flow-cytometry analysis (D) Immunofluorescence. (E) S100A7 binding to RAGE as determined by ELISA using RAGE-Fc or EGFR-Fc proteins. Graph shows mean ± SEM of three independent experiments. (F) 1mg of cell lysates from S100A7-overexpressing MDA-MB-231 cells were subjected to immunoprecipitation with IgG or S100A7 and probed with anti-RAGE antibody. (G) MDA-MB-231 cells and (H) SCP2 cells were pretreated with RAGE neutralizing antibody (naRAGE) or control IgG (10μg/ml) for 30 min before being subjected to recombinant S100A7 (SA7, 50ng/ml)-induced migration. (I) MDA-MB-231 and (J) SCP2 cells were pretreated with soluble RAGE (sRAGE) before stimulation with S100A7 at the indicated times. Cell lysates (50 μg) were subjected to Western blot using phospho-ERK (P-ERK) and total ERK (T-ERK). (K) MDA-MB-231 cells were transfected with either wild-type or NF-kB plasmid for 24h, stimulated with S100A7 (100ng/ml) or IgG (10μg) or naRAGE (10μg) for additional 24h, lysed and analyzed for luciferase activity. Renilla luciferase vector served as internal control. Graphs represent mean ± SD for each experiment repeated three times with similar results.*, p<0.05 and **, p<0.01.
Blockade of RAGE inhibits mammary tumor growth and metastasis in inducible MMTV-mS100a7a15 mice
In order to analyze the relevance of RAGE/mS100a7a15 in a mammary tumor growth and metastasis model, we employed a MVT-1 syngeneic orthotopic model which recapitulates the stages of human primary tumors. We injected MVT-1 cells into inducible MMTV-mS100a7a15 transgenic mice and blocked RAGE with neutralizing RAGE antibody. Mice were treated with doxycycline (Dox)-chow (1g/kg) one week before MVT-1 injection to switch on the expression of mS100a7a15. When tumors grew as large as 100mm3, the Dox-treated group was given naRAGE (20 μg/mouse) or IgG (20μg/mouse) intraperitoneally three times a week for 20 days. MMTV-mS100a7a15 mice fed with normal chow were used as a negative control. Inducible mice treated with naRAGE showed reduced tumor progression compared to the IgG treated group (Figure 4A-B-C). In order to determine if a blocking ligand of RAGE inhibits mammary tumor progression, we used soluble RAGE (sRAGE). Mice were fed with Dox-chow one week before the injection of MVT-1 cells into the #4 mammary gland. After day 1 of Dox induction, mice were injected intra-peritoneally with murine sRAGE (2μg/ mouse) or PBS. MMTV-mS100a7a15 mice who received normal chow served as a negative control. As shown in Figure 4D-E-F, sRAGE treatment significantly reduced tumor progression in the Dox-induced group as compared to the PBS Dox-induced group. Furthermore, we observed that blocking RAGE or mS100a7a15 substantially decreased proliferation and angiogenesis in MVT-1-derived tumors obtained from inducible MMTV-mS100a7a15 (Figure 4G-H). Next, we investigated whether RAGE inhibition reduces surface lung metastases in inducible MMTV-mS100a7a15 mice. We observed a significant decrease in surface lung metastases in the mice treated with naRAGE (Figure 4I-J) or sRAGE (Figure 4K-L) in MVT-1-derived tumors obtained from inducible MMTV-mS100a7a15 mice as compared to control groups. Taken together, these results suggest that RAGE plays an important role in mS100a7a15-induced breast cancer progression and metastasis.
Figure 4. Blockade of RAGE inhibits mammary tumor growth and metastasis in MMTV-mS100a7a15 inducible mice.
(A) MVT-1 (1× 105) cells were injected into the mammary gland of doxycycline treated (+Dox) or untreated (-Dox) MMTV-mS100a7a15 mice (n=5). Dox treated mice (n=10) were injected intraperitoneally with RAGE neutralizing antibody (naRAGE) or control IgG (20 μg/mouse) for every alternate day and tumor volume was measured every week. (B) After 28 days, the tumors were excised. (C) Representative photograph of tumors dissected from different experimental groups. (D) MVT-1 (1× 105) cells were injected into the mammary gland of +Dox or -Dox MMTV-mS100a7a15 mice (n=5). Dox treated MMTV-mS100a7a15 mice (n=10) were either treated with sRAGE (SR, 10μg/mouse) or PBS for every alternate day and tumor volume was measured every week. (E) After 28 days, the tumors were excised. (F) Representative photograph of tumors dissected from different experimental groups. IHCs were performed for CD31 and Ki67 in tumors from Dox–induced MMTV-mS100a7a15 mice that were either treated with (G) naRAGE or control IgG or (H) SR. Data represent mean ± SD of three independent experiments. Scale bar: 100μm. *, p<0.05 and **, p<0.01. (I) Representative photograph of lungs dissected from naRAGE treated groups. Right panel showed H&E staining of metastatic deposits. (J) Bar graph showing the number of metastatic nodules on the lungs (18). (K) Representative photograph of lungs dissected from SR treated groups. Right panel showed H&E staining of metastatic deposits. (L) Bar graph showing the number of metastatic nodules on the lungs. Data represent mean ± SD per experimental group. Scale bar: 20μm. *, p<0.05 and **, p<0.01.
RAGE/S100A7-axis modulates the tumor microenvironment
Macrophages, especially M2-TAMs, have been shown to enhance tumor growth and metastasis (21-22,38-39). In order to identify the molecular mechanism of RAGE-mediated breast tumor growth and metastasis, we analyzed macrophage recruitment in the PyMT-derived tumors of RAGE+/+ and RAGE−/− mice. As shown in Figure S2, F4/80/Arg1-positive macrophages were substantially reduced in RAGE−/− PyMT derived tumors as compared to RAGE+/+ tumors. Next, we observed that mS100a7a15-overexpressing mice significantly increased the recruitment of F4/80+/Arg-1 macrophages and RAGE blockage by naRAGE-treated significantly reduced the recruitment of F4/80+/Arg-1 macrophages compared to IgG control, as analyzed by IHC (Figure 5A). In addition, we observed substantial decrease in the recruitment of M2 macrophages in MVT-1 derived tumors pretreated with sRAGE in inducible mice compared to PBS treated mice (Figure 5B). We also observed reduced CD11b/F4/80+ TAMs by FACS (Figure 5C). We further showed that decreased expression of iNOS (M1 marker) in primary tumors compared to IgG control (Figure 5A-B). We also analyzed the infiltrations of macrophages in the lung tissues and observed reduced expression of F4/80+ macrophages and Arg-1 expression in naRAGE or sRAGE treated MMTV-mS100a7a15 inducible mice when compared to control mice (Figure S3). These studies suggest that blockade of RAGE in mS100a7a15-transgenic mice inhibits tumor growth and metastasis through inhibition of M2 macrophage recruitment.
Figure 5. RAGE/mS100a7a15 regulates tumor associated macrophage recruitment.
(A) Representative IHC staining of macrophage markers (F4/80+, Arg-1 and iNOS) in MVT-1 tumors derived from naRAGE or IgG-treated Dox-induced (+Dox) or uninduced (-Dox) MMTV-mS100a7a15 mice. (B) Intratumoral macrophages are highlighted with IHC for macrophage markers in MVT-1 tumors derived from sRAGE (SR) or PBS treated Dox-induced or uninduced MMTV-mS100a7a15 mice. Scale bar: 100μm. (C-D) Quantitative analyses of F4/80+ macrophages by FACS in MVT-1 tumors derived from naRAGE or IgG-treated +Dox or –Dox MMTV-mS100a7a15 mice.
Since macrophage recruitment to primary tumors plays an important role in promoting S100A7/mS100a7a15-induced metastasis (18), we wanted to know whether recombinant mS100a7a15- might affect macrophage activity in a RAGE dependent manner. We analyzed the migration of RAW264.7 (RAW), a macrophage cell line, in the presence of mS100a7a15 recombinant protein. We showed that mS100a7a15 significantly enhanced the migration of RAW and that pretreatment with naRAGE significantly abrogated mS100a7a15-induced migration compared to IgG control (Figure 6A). Though S100A7 has been shown to regulate MMPs in cancer cells (17), its role in the macrophage is not known. We analyzed MMP9 secretion in the presence of mS100a7a15 with or without sRAGE treatment. mS100a7a15-induced MMP9 secretion was enhanced in RAW cells and this effect was diminished in the presence of sRAGE (Figure 6B). Using double immunofluorescence, we also observed enhanced recruitment of MMP9-expressing F4/80+ macrophages in the MVT-1 tumors of MMTV-mS100a7a15 inducible mice compared to noninducible mice. The recruitment of MMP9+/F4/80+ macrophages was substantially decreased in the naRAGE treated group compared to IgG treated MMTV-mS100a7a15 inducible mice (Figure 6C). We further analyzed the molecular mechanism of the mS100a7a15-induced migration of RAW cells. As shown in Figure 6D&E, mS100a7a15-induced ERK activation was significantly reduced in the presence of sRAGE treatment. Furthermore, our double immunofluorescence analyses showed that RAGE ablation or its blocking by naRAGE or sRAGE substantially reduced F4/80+ macrophages (Figure 6C). These studies suggest that RAGE receptor regulates mS100a7a15-induced ERK activation in macrophages.
Figure 6. RAGE/mS100a7a15 regulates MMP9+ macrophages.
(A) RAGE expression was analyzed on RAW cells by FACS. (B) RAW cells were subjected to mS100a7a15 (100 ng/ml)-induced migration in presence of murine RAGE neutralizing (naRAGE) or control IgG antibodies. (C) RAW cells were stimulated with mS100a7a15 (100ng/ml) in the presence or absence of sRAGE (SR, 50 ng/ml) for 24h. Conditioned media was analyzed for MMP activity. (C lower panel) Quantification (D) Tumors excised from (−Dox) MMTV-mS100a7a15 mice were subjected to double immunofluorescence for MMP9 (green), F4/80 (red) or DAPI (blue). (E) RAW cells were pretreated with naRAGE or IgG antibodies for 1h, stimulated with mS100a7a15 (SA15, 100ng/ml) and subjected to WB for P-ERK. (E) Quantification of Western blots. Data represent mean ± SD per experimental group. Scale bar: 100μm.*, p<0.05 and **, p<0.01.
Discussion
Emerging data have implicated importance of RAGE in the pathogenesis of various human disorders including cancers (2,10). The interactions between RAGE and its ligands trigger the activation of MAPK, JAK/STAT, and NF-κB in prostate cancer (1-2). In this work, we identified that RAGE plays a critical role in promoting breast cancer growth and metastasis. We documented that RAGE is highly expressed in human TNBC and murine breast cancer cell lines and weakly expressed in low metastatic ER+ cells. However, there are some conflicting reports regarding the expression of RAGE in the MCF7 cell line (40-41). It could be due to the difference in the techniques which were employed. We detected RAGE expression by FACS analysis. However, other studies detected RAGE expression by Western blot analysis. There is possibility that detected RAGE by Western blot could be a truncated or soluble form of RAGE. In addition, we showed that RAGE is preferentially expressed in invasive and metastatic tumor deposits. This is consistent with a recent report which demonstrated that high RAGE expression was observed in lymph node and distant metastases patients samples (42). Moreover, we observed that RAGE expression was associated with poor prognosis in breast cancer. We also characterized the role of RAGE in breast cancer progression and metastasis. Using a genetic approach, we showed that RAGE ablation significantly reduced PyMT cell-derived tumor growth while blocking RAGE with neutralizing antibodies inhibits breast cancer visceral metastasis in preclinical mouse models. It has been shown recently that RAGE knockdown by siRNA significantly inhibited tumorigenic potential of MDA-MB231 cell line (40). RAGE ablation in a triple transgenic model of spontaneous pancreatic cancer has also been shown to delay pancreatic cancer development (9,43).
Moreover, we demonstrated that RAGE binding to S100A7 enhanced RAGE expression in breast cancer cells. In addition, we provide evidence that S100A7 co-immuno-precipitated with RAGE in S100A7-overexpressing MDA-MB-231 cells. Our bio-informatics data also showed that RAGE is co-overexpressed with S100A7 in human breast cancer tissue. Proinflammatory RAGE ligands, such as S100B, S100A4, and S100A8/A9, have been shown to enhance RAGE expression, and continuous activation of RAGE fuels an inflammatory milieu at the tumor microenvironment (2,44). Our data suggests that S100A7 enhances activation of NF-κB through RAGE activation. We also showed that blocking RAGE or inhibiting S100A7/mS100a7a15 binding to RAGE by soluble RAGE (sRAGE) inhibits breast cancer cell migration and ERK activation. The sRAGE acts as a decoy that prevents ligands from interacting with the cell surface receptor. The application of sRAGE in vitro and in vivo resulted in an effective blockade of RAGE, in accordance with this decoy mechanism, in a range of animal models (11). It is well documented that S100A8/A9-RAGE axis plays a significant role in breast and colon cancer growth and metastasis by modulating its downstream targets such as ERK1/2 (P44/p42), mitogen activated protein kinases (MAPK), and NF-κB signaling pathways (7,42,45).
In the current study, we have shown that RAGE deficiency in the host reduced breast cancer growth by decreasing recruitment of tumor associated macrophages and tumor angiogenesis. We have shown previously that mS100a7a15 overexpression in mammary glands enhanced mammary tumor growth metastasis through macrophage recruitment (18). To further analyze molecular mechanism of these effects, we blocked RAGE activation in MMTV-mS100a7a15 inducible model by naRAGE or sRAGE.Blocking RAGE reduced macrophage recruitment into the MVT1 derived tumors in the MMTV-mS100a7a15 inducible model. Our study further revealed that blocking the RAGE/mS100a7a15 axis inhibits M2-marker arginase expression. M2 polarized TAMs are known to drive tumor progression by stimulating angiogenesis and metastasis (21,38,46). We did not observe a significant change in CD4/CD3/CD8-positive T-cells and other immune cells such as NK cells, as detected by FACS in MVT-1 derived tumors obtained from MMTV-mS100a7a15 inducible mice treated with RAGE neutralizing antibody or sRAGE.
We showed that blocking of RAGE inhibits mS100a7a15-induced recruitment and MMP9 activation in macrophages. MMP9 has been shown to degrade the ECM and release growth factors to enhance angiogenesis (47-48). Furthermore, it has been shown that MMP9 induction by primary tumors in macrophages and the lung endothelium promotes metastasis, especially to lung (48-49).
In summary, our study shows that RAGE is highly expressed in basal-type breast cancer, especially TNBC, and is preferentially expressed in invasive and lymph node metastasis tissues. Elucidation of the molecular mechanism behind enhanced breast cancer growth and metastasis shows that this is likely due to binding of RAGE to S100A7. In turn, the RAGE/S100A7 axis is responsible for enhanced recruitment of MMP9 positive TAMs. We have also shown that RAGE neutralizing antibody and soluble RAGE, significantly decrease tumor growth and metastasis in an inducible mS100a7a15 transgenic mouse model. This data implies that the RAGE/S100A7 signaling axis could be used to inhibit TNBC breast cancer growth and metastasis. Furthermore, these studies demonstrate that RAGE could be used as a novel biomarker and that neutralizing antibodies/soluble RAGE could be used to develop novel therapeutic strategies against TNBC.
Supplementary Material
Precis.
Findings offer robust evidence for key contibutions of the RAGE/S100A7 signaling axis in conditioning an inflammatory microenvironment that drives aggressive breast cancers, including deadly triple-negative tumors that occur more commonly in pre-menopausal women.
Figure 7. Schematic representation of RAGE-mediated S100A7-induced signaling that regulates breast cancer growth and metastasis.
Epithelial cells release S100A7/mS100a7a15 which binds to RAGE and activates signaling cascades that recruit TAMs to the tumor stroma. TAMs in turn enhance growth and metastasis by secreting growth factor, chemokines/cytokines, and MMPs. Blocking of RAGE/S100A7-axis by sRAGE or naRAGE may reduce breast tumor growth and metastasis, especially to lungs.
Acknowledgements
We thank Kristin Kovach for assistance with immunohistochemistry. We thank Drs. Z. Qamri, A. Sneh, D. Chakroborty and G. Amponsah for technical assistance and S. Adamovich for critical reading of the manuscript.
Grant Support: This work was supported by grants from NIH (CA109527 and CA153490) and Department of Defense to RKG. NAW, DKA and HZ were supported by Pelotonia Fellowship from the Comprehensive Cancer Center, OSU.
Abbreviations
- S100A7
Psoriasin
- TAM
tumor associated macrophage
- MMP9
metalloproteinase 9
Footnotes
The authors declare that they have no potential conflicts of interest.
Authors' Contributions
Conception, design, interpretation and manuscript writing and revision: MWN, NAW, RKG
Data acquisition, statistical and computational analysis and technical support: MWN, NAW, DKA, CAP, JR, ME, HZ, LP, XZ, KS, MO, CS, WEC, RKG.
Study supervision: RKG
References
- 1.Sparvero LJ, Asafu-Adjei D, Kang R, Tang D, Amin N, Im J, et al. RAGE (Receptor for Advanced Glycation Endproducts), RAGE ligands, and their role in cancer and inflammation. J Transl Med. 2009;7:17. doi: 10.1186/1479-5876-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rojas A, Figueroa H, Morales E. Fueling inflammation at tumor microenvironment: the role of multiligand/RAGE axis. Carcinogenesis. 2010;31(3):334–41. doi: 10.1093/carcin/bgp322. [DOI] [PubMed] [Google Scholar]
- 3.de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13(6):607–15. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 4.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–37. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 7.Gebhardt C, Riehl A, Durchdewald M, Nemeth J, Furstenberger G, Muller-Decker K, et al. RAGE signaling sustains inflammation and promotes tumor development. J Exp Med. 2008;205(2):275–85. doi: 10.1084/jem.20070679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heijmans J, Buller NV, Hoff E, Dihal AA, van der Poll T, van Zoelen MA, et al. Rage signalling promotes intestinal tumourigenesis. Oncogene. 2013;32(9):1202–6. doi: 10.1038/onc.2012.119. [DOI] [PubMed] [Google Scholar]
- 9.Kang R, Loux T, Tang D, Schapiro NE, Vernon P, Livesey KM, et al. The expression of the receptor for advanced glycation endproducts (RAGE) is permissive for early pancreatic neoplasia. Proc Natl Acad Sci U S A. 2012;109(18):7031–6. doi: 10.1073/pnas.1113865109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riehl A, Nemeth J, Angel P, Hess J. The receptor RAGE: Bridging inflammation and cancer. Cell Commun Signal. 2009;7:12. doi: 10.1186/1478-811X-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taguchi A, Blood DC, del Toro G, Canet A, Lee DC, Qu W, et al. Blockade of RAGE- amphoterin signalling suppresses tumour growth and metastases. Nature. 2000;405(6784):354–60. doi: 10.1038/35012626. [DOI] [PubMed] [Google Scholar]
- 12.Turovskaya O, Foell D, Sinha P, Vogl T, Newlin R, Nayak J, et al. RAGE, carboxylated glycans and S100A8/A9 play essential roles in colitis-associated carcinogenesis. Carcinogenesis. 2008;29(10):2035–43. doi: 10.1093/carcin/bgn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Zhang L, Zhang IY, Chen X, Da Fonseca A, Wu S, et al. S100B promotes glioma growth through chemoattraction of myeloid-derived macrophages. Clin Cancer Res. 2013;19(14):3764–75. doi: 10.1158/1078-0432.CCR-12-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf R, Howard OM, Dong HF, Voscopoulos C, Boeshans K, Winston J, et al. Chemotactic activity of S100A7 (Psoriasin) is mediated by the receptor for advanced glycation end products and potentiates inflammation with highly homologous but functionally distinct S100A15. J Immunol. 2008;181(2):1499–506. doi: 10.4049/jimmunol.181.2.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolf R, Mascia F, Dharamsi A, Howard OM, Cataisson C, Bliskovski V, et al. Gene from a psoriasis susceptibility locus primes the skin for inflammation. Sci Transl Med. 2010;2(61):61ra90. doi: 10.1126/scitranslmed.3001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Haddad S, Zhang Z, Leygue E, Snell L, Huang A, Niu Y, et al. Psoriasin (S100A7) expression and invasive breast cancer. Am J Pathol. 1999;155(6):2057–66. doi: 10.1016/S0002-9440(10)65524-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sneh A, Deol YS, Ganju A, Shilo K, Rosol TJ, Nasser MW, et al. Differential role of psoriasin (S100A7) in estrogen receptor alpha positive and negative breast cancer cells occur through actin remodeling. Breast Cancer Res Treat. 2013;138(3):727–39. doi: 10.1007/s10549-013-2491-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasser MW, Qamri Z, Deol YS, Ravi J, Powell CA, Trikha P, et al. S100A7 enhances mammary tumorigenesis through upregulation of inflammatory pathways. Cancer Res. 2012;72(3):604–15. doi: 10.1158/0008-5472.CAN-11-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.West NR, Watson PH. S100A7 (psoriasin) is induced by the proinflammatory cytokines oncostatin-M and interleukin-6 in human breast cancer. Oncogene. 2010;29(14):2083–92. doi: 10.1038/onc.2009.488. [DOI] [PubMed] [Google Scholar]
- 20.Hartman ZC, Poage GM, den Hollander P, Tsimelzon A, Hill J, Panupinthu N, et al. Growth of triple-negative breast cancer cells relies upon coordinate autocrine expression of the proinflammatory cytokines IL-6 and IL-8. Cancer Res. 2013;73(11):3470–80. doi: 10.1158/0008-5472.CAN-12-4524-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4(1):71–8. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 22.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122(3):787–95. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biswas SK, Lewis CE. NF-kappaB as a central regulator of macrophage function in tumors. J Leukoc Biol. 2010;88(5):877–84. doi: 10.1189/jlb.0310153. [DOI] [PubMed] [Google Scholar]
- 24.Gasparini P, Cascione L, Fassan M, Lovat F, Guler G, Balci S, et al. microRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget. 2014;5(5):1174–84. doi: 10.18632/oncotarget.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shilo K, Dracheva T, Mani H, Fukuoka J, Sesterhenn IA, Chu WS, et al. Alpha- methylacyl CoA racemase in pulmonary adenocarcinoma, squamous cell carcinoma, and neuroendocrine tumors: expression and survival analysis. Arch Pathol Lab Med. 2007;131(10):1555–60. doi: 10.5858/2007-131-1555-MCRIPA. [DOI] [PubMed] [Google Scholar]
- 26.Hernandez JL, Padilla L, Dakhel S, Coll T, Hervas R, Adan J, et al. Therapeutic targeting of tumor growth and angiogenesis with a novel anti-S100A4 monoclonal antibody. PLoS One. 2013;8(9):e72480. doi: 10.1371/journal.pone.0072480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9(2):121–32. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 28.Minn AJ, Kang Y, Serganova I, Gupta GP, Giri DD, Doubrovin M, et al. Distinct organ- specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Invest. 2005;115(1):44–55. doi: 10.1172/JCI22320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolford CC, McConoughey SJ, Jalgaonkar SP, Leon M, Merchant AS, Dominick JL, et al. Transcription factor ATF3 links host adaptive response to breast cancer metastasis. J Clin Invest. 2013;123(7):2893–906. doi: 10.1172/JCI64410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qamri Z, Preet A, Nasser MW, Bass CE, Leone G, Barsky SH, et al. Synthetic cannabinoid receptor agonists inhibit tumor growth and metastasis of breast cancer. Mol Cancer Ther. 2009;8(11):3117–29. doi: 10.1158/1535-7163.MCT-09-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prasad A, Paruchuri V, Preet A, Latif F, Ganju RK. Slit-2 induces a tumor-suppressive effect by regulating beta-catenin in breast cancer cells. J Biol Chem. 2008;283(39):26624–33. doi: 10.1074/jbc.M800679200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iorns E, Drews-Elger K, Ward TM, Dean S, Clarke J, Berry D, et al. A new mouse model for the study of human breast cancer metastasis. PLoS One. 2012;7(10):e47995. doi: 10.1371/journal.pone.0047995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lasfargues EY, Coutinho WG, Redfield ES. Isolation of two human tumor epithelial cell lines from solid breast carcinomas. J Natl Cancer Inst. 1978;61(4):967–78. [PubMed] [Google Scholar]
- 34.Lee TH, Seng S, Sekine M, Hinton C, Fu Y, Avraham HK, et al. Vascular endothelial growth factor mediates intracrine survival in human breast carcinoma cells through internally expressed VEGFR1/FLT1. PLoS Med. 2007;4(6):e186. doi: 10.1371/journal.pmed.0040186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang TC, Cardiff RD, Zukerberg L, Lees E, Arnold A, Schmidt EV. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature. 1994;369(6482):669–71. doi: 10.1038/369669a0. [DOI] [PubMed] [Google Scholar]
- 36.Weinstat-Saslow D, Merino MJ, Manrow RE, Lawrence JA, Bluth RF, Wittenbel KD, et al. Overexpression of cyclin D mRNA distinguishes invasive and in situ breast carcinomas from non-malignant lesions. Nat Med. 1995;1(12):1257–60. doi: 10.1038/nm1295-1257. [DOI] [PubMed] [Google Scholar]
- 37.Bierhaus A, Schiekofer S, Schwaninger M, Andrassy M, Humpert PM, Chen J, et al. Diabetes-associated sustained activation of the transcription factor nuclear factor-kappaB. Diabetes. 2001;50(12):2792–808. doi: 10.2337/diabetes.50.12.2792. [DOI] [PubMed] [Google Scholar]
- 38.Lin EY, Pollard JW. Tumor-associated macrophages press the angiogenic switch in breast cancer. Cancer Res. 2007;67(11):5064–6. doi: 10.1158/0008-5472.CAN-07-0912. [DOI] [PubMed] [Google Scholar]
- 39.Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol. 2010;22(2):231–7. doi: 10.1016/j.coi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 40.Radia AM, Yaser AM, Ma X, Zhang J, Yang C, Dong Q, et al. Specific siRNA Targeting Receptor for Advanced Glycation End Products (RAGE) Decreases Proliferation in Human Breast Cancer Cell Lines. Int J Mol Sci. 2013;14(4):7959–78. doi: 10.3390/ijms14047959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lata K, Mukherjee TK. Knockdown of receptor for advanced glycation end products attenuate 17alpha-ethinyl-estradiol dependent proliferation and survival of MCF-7 breast cancer cells. Biochim Biophys Acta. 2014;1840(3):1083–91. doi: 10.1016/j.bbagen.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 42.Yin C, Li H, Zhang B, Liu Y, Lu G, Lu S, et al. RAGE-binding S100A8/A9 promotes the migration and invasion of human breast cancer cells through actin polymerization and epithelial-mesenchymal transition. Breast Cancer Res Treat. 2013;142(2):297–309. doi: 10.1007/s10549-013-2737-1. [DOI] [PubMed] [Google Scholar]
- 43.DiNorcia J, Lee MK, Moroziewicz DN, Winner M, Suman P, Bao F, et al. RAGE gene deletion inhibits the development and progression of ductal neoplasia and prolongs survival in a murine model of pancreatic cancer. J Gastrointest Surg. 2012;16(1):104–12. doi: 10.1007/s11605-011-1754-9. discussion 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bierhaus A, Humpert PM, Morcos M, Wendt T, Chavakis T, Arnold B, et al. Understanding RAGE, the receptor for advanced glycation end products. J Mol Med (Berl) 2005;83(11):876–86. doi: 10.1007/s00109-005-0688-7. [DOI] [PubMed] [Google Scholar]
- 45.Ichikawa M, Williams R, Wang L, Vogl T, Srikrishna G. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol Cancer Res. 2011;9(2):133–48. doi: 10.1158/1541-7786.MCR-10-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sica A, Allavena P, Mantovani A. Cancer related inflammation: the macrophage connection. Cancer Lett. 2008;267(2):204–15. doi: 10.1016/j.canlet.2008.03.028. [DOI] [PubMed] [Google Scholar]
- 47.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Kempen LC, Coussens LM. MMP9 potentiates pulmonary metastasis formation. Cancer Cell. 2002;2(4):251–2. doi: 10.1016/s1535-6108(02)00157-5. [DOI] [PubMed] [Google Scholar]
- 49.Deng J, Liu Y, Lee H, Herrmann A, Zhang W, Zhang C, et al. S1PR1-STAT3 signaling is crucial for myeloid cell colonization at future metastatic sites. Cancer Cell. 2012;21(5):642–54. doi: 10.1016/j.ccr.2012.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Network TCGA Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.