Abstract
Studies of genetic variation can clarify the role of geography and spatio-temporal variation of climate in shaping demography, particularly in temperate zone tree species with large latitudinal ranges. Here, we examined genetic variation in narrowleaf cottonwood, Populus angustifolia, a dominant riparian tree. Using multi-locus surveys of polymorphism in 363 individuals across the species' 1800 km latitudinal range, we found that, first, P. angustifolia has stronger neutral genetic structure than many forest trees (simple sequence repeat (SSR) FST=0.21), with major genetic groups corresponding to large apparent geographical barriers to gene flow. Second, using SSRs and putatively neutral sequenced loci, coalescent simulations indicated that populations diverged before the last glacial maximum (LGM), suggesting the presence of population structure before the LGM. Third, the LGM and subsequent warming appear to have had different influences on each of these distinct populations, with effective population size reduction in the southern extent of the range but major expansion in the north. These results are consistent with the hypothesis that climate and geographic barriers have jointly affected the demographic history of P. angustifolia, and point the importance of both factors as being instrumental in shaping genetic variation and structure in widespread forest trees.
Introduction
Widespread, ecologically dominant forest tree species provide excellent opportunities to explore the relative influences of climate and geography on demographic history and selection (Savolainen et al., 2007). Across their broad ranges, such species span large environmental gradients, and populations are often locally adapted. For example, growth and phenology traits are moderately to strongly heritable and display climatic clines (Howe et al., 2003; Savolainen et al., 2007). Furthermore, repeated Quaternary glacial cycles have had a pronounced effect, seen in both the past and current distribution of species, even in areas that were free of ice during the last glacial maximum (LGM, ca 18 000 years ago; Frenzel et al., 1992; Jackson and Weng, 1999; Hu et al., 2009). These climatic shifts impacted population size, subdivision and migration, leaving genetic signatures in genome-wide patterns of polymorphism and influencing patterns of natural selection (Ingvarsson, 2008; Keller et al., 2010, 2011; Ma et al., 2010; Levsen et al., 2012; Cushman et al., 2014; Evans et al., 2014; Zhou et al., 2014).
Investigating patterns of diversity across putatively neutral and potentially adaptive genes controlling ecologically important traits can provide insight into the importance of neutral demographic and selective forces in forest trees (Ma et al., 2010), identify potential targets for tree breeding programs (Howe et al., 2003) and aid conservation practices by clarifying the role of climate change as a driver of evolution (Savolainen et al., 2007; Alberto et al., 2013). In areas covered by ice sheets during the LGM, several studies have found evidence of recent expansion. Ingvarsson (2008) and Ma et al. (2010) found support for models of population bottlenecks and expansion in P. tremula, as did Keller et al. (2010) in a sample of P. balsamifera, and Zhou et al. (2014) in P. trichocarpa. Outside of regions covered by Pleistocene ice, Eckert et al. (2010) found evidence of expansion from multiple refugia for Pinus taeda based on patterns of population structure. Although these studies have found evidence of expansion since the LGM, warming and drying of southern range portions could reduce regional suitability for some species, leading to population contraction. If this is the case, then local dynamics could vary across a species' range after the LGM, which has yet to be inferred from patterns of genetic variation.
Narrowleaf cottonwood, Populus angustifolia James (Salicaceae), is well-suited to test hypotheses regarding the effect of climate change on temperate species across regions that have warmed and dried considerably since the LGM. This tree is native to North America, diploid, dioecious, wind pollinated and dispersed by wind and water. It is relatively flood tolerant, reproduces sexually and vegetatively, and occurs in riparian habitats in intermountain valleys and along the Rocky Mountains, from southern Arizona northward to southern Alberta, Canada (Cooke and Rood, 2007). Climate and photoperiod vary strongly along this latitudinal gradient, with mean annual temperature ranging from 5 to 13 °C. Narrowleaf cottonwood hybridizes with other species of Populus throughout its range and where species co-occur, it grows at elevations higher than that of P. deltoides or P. fremontii, but lower than that of P. trichocarpa or P. balsamifera, and typically in relatively small, disjunct woodlands separated by river drainages (Floate, 2004; Cooke and Rood, 2007).
Here we examine patterns of diversity at putatively neutral and coding sequences to examine current population structure and infer past demographic events since the LGM. We are specifically addressing the following three questions: (1) what is the role of geographic barriers in genetically structuring populations of P. angustifolia? We used neutral genetic markers (SSRs and sequenced loci) to address this question. (2) Did the LGM (ca 18 000 years ago) and subsequent warming influenced P. angustifolia populations? We used two distinct demographic analyses, to test whether population divergences in the north or south of the range coincided with the LGM, and whether population sizes changed subsequent to the LGM. (3) Do vegetative phenology candidate genes show patterns of nucleotide variation consistent with divergent selection? The candidate genes we tested included two phytochrome B (PHYB) genes and an APETALA 2/ethylene-responsive element-binding factor (AP2/ERF) family transcription factor. In Populus, phytochromes are strongly implicated in the photoperiodic control of dormancy (Howe et al., 2003; Ma et al., 2010). Phytochromes are light-sensing proteins that respond to red and far-red light cues, and phytochrome B2 (PHYB2) has been associated with daylength-driven budset in P. tremula (Ma et al., 2010). AP2/ERF is a large family of 200 genes in Populus that are involved in a number of stress responses (Zhuang et al., 2008) and potentially in dormancy induction (Rohde et al., 2007). In P. euphratica, expression of an AP2/ERF locus is induced by cold and drought (Chen et al., 2009). Ethylene-responsive factor 61 (ERF61=EBB1=PtERF-B1-2) contains a typical AP2/ERF-binding domain (Zhuang et al., 2008), and has been implicated in date of leaf flush in Populus based on activation tagging (Yordanov et al., 2014). Because of their role in tree development, we hypothesized that these phenology candidate genes would show evidence of divergent selection.
Materials and methods
Plant materials
In January–February 2009, we collected branch cuttings from 447 trees from the drainages of nine rivers, spanning ca 1800 km across the full latitudinal range of P. angustifolia (Figure 1a). These collections represent a strong latitudinal and climatic cline, and also cover large geographic disjunctions in the range of P. angustifolia (Geographical collections in the online Supplementary Information). We collected leaf material from the rooted, potted cuttings during September 2009, and extracted total genomic DNA from ca 6 mg of dried leaf tissue using DNeasy 96 Plant Kits (Qiagen, Valencia, CA, USA).
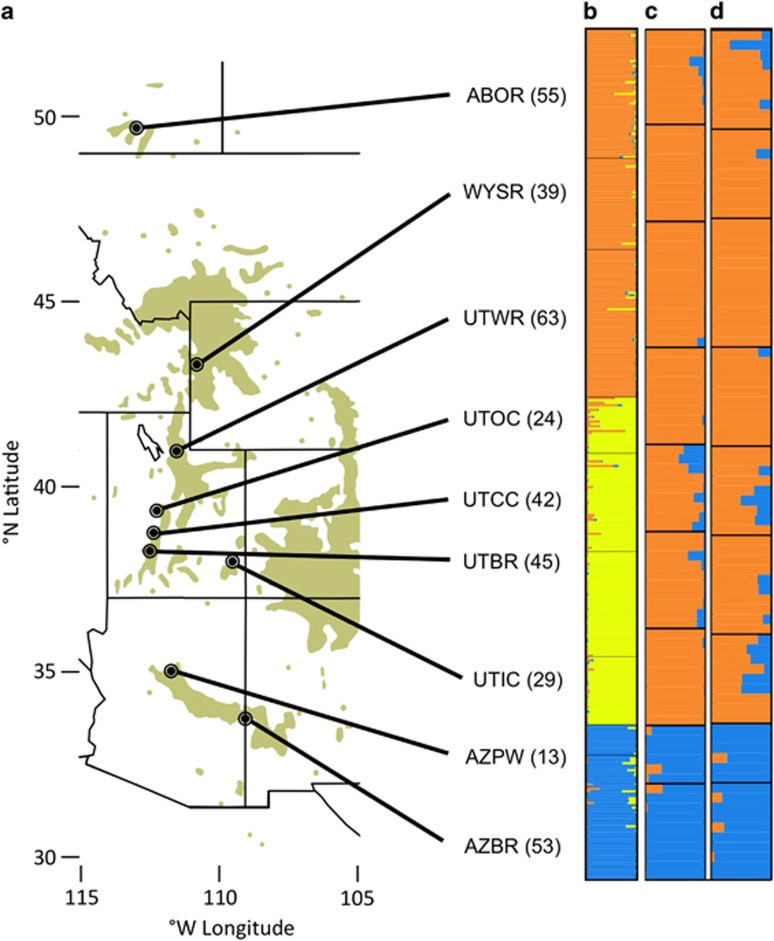
Figure 1.
Study populations and evidence for population substructure. (a) Populus angustifolia sample locations. Population codes: Oldman River, AB (ABOR); Snake River, WY (WYSR); Weber River, UT (UTWR); Oak Creek, UT (UTOC); Indian Creek, UT (UTIC); Corn Creek, UT (UTCC); Beaver River, UT (UTBR); Pumphouse Wash, AZ (AZPW); and Blue River, AZ (AZBR). Numbers in parentheses represent sample sizes within each population. (b) K=3 STRUCTURE barplot for 24 SSRs. (c) K=2 STRUCTURE barplot for random sequenced loci SNPs. (d) K=2 STRUCTURE barplot for candidate gene SNPs.
PCR and sequencing methods
To investigate neutral genetic variation, we used a combination of SSRs and sequenced loci. We chose 24 SSR loci (Supplementary Table 1) from a publically available list identified by the Populus trichocarpa Genome Project (Tuskan et al., 2004; http://www.ornl.gov/sci/ipgc/ssr_resource.htm). These loci have reliable repeat motifs, most are tri- or tetranucleotide repeats, are easily amplified, and are distributed throughout the genome. PCR amplifications were carried out in 18 μl volume reactions, with 1.5 μl (10 ng μl−1) template DNA, 0.17–0.22 μl of 10 μM primer, 0.11 mM dNTPs, 0.75 U Taq polymerase, 1.1 × PCR Buffer and 2.78 mM MgCl2. Cycling conditions consisted of the following: 1 cycle at 95 °C (5 min); 9 cycles of touchdown at 95 °C (15 s), 58 °C (15 s, decreasing 1 °C each cycle), 72 °C (30 s); 20 cycles at 95 °C (15 s), 50 °C (15 s), 72 °C (30 s); and 1 cycle at 72 °C (3 min). Forward primers were end-labeled with FAM, HEX, NED, PET or VIC fluorescent dye (Applied Biosystems (ABI), Foster City, CA, USA) and PCR products were analyzed on an ABI 3730 × l automated sequencer (ABI) using Genescan LIZ 500 or LIZ 600 internal size standard (ABI). Alleles were sized and scored using Genemapper v4.0 (ABI), and all genotypes were manually checked for accuracy.
After removing clonal ramets and putative interspecific hybrids (Clone Identification and Hybridization in the online Supplementary Information), our final SSR analyses were performed using 363 unique P. angustifolia individuals (Figure 1).
In a subset of 6–12 individuals per population, we sequenced 10 putatively neutral control loci to complement the neutral SSR data set for our demographic analyses. We also sequenced the phenology candidate genes PHYB1 (>8 kb), PHYB2 (>7 kb) and ERF61 (>2 kb) to test our hypothesis regarding divergent selection (Supplementary Tables 2–4). These sequenced regions spanned introns, exons, and flanking noncoding regions. The 10 control loci were randomly chosen as a subset of those used by Olson et al. (2010) to allow direct comparison of our data from P. angustifolia to those from P. balsamifera. In that study, which examined genome-wide polymorphism patterns, but not demography, samples spanned subpopulations across the entire range of P. balsamifera, and were therefore similar to our collections.
Sequenced loci (Supplementary Tables 2–4) were amplified in 20 μl volumes, with 15 ng template DNA, 0.2 mM dNTPs, 0.4 μM each primer, 0.8 U Taq polymerase, 1 × PCR buffer and 2 mM MgCl2. Cycling conditions consisted of 1 cycle at 95° (5 min); 30 cycles at 95° (15 s), 60° (30 s), 72° (90 s); and 1 cycle at 72° (7 min). We directly sequenced PCR products in both directions using ABI Big Dye Terminator v3.1 chemistry on an ABI 3730 × l sequencer. We compiled sequences for each individual using Seqman (DNAStar, Lasergene, Madison, WI, USA) and manually checked all variants, including single-nucleotide polymorphisms (SNPs) and insertion-deletion (indel) polymorphisms. We aligned all sequences at each locus using Clustal X (Larkin et al., 2007), manually checked the alignments and determined the gametic phase of haplotypes using PHASE v. 2.1 (Stephens et al., 2001) as implemented in DnaSP v.5 (Librado and Rozas, 2009) using 6000 steps through the Gibbs chain, with a 1000-step burnin period.
Neutral population structure and polymorphism
To examine patterns of genetic diversity across our collection, we first estimated standard polymorphism indices using DnaSP, including Watterson's θ, nucleotide polymorphism and divergence from P. trichocarpa. We tested for evidence of non-neutral evolution by estimating Tajima's D in DnaSP for each locus within each population and over all populations (Supplementary Table 4) and performing 10 000 neutral coalescent simulations, as implemented in DnaSP conditioned on Watterson's θ. To assess neutral population structure, we performed an analysis of molecular variance (AMOVA) among source rivers using the SSR data and the control loci SNPs independently, implemented using Arlequin v.3.1 (Excoffier and Lischer, 2010). We estimated 95% confidence intervals (CIs) using 20 000 bootstrap replicates, resampled over loci and tested the correlation between pairwise FST values and geographic distances among populations using a Mantel test with Arlequin using 1000 permutations. Similarly, we tested the correlation of FST estimates between the marker types using a Mantel test.
To examine genetic groupings in the 363 unique P. angustifolia individuals (using SSRs) or 86 individuals (sequenced loci, control and candidate genes separately), we used the program STRUCTURE 2.3.3 (Falush et al., 2003). We used the admixture model with correlated allele frequencies, running 10 iterations each with 15 000 burnin steps, followed by 20 000 steps through the MCMC chain for each of K=1 to K=10. Visual inspection of chains indicated that convergence was reached and the chains had mixed well. To choose the best K, the ΔK statistic (Evanno et al., 2005) was estimated using Structure Harvester (Earl and VonHoldt, 2012; Supplementary Figure 1). CLUMPP (Jakobsson and Rosenberg, 2007) was used to combine the results of the 10 replicate runs, and DISTRUCT (Rosenberg, 2004) was used for visualization.
Based on the findings of STRUCTURE groupings, we performed hierarchical AMOVA using Arlequin, with river of origin nested within STRUCTURE group. We did this for K=2 groups using all rivers, and K=3 groups excluding the UTOC river, which appears admixed using the SSR data. This analysis tested whether the large regional genetic groupings (STRUCTURE groups) account for most of the differentiation, or whether rivers remained differentiated from one another within these groups.
Demographic history
We performed two different analyses to investigate different aspects of demographic history. The goal of the first was to determine if population structure in P. angustifolia existed before the LGM. We used IMa2 (Hey, 2010) to simultaneously estimate divergence times, effective population sizes (Ne) and migration rates among three regional groupings of populations (see results from the STRUCTURE analysis): a northern region (ABOR, WYSR, UTWR), a central region (UTCC, UTBR, UTIC) and a southern region (AZPW, AZBR). We estimated these parameters using the combined SSR and control loci SNP data set for two different population divergence models: first using a model in which the northern population diverged before a central/southern divergence, and second using a model in which the southern population diverged before a northern/central divergence (Supplementary Figure 2 and IMa2 Analysis of Demographic History in the online Supplementary Information). These divergence models were based on the geographical orientation of the species and the major groupings identified in STRUCTURE. We did not test a model in which the central population diverged before a northern/southern divergence because of the strong latitudinal orientation of the populations (Figure 1). For each model, we used seven independent instances, each with randomly generated initial parameter values, with a total of 243 050 (northern divergence first) or 53 258 (southern divergence first) sampled genealogies after burnin periods of 106 samples. Using multiple independent instances of the model, different starting parameter values, increases confidence in the posterior estimates of the model. Both models had similarly variable posterior credible intervals, and the independent runs generally converged; therefore, we were confident that both had sufficiently searched the parameter space. For scaling the genetic parameters to individuals and years, we used a generation time of 15 years and mutation rate per base per year (μ) of 2.5 × 10−9 (Tuskan et al., 2006), similar to other studies in Populus (Ingvarsson, 2008; Keller et al., 2010, 2011; Levsen et al., 2012).
Our second analysis assessed patterns of recent population growth and decline after the LGM, which has been linked to Ne bottlenecks and subsequent expansion of P. trichocarpa (Zhou et al., 2014). We therefore used the program MIGRATE (Beerli, 2006) to estimate Ne and all pairwise migration rates, as well as Ne changes through time for each of the three STRUCTURE groups. We used the same set of three groupings as with the IMa2 analyses described above and the 10 sequenced control loci. We used uniform priors for both effective population size (θ=4Neμ) and mutation-scaled migration (M=m/μ) with ranges (0, 0.15) and (0, 5000), respectively. We used six heated chains with temperatures of 1, 100, 1000, 5000, 10 000 and 100 000; a 10 000-step burnin; and a collection period of 48 750 000 steps, sampling only every 65th step for a total of 750 000 recorded steps. We examined changes of Ne through time with the Bayesian skyline plots in MIGRATE, as was done for P. trichocarpa (Zhou et al., 2014). We used the same estimates of generation time and μ as above to scale parameters to demographic units.
Tests of selection
We first performed the same set of analyses using the candidate gene SNPs as we performed with the control loci SNPs, including STRUCTURE, AMOVA and Mantel tests. We then compared patterns of differentiation for candidate genes to the putatively neutral control genes using both FST and the among population component of π (πT-S) as our estimate of among-population genetic differentiation (Charlesworth, 1998). We also tested for differences in the rate of accumulation of silent polymorphisms within and between species across multiple loci using the Hudston-Kreitman-Aguadé (HKA) test (Wright and Charlesworth, 2004) test and for differences in the rate of synonymous and nonsynonymous mutation using the McDonald–Kreitman tests (McDonald and Kreitman, 1991) using DnaSP.
Results
Diversity and polymorphism
We genotyped all individuals at 24 SSR loci and sequenced >22.8 kb in more than 80 individuals (Supplementary Table 1). FIS was, on average, weakly positive in most populations, unsurprising given the clonal nature and isolated habitat of P. angustifolia (Supplementary Table 1). Indels were found in all three phenology genes, including two polymorphic deletions within the coding region of ERF61, deleting one and three amino acids, respectively. A premature stop codon was found in the third exon of PHYB1 based on the P. trichocarpa reference genome sequence (Tuskan et al., 2006). Because this mutation was fixed in all sequenced P. angustifolia, diversity measures were estimated using this premature stop codon as the end of the coding region. The rate of nonsynonymous to synonymous substitutions and divergence from P. trichocarpa remained «1 (Supplementary Table 4), indicating that the truncated locus may still be under purifying selection, particularly given that it is fixed within the species. The relatively old divergence times among populations and limited migration (see below) suggest that there has been sufficient time to accumulate nonsynonymous polymorphisms if the locus were a pseudogene. A total of 346 SNPs were found across all loci, or approximately 1 SNP per 66 bp (Supplementary Table 4). Average overall nucleotide diversity (π) was 0.00381 (θ=0.00335), and somewhat lower in each of the individual subpopulations (Supplementary Tables 4 and 5). Overall diversity was similar to P. balsamifera (π=0.00334; θ=0.00335). Estimates of nonsynonymous diversity tended to be lower than those of silent (synonymous+noncoding) diversity (Supplementary Table 4). Only one locus showed any evidence of deviation from the expected Tajima's D (P<0.01), and only 3 of the 10 control loci were positive overall (Supplementary Table 4).
Population structure
Two lines of evidence demonstrated strong population structure in P. angustifolia. First, our STRUCTURE analyses clearly supported the presence of multiple groups, representing the highest hierarchical level of differentiation identified in our sample (Figures 1b–d and Supplementary Figure 1). Patterns differed somewhat for different sets of loci. The SSRs showed clear evidence of three groups, corresponding to major geographical regions: (1) the southern range along the Mogollon Rim in Arizona, (2) the center of the range in the central and southern Wasatch Mountains in Utah and (3) the northern Wasatch and Rocky Mountains. There were clear genetic discontinuities among these regions, with only collections from Oak Creek, Utah, exhibiting evidence of overlap. Within these three regional genetic groupings, we found genetic structure primarily among rivers (Supplementary Figure 3). The results from control locus and candidate gene SNPs were somewhat different (Figures 1c and d). For both sets of SNPs, the southern grouping was divergent, but there was little distinction between the northern and central groupings.
Second, we found strong overall differentiation among a priori defined populations using SSRs (FST (95% CI)=0.21 (0.16–0.26)), as well as using control locus SNPs (FST (95% CI)=0.27 (0.21–0.33)). Patterns of pairwise SNP FST were correlated to those found using SSR loci (Mantel R=0.86, P<0.001). FST (95% CI) of the candidate genes was 0.29 (0.24–0.34) for PHYB1, 0.25 (0.19–0.31) for PHYB2 and 0.41 (0.31–0.51) for ERF61. Mantel tests of pairwise FST between phenology candidate genes and SSRs were significant (all Mantel R⩾0.54, P⩽0.003), indicating that the phenology candidate genes generally followed patterns similar to those of neutral loci.
Finally, a weak (SSR Mantel R=0.31) but nonsignificant (P=0.15) relationship existed between pairwise geographic distance and genetic differentiation. SNPs showed similar patterns (all candidate gene Mantel R>0.22, control loci Mantel R=0.36, all P>0.08). Despite this lack of significant geographic signal, the AMOVA at the source river level showed that greater differentiation occurred between populations from different STRUCTURE groups versus populations from the same group, although many pairs of populations within regions were still significantly differentiated (for example, pairwise FST>0.1, P<0.001, Table 1). Our hierarchical AMOVA demonstrated that a large fraction of the variance results from among regional genetic group variation. However, a significant proportion of the variance remains among rivers within these groups (Table 2), which were clearly latitudinally oriented (Table 1, Figure 1 and Supplementary Figures 2 and 3).
Table 1. Pairwise population differentiation (FST) among the nine collection locations of P. angustifolia using 24 SSR loci (below diagonal) and control sequenced loci (above diagonal).
| ABOR | WYSR | UTWR | UTOC | UTCC | UTBR | UTIC | AZPW | AZBR | |
|---|---|---|---|---|---|---|---|---|---|
| ABOR | — | 0.02* | 0.08 | 0.13 | 0.27 | 0.30 | 0.21 | 0.39 | 0.39 |
| WYSR | 0.09 | — | 0.01* | 0.06 | 0.25 | 0.28 | 0.15 | 0.46 | 0.45 |
| UTWR | 0.09 | 0.01* | — | 0.01 | 0.19 | 0.19 | 0.12 | 0.41 | 0.42 |
| UTOC | 0.14 | 0.14 | 0.13 | — | 0.19 | 0.19 | 0.12 | 0.43 | 0.44 |
| UTCC | 0.13 | 0.15 | 0.16 | 0.11 | — | 0.00* | 0.24 | 0.35 | 0.38 |
| UTBR | 0.14 | 0.18 | 0.19 | 0.10 | 0.06 | — | 0.26 | 0.38 | 0.39 |
| UTIC | 0.14 | 0.24 | 0.24 | 0.18 | 0.18 | 0.16 | — | 0.45 | 0.46 |
| AZPW | 0.35 | 0.41 | 0.39 | 0.32 | 0.31 | 0.33 | 0.35 | — | 0.06 |
| AZBR | 0.32 | 0.39 | 0.36 | 0.28 | 0.29 | 0.28 | 0.29 | 0.15 | — |
Population abbreviations are as in legend of Figure 1.
*P>0.01.
Table 2. Mode (95% posterior credible intervals) of demographic parameters for the three subpopulations (SSR STRUCTURE groups) of P. angustifolia estimated in IMa2, using two different divergence models (top) and MIGRATE (bottom).
| Southern divergence first | Northern divergence first | ||
|---|---|---|---|
|
Divergence time (years) |
Divergence time (years) |
||
| Northern-Central | 555 463 (378 359–1 489 286) | Southern-Central | 334 805 (77 263–1 158 941) |
| Southern—Northern/Central | 3 002 723 (1 972 298–10 552 195) | Northern—Southern/Central | 437 822 (386 314–15 376 999) |
|
Population size (Ne, individuals) |
Population size (Ne, individuals) |
||
| Northern | 16 503 (12 478–22 943) | Northern | 15 728 (12 869–21 448) |
| Central | 18 113 (12 283–25 358) | Central | 15 728 (10 009–21 448) |
| Southern | 10 063 (7648–15 698) | Southern | 10 009 (7149–12 869) |
| Northern/Central Ancestor | 56 781 (20 528–675 143) | Southern/Central Ancestor | 55 764 (38 606–2 732 158) |
| P. angustifolia Ancestor | 804 483 (318 412–799 653) | P. angustifolia Ancestor | 132 976 (118 678–2 807 082) |
|
Migration=2Nimij |
Migration=2Nimij |
||
| Northern>Central | 0.756 (0.387–1.769) | Northern>Central | 0.485 (0.169–1.272) |
| Central>Northern | 1.050 (0.320–1.905) | Central>Northern | 0.439 (0.051–1.316) |
| Northern>Southern | 0.011 (0.002–0.143) | Northern>Southern | 0.025 (0.003–0.248) |
| Southern>Northern | 0.046 (0.008–0.331) | Southern>Northern | 0.018 (0.002–0.265) |
| Central>Southern | 0.098 (0.005–0.287) | Central>Southern | 0.043 (0.004–0.362) |
| Southern>Central | 0.121 (0.017–0.405) | Southern>Central | 0.009 (0.003–0.367) |
| Southern>Northern/Central Ancestor | 0.079 (0.004–0.473) | Northern>Southern/Central Ancestor | 0.037 (0.003–0.334) |
| Northern/Central Ancestor>Southern | 0.002 (0.003–14.96) | Southern/Central Ancestor>Northern | 0.408 (0.049–129.727) |
2Nimij represents the rate at which migrants from population j move to population i (from i to j in the reverse-time coalescent).
Demographic history
Our demographic history analysis using IMa2 resulted in three main findings (Table 2, Figures 2 and 3). First, the modal estimates and 95% credible intervals of all divergences clearly predate the LGM (ca 18 000 years ago). This was true for both the northern divergence first model as well as the southern divergence first model (Figures 2a and 3a). The credible intervals were quite large, however, indicating little confidence in either the order of divergence or the exact time.
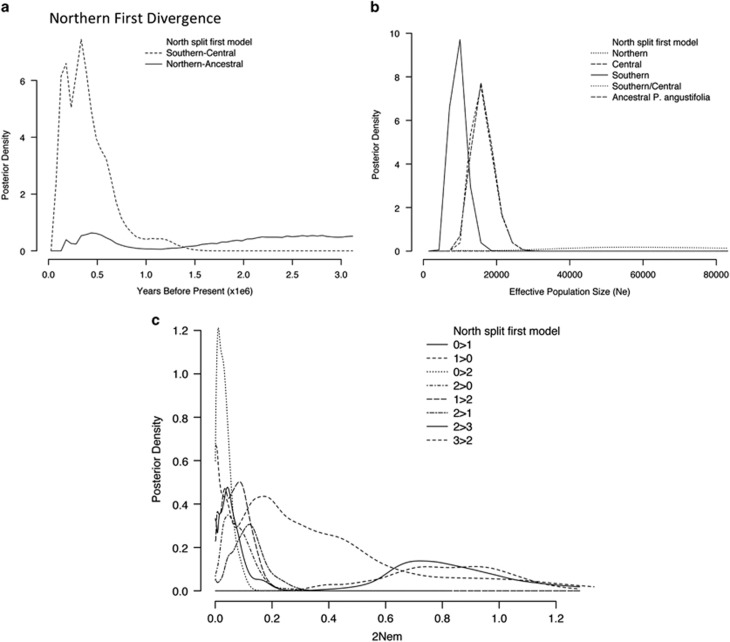
Figure 2.
IMa2 demographic analyses in three populations of P. angustifolia as estimated in IMa2 using a population model in which the northern group diverged before a southern-central divergence. Posterior densities of (a) population splitting times, (b) effective population sizes and (c) population migration rates (2Nem). In a, x-axis has been truncated to 3 million years.
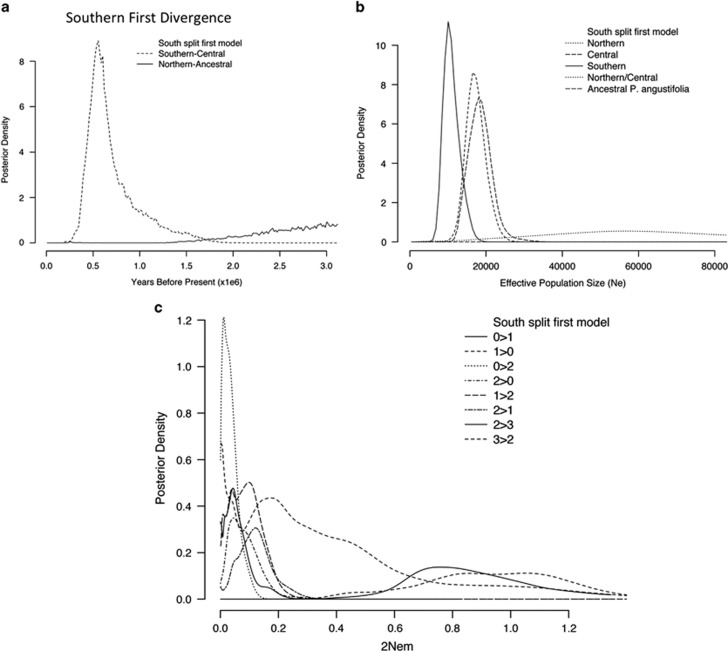
Figure 3.
IMa2 demographic analyses in three populations of P. angustifolia as estimated in IMa2 using a population model in which the southern group diverged before a northern-central divergence. Posterior densities of (a) population splitting times, (b) effective population sizes and (c) population migration rates (2Nem). In a, x-axis has been truncated to 3 million years.
Second, the extant effective population size estimates were all considerably smaller than the ancestral population estimates (Table 3, Figures 2b and 3b). Posterior distributions of ancestral population sizes plateaued. However, both had strong modal peaks that indicated much larger ancestral effective population sizes than the estimates for extant populations. Furthermore, Ne of the northern and central groupings was substantially larger than the southern population. These patterns remained, regardless of the divergence model used.
Table 3. Median (95% posterior credible intervals) of effective population size (N e) and migration (2N i m ij ) rate for the three subpopulations (SSR STRUCTURE groups) of P. angustifolia estimated in MIGRATE.
|
Population size (Ne, individuals) | |
| Northern | 10 933 (7–27 467) |
| Central | 4667 (7–24 400) |
| Southern | 333 (7–18 133) |
| Migration=2Nimij | |
| Southern>Northern | 0.007 (0–0.014) |
| Central>Northern | 0.22 (0.063–1.895) |
| Northern>Southern | 0.125 (0–0.525) |
| Central>Southern | 0.036 (0–0.247) |
| Northern>Central | 0.408 (0–1.72) |
| Southern>Central | 0.002 (0–0.009) |
Third, migration among populations appeared to be largely limited to gene exchange between the current northern and central populations (Table 3, Figures 2c and 3c). Extensive unidirectional gene flow was inferred from the central/southern ancestor to the northern population in the northern divergence first model.
Our analysis using MIGRATE indicated that both the northern and central groupings are substantially larger than the southern, consistent with the IMa2 analysis (Table 3). The long-term Ne estimate of the central group was smaller than the northern; however, the 95% credible intervals substantially overlapped. Patterns of migration mirrored those found with IMa2, indicating little migration of the northern or central group with the southern population.
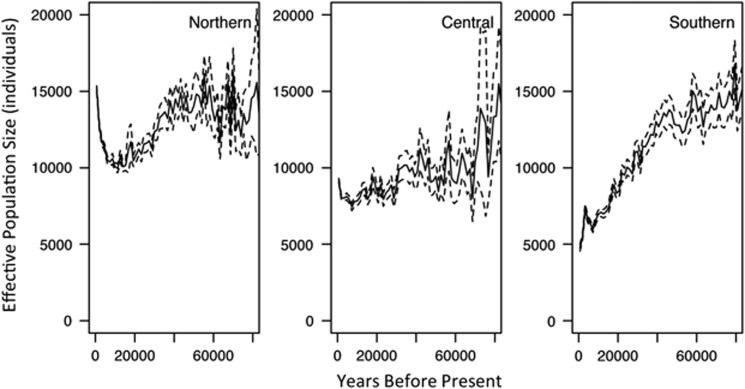
Patterns of Ne over time, however, revealed strikingly different trends among the three groupings (Figure 4). Scaled to demographic units, before 80 000 years ago all three populations were of similar effective size (ca 15 000 individuals). Although strong declines in the northern and southern groups began roughly 40 000 years ago, the southern population continued this decline even more rapidly in the very recent past. The northern group apparently rebounded beginning approximately 20 000 years ago and continues to expand. The central population remained relatively constant, with possible recent growth.
Figure 4.
Bayesian skyline plot from MIGRATE analysis of effective population size (Ne) through time, scaled to real demographic units for each of the three genetic subpopulations identified in Figure 1b. Solid line indicates the average, whereas dotted lines represent 2 × the standard deviation.
Selection in phenology candidate genes
Nonsynonymous and silent diversity estimates differed (Supplementary Table 4), but there was no indication that the rate of evolution differed among genes when only phenology candidates were compared with control loci, together or individually (all HKA P>0.3). In addition, the ratio of polymorphism to divergence at synonymous to nonsynonymous sites gave no indication for departures from neutrality (McDonald–Kreitman test, P>0.1 for all loci). Similarly, observed πT-S for candidate genes were similar to estimates using control loci (Supplementary Tables 4 and 5). Overall patterns of structure were similar to SSRs and control loci, as indicated by the strong Mantel correlation of FST estimates between candidate gene SNPs with both SSRs and control locus SNPs (all Mantel R⩾0.54, P⩽0.003).
Discussion
Population structure
Compared with its congeners (reviewed by Slavov and Zhelev, 2010), genetic structure across the range of P. angustifolia is much stronger than expected. P. angustifolia is dioecious with putatively long-distance dispersal of pollen and seed, traits that are expected to minimize population structure (Hamrick and Godt, 1996), as has been shown in other species (for example, Namroud et al., 2008). Three factors could explain this result. First, P. angustifolia is a riparian species that grows primarily in isolated, montane canyons throughout much of its range. In the most northern populations, it is even more limited in distribution, being elevationally restricted between two other Populus species (Floate, 2004; Cooke and Rood, 2007). This discontinuous range at both broad and local scales could be limiting gene flow, even between adjacent rivers (Table 1 and Supplementary Figure 3).
Second, the range of P. angustifolia spans several major geographic features. One of these is the unsuitable arid lands that separate the genetically distinct populations on the Mogollon Rim, Arizona, from those growing in Utah (Figure 1). A second division occurs between the northern and central Wasatch Mountains, Utah. This corresponds to the northern and central STRUCTURE groups found using SSRs. Weaker but still highly significant structure is observed with the SNPs between these two areas (for example, FST between UTWR and UTCC is 0.16 and 0.19 for SSRs and control loci SNPs, respectively). The apparent area of overlap or admixture in the Oak Creek, Utah, area may result from recent gene flow or may be resolved with more loci in future studies. The difference in STRUCTURE groupings between SSRs and sequenced loci (K=3 vs K=2, respectively), may be due to the smaller number of loci or mutation rate of nucleotide substitutions compared with SSRs. However, similar patterns were found using hierarchical AMOVA and pairwise FST analyses, suggesting that structure exists at large regional scales as well as among individual rivers, a result seen in other Populus species (Cushman et al., 2014; Evans et al., 2014).
Although overall structure is relatively weak in many forest trees (for example, Namroud et al., 2008), differentiation of populations from range-wide collections tends to be stronger than that of more local collections (Ingvarsson, 2005; Keller et al., 2010; Slavov et al., 2012; Cushman et al., 2014; Evans et al., 2014; Zhou et al., 2014). For instance, Heuertz et al. (2006) found that across large geographic distances, Picea abies is strongly differentiated across large geographic barriers, with FST estimates approaching those that we found among genetic groupings. Therefore, even though many forest trees have extensive potential for long-distance dispersal (Savolainen et al., 2007), they can still develop strong neutral genetic structure across large spatial scales and geographic barriers. This is supported by the estimates of migration among groups, with the southern population being the most isolated. Third, the northern and southern regions of the range would have experienced very different environmental conditions during Quaternary glacial cycles (Frenzel et al., 1992), which likely resulted in contrasting demographic effects, as discussed next.
Polymorphism and demographic history
Polymorphism levels in P. angustifolia were high, as is expected for forest trees (Hamrick and Godt, 1996). Patterns of polymorphism were generally consistent with purifying selection and similar between phenology candidate genes and control loci, indicating that these genes, on the whole, were not affected by strong divergent selection. These findings are similar to a recent study of P. balsamifera photoperiodic genes, including PHYB1 and PHYB2, in which most loci showed no evidence of stronger differentiation than that expected under a selectively neutral demographic model (Keller et al., 2011). The strong genome-wide latitudinal clines in P. angustifolia, which covary with the expected patterns of selection in climate-related traits, make identifying signatures of selection difficult.
The three P. angustifolia groups that we detected appear to have similar long-term effective population sizes. This finding is surprising given the much smaller census size in the southern part of the range where the species is primarily restricted to small numbers of individuals along fewer perennial streams, compared with extensive gallery forests along major waterways in the north. Nevertheless, estimates of π and extant Ne estimates obtained using IMa2 and MIGRATE were qualitatively similar among the three groups (Tables 2 and 3, and Supplementary Table 5). The northern group had higher estimated Ne than the southern group, but 95% credible intervals largely overlapped, indicating that the long-term Ne is similar. Contemporary migration appears to occur mostly between the Northern and Central populations (Table 3).
The fact that ancestral P. angustifolia populations were larger than extant subpopulations could reflect bottlenecks after these subpopulations diverged, similar to such bottlenecks in two other Populus species (Ingvarsson, 2008; Keller et al., 2011). The overall estimate of nucleotide diversity was slightly higher than that of P. balsamifera (Table 4), suggesting that the long-term effective population sizes of these two species are similar. This fact is surprising given that the range of P. balsamifera is much larger than that of P. angustifolia (Cooke and Rood, 2007), but P. balsamifera likely experienced a range-wide bottleneck after the LGM (Keller et al., 2010). These Pleistocene glacial cycles are also hypothesized to have driven the divergence and allopatric distributions of P. trichocarpa and P. balsamifera (Breen et al., 2012; Levsen et al., 2012).
Table 4. Hierarchical AMOVA results using the K=2 or K=3 STRUCTURE groupings, for the SSRs, the control sequenced loci and the candidate genes separately.
| Data set | Source | K=2 Genetic groups |
K=3 Genetic groups |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Rivers within groups |
Rivers within groups, UTOC removed |
||||||||||||||
| df | Sum of Squares | σ2 | % | F-statistic | P | df | Sum of Squares | σ2 | % | F-statistic | P | ||||
| SSRs | Among groups | 1 | 431.75 | 1.61 | 20.75 | FCT | 0.207 | 0.033 | 2 | 642.79 | 1.23 | 17.4 | FCT | 0.174 | 0.004 |
| Among rivers within groups | 7 | 508.36 | 0.86 | 11.11 | FSC | 0.140 | <0.001 | 5 | 256.56 | 0.57 | 8.07 | FSC | 0.098 | <0.001 | |
| Among individuals within rivers | 354 | 2006.65 | 0.38 | 4.89 | FIS | 0.072 | <0.001 | 331 | 1881.58 | 0.41 | 5.75 | FIS | 0.077 | <0.001 | |
| Within individuals | 363 | 1782 | 4.91 | 63.24 | FIT | 0.367 | <0.001 | 339 | 1651 | 4.87 | 68.78 | FIT | 0.312 | <0.001 | |
| Total | 725 | 4728.76 | 7.76 | 677 | 4431.94 | 7.08 | |||||||||
| Random Loci | Among groups | 1 | 189.73 | 3.13 | 38.77 | FCT | 0.388 | 0.023 | 2 | 150.18 | 1.30 | 30.72 | FCT | 0.307 | 0.004 |
| Among rivers within groups | 7 | 198.43 | 1.24 | 15.38 | FSC | 0.251 | <0.001 | 5 | 50.47 | 0.38 | 8.94 | FSC | 0.129 | <0.001 | |
| Among individuals within rivers | 79 | 303.22 | 0.13 | 1.67 | FIS | 0.036 | 0.201 | 70 | 190.68 | 0.17 | 3.93 | FIS | 0.065 | 0.096 | |
| Within individuals | 88 | 314 | 3.57 | 44.19 | FIT | 0.558 | <0.001 | 78 | 186.5 | 2.39 | 56.42 | FIT | 0.436 | <0.001 | |
| Total | 175 | 1005.38 | 8.08 | 155 | 577.8 | 4.24 | |||||||||
| Candidate Genes | Among groups | 1 | 597.41 | 10.30 | 38.5 | FCT | 0.385 | 0.036 | 2 | 272.52 | 2.38 | 25.19 | FCT | 0.252 | 0.009 |
| Among rivers within groups | 7 | 447.29 | 2.49 | 9.32 | FSC | 0.152 | <0.001 | 5 | 92.96 | 0.59 | 6.25 | FSC | 0.084 | <0.001 | |
| Among individuals within rivers | 78 | 1193.84 | 1.35 | 5.03 | FIS | 0.096 | 0.0156 | 69 | 502.70 | 0.81 | 8.59 | FIS | 0.125 | 0.046 | |
| Within individuals | 87 | 1097.5 | 12.61 | 47.15 | FIT | 0.528 | <0.001 | 77 | 436 | 5.66 | 59.96 | FIT | 0.400 | <0.001 | |
| Total | 173 | 3336.05 | 26.76 | 153 | 1304.18 | 9.44 | |||||||||
Abbreviation: AMOVA, analysis of molecular variance.
P-values based on 1000 permutations.
As areas outside of the maximal extent of the ice sheets were impacted by altered climate during glacial cycles (Frenzel et al., 1992; Jackson and Weng, 1999; Hu et al., 2009), it is likely that populations throughout the range of P. angustifolia were altered by these cycles as well. The estimated times of divergence significantly predate the LGM, indicating that the current genetic groupings existed before the last major glacial period, regardless of the divergence model used (Table 2). Interglacial periods would have restricted southern populations to isolated pockets, but during glacial maxima, northern populations would have been restricted. This is consistent with our findings of the change in Ne through time, which showed a continuing decline in the southern region, a drastic expansion in the north, and a moderate recent increase in the center after the LGM, roughly 18 000 years ago (Figure 4). The negative Tajima's D estimates for many loci (Supplementary Table 4) also support a recent expansion. Other North American tree species support the hypothesis of impacts of past climate on population sizes and distributions. In a latitudinally oriented study of Picea sitchensis, Holliday et al. (2010) found support for an historical bottleneck and expansion, and also found negative Tajima's D, as did a study of Picea aibes in Eurasia (Heuertz et al., 2006). In P. trichocarpa of the central and northern Pacific Northwest, populations have expanded rapidly since the LGM (Zhou et al., 2014). Quaternary climate changes have also been hypothesized to have driven the decline of a now-extinct species of Picea in the southeastern United State, far outside of the ice sheet distribution (Jackson and Weng, 1999). Thus, current P. angustifolia genetic subdivision likely reveals the existence of multiple Pleistocene populations, similar to hypotheses about P. trichocarpa (Slavov et al., 2012), P. balsamifera (Breen et al., 2012) and Pinus taeda (Eckert et al., 2010). Among these P. angustifolia subpopulations, however, Ne changes in the recent past reflect drastically different impacts of western North America climate.
Conclusions
Our surveys of polymorphism throughout the latitudinal range of P. angustifolia demonstrate that geographical barriers and climate fluctuations jointly shape genetic variation and structure in this species. The intricate geography and climatic history, combined with the likely existence of Pleistocene population structure, appears to lead to distinct dynamics across its range, as seen in comparisons between the northern and southern subpopulations. For species with strong latitudinal distributions and isolated marginal populations (for example, many plant species along the coastal ranges of North America and along the Rocky and Appalachian Mountains), such dynamics may be common (Alberto et al., 2013). For example, complex alternative hypotheses have been proposed to explain genetic patterns found in Pacific Northwest species involving refugia, vicariance, recent colonization and growth (Brunsfeld et al., 2001). Paradoxically, the strong signal in neutral loci left by these processes makes it particularly difficult to test whether divergent selection has occurred in candidate genes. Despite this, understanding neutral demographic processes is important, because they can aid our understanding of current climate change as a driver of evolution (Alberto et al., 2013), and help identify genetically unique populations that may warrant special management consideration. This is particularly relevant for a species like P. angustifolia, which in the southern part of the range is narrowly distributed in isolated drainages and potentially threatened by rapidly changing climate (Gitlin et al., 2006).
Data archiving
We deposited all sequences (control and phenology candidates) in GenBank (Accessions JX115218-JX117417). SSR genotypes and geographical locations of collections are deposited in Dryad Digital Repository (http://doi.org/10.5061/dryad.82kv2).
Acknowledgments
We thank Deb Ball, Brad Blake, Victor Busov, Karen Gill, Basil Jefferies, Sobadini Kaluthota, Karla Kennedy, Mindie Lipphardt, Tamara Max, Julie Nielsen, Ian Ouwerkerk, Phil Patterson, David Pearce, David Smith, Amy Whipple, Matt Zinkgraf and the Cottonwood Ecology Group for help in the field, laboratory, greenhouse and discussion. We thank the Apache-Sitgreaves, Fishlake and Bridger-Teton National Forests; Monticello BLM; Weber Basin Water Conservancy District; AZ Game & Fish and UT Division of Wildlife Resources. We also thank the anonymous reviewers for many helpful comments on previous versions of the manuscript. Research was supported by NSF-FIBR DEB-0425908 to TGW, GJA and SPD; a Science Foundation Arizona Graduate Student Fellowship, NSF IGERT and Doctoral Dissertation Improvement Grant to LME and Northern Arizona University.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Alberto FJ, Aitken SN, Alía R, González-Martínez SC, Hänninen H, Kremer A, et al. Potential for evolutionary responses to climate change—evidence from tree populations. Glob Chang Biol. 2013;19:1645–1661. doi: 10.1111/gcb.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P. Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics. 2006;22:341–345. doi: 10.1093/bioinformatics/bti803. [DOI] [PubMed] [Google Scholar]
- Breen AL, Murray DF, Olson MS. Genetic consequences of glacial survival: the late Quaternary history of balsam poplar (Populus balsamifera L.) in North America. J Biogeogr. 2012;39:918–928. [Google Scholar]
- Brunsfeld S, Sullivan J, Soltis D, Soltis P.2001Comparative phylogeography of north- western North America: a synthesisIn: Silvertown J, Antonovics J (eds)Integrating Ecological and Evolutionary Processes in a Spatial Context Blackwell Publishing, Ltd.: Oxford; 319–339. [Google Scholar]
- Charlesworth B. Measures of divergence between populations and the effect of forces that reduce variability. Mol Biol Evol. 1998;15:538–543. doi: 10.1093/oxfordjournals.molbev.a025953. [DOI] [PubMed] [Google Scholar]
- Chen J, Xia X, Yin W. Expression profiling and functional characterization of a DREB2-type gene from Populus euphratica. Biochem Biophys Res Commun. 2009;378:483–487. doi: 10.1016/j.bbrc.2008.11.071. [DOI] [PubMed] [Google Scholar]
- Cooke JEK, Rood SB. Trees of the people: the growing science of poplars in Canada and worldwide. Can J Bot. 2007;85:1103–1110. [Google Scholar]
- Cushman S, Max T, Meneses N, Evans LM, Ferrier S, Honchak B, et al. Landscape genetic connectivity in a riparian foundation tree is jointly driven by climatic gradients and river networks. Ecol Appl. 2014;24:1000–1014. doi: 10.1890/13-1612.1. [DOI] [PubMed] [Google Scholar]
- Earl DA, VonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–361. [Google Scholar]
- Eckert AJ, van Heerwaarden J, Wegrzyn JL, Nelson CD, Ross-Ibarra J, González-Martínez SC, et al. Patterns of population structure and environmental associations to aridity across the range of loblolly pine (Pinus taeda L., Pinaceae) Genetics. 2010;185:969–982. doi: 10.1534/genetics.110.115543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Evans LM, Slavov GT, Rodgers-Melnick E, Martin J, Ranjan P, Muchero W, et al. Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations. Nat Genet. 2014;46:1089–1096. doi: 10.1038/ng.3075. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure ising multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floate K. Extent and patterns of hybridization among the three species of Populus that constitute the riparian forest of southern Alberta, Canada. Can J Bot. 2004;82:253–264. [Google Scholar]
- Frenzel BB, Pecsi M, Velichko AA. Atlas of Paleoclimates and Paleoenvironments of the Northern Hemisphere: Late Pleistocene—Holocene. Geographical Research Institute, Hungarian Academy of Sciences: Budapest, Hungary; 1992. [Google Scholar]
- Gitlin AR, Sthultz CM, Bowker Ma, Stumpf S, Paxton KL, Kennedy K, et al. Mortality gradients within and among dominant plant populations as barometers of ecosystem change during extreme drought. Conserv Biol. 2006;20:1477–1486. doi: 10.1111/j.1523-1739.2006.00424.x. [DOI] [PubMed] [Google Scholar]
- Hamrick J, Godt M. Effects of life history traits on genetic diversity in plant species. Philos Trans R Soc London B. 1996;351:1291–1298. [Google Scholar]
- Heuertz M, De Paoli E, Källman T, Larsson H, Jurman I, Morgante M, et al. Multilocus patterns of nucleotide diversity, linkage disequilibrium and demographic history of Norway spruce [Picea abies (L.) Karst] Genetics. 2006;174:2095–2105. doi: 10.1534/genetics.106.065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J. Isolation with migration models for more than two populations. Mol Biol Evol. 2010;27:905–920. doi: 10.1093/molbev/msp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday Ja, Yuen M, Ritland K, Aitken SN. Postglacial history of a widespread conifer produces inverse clines in selective neutrality tests. Mol Ecol. 2010;19:3857–3864. doi: 10.1111/j.1365-294X.2010.04767.x. [DOI] [PubMed] [Google Scholar]
- Howe GT, Aitken SN, Neale DB, Jermstad KD, Wheeler NC, Chen THH. From genotype to phenotype: unraveling the complexities of cold adaptation in forest trees. Can J Bot. 2003;1266:1247–1266. [Google Scholar]
- Hu FS, Hampe A, Petit RJ. Paleoecology meets genetics: deciphering past vegetational dynamics. Front Ecol Environ. 2009;7:371–379. [Google Scholar]
- Ingvarsson PK. Nucleotide polymorphism and linkage disequilibrium within and among natural populations of European aspen (Populus tremula L., Salicaceae) Genetics. 2005;169:945–953. doi: 10.1534/genetics.104.034959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson PK. Multilocus patterns of nucleotide polymorphism and the demographic history of Populus tremula. Genetics. 2008;180:329–340. doi: 10.1534/genetics.108.090431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson ST, Weng C. Late quaternary extinction of a tree species in eastern North America. Proc Natl Acad Sci USA. 1999;96:13847–13852. doi: 10.1073/pnas.96.24.13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Keller S, Levsen N, Ingvarsson P. Local selection across a latitudinal gradient shapes nucleotide diversity in Balsam Poplar, Populus balsamifera L. Genetics. 2011;188:941–952. doi: 10.1534/genetics.111.128041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller SR, Olson MS, Silim S, Schroeder W, Tiffin P. Genomic diversity, population structure, and migration following rapid range expansion in the Balsam Poplar, Populus balsamifera. Mol Ecol. 2010;19:1212–1226. doi: 10.1111/j.1365-294X.2010.04546.x. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Levsen ND, Tiffin P, Olson MS. Pleistocene speciation in the genus Populus (Salicaceae) Syst Biol. 2012;61:401–412. doi: 10.1093/sysbio/syr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Ma X-F, Hall D, Onge KRS, Jansson S, Ingvarsson PK. Genetic differentiation, clinal variation and phenotypic associations with growth cessation across the Populus tremula photoperiodic pathway. Genetics. 2010;186:1033–1044. doi: 10.1534/genetics.110.120873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Namroud M-C, Beaulieu J, Juge N, Laroche J, Bousquet J. Scanning the genome for gene single nucleotide polymorphisms involved in adaptive population differentiation in white spruce. Mol Ecol. 2008;17:3599–3613. doi: 10.1111/j.1365-294X.2008.03840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MS, Robertson AL, Takebayashi N, Silim S, Schroeder WR, Tiffin P. Nucleotide diversity and linkage disequilibrium in balsam poplar (Populus balsamifera. New Phytol. 2010;186:526–536. doi: 10.1111/j.1469-8137.2009.03174.x. [DOI] [PubMed] [Google Scholar]
- Rohde A, Ruttink T, Hostyn V, Sterck L, Van Driessche K, Boerjan W. Gene expression during the induction, maintenance, and release of dormancy in apical buds of poplar. J Exp Bot. 2007;58:4047–4060. doi: 10.1093/jxb/erm261. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. distruct: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- Savolainen O, Pyhäjärvi T, Knurr Ti. Gene flow and local adaptation in trees. Annu Rev Ecol Evol Syst. 2007;38:595–619. [Google Scholar]
- Slavov GT, DiFazio SP, Martin J, Schackwitz W, Muchero W, Rodgers-Melnick E, et al. Genome resequencing reveals multiscale geographic structure and extensive linkage disequilibrium in the forest tree Populus trichocarpa. New Phytol. 2012;196:713–725. doi: 10.1111/j.1469-8137.2012.04258.x. [DOI] [PubMed] [Google Scholar]
- Slavov GT, Zhelev P.2010Salient biological features, systematics, and genetic variation of populusIn: Jansson S, Bhalerao R, Groover A (eds)Genetics and Genomics of Populus Springer: New York: New York, NY; 15–38. [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Gunter LE, Yang ZK, Yin T, Sewell MM, Difazio SP. Characterization of microsatellites revealed by genomic sequencing of Populus trichocarpa. Can J For Res. 2004;34:85–93. [Google Scholar]
- Wright SI, Charlesworth B. The HKA test revisited: a maximum-likelihood-ratio test of the standard neutral model. Genetics. 2004;168:1071–1076. doi: 10.1534/genetics.104.026500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordanov YS, Ma C, Strauss SH, Busov VB. EARLY BUD-BREAK 1 (EBB1) is a regulator of release from seasonal dormancy in poplar trees. Proc Natl Acad Sci USA. 2014;111:10001–10006. doi: 10.1073/pnas.1405621111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Bawa R, Holliday J. Exome resequencing reveals signatures of demographic and adaptive processes across the genome and range of black cottonwood (Populus trichocarpa. Mol Ecol. 2014;23:2486–2499. doi: 10.1111/mec.12752. [DOI] [PubMed] [Google Scholar]
- Zhuang J, Cai B, Peng R-H, Zhu B, Jin X-F, Xue Y, et al. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem Biophys Res Commun. 2008;371:468–474. doi: 10.1016/j.bbrc.2008.04.087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.