Abstract
Small heat shock proteins (sHsps) are virtually ubiquitous molecular chaperones that can prevent the irreversible aggregation of denaturing proteins. To maintain protein homeostasis, sHsps complex with a variety of nonnative proteins in an ATP-independent manner and, in the context of the stress response, form a first line of defense against protein aggregation. In vertebrates they act to maintain the clarity of the eye lens, and in humans sHsp mutations are linked to myopathies and neuropathies. Although found in all domains of life, sHsps are quite diverse and have evolved independently in metazoans, plants and fungi. sHsp monomers range in size from approximately 12 to 42 kDa and are defined by a conserved β-sandwich α-crystallin domain, flanked by variable N- and C-terminal sequences. Most sHsps form large oligomeric ensembles with a broad distribution of different, sphere- or barrel like oligomers, with the size and structure of the oligomers dictated by features of the N- and C-termini. The activity of sHsps is regulated by mechanisms that change the equilibrium distribution in tertiary features and/or quaternary structure of the sHsp ensembles. Cooperation and/or coassembly between different sHsps in the same cellular compartment adds an underexplored level of complexity to sHsp structure and function.
Keywords: protein aggregation, molecular chaperones, stress response, protein folding, α-crystallin
Evolution of sHsps
In the course of evolution, a network of proteins arose to protect cells against stress conditions (e.g. heat, cold, oxidative stress) (cf. [1]). A prominent group of these stress proteins are the molecular chaperones, which comprise several families classified according to their molecular mass and evolutionary history [2]. Small heat shock proteins (sHsps), which are present in all three domains of life, are the least conserved of the molecular chaperones [3, 4]. Among the most well-studied members of the sHsp family are the two α-crystallins, αA (or HspB4) and αB (or HspB5), which share 60% amino acid identity, and account for over 30% of protein in the vertebrate eye lens. In the lens they act to maintain the solubility of other lenticular proteins, preventing aggregate-induced light scattering [5, 6]. The clinical importance of αB is highlighted by its additional expression in many other tissues [7] and the fact that αB mutations are linked to myopathies [8]. Furthermore, αB accumulates in plaques of amyloid proteins/peptides that are correlated with neurological disorders such as Alzheimer's, Creutzfeld-Jacob, as well as other diseases [8]. The link of sHsps to human disease, along with the fact that sHsps are expressed during stress and specific stages of development in other organisms, indicates the importance of this virtually ubiquitous group of molecular chaperones.
sHsp primary structure can be dissected into a non-conserved N-terminal sequence (NTS) of variable length, a conserved α-crystallin domain (ACD) and a non-conserved short C-terminal sequence (CTS) (Fig. 1A) [3, 9]. The ACD (or Hsp20 domain, PF00011) represents the conserved signature motif of sHsps. Phylogenetic analyses indicate that sHsps were already present in the last common ancestor of pro- and eukaryotes [3, 10, 11]. Prokaryotes contain usually only one or two sHsps [3, 11, 12]. However, a few, mostly pathogenic bacteria do not encode sHsps, and some, often symbiotic bacteria, have as many as 12 sHsp genes [3, 12, 13].
Figure 1.
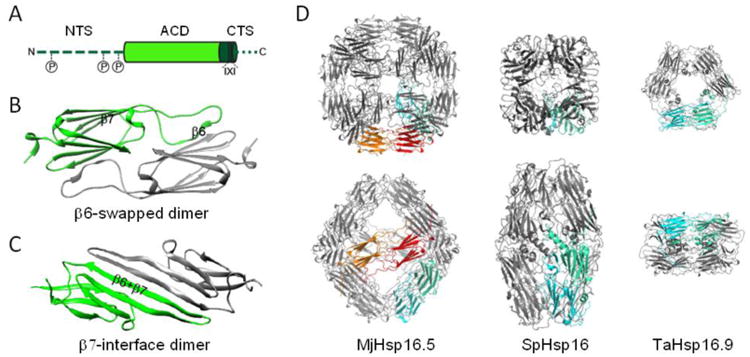
(A) Domain organization of sHsps. N-terminal sequence (NTS; dark green, dashed line), α-crystallin domain (ACD; light green), C-terminal sequence (CTS; dark green with the conserved I-X-I motive in cylinder form, and remainder as a dotted line). As indicated, up to three phosphorylation sites exist in the NTS of some sHsps as discussed in the text. (B) Structure of a β6-swapped dimer of the ACD of M. jannaschii Hsp16.5 (X-ray crystallography, PDB: 1SHS; [45]). The ACDs of individual protomers are colored green and gray. (C) Structure of a β7-interface dimer of the ACD of human αB-crystallin (solid state NMR, PDB: 2KLR; [49]). (D) To scale comparison of the three available oligomeric crystal structures of sHsps. One dimeric building block is marked in green-cyan to highlight the variable interconnections of the dimers in the respective structures. MjHsp16.5; M. jannashii Hsp16.5 representing a 24mer [45]. The orange-red highlighted dimeric building block additionally highlights the equatorial protein axis forming an octahedron. SpHsp16; Schizosaccharomyces pombe Hsp16 representing a 16mer ellipsoid composed of two half-spheres of four dimers [46]. TaHsp16.9; Tritium aestivum (wheat) Hsp16.9 representing a 12mer of two stacked rings [40].
In comparison, in most multicellular eukaryotes the number of sHsps is significantly larger [3, 14]. For example, in addition to αA- and αB-crystallin there are eight other sHsps encoded in the human genome, 16 sHsps are present in Caenorhabditis elegans, and in higher plants there are at least 12 sHsp gene families, with multiple family members bringing the total number of plant sHsp genes in any one species to 20 or more [3, 15-17].
A number of studies have considered the evolutionary trajectory of sHsps, both as a superfamily and within individual domains of life [4, 10, 11, 18-20]. From a comprehensive analysis of 8714 sHsp sequences, Kriehuber et al. [3] were able to conclude that the sHsp signature ACD has evolved independently of the flanking NTS and CTS. A phylogeny of the ACD reflects speciation events, with sHsps from different phyla clustering together on specific branches of the evolutionary tree, while analysis of the NTS and CTS shows no such relationship. In addition, bacterial sHsps are spread into several branches of the tree, even when only considering the ACD, seemingly reflecting a functional diversification of these sequences. Notably, all metazoan sHsps belong to a distinct clade and appear to have evolved from a single ancestral gene by repeated duplications. Similarly, the gene families of higher plant sHsps have arisen by duplication and divergence, which has also included acquisition of specific targeting signals to direct the encoded sHsps to intracellular organelles [17]. In this regard higher plants are unique compared to other eukaryotes, in that sHsps are found not only in the cytosol, but in virtually every membrane bound compartment – chloroplasts, mitochondria, endoplasmic reticulum (ER), peroxisomes, and the nucleus [17, 18, 21-23.Drosophila and Toxoplasma gondii, in which an sHsp is found in mitochondria, are up to now the only other eukaryotes known to have other organelle-localized sHsps [24-26]. Shuttling of cytosolic sHsps into the nucleus is observed under certain conditions in virtually all eukaryotes, but this behavior is distinct from the nuclear-targeted sHsp in plants, which possesses a canonical nuclear-targeting signal. The sessile lifestyle of plants after their movement to land may have driven the evolution of these chaperones to provide protection of proteins throughout the cell. Indeed, there is no evidence for organelle-targeted sHsps in algae [17, 21]. While the presence of sHsps in mitochondria, chloroplasts and the ER may appear similar to the existence of Hsp70/DnaK homologues in the same compartments [27, 28], their evolution is dramatically different. The Hsp70/DnaK homologues were present in the common ancestor of eukaryotes (or of plants in the case of chloroplasts) [29, 30], while the mitochondrial, chloroplast and ER sHsps arose from cytosolic sHsp genes in the plant lineage [17, 18]. The continuing evolution of sHsps is also readily seen in plants. Monocots and eudicots, which diverged on the order of 200 million years ago, have unique cytosolic sHsps, and unique sHsps are found in even more recently diverged taxonomic groups [17, 31]. Altogether, the sHsps appear to have evolved with a flexibility unique to this family of chaperones.
sHsp Structure
A striking feature of most sHsps, which has been considered important for their function, is their ability to assemble into oligomers. The majority of sHsps are found as large, often polydisperse ensembles typically ranging from 12 to greater than 32 subunits, although some dimeric sHsps have now been described [32-36]. The oligomeric sHSPs are all built from an underlying dimeric structure, with a hierarchal arrangement mediated by different protein domains [14, 34, 36, 37]. The ACD, for which there is a growing number of structures [38], is on average 94 amino acids long [3] and forms a compact β-sandwich similar to the immunoglobulin fold, but with a different topology that is identical to that of the Hsp90 cochaperone p23 (Fig. 1B/C). The β-sandwich is composed of two anti-parallel sheets of three and four β-strands, connected by a short inter-domain loop (Fig. 1) [34, 39, 40]. 3D structures of isolated ACDs demonstrate that they usually form stable dimers (Fig. 1B/C), but that the ACD alone is not sufficient for oligomer formation [41-44], although the dimer is the basic building block and the first level of structural order (Fig. 1) [34]. Dimerization of plant, yeast, archaeal and bacterial sHsps characterized to date occurs via reciprocal swapping of the β6 strands into the β-sandwich of the neighboring monomer (Fig. 1B) [33, 40, 45, 46]. This dimer structure has commonly been called a “bacterial” type of dimer, but should more appropriately be called a “β6-swapped dimer”, as higher eukaryotes (eg. wheat Hsp16.9; 1GME) also show this conformation. A second type of dimer structure has been observed and referred to as the “metazoan” type, which is found for example in the α-crystallins [34, 47]. In this type of dimer, the β6- and β7-strands are fused into an elongated strand that forms the dimer interface with its counterpart from the neighboring monomer in an anti-parallel orientation (Fig. 1C). We suggest that this dimer structure should be called a “β7-interface dimer”, as this is more descriptive, and because it remains unclear if this organization will be limited to metazoans.
The variable NTS and CTS that flank the ACD are essential for assembly of the higher order oligomers. Most importantly, the CTS contains a conserved I-x-I motif that is involved in the association of sHsp dimers into oligomers (Fig. 1) [34, 36, 48]. Due to the limited number of high resolution oligomeric sHsp structures, it is still enigmatic which inter-subunit interactions are responsible for the formation of the defined, higher-order oligomeric species. The emerging picture is that residues in all three regions of the sHsp are required for oligomerization. While the ACD forms the basic dimeric building block, both flanking regions contribute to the assembly process. Binding of the CTS I-x-I motif into the hydrophobic groove formed by the β4- and β8-strands of the ACD of a neighboring monomer forms tetramers or hexamers that then further associate into oligomers through contacts within the NTS [34, 39, 40, 45, 46, 49, 50]. However, currently only three complete, oligomeric crystal structures and one pseudo-atomic model derived from a cryo EM structure are available (Fig. 1) [40, 45, 46, 50]. All other 3D structures only represent isolated ACDs or parts of the oligomers. Furthermore, all available structures lack complete information on the structure of the NTS, as all or some are unresolved in the crystal structures, leading to the suggestion that the NTS might be, at least in part, intrinsically disordered [51]. Alternatively, the NTS may include highly dynamic structural elements that fluctuate between different contacts and positions within the oligomer [52]. In this context, it remains possible that the available crystal structures represent only a snapshot of a single stable conformer of what is otherwise a highly dynamic set of conformers. Thus, a combination of different techniques, including crystallography, cryo EM and NMR, as well as EPR and native mass spectrometry, is needed to understand the full, dynamic ensemble of sHsp oligomers and conformers. It will be highly interesting to define the sHsp structural features that mediate the dynamic nature of the oligomer and the variability of subunit interactions.
The hierarchal oligomerization principle of dimers assembling through CTS and NTS contacts seems to be conserved among sHsps and allows the total number of subunits in the oligomers, as well as the geometry of the oligomer, to be modulated by variations, especially in the NTS. This is highlighted by studies on a variant of Hsp16.5 from Methanocaldococcus jannaschii [53] where, the insertion of a 14 amino acid sequence (from the NTS of human Hsp27) adjacent to the ACD of Hsp16.5 resulted in an increase of the number of subunits in the oligomer from 24 to 48 subunits.
Another key feature of sHsps is their tendency to populate a range of oligomeric states at equilibrium [35, 36, 50, 54]. The oligomers constantly exchange subunits and are thus polydisperse and dynamic ensembles (indicated schematically in Fig. 2). The degree of structural plasticity and heterogeneity appears, however, variable for different members of the family [36]. Conditions that destabilize interactions at subunit interfaces lead to an enhanced rate of subunit dissociation, concomitant with enhanced subunit exchange and an increase in smaller ensembles. The ability to exist in a balance between different oligomer populations is correlated with the regulation of the chaperone activity of sHsps [35, 36].
Figure 2.
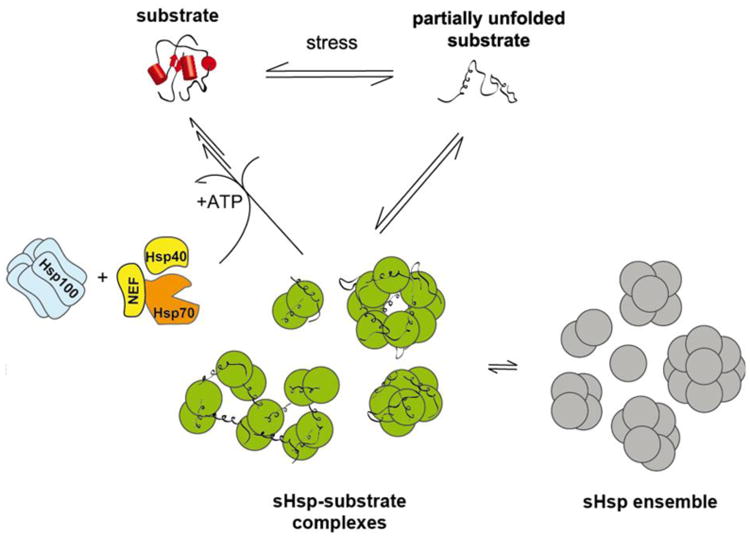
Model for the chaperone function of sHsps. Under stress conditions when substrate proteins are destabilized and begin to unfold, sHsps bind these partially unfolded substrates in an energy-independent manner and keep them in a folding-competent state. The physiologic ensemble of sHsp oligomers (grey) are activated (green) by a shift to a higher content of smaller species (often dimers). The substrate is stabilized by this activated ensemble of sHsps (green) and may reactivate spontaneously or is captured in stable sHsp/substrate complexes (of still enigmatic organization). Bound substrates are subsequently refolded by the ATP-dependent Hsp70 chaperone system (composed of Hsp70, Hsp40 and a nucleotide exchange factor; NEF) and may involve the Hsp100/ClpB chaperone system in cells and cellular compartments where it is found.
Model for sHsp chaperone activity
It has been shown for many sHsps from different species that they can act as molecular chaperones by binding denaturing proteins and preventing them from irreversible aggregation in an ATP-independent fashion (Fig. 2) [4, 55-62]. sHsps fulfill their task as molecular chaperones by stabilizing early unfolding intermediates of aggregation prone proteins, arising as a result of diverse stress conditions (e.g. temperature, oxidative stress). sHsps must be present during the time in which substrate is unfolding; they cannot rescue already unfolded and aggregated substrates. Some early unfolding intermediates may dissociate from the sHsp and refold spontaneously [33, 35, 61, 63, 64]. However, the identity of the substrate, the degree to which it is unfolded, and the specific properties of the sHsp determine the stability of the interaction, and some sHsp-substrate complexes appear essentially irreversible (Fig. 3) [14, 36, 37, 54, 57, 65]. Analyses by electron microscopy [60, 62, 66] and mass spectrometry [54] have revealed that the sHsp-substrate complexes are a discrete ensemble of soluble species that are larger than the substrate-free sHsp oligomers. Although there are no resolved structures of a sHSP-substrate complex, these species convey the impression that they have re-assembled from a dissociated form of the sHsps oligomers, presumably dimers, which re-associate to a new oligomeric form containing the bound substrate. Thus, within the protein homeostasis network of the cell, sHsps can function as a buffer system to bind unfolding proteins upon stress, protecting them from irreversible aggregation (Fig. 2). In vitro experiments revealed that the non-native protein trapped in sHsp/substrate complexes can be released and refolded in the presence of additional ATP-dependent chaperones (Fig. 2). In mammalian cells and in plants, the Hsp70/Hsp40 system is required for refolding of substrate proteins bound to sHsps [67-70]. Similarly, in bacteria such as E. coli, sHsp-bound non-native proteins are transferred to the DnaK-DnaJ-GrpE chaperone system and subsequently reactivated [33, 71-73]. However, this reactivation mechanism appears to depend on the ratio of sHsp to substrate, that is, the Hsp70/Hsp40 system is effective in refolding substrate proteins only if soluble sHsp/substrate complexes form, which requires that sHsps are present at stoichiometric or excess concentrations compared to substrate. At excess levels of substrate, sHsps are incorporated into amorphous aggregates of the substrate protein (Fig. 2). Refolding of substrates from these large, sHsp-containing aggregates requires a mechanism involving not only DnaK-DnaJ-GrpE, but also the protein disaggregase ClpB in E. coli [33, 71, 73-75]. This mechanism is conserved from bacteria to lower eukaryotes, and is also found in higher plants, involving the Hsp100 family members ClpB in bacteria, Hsp104 in S. cerevisiae, and Hsp101 in plants [71, 74-76].
Figure 3.
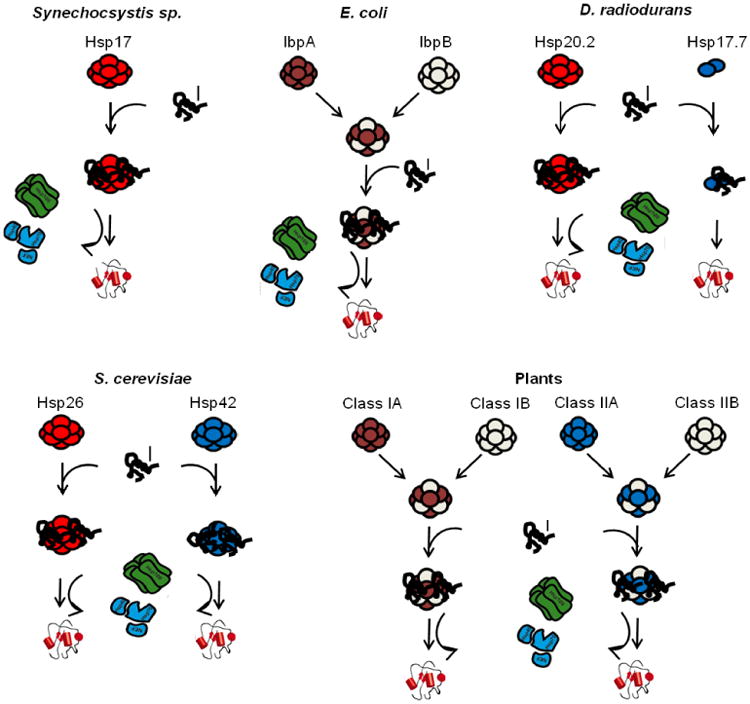
Comparison of types of interactions seen for different, cytosolic sHsp systems from bacteria to higher eukaryotes. The illustrated oligomers and sHsp-substrate complexes symbolize ensembles of oligomers as in Fig. 2. In a number of bacteria (e.g. Synechocystis sp. 6803) there is only a single sHsp (Hsp16.6) that is essential for heat tolerance and acts according to the mechanism described in Fig. 2 [96]. In other bacteria, such as E. coli, there are two (IbpA and IbpB) or more sHsps that form hetero-oligomers and function cooperatively [73, 128]. In yet other bacteria, like D. radiodurans, there are sHsps that work in parallel, independently of each other [33, 128]. In lower eukaryotes like baker`s yeast there are also two, oligomeric sHsps that act independently in parallel. In higher plants there are multiple sHsps classes, and each can have multiple members. Commonly members of the classes are oligomeric and from hetero-oligomers only within members of the same class [36]. The individual classes act in parallel, following in principle the general model (Fig. 2) with the exception that the Hsp100 chaperone system is not found in eukaryotes outside of plants, yeasts and parasitic protozoans. Variations in the spectrum of sHsps in other eukaryotes determine the extent to which sHsp coassembly occurs and the number of independent, parallel sHsp pathways that may be present.
Substrate recognition by sHsps
It is still enigmatic how sHsps recognize denaturing protein substrates. Studies using proteomic approaches in different organisms have identified a significant number of cytosolic proteins associated with, or maintained soluble by sHsps under heat shock conditions [33, 63, 77-79]. Overall, the spectrum of proteins identified as potential in vivo substrates of sHsps during stress indicates that sHsps are promiscuous, although, a preference for translation-related proteins (e.g. ribosomal proteins, translation factors and amino-acyl tRNA synthases) and for metabolic enzymes has been observed in bacteria [33, 78, 80]. Nevertheless, it remains unclear which regions of substrate proteins are bound by sHsps and whether common recognition motifs exist. In terms of the ratio of sHsp to substrate required for substrate protection, sHsps are typically less effective in suppressing the aggregation of larger proteins, indicating that the interaction depends on the mass ratio rather than on the molar ratio, which hints to a charge- and/or hydrophobicity-driven capturing of substrates [36]. While recognition motifs on the sHsps that are involved in substrate interaction are still enigmatic, the emerging picture is that multiple binding sites throughout the protein act together, presumably with a different constellation of sites binding different substrates. Early studies of the incorporation of hydrophobic dyes suggested that substrates bind to short segments in the NTS [67, 81]. Recent evidence from cross-linking experiments and analyses by mass spectrometry or using peptide libraries support this conclusion [82-85]. Molecular dynamics simulations of the NTS in the dimeric form of wheat Hsp16.9 (1GME) (and the homologous pea Hsp18.1) described solvent exposed hydrophobic surface patches on the NTS ranging from 800 to 1700 Å2 that would be available to interact with hydrophobic regions of unfolding substrates [52]. Altogether, there is extensive support for a major role of the NTS in substrate binding.
Other work, however, clearly indicates the ACD and CTS are also involved in substrate interactions. An exchange of the highly conserved G in the AxxxGVL motif of HspH from Bradyrhizobium japonicum specifically impaired chaperone activity without interfering with other properties of the protein, supporting the involvement of the ACD [86-88]. Analysis of chimeric sHsps with different efficiencies of substrate protection also supported a role for the ACD with specific substrates [89]. Additionally, mutations of amino acid residues in the CTS of αB-crystallin affect chaperone activity and indicate that the flexibility of the extended C-terminus of these vertebrate proteins is necessary for substrate recognition or sHsp-substrate complex solubility [90, 91]. The I-x-I motif binds a hydrophobic groove between the β4- and β8-strands within the ACD of the adjacent monomer, which is a potential binding pocket for unfolded proteins as these hydrophobic patches are exposed when the C-terminal contacts dissociate during oligomer disassembly [40, 44, 91, 92]. However, few interactions of the ACD with substrates have been detected in cross-linking experiments, and the β4-β8 hydrophobic groove presents much less accessible hydrophobic surface (<300 Å2) for substrate binding than the NTS [52, 83, 84].
Altogether, substrate binding seems to be accomplished primarily by the non-conserved, variable sequences outside the ACD, which may explain variations in the substrate specificity of different sHsps. Furthermore, as discussed above, independent evolution of the NTS and CTS [3], would suggest it is likely that there are variations in the profile of substrates recognized by sHsps from evolutionally distant species.
The few studies that have used crosslinking to investigate sHsp-binding sites on model substrates indicated that a limited number of substrate sequence segments are bound to the sHsps [85, 93, 94]. No crosslinks have been detected to regions or sequence stretches comprising the interior of a native, folded substrate. These findings are consistent with the model that during stress, substrate proteins in the cell are bound as early unfolding intermediates, rather than as more fully unfolded peptide chains, as only regions presumably exposed early during unfolding bind to the sHsps [83, 85, 93]. This is further evident in experiments using a library of fluorescently labeled T4-lysozyme variants with different thermodynamic stabilities [61, 65, 95]. In these studies, already very weakly destabilized T4-lysozyme variants (by 5 kcal/mol of free energy of unfolding) bound to human Hsp27 (HspB1) or αA-crystallin (HspB4) and the binding affinity increased with stronger destabilization of the T4 lysozyme. Experiments have also been performed to examine the structure of malate dehydrogenase bound to two different sHsps using amide hydrogen-deuterium exchange coupled with mass spectrometry [85]. Data revealed that MDH retained significant core structure, again supporting interaction with an early unfolding intermediate.
Regulation of sHsp activity
For efficient recognition and binding of substrate proteins the majority of sHsps requires a shift in the equilibrium of the oligomeric ensemble from an “inactive state” comprising a high fraction of large oligomers, to an ensemble weighted toward smaller species, representing the “active state” [14, 34-37, 65]. This is necessary, because as described above, substrate binding interactions are thought in a large extent to involve the NTS, which is primarily sequestered within the oligomer. The oligomer is then essentially a storage form that must be regulated to expose substrate binding sites. Support for activation of sHsps by dissociation, in particular to a dimeric form, also comes from studies of atypical sHsps that appear to exist solely as dimers in the native state. A dimeric sHsp from Arabidopsis thaliana, Hsp18.5, is active as a chaperone, binds denaturing model substrates, and forms stable, large sHsp-substrate complexes as seen for its oligomeric relatives [32]. A further example from Deinocaldococcus radiodurans, is dimeric Hsp17.7, which effectively protects substrate in vitro, but interacts with substrates only transiently without forming stable sHsp/substrate complexes [33].
How is the activity of oligomeric sHsps regulated? Of course the expression of many sHsps is dramatically elevated by heat and other stresses, increasing the availability of these chaperones when needed during stress. However, along with the activity of the many sHsps that are present through the life of certain cells, the activity of the oligomeric sHsps that accumulate during stress must be regulated. Four different regulatory mechanisms are recognized that shift the oligomeric equilibrium to the proposed active state consisting of dimers or other small oligomers: first, the presence of unfolded or partially folded substrates; second, changes in the environmental temperature; third, phosphorylation or other post-translational modifications; and fourth, the formation of hetero-oligomers [35].
The first regulatory mechanism establishes sHsps as the initial line of defense in the cell, ensuring the stability of the proteome under physiological and stress conditions. The dynamic assembly/disassembly of sHsp oligomers allows potential substrate binding sites, which are sequestered in the oligomers, to become exposed [65, 95-98]. As there will always be some population of dissociated sHsp subunits (primarily dimers) in the ensemble, a small number of binding sites for unfolding substrate proteins are always available. The presence of unfolding substrate proteins that interact with dissociated sHsp subunits would then lead to a shift in the equilibrium of the sHsp ensemble towards the more active species. Thus, sHsp ensemble dynamics act as a sensing mechanism that monitors the presence of non-native proteins in the cellular environment [65, 99, 100].
Many studies have demonstrated that heat stress temperature (or more generally, a stressor itself) is a general trigger activating sHsps [35, 36]. Interestingly, the temperature range of activation reflects the physiological temperature of the respective organism. For example, mesophilic yeast Hsp26 is activated in a temperature range from 20°C to 43°C, Hsp16.5 from the hyperthermophile M. jannaschii is activated from 60°C to 95°C, human Hsp27 (HspB1) is activated from 37°C to 42°C, and sHsps from fish living below 15°C are activated at ∼10°C-25°C [35, 62, 101-103]. The shift from physiological to heat stress temperatures, which usually requires only a few degree increase, provides sufficient activation energy to shift the sHsp ensemble equilibrium towards enhanced dissociation. Temperature stress itself as an activation mechanism is additionally important at the level of translation control. mRNAs encoding sHsps from mesophilic and thermophilic cyanobacteria, as well as in Pseudomonas putida, contain “RNA thermometers” (RNAT), which are internal hairpins in the 5′-untranslated region that inhibit translation at physiological temperatures [104, 105]. At heat-stress temperatures the RNATs melt, allowing effective translation of the sHsp mRNA. Thus, at least bacteria seem to control the activity and level of sHsps by stress-dependent transcription, RNAT dependent translation, and temperature-induced dissociation/activation of the protein oligomers.
Similar to the presence of substrate, the temperature stimulus represents a very effective and rapid trigger for the activation of sHsps when the level of unfolded proteins increases within a cell. It is tempting to speculate that temperature activation of sHsps is one of the oldest (in terms of evolution) regulatory mechanisms. It allows organisms to rapidly stabilize the proteome without the need for new protein synthesis.
The regulation of sHsp chaperone activity by phosphorylation or more generally, by post-translational modifications, appears to be specific to eukaryotes. With the exception of αA-crystallin (HspB4), all other human sHsps are regulated by serine phosphorylation in response to stress [7, 106-108]. Human Hsp27 (HspB1), for example, possesses three phosphorylation sites, S15, S78 and S82, whose modification via a MAP-kinase cascade leads to eight possible isoforms [108]. sHsp phosphorylation, similar to high temperature and the presence of unfolded proteins, usually shifts the distribution of the sHsp ensemble towards smaller species by dissociation of the larger oligomers, although exceptions are also observed [7, 35]. The smaller species are enriched in tetramers and hexamers reflecting the fact that the assembly process comprises multiple equilibria that are differentially affected by phosphorylation [79, 109]. For example, phosphorylation by MAPKAP2/3 kinase of human Hsp27 (HspB1) leads to an enrichment of tetramers that further dissociate into dimers [68, 109-111]. Similarly, studies using phosphorylation-mimicking variants of αB-crystallin reveal an oligomer ensemble mainly consisting of 12-mers, hexamers and dimers [79, 112]. Predominance of these species indicates that phosphorylation primarily affects N-terminal contacts in the oligomer, which is also in accordance with the location of the phosphorylation sites (Fig. 1A). Mechanistically, the negative charges incorporated upon phosphorylation are predicted to destabilize subunit interfaces. In a pseudo-atomic model of αB-crystallin, the three phosphorylation sites are found in the same region indicating subunit interactions could be destabilized by increasing phosphorylation (and hence the degree of negative charge) in a titratable manner [50, 79].
It should also be mentioned that non-mammalian sHsps can be phosphorylated. Phosphorylated species of Hsp22 from maize mitochondria [113] and yeast Hsp26 have been described [114-116]. However, it remains unknown whether phosphorylation of non-mammalian sHsps has a similar impact on their activity and substrate interactions as is the case for mammalian sHsps. Additionally, other post-translational modifications including deamination, oxidation, glycation or the attachment of methylglyoxal, have been described to have regulatory influence on the chaperone activity of sHsps, primarily for the vertebrate α-crystallins [117-120].
The dynamic sHsp oligomer structure, which constantly exchanges subunits, provides the possibility to form hetero-oligomers with other sHsps present in the same cellular compartment, and represents a further mechanism by which sHsp activity can be modulated. The potential to form sHsp hetero-oligomers varies widely between organisms depending on their constellation of sHsps and the compatibility of the sHsps for co-assembly. This variation in the regulatory potential of hetero-oligomer formation is already evident in bacteria. A number of bacteria encode only a single sHsp, commonly existing as a large oligomer. This is the case for the cyanobacterium Synechocystis sp. PCC6803 (Hsp16.6), Xanthomonas campestris (HspA) and the cyst forming bacterium Azotobacter vinelandii (Hsp20). Notably, in these bacteria Hsp16.6 and HspA are essential for thermotolerance [96, 121], and Hsp20 is essential for cyst desiccation resistance [122]. In other bacteria, such as E. coli, there are two sHsps, one of which (IbpA in E. coli) has low chaperone activity alone, but which acts to enhance chaperone activity of the second sHsp (IbpB in E. coli) when they form a hetero-oligomer (Fig. 3). This observation suggests that one sHsp can modulate the activity of a “partner” sHsp [73]. However, it is unclear if the E. coli “two component sHsp system” can be generalized. In bacteria encoding such a two component sHsp system, the sHsps are either near relatives (located in the same branches of the phylogenetic tree, e.g. E. coli) or more distantly related (located in different branches) [3, 33]. In the latter type of a two component sHsp system, studied in Deinococcus radiodurans in vitro the two sHsps act independently, but in parallel, without hetero-oligomer formation, and they differ substantially in their quaternary structure and function within the chaperone network (Fig. 3) [33]. One of the sHsps, Hsp17.7, is dimeric and an active chaperone as noted above, while the other one, Hsp20.2 is predominantly oligomeric. Hsp20.2 cooperates with ATP-dependent chaperones, while Hsp17.7 appears to keep substrates in a refolding-competent state solely by transient interactions (Fig. 3) [33].
In the context of regulation by hetero-oligomer formation, it is of special interest that not all sHsps found in the same compartment form hetero-oligomers. The sHsps from the soybean symbiont Bradyrhizobium japonicum sort into two classes, where hetero-oligomerization is restricted to the respective class [58]. Similarly Pseudomonas putida has a tricistronic operon of three sHsps (hspX, hspY and hspZ) and a separately encoded ibpA gene that cooperate, but seem to form no (or only weak) hetero-oligomers [105]. This tricistronic operon is conserved in bacteria that are metabolically related to P. putida, suggesting that several, independently acting sHsps might be another common scenario in bacteria. Similarly, in lower eukaryotes like baker s yeast, two sHsps, Hsp26 and Hsp42, act independently and do not form hetero-oligomers (Fig. 3) [63, 123, 124].
In higher eukaryotes the potential for regulation by hetero-oligomerization, as well as the existence of many independently acting sHsps is further evident. In humans (and other vertebrates), specific sHsps are found hetero-oligomerized in vivo (e.g. human αA- and αB-crystallin in the eye lens), and may also act in parallel as homo-oligomers [7, 125, 126]. In higher plants both scenarios are well-developed. In angiosperms (flowering plants) there are at least six sub-families of sHsps, termed Class I to VI that can be found in the cytosol [17, 36]. To date, none of the members of these different sHsp classes have been shown to co-assemble into hetero-oligomers [32, 127], and so presumably act in parallel. The evolutionary history of the class I and II proteins further supports a parallel role, as they are both present in mosses, indicating that they diverged following gene duplication over 400 million years ago [19]. However, at the same time, the class I group of plant cytosolic sHsps has itself undergone extensive gene duplication in many species, and all members of the class readily form hetero-oligomers offering extensive possibilities for subtle modification of substrate interactions [32, 40, 54]. The extent to which hetero-oligomerization may regulate sHsp function in higher eukaryotes remains to be explored.
Summary
In summary, the sHsps are virtually ubiquitous molecular chaperones that have evolved independently in the different branches of life. While they all share a conserved structural domain, the ACD, their divergent NTS and CTS dictate differences in their overall structure, and likely in their substrate interactions and roles within the cell. With some exceptions, sHsps assemble into large oligomers that are built from either a β6-swapped dimer or a β7-interface dimer. sHsp oligomers from some species form monodisperse oligomers, but many are polydisperse, and all exist in a dynamic equilibrium in which subunits exchange between oligomers. Increasing evidence points to the critical role of oligomer dynamics in the ability of sHsps to bind denaturing substrates and to maintain substrates in a soluble and folding competent state. There is still some way to go before we understand the precise details of substrate recognition by sHsps or the potential mechanistic differences between diverse family members. Cooperation between different sHsps in the same cell, either acting in parallel or as hetero-oligomers, adds another level of complexity to defining sHsp function and mechanism. The requirement of sHsps for stress tolerance, their dramatic diversification and the links between sHsp mutations and different human diseases all reinforce the importance of further studies of these enigmatic molecular chaperones.
Research Highlights.
A signature of all sHsps is a core α-crystallin domain which has a β-sandwich structure
Most sHsps form large oligomers built in different geometries from a dimeric substructure
sHsps can bind and stabilize diverse nonnative proteins to maintain protein homeostasis
sHsps evolved independently in different eukaryotes, potentially reflecting diverse functions
Oligomeric sHsps exist as dynamic ensembles that are likely critical to their chaperone function
Acknowledgments
The authors thank the Deutsche Forschungsgemeinschaft (SFB 1035) for financial support to M.H. and Tilly Fleckenstein for help with illustrations. E.V. acknowledges major support for studies of sHsps from the US National Institutes of Health (RO1 GM42762), as well as previous grants from the US Department of Agriculture (NRICGP 99-351007618), the National Science Foundation (IBN-0213128), and current faculty start-up funds from the Massachusetts Life Sciences Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40:253–66. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–51. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Kriehuber T, Rattei T, Weinmaier T, Bepperling A, Haslbeck M, Buchner J. Independent evolution of the core domain and its flanking sequences in small heat shock proteins. FASEB J. 2010;24:3633–42. doi: 10.1096/fj.10-156992. [DOI] [PubMed] [Google Scholar]
- 4.De Jong WW, Leunissen JA, Voorter CE. Evolution of the alpha-crystallin/small heat-shock protein family. Mol Biol Evol. 1993;10:103–26. doi: 10.1093/oxfordjournals.molbev.a039992. [DOI] [PubMed] [Google Scholar]
- 5.Horwitz J. Alpha-crystallin. Exp Eye Res. 2003;76:145–53. doi: 10.1016/s0014-4835(02)00278-6. [DOI] [PubMed] [Google Scholar]
- 6.Slingsby C, Wistow GJ, Clark AR. Evolution of crystallins for a role in the vertebrate eye lens. Protein Sci. 2013;22:367–80. doi: 10.1002/pro.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arrigo AP. Human small heat shock proteins: protein interactomes of homo- and hetero-oligomeric complexes: an update. FEBS Lett. 2013;587:1959–69. doi: 10.1016/j.febslet.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Sun Y, MacRae TH. The small heat shock proteins and their role in human disease. FEBS J. 2005;272:2613–27. doi: 10.1111/j.1742-4658.2005.04708.x. [DOI] [PubMed] [Google Scholar]
- 9.Caspers GJ, Leunissen JA, De Jong WW. The expanding small heat-shock protein family, and structure predictions of the conserved “alpha-crystallin domain”. J Mol Evol. 1995;40:238–48. doi: 10.1007/BF00163229. [DOI] [PubMed] [Google Scholar]
- 10.Waters ER, Lee GJ, Vierling E. Evolution, structure and function of the small heat shock proteins in plants. J Exp Bot. 1996;47:325–38. [Google Scholar]
- 11.Kappe G, Leunissen JA, De Jong WW. Evolution and diversity of prokaryotic small heat shock proteins. Prog Mol Subcell Biol. 2002;28:1–17. doi: 10.1007/978-3-642-56348-5_1. [DOI] [PubMed] [Google Scholar]
- 12.Narberhaus F. Alpha-crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiol Mol Biol Rev. 2002;66:64–93. doi: 10.1128/MMBR.66.1.64-93.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arrigo AP. Small stress proteins: Chaperones that act as regulators of intracellular redox state and programmed cell death. Biol Chem. 1998;379:19–26. [PubMed] [Google Scholar]
- 14.Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12:842–6. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- 15.Kappe G, Franck E, Verschuure P, Boelens WC, Leunissen JAM, De Jong WW. The human genome encodes 10 alpha-crystallin-related small heat shock proteins: HspB1-10. Cell Stress Chaperones. 2003;8:53–61. doi: 10.1379/1466-1268(2003)8<53:thgecs>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scharf KD, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins) Cell Stress Chaperones. 2001;6:225–37. doi: 10.1379/1466-1268(2001)006<0225:tefoat>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waters ER. The evolution, function, structure, and expression of the plant sHSPs. J Exp Bot. 2013;64:391–403. doi: 10.1093/jxb/ers355. [DOI] [PubMed] [Google Scholar]
- 18.Waters ER, Vierling E. Chloroplast small heat shock proteins: evidence for atypical evolution of an organelle-localized protein. Proc Natl Acad Sci USA. 1999;96:14394–9. doi: 10.1073/pnas.96.25.14394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waters ER, Vierling E. The diversification of plant cytosolic small heat shock proteins preceded the divergence of mosses. Mol Biol Evol. 1999;16:127–39. doi: 10.1093/oxfordjournals.molbev.a026033. [DOI] [PubMed] [Google Scholar]
- 20.De Jong WW, Caspers GJ, Leunissen JA. Genealogy of the alpha-crystallin--small heat-shock protein superfamily. Int J Biol Macromol. 1998;22:151–62. doi: 10.1016/s0141-8130(98)00013-0. [DOI] [PubMed] [Google Scholar]
- 21.Waters ER, Rioflorido I. Evolutionary analysis of the small heat shock proteins in five complete algal genomes. J Mol Evol. 2007;65:162–74. doi: 10.1007/s00239-006-0223-7. [DOI] [PubMed] [Google Scholar]
- 22.Ma CL, Haslbeck M, Babujee L, Jahn O, Reumann S. Identification and characterization of a stress-inducible and a constitutive small heat-shock protein targeted to the matrix of plant peroxisomes. Plant Physiol. 2006;141:47–60. doi: 10.1104/pp.105.073841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siddique M, Gernhard S, von Koskull-Doring P, Vierling E, Scharf KD. The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones. 2008;13:183–97. doi: 10.1007/s12192-008-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrow G, Inaguma Y, Kato K, Tanguay RM. The small heat shock protein Hsp22 of Drosophila melanogaster is a mitochondrial protein displaying oligomeric organization. J Biol Chem. 2000;275:31204–10. doi: 10.1074/jbc.M002960200. [DOI] [PubMed] [Google Scholar]
- 25.Michaud S, Morrow G, Marchand J, Tanguay RM. Drosophila small heat shock proteins: cell and organelle-specific chaperones? Prog Mol Subcell Biol. 2002;28:79–101. doi: 10.1007/978-3-642-56348-5_5. [DOI] [PubMed] [Google Scholar]
- 26.de Miguel N, Echeverria PC, Angel SO. Differential subcellular localization of members of the Toxoplasma gondii small heat shock protein family. Eukaryot Cell. 2005;4:1990–7. doi: 10.1128/EC.4.12.1990-1997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sung DY, Vierling E, Guy CL. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 2001;126:789–800. doi: 10.1104/pp.126.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renner T, Waters ER. Comparative genomic analysis of the Hsp70s from five diverse photosynthetic eukaryotes. Cell Stress Chaperones. 2007;12:172–85. doi: 10.1379/CSC-230R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta RS. Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev. 1998;62:1435–91. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- 31.Waters ER, Aevermann BD, Sanders-Reed Z. Comparative analysis of the small heat shock proteins in three angiosperm genomes identifies new subfamilies and reveals diverse evolutionary patterns. Cell Stress Chaperones. 2008;13:127–42. doi: 10.1007/s12192-008-0023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basha E, Jones C, Blackwell AE, Cheng G, Waters ER, Samsel KA, et al. An unusual dimeric small heat shock protein provides insight into the mechanism of this class of chaperones. J Mol Biol. 2013;425:1683–96. doi: 10.1016/j.jmb.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bepperling A, Alte F, Kriehuber T, Braun N, Weinkauf S, Groll M, et al. Alternative bacterial two-component small heat shock protein systems. Proc Natl Acad Sci USA. 2012;109:20407–12. doi: 10.1073/pnas.1209565109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delbecq SP, Klevit RE. One size does not fit all: the oligomeric states of alphaB crystallin. FEBS Lett. 2013;587:1073–80. doi: 10.1016/j.febslet.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haslbeck M, Weinkauf S, Buchner J. Regulation of the chaperone function of small Hsps. In: Tanguay RM, Hightower LE, editors. The Small HSP World. Cham: Springer International Publishing AG; in press. [Google Scholar]
- 36.Basha E, O'Neill H, Vierling E. Small heat shock proteins and alpha-crystallins: dynamic proteins with flexible functions. Trends Biochem Sci. 2012;37:106–17. doi: 10.1016/j.tibs.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garrido C, Paul C, Seigneuric R, Kampinga HH. The small heat shock proteins family: the long forgotten chaperones. Int J Biochem Cell Biol. 2012;44:1588–92. doi: 10.1016/j.biocel.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 38.Santhanagopalan I, Basha E, Ballard KN, Bopp NE, Vierling E. Model Caperones: small Heat Shock Proteins from Plants. In: Tanguay RM, Hightower LE, editors. The Small HSP World. Cham: Springer International Publishing AG; in press. [Google Scholar]
- 39.Van Montfort R, Slingsby C, Vierling E. Structure and function of the small heat shock protein/alpha-crystallin family of molecular chaperones. Adv Protein Chem. 2002;59:105–56. doi: 10.1016/s0065-3233(01)59004-x. [DOI] [PubMed] [Google Scholar]
- 40.van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol. 2001;8:1025–30. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- 41.Bagneris C, Bateman OA, Naylor CE, Cronin N, Boelens WC, Keep NH, et al. Crystal structures of alpha-crystallin domain dimers of alphaB-crystallin and Hsp20. J Mol Biol. 2009;392:1242–52. doi: 10.1016/j.jmb.2009.07.069. [DOI] [PubMed] [Google Scholar]
- 42.Baranova EV, Weeks SD, Beelen S, Bukach OV, Gusev NB, Strelkov SV. Three-Dimensional Structure of alpha-Crystallin Domain Dimers of Human Small Heat Shock Proteins HSPB1 and HSPB6. J Mol Biol. 2011 doi: 10.1016/j.jmb.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 43.Clark AR, Naylor CE, Bagneris C, Keep NH, Slingsby C. Crystal structure of R120G disease mutant of human alphaB-crystallin domain dimer shows closure of a groove. J Mol Biol. 2011;408:118–34. doi: 10.1016/j.jmb.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laganowsky A, Benesch JL, Landau M, Ding L, Sawaya MR, Cascio D, et al. Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function. Protein Sci. 2010;19:1031–43. doi: 10.1002/pro.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim KK, Kim R, Kim SH. Crystal structure of a small heat-shock protein. Nature. 1998;394:595–9. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- 46.Hanazono Y, Takeda K, Oka T, Abe T, Tomonari T, Akiyama N, et al. Nonequivalence observed for the 16-meric structure of a small heat shock protein, SpHsp16.0, from Schizosaccharomyces pombe. Structure. 2013;21:220–8. doi: 10.1016/j.str.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 47.Jehle S, van RB, Stout JR, Noguchi SM, Falber K, Rehbein K, et al. alphaB-crystallin: a hybrid solid-state/solution-state NMR investigation reveals structural aspects of the heterogeneous oligomer. J Mol Biol. 2009;385:1481–97. doi: 10.1016/j.jmb.2008.10.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J, Feige MJ, Franzmann TM, Bepperling A, Buchner J. Regions outside the alpha- crystallin domain of the small heat shock protein Hsp26 are required for its dimerization. J Mol Biol. 2010;398:122–31. doi: 10.1016/j.jmb.2010.02.022. [DOI] [PubMed] [Google Scholar]
- 49.Jehle S, Vollmar BS, Bardiaux B, Dove KK, Rajagopal P, Gonen T, et al. N-terminal domain of alphaB-crystallin provides a conformational switch for multimerization and structural heterogeneity. Proc Natl Acad Sci USA. 2011;108:6409–14. doi: 10.1073/pnas.1014656108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Braun N, Zacharias M, Peschek J, Kastenmuller A, Zou J, Hanzlik M, et al. Multiple molecular architectures of the eye lens chaperone alphaB-crystallin elucidated by a triple hybrid approach. Proc Natl Acad Sci USA. 2011;108:20491–6. doi: 10.1073/pnas.1111014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804:1231–64. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patel S, Vierling E, Tama F. Replica exchange molecular dynamics simulations provide insight into substrate recognition by small heat shock proteins. Biophys J. 2014;106:2644–55. doi: 10.1016/j.bpj.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McHaourab HS, Lin YL, Spiller BW. Crystal structure of an activated variant of small heat shock protein Hsp16.5. Biochemistry. 2012;51:5105–12. doi: 10.1021/bi300525x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stengel F, Baldwin AJ, Painter AJ, Jaya N, Basha E, Kay LE, et al. Quaternary dynamics and plasticity underlie small heat shock protein chaperone function. Proc Natl Acad Sci USA. 2010;107:2007–12. doi: 10.1073/pnas.0910126107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ingolia TD, Craig EA. Four small Drosophila heat shock proteins are related to each other and to mammalian alpha-crystallin. Proc Natl Acad Sci USA. 1982;79:2360–4. doi: 10.1073/pnas.79.7.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jakob U, Gaestel M, Engel K, Buchner J. Small heat shock proteins are molecular chaperones. J Biol Chem. 1993;268:1517–20. [PubMed] [Google Scholar]
- 57.Horwitz J. Alpha-crystallin can function as a molecular chaperone. Proc Natl Acad Sci USA. 1992;89:10449–53. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Studer S, Narberhaus F. Chaperone activity and homo- and hetero-oligomer formation of bacterial small heat shock proteins. J Biol Chem. 2000;275:37212–8. doi: 10.1074/jbc.M004701200. [DOI] [PubMed] [Google Scholar]
- 59.Lee GJ, Pokala N, Vierling E. Structure and in vitro molecular chaperone activity of cytosolic small heat shock proteins from pea. J Biol Chem. 1995;270:10432–8. doi: 10.1074/jbc.270.18.10432. [DOI] [PubMed] [Google Scholar]
- 60.Basha E, Lee GJ, Demeler B, Vierling E. Chaperone activity of cytosolic small heat shock proteins from wheat. Eur J Biochem. 2004;271:1426–36. doi: 10.1111/j.1432-1033.2004.04033.x. [DOI] [PubMed] [Google Scholar]
- 61.Mchaourab HS, Dodson EK, Koteiche HA. Mechanism of chaperone function in small heat shock proteins. Two-mode binding of the excited states of T4 lysozyme mutants by alphaA-crystallin. J Biol Chem. 2002;277:40557–66. doi: 10.1074/jbc.M206250200. [DOI] [PubMed] [Google Scholar]
- 62.Haslbeck M, Walke S, Stromer T, Ehrnsperger M, White HE, Chen S, et al. Hsp26: a temperature-regulated chaperone. EMBO J. 1999;18:6744–51. doi: 10.1093/emboj/18.23.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haslbeck M, Braun N, Stromer T, Richter B, Model N, Weinkauf S, et al. Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J. 2004;23:638–49. doi: 10.1038/sj.emboj.7600080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McHaourab HS, Godar JA, Stewart PL. Structure and mechanism of protein stability sensors: chaperone activity of small heat shock proteins. Biochemistry. 2009;48:3828–37. doi: 10.1021/bi900212j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mchaourab HS, Godar JA, Stewart PL. Structure and mechanism of protein stability sensors: chaperone activity of small heat shock proteins. Biochemistry. 2009;48:3828–37. doi: 10.1021/bi900212j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stromer T, Ehrnsperger M, Gaestel M, Buchner J. Analysis of the interaction of small heat shock proteins with unfolding proteins. J Biol Chem. 2003;278:18015–21. doi: 10.1074/jbc.M301640200. [DOI] [PubMed] [Google Scholar]
- 67.Lee GJ, Roseman AM, Saibil HR, Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 1997;16:659–71. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ehrnsperger M, Graber S, Gaestel M, Buchner J. Binding of non-native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 1997;16:221–9. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stege GJ, Li GC, Li L, Kampinga HH, Konings AW. On the role of hsp72 in heat-induced intranuclear protein aggregation. Int J Hyperthermia. 1994;10:659–74. doi: 10.3109/02656739409022446. [DOI] [PubMed] [Google Scholar]
- 70.Lee GJ, Vierling E. A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol. 2000;122:189–98. doi: 10.1104/pp.122.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mogk A, Deuerling E, Vorderwulbecke S, Vierling E, Bukau B. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol. 2003;50:585–95. doi: 10.1046/j.1365-2958.2003.03710.x. [DOI] [PubMed] [Google Scholar]
- 72.Veinger L, Diamant S, Buchner J, Goloubinoff P. The small heat-shock protein IbpB from Escherichia coli stabilizes stress-denatured proteins for subsequent refolding by a multichaperone network. J Biol Chem. 1998;273:11032–7. doi: 10.1074/jbc.273.18.11032. [DOI] [PubMed] [Google Scholar]
- 73.Ratajczak E, Zietkiewicz S, Liberek K. Distinct activities of Escherichia coli small heat shock proteins IbpA and IbpB promote efficient protein disaggregation. J Mol Biol. 2009;386:178–89. doi: 10.1016/j.jmb.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 74.Haslbeck M, Miess A, Stromer T, Walter S, Buchner J. Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J Biol Chem. 2005;280:23861–8. doi: 10.1074/jbc.M502697200. [DOI] [PubMed] [Google Scholar]
- 75.Cashikar AG, Duennwald ML, Lindquist SL. A chaperone pathway in protein disaggregation: Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J Biol Chem. 2005 doi: 10.1074/jbc.M502854200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee U, Wie C, Escobar M, Williams B, Hong SW, Vierling E. Genetic analysis reveals domain interactions of Arabidopsis Hsp100/ClpB and cooperation with the small heat shock protein chaperone system. Plant Cell. 2005;17:559–71. doi: 10.1105/tpc.104.027540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Basha E, Lee GJ, Breci LA, Hausrath AC, Buan NR, Giese KC, et al. The identity of proteins associated with a small heat shock protein during heat stress in vivo indicates that these chaperones protect a wide range of cellular functions. J Biol Chem. 2004;279:7566–75. doi: 10.1074/jbc.M310684200. [DOI] [PubMed] [Google Scholar]
- 78.Fu X, Shi X, Yan L, Zhang H, Chang Z. In vivo substrate diversity and preference of small heat shock protein IbpB as revealed by using a genetically incorporated photo-cross-linker. J Biol Chem. 2013;288:31646–54. doi: 10.1074/jbc.M113.501817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peschek J, Braun N, Rohrberg J, Back KC, Kriehuber T, Kastenmuller A, et al. Regulated structural transitions unleash the chaperone activity of alphaB-crystallin. Proc Natl Acad Sci USA. 2013;110:E3780–9. doi: 10.1073/pnas.1308898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fu X. Chaperone function and mechanism of small heat-shock proteins. Acta Biochimi Bbiophys Sin. 2014;46:347–56. doi: 10.1093/abbs/gmt152. [DOI] [PubMed] [Google Scholar]
- 81.Sharma KK, Kumar GS, Murphy AS, Kester K. Identification of 1,1′-bi(4-anilino) naphthalene-5,5′-disulfonic acid binding sequences in alpha-crystallin. J Biol Chem. 1998;273:15474–8. doi: 10.1074/jbc.273.25.15474. [DOI] [PubMed] [Google Scholar]
- 82.Ghosh JG, Shenoy AK, Jr, Clark JI. Interactions between important regulatory proteins and human alphaB crystallin. Biochemistry. 2007;46:6308–17. doi: 10.1021/bi700149h. [DOI] [PubMed] [Google Scholar]
- 83.Ahrman E, Lambert W, Aquilina JA, Robinson CV, Emanuelsson CS. Chemical cross-linking of the chloroplast localized small heat-shock protein, Hsp21, and the model substrate citrate synthase. Protein Sci. 2007;16:1464–78. doi: 10.1110/ps.072831607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jaya N, Garcia V, Vierling E. Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc Natl Acad Sci USA. 2009;106:15604–9. doi: 10.1073/pnas.0902177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cheng G, Basha E, Wysocki VH, Vierling E. Insights into small heat shock protein and substrate structure during chaperone action derived from hydrogen/deuterium exchange and mass spectrometry. J Biol Chem. 2008;283:26634–42. doi: 10.1074/jbc.M802946200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lentze N, Aquilina JA, Lindbauer M, Robinson CV, Narberhaus F. Temperature and concentration-controlled dynamics of rhizobial small heat shock proteins. Eur J Biochem. 2004;271:2494–503. doi: 10.1111/j.1432-1033.2004.04180.x. [DOI] [PubMed] [Google Scholar]
- 87.Lentze N, Narberhaus F. Detection of oligomerisation and substrate recognition sites of small heat shock proteins by peptide arrays. Biochem Biophys Res Commun. 2004;325:401–7. doi: 10.1016/j.bbrc.2004.10.043. [DOI] [PubMed] [Google Scholar]
- 88.Hochberg GK, Ecroyd H, Liu C, Cox D, Cascio D, Sawaya MR, et al. The structured core domain of alphaB-crystallin can prevent amyloid fibrillation and associated toxicity. Proc Natl Acad Sci USA. 2014;111:E1562–70. doi: 10.1073/pnas.1322673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Basha E, Friedrich KL, Vierling E. The N-terminal arm of small heat shock proteins is important for both chaperone activity and substrate specificity. J Biol Chem. 2006;281:39943–52. doi: 10.1074/jbc.M607677200. [DOI] [PubMed] [Google Scholar]
- 90.Treweek TM, Ecroyd H, Williams DM, Meehan S, Carver JA, Walker MJ. Site-directed mutations in the C-terminal extension of human alphaB-crystallin affect chaperone function and block amyloid fibril formation. PloS One. 2007;2:e1046. doi: 10.1371/journal.pone.0001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Treweek TM, Rekas A, Walker MJ, Carver JA. A quantitative NMR spectroscopic examination of the flexibility of the C-terminal extensions of the molecular chaperones, alphaA- and alphaB-crystallin. Exp Eye Res. 2010;91:691–9. doi: 10.1016/j.exer.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 92.Delbecq SP, Jehle S, Klevit R. Binding determinants of the small heat shock protein, alphaB-crystallin: recognition of the ‘I×I’ motif. EMBO J. 2012;31:4587–94. doi: 10.1038/emboj.2012.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ahrman E, Gustavsson N, Hultschig C, Boelens WC, Emanuelsson CS. Small heat shock proteins prevent aggregation of citrate synthase and bind to the N-terminal region which is absent in thermostable forms of citrate synthase. Extremophiles. 2007;11:659–66. doi: 10.1007/s00792-007-0080-3. [DOI] [PubMed] [Google Scholar]
- 94.Santhoshkumar P, Sharma KK. Identification of a region in alcohol dehydrogenase that binds to alpha-crystallin during chaperone action. Biochim Biophys Acta. 2002;1598:115–21. doi: 10.1016/s0167-4838(02)00356-4. [DOI] [PubMed] [Google Scholar]
- 95.Shashidharamurthy R, Koteiche HA, Dong J, Mchaourab HS. Mechanism of chaperone function in small heat shock proteins: dissociation of the HSP27 oligomer is required for recognition and binding of destabilized T4 lysozyme. J Biol Chem. 2005;280:5281–9. doi: 10.1074/jbc.M407236200. [DOI] [PubMed] [Google Scholar]
- 96.Giese KC, Vierling E. Changes in oligomerization are essential for the chaperone activity of a small heat shock protein in vivo and in vitro. J Biol Chem. 2002;277:46310–8. doi: 10.1074/jbc.M208926200. [DOI] [PubMed] [Google Scholar]
- 97.Yang H, Huang S, Dai H, Gong Y, Zheng C, Chang Z. The Mycobacterium tuberculosis small heat shock protein Hsp16.3 exposes hydrophobic surfaces at mild conditions: conformational flexibility and molecular chaperone activity. Protein Sci. 1999;8:174–9. doi: 10.1110/ps.8.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lindner RA, Kapur A, Mariani M, Titmuss SJ, Carver JA. Structural alterations of alpha-crystallin during its chaperone action. Eur J Biochem. 1998;258:170–83. doi: 10.1046/j.1432-1327.1998.2580170.x. [DOI] [PubMed] [Google Scholar]
- 99.Shi J, Koteiche HA, McDonald ET, Fox TL, Stewart PL, McHaourab HS. Cryoelectron microscopy analysis of small heat shock protein 16.5 (Hsp16.5) complexes with T4 lysozyme reveals the structural basis of multimode binding. J Biol Chem. 2013;288:4819–30. doi: 10.1074/jbc.M112.388132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Benesch JL, Ayoub M, Robinson CV, Aquilina JA. Small heat shock protein activity is regulated by variable oligomeric substructure. J Biol Chem. 2008;283:28513–7. doi: 10.1074/jbc.M804729200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Haslbeck M, Kastenmuller A, Buchner J, Weinkauf S, Braun N. Structural dynamics of archaeal small heat shock proteins. J Mol Biol. 2008;378:362–74. doi: 10.1016/j.jmb.2008.01.095. [DOI] [PubMed] [Google Scholar]
- 102.Ehrnsperger M, Lilie H, Gaestel M, Buchner J. The dynamics of Hsp25 quaternary structure. Structure and function of different oligomeric species. J Biol Chem. 1999;274:14867–74. doi: 10.1074/jbc.274.21.14867. [DOI] [PubMed] [Google Scholar]
- 103.Posner M, Kiss AJ, Skiba J, Drossman A, Dolinska MB, Hejtmancik JF, et al. Functional validation of hydrophobic adaptation to physiological temperature in the small heat shock protein alphaA-crystallin. PloS one. 2012;7:e34438. doi: 10.1371/journal.pone.0034438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cimdins A, Klinkert B, Aschke-Sonnenborn U, Kaiser FM, Kortmann J, Narberhaus F. Translational control of small heat shock genes in mesophilic and thermophilic cyanobacteria by RNA thermometers. RNA Biol. 2014;11 doi: 10.4161/rna.28648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Krajewski SS, Joswig M, Nagel M, Narberhaus F. A tricistronic heat shock operon is important for stress tolerance of Pseudomonas putida and conserved in many environmental bacteria. Environ Microbiol. 2014;16:1835–53. doi: 10.1111/1462-2920.12432. [DOI] [PubMed] [Google Scholar]
- 106.Ito H, Iida K, Kamei K, Iwamoto I, Inaguma Y, Kato K. AlphaB-crystallin in the rat lens is phosphorylated at an early post-natal age. FEBS Lett. 1999;446:269–72. doi: 10.1016/s0014-5793(99)00242-2. [DOI] [PubMed] [Google Scholar]
- 107.van den IJssel PR, Overkamp P, Bloemendal H, De Jong WW. Phosphorylation of alphaB-crystallin and HSP27 is induced by similar stressors in HeLa cells. BiochemBiophysResCommun. 1998;247:518–23. doi: 10.1006/bbrc.1998.8699. [DOI] [PubMed] [Google Scholar]
- 108.Gaestel M. sHsp-phosphorylation: enzymes, signaling pathways and functional implications. Prog Mol Subcell Biol. 2002;28:151–69. doi: 10.1007/978-3-642-56348-5_8. [DOI] [PubMed] [Google Scholar]
- 109.Rogalla T, Ehrnsperger M, Preville X, Kotlyarov A, Lutsch G, Ducasse C, et al. Regulation of Hsp27 oligomerization, chaperone function, and protective activity against oxidative stress/tumor necrosis factor alpha by phosphorylation. J Biol Chem. 1999;274:18947–56. doi: 10.1074/jbc.274.27.18947. [DOI] [PubMed] [Google Scholar]
- 110.Hayes D, Napoli V, Mazurkie A, Stafford WF, Graceffa P. Phosphorylation dependence of hsp27 multimeric size and molecular chaperone function. J Biol Chem. 2009;284:18801–7. doi: 10.1074/jbc.M109.011353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kato K, Hasegawa K, Goto S, Inaguma Y. Dissociation as a result of phosphorylation of an aggregated form of the small stress protein, hsp27. J Biol Chem. 1994;269:11274–8. [PubMed] [Google Scholar]
- 112.Ito H, Kamei K, Iwamoto I, Inaguma Y, Nohara D, Kato K. Phosphorylation-induced change of the oligomerization state of alpha B-crystallin. J Biol Chem. 2001;276:5346–52. doi: 10.1074/jbc.M009004200. [DOI] [PubMed] [Google Scholar]
- 113.Lund AA, Rhoads DM, Lund AL, Cerny RL, Elthon TE. In vivo modifications of the maize mitochondrial small heat stress protein, HSP22. J Biol Chem. 2001;276:29924–9. doi: 10.1074/jbc.M103373200. [DOI] [PubMed] [Google Scholar]
- 114.Bentley NJ, Fitch IT, Tuite MF. The small heat-shock protein Hsp26 of Saccharomyces cerevisiae assembles into a high molecular weight aggregate. Yeast. 1992;8:95–106. doi: 10.1002/yea.320080204. [DOI] [PubMed] [Google Scholar]
- 115.Bodenmiller B, Wanka S, Kraft C, Urban J, Campbell D, Pedrioli PG, et al. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal. 2010;3:rs4. doi: 10.1126/scisignal.2001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nature Biotech. 2002;20:301–5. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 117.Gupta R, Srivastava OP. Deamidation affects structural and functional properties of human alphaA-crystallin and its oligomerization with alphaB-crystallin. J Biol Chem. 2004;279:44258–69. doi: 10.1074/jbc.M405648200. [DOI] [PubMed] [Google Scholar]
- 118.Chen SJ, Sun TX, Akhtar NJ, Liang JJ. Oxidation of human lens recombinant alphaA-crystallin and cysteine-deficient mutants. J Mol Biol. 2001;305:969–76. doi: 10.1006/jmbi.2000.4348. [DOI] [PubMed] [Google Scholar]
- 119.Chalova AS, Sudnitsyna MV, Semenyuk PI, Orlov VN, Gusev NB. Effect of disulfide crosslinking on thermal transitions and chaperone-like activity of human small heat shock protein HspB1. Cell Stress Chaperones. 2014;19:963–72. doi: 10.1007/s12192-014-0520-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Satish Kumar M, Mrudula T, Mitra N, Bhanuprakash Reddy G. Enhanced degradation and decreased stability of eye lens alpha-crystallin upon methylglyoxal modification. Exp Eye Res. 2004;79:577–83. doi: 10.1016/j.exer.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 121.Lin CH, Lee CN, Lin JW, Tsai WJ, Wang SW, Weng SF, et al. Characterization of Xanthomonas campestris pv. campestris heat shock protein A (HspA), which possesses an intrinsic ability to reactivate inactivated proteins. Appl Microbiol Biotechnol. 2010;88:699–709. doi: 10.1007/s00253-010-2776-z. [DOI] [PubMed] [Google Scholar]
- 122.Cocotl-Yanez M, Moreno S, Encarnacion S, Lopez-Pliego L, Castaneda M, Espin G. A small heat-shock protein (Hsp20) regulated by RpoS is essential for cyst desiccation resistance in Azotobacter vinelandii. Microbiology. 2014;160:479–87. doi: 10.1099/mic.0.073353-0. [DOI] [PubMed] [Google Scholar]
- 123.Duennwald ML, Echeverria A, Shorter J. Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol. 2012;10:e1001346. doi: 10.1371/journal.pbio.1001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Specht S, Miller SB, Mogk A, Bukau B. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol. 2011;195:617–29. doi: 10.1083/jcb.201106037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mymrikov EV, Seit-Nebi AS, Gusev NB. Heterooligomeric complexes of human small heat shock proteins. Cell Stress Chaperones. 2012;17:157–69. doi: 10.1007/s12192-011-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mymrikov EV, Seit-Nebi AS, Gusev NB. Large potentials of small heat shock proteins. Physiol Rev. 2011;91:1123–59. doi: 10.1152/physrev.00023.2010. [DOI] [PubMed] [Google Scholar]
- 127.Basha E, Jones C, Wysocki V, Vierling E. Mechanistic differences between two conserved classes of small heat shock proteins found in the plant cytosol. J Biol Chem. 2010;285:11489–97. doi: 10.1074/jbc.M109.074088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Haslbeck M. Small heat shock proteins in bacteria. In: De Bruijn FJ, editor. Stress and Enviromental Control of Gene Expression in Bacteria. Hoboken: John Wiley & Sons, Inc.; in press. [Google Scholar]


