Abstract
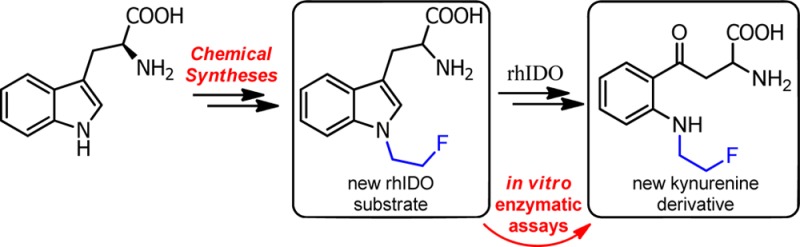
Indoleamine 2,3-dioxygenase (hIDO) is an enzyme that catalyzes the oxidative cleavage of the indole ring of l-tryptophan through the kynurenine pathway, thereby exerting immunosuppressive properties in inflammatory and tumoral tissues. The syntheses of 1-(2-fluoroethyl)-tryptophan (1-FETrp) and 1-((1-(2-fluoroethyl)-1H-1,2,3-triazol-4-yl)methyl)-tryptophan, two N1-fluoroalkylated tryptophan derivatives, are described here. In vitro enzymatic assays with these two new potential substrates of hIDO show that 1-FETrp is a good and specific substrate of hIDO. Therefore, its radioactive isotopomer, 1-[18F]FETrp, should be a molecule of choice to visualize tumoral and inflammatory tissues and/or to validate new potential inhibitors.
Keywords: Tryptophan; indoleamine 2,3-dioxygenase; kynurenine; 1-(2-fluoroethyl)-tryptophan; tryptophan 2,3-dioxygenase; positron emission tomography
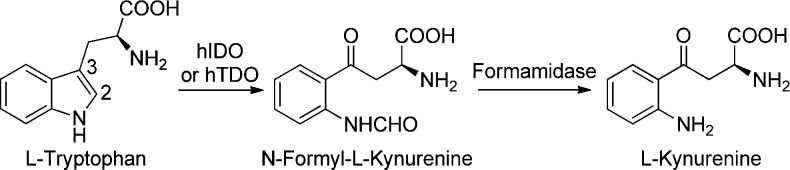
l-Tryptophan (Trp), the least abundant of the essential amino acids, is used in three biological processes: (i) the serotonin pathway, which converts only tiny amounts (about 1%) of dietary Trp into serotonin, (ii) protein synthesis, and (iii) the kynurenine pathway, metabolizing more than 95% of Trp and leading to the biosynthesis of kynurenine (KYN), nicotinamide adenine dinucleotide (NAD+), kynurenic acid (KYNA), 3-hydroxyanthranilic acid (3-HAA), etc.1−4 The initial and rate-limiting step of the kynurenine pathway requires the involvement of indoleamine 2,3-dioxygenase (hIDO), or tryptophan 2,3-dioxygenase (hTDO; mainly expressed in liver). These structurally different heme-containing enzymes catalyze the C2–C3 oxidative cleavage of the pyrrole ring of the indole nucleus of Trp, yielding to the N-formyl-l-kynurenine (Scheme 1).5−8 hIDO expression is strongly stimulated by interferon-γ (IFN-γ) and also, to some extent, by cytokines such as interleukins (IL-1β, IL-2) and tumor necrosis factor (TNF-α).6,9−15
Scheme 1. The Two First Steps of the Kynurenine Pathway.
l-Kynurenine, the key intermediate of the kynurenine pathway, is formed from l-tryptophan in two steps catalyzed by indoleamine 2,3-dioxygenase (or tryptophan 2,3-dioxygenase) and formamidase, respectively.
hIDO is mainly and highly expressed (i) in placenta, preventing the rejection of allogeneic fetuses,5 (ii) in inflammatory tissues,11,12 and (iii) in human tumors such as mammary, prostatic, colorectal, pancreatic, cervical, and endometrial carcinomas.4,5,11−15 In these various tissues, this high expression of hIDO creates a depletion of tryptophan and an accumulation of immunosuppressive tryptophan catabolites, both of which result in a local suppression of the immune response by blocking T-lymphocyte proliferation in the G1 phase of the cell cycle.1,5,7,16,17
Recently, in order to improve cancer immunotherapy and to counteract the local immunosuppression that protects malignancies from endogenous and drug-induced destruction, a number of hIDO inhibitors have been developed.18 To facilitate (i) the preclinical and clinical development and validation of such compounds and (ii) the clinical detection of hIDO expressing cells in cancer imaging, the synthesis (and the radiosynthesis) of a PET imaging tracer could prove to be a highly desirable and valuable tool.
[α/β-11C]-l-Tryptophan,19,20 5-hydroxy-[α/β-11C]-l-tryptophan,20,21 and α-[11C]methyl-l-tryptophan ([11C]AMT)15,21−23 are radiotracers used essentially in positron emission tomography (PET) studies of the in vivo transport and brain serotonin metabolism of l-tryptophan in health and disease.
Inasmuch as following the metabolization of a radioactive substrate of hIDO through the kynurenine pathway could highlight the presence of this enzyme, the introduction into tryptophan of a radionucleide with a longer half-life time, such as [18F]fluorine (t1/2 = 109.7 min), seems to be better adapted than [11C]carbon (t1/2 = 20.3 min) (in terms of [18F]fluorine production, radiotracer synthesis, biological studies over longer periods, etc.).
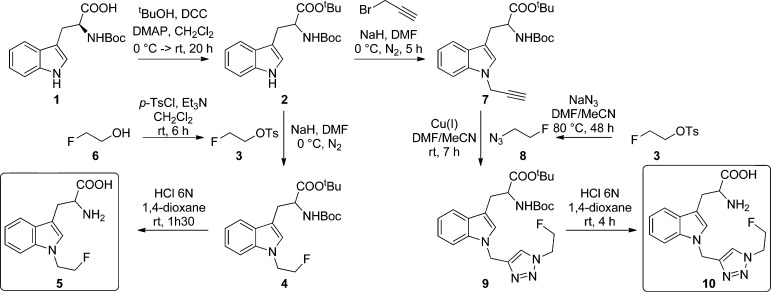
In order to achieve these objectives, two molecules alkylated on the nitrogen atom of the indole ring, 1-(2-fluoroethyl)-tryptophan (5) and 1-((1-(2-fluoroethyl)-1H-1,2,3-triazol-4-yl)methyl)-tryptophan (10) (Scheme 2), were synthesized using two different approaches (including a “Click” reaction), both easily transposable for the radiosynthesis.
Scheme 2. General Scheme for the Syntheses of 1-(2-Fluoroethyl)-tryptophan (5) and 1-((1-(2-Fluoroethyl)-1H-1,2,3-triazol-4-yl)methyl)-tryptophan (10).
The synthesis of the key intermediate 2, used for those of 5 and 10 (Scheme 2), involved the protection of the carboxylic acid function of the commercially available Boc-l-tryptophan 1 with tert-butyl ester (27%). The synthesis of 5, allowing the introduction in two steps of a fluoroethyl group, one of the smallest monofluoroalkyl chains, required the N1-alkylation of 2 with 1-fluoro-2-tosyloxyethane (3) (1.1 equiv; synthesized beforehand from 2-fluoroethanol 6 (95%)) with sodium hydride (2.5 equiv) at 0 °C in DMF. These conditions were crucial to minimize side reactions while maximizing the yield of the N1-alkylation (52%). Hydrolysis of the intermediate 4 in hydrochloric acid (6 N) and 1,4-dioxane (1/1 (v/v)) then gave 1-(2-fluoroethyl)-tryptophan as its hydrochloride salt (5·HCl; 93%). The enantiomeric excess was determined for 5 by a chiral HPLC analysis. A racemization of the alpha-amino acid product was observed. However, our method using sodium hydride offered a better yield (52%) compared with that recently obtained by Sun et al. (NaOH, 30%). Moreover, epimerization was not evaluated in this paper.24
The synthesis of 10 began with the N1-alkylation of 2 with propargyl bromide (5 equiv) and sodium hydride (5 equiv) at 0 °C for 5 h in DMF (93%). This result was in good accordance with that obtained by Schultz et al.25 The 1,2,3-triazole ring was formed by a Huisgen 1,3-dipolar cycloaddition (also known as a Click reaction), usually catalyzed by copper(I) salt (directly introduced or generated in situ) in conjunction with an added organic or inorganic base.26−28 This synthesis allowed an easy introduction of the fluorine, leading to a longer and more rigid monofluorinated chain, due to a triazole ring formation between 7 and the fluorinated prosthetic group 8 (synthesized beforehand in one step from 3 and sodium azide), at room temperature for 7 h. The reusable polymer-supported Amberlyst A-21·CuI, which associates an organic base and Cu(I) salt, should facilitate Cu(I) elimination.26 However, even though the yields obtained were good (73%), after an extended stirring, copper was partially released into the solution. Copper(I) iodide offered the desired product but with a small amount of 5-iodo-1,2,3-triazole derivative as an unwanted side-product. The Hünig’s base and tetrakis(acetonitrile)copper(I) hexafluorophosphate (Cu(CH3CN)4PF6), which is much more soluble than CuI,28 gave higher yields (81%) while limiting the number of side-products. Nevertheless, an inert atmosphere is essential to limit the formation of the 5-hydroxy-1,2,3-triazole analogue. Finally, hydrolysis of 9 in hydrochloric acid (6 N) and 1,4-dioxane (1/1 (v/v)) offered 1-((1-(2-fluoroethyl)-1H-1,2,3-triazol-4-yl)methyl)-tryptophan as its hydrochloride salt (10·HCl) (83%). A complete racemization of the alpha-amino acid product was also observed.
In vitro enzymatic assays with 5 and 10 were performed in order to characterize the affinity and the specificity of these new potential substrates for recombinant human indoleamine 2,3-dioxygenase (rhIDO) (Scheme 1; see Supporting Information for experimental details). This reaction has been widely studied, and substrates have been characterized by kinetic constants determined through the use of a range of analytical methods, observing either the formation of N-formyl-l-kynurenine derivatives (direct method)29,30 or an increase in the concentration of l-kynurenine analogues after incubation with perchloric acid (indirect method).31−33
The formation of the product, N-formyl-l-kynurenine, was followed by measuring its absorbance at 321 nm.30−32 Nevertheless, as mentioned briefly by Chauhan et al.,31 the UV-absorbance of the N-formyl-l-kynurenine derivative decreased significantly when the nitrogen in the substrate was alkylated, as for 1-Me-l-Trp (12). This decrease was also observed here for 1-FETrp (5), where the low UV-absorption of the product did not allow the study of the reaction by this method. An indirect method was therefore investigated. Moreover, it was observed that N-alkylated-N-formyl-kynurenine derivatives were hydrolyzed into N-alkylated-kynurenine analogues much more slowly (for at least 30 min in HClO4). The substrates and their products were separated by HPLC or UPLC and detected by UV (method A, less sensitive; [S] ≥ 200 μM) or by fluorescence detection (method B; used to detect only the substrates; [S] ≥ 2 μM).34
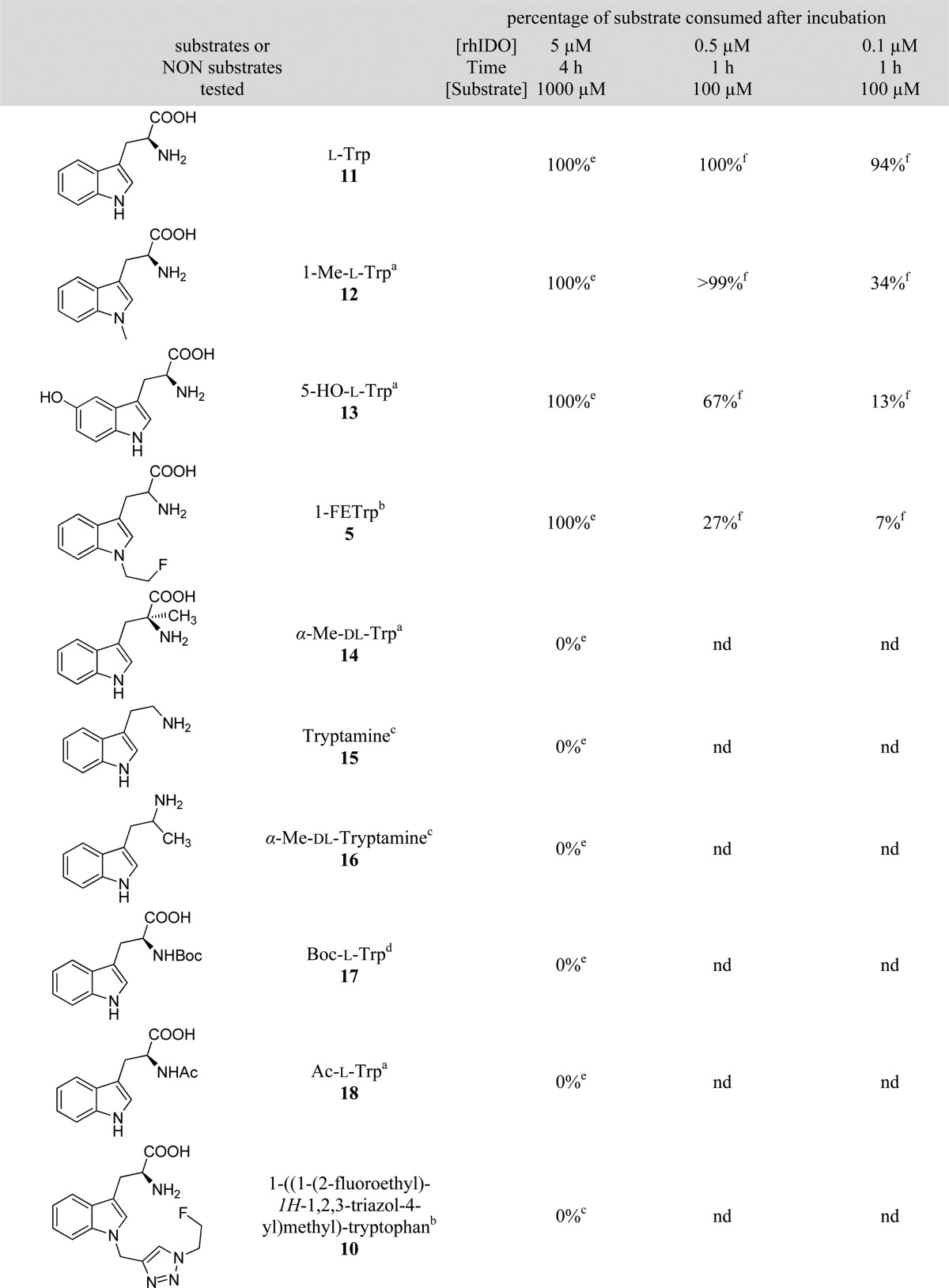
The oxidative conditions of the enzymatic reaction were first improved to allow a complete oxidation of the natural substrate: l-tryptophan (11) (Table 1). Under these optimal conditions, 12, 13, and 5 were also completely oxidized. These results were in contrast to those found with the following six compounds: 10 (which probably had a too long and rigid substituent to fit adequately into the catalytic cavity of the enzyme); α-Me-dl-Trp (14); tryptamine (15); α-Me-dl-tryptamine (16); Boc-l-Trp (17); and Ac-l-Trp (18) (whose NH3+ group is protected and no longer available for H-bonding interaction with the catalytic center facilitating the ring-opening reaction7). These six compounds were therefore found not to be substrates of this enzyme. In our experimental conditions, the opening of the indole ring was also not observed for α-Me-dl-Trp (AMT, 14). Thus, our results showed that AMT was not metabolized by rhIDO through the kynurenine pathway, contrary to the findings reported in the literature.35
Table 1. Enzymatic Assays Carried out under Different Conditions with Some Substrates and NON Substrates of rhIDO.
From Aldrich.
Racemate.
Acros Organics.
Iris Biotech Gmbh.
Percentage determined by UV detection method.
Percentage obtained by fluorescence detection method. All assays were repeated two to three times. nd: not determined.
The incubation conditions (substrate concentration, enzyme concentration, and time) were then changed in order to compare the substrates. Compound 5 was identified as a good substrate of this enzyme, but it was found to be oxidized more slowly than 5-HO-l-Trp (around 2 times), 1-Me-l-Trp (around 4 times), and l-Trp, which is known as the best substrate for rhIDO (Table 1).
The fluorescence method was used to determine the initial rates, allowing the determination of the Km and kcat values. An inhibition by the substrate was observed for l-Trp and 1-Me-l-Trp, as described in the literature,30 but not for 5-HO-l-Trp and 1-FETrp under the conditions studied. However, even though the concentration of rhIDO was reduced as much as possible, the reaction was still too fast to allow an accurate determination of the initial rates. Nonetheless, these results still allowed an estimation of the Km value (70 ± 30 μM for 1-FETrp).
Because of the difficulties involved in determining accurately these kinetic constants, the kcat/Km ratio, a second order constant, characterizing the reaction rate for substrate concentrations smaller than Km (values summarized in Table 2), was determined via method B.29,30 The oxidation rate of 5 was smaller than for 13 (around 3 times), 12 (6 times), and 11 (360 times) (Table 2; Supporting Information), suggesting that, having a more sterically hindered structure than l-Trp, 5 has lesser conformational freedom in the catalytic cavity, reducing the flexibility of its complex with rhIDO and undoubtedly conducing to a slower enzymatic reaction.
Table 2. In vitro Enzymatic Assays Performed with rhIDO (37 °C, pH 6.5).
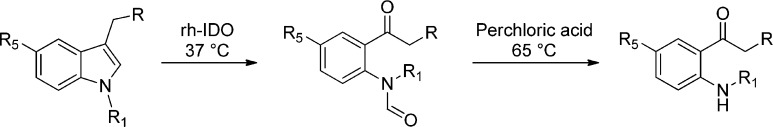
HPLC and UPLC analyses were performed, and the two product peaks appearing on the chromatograms for 11, 12, 13 (only one peak), and 5 (see Supporting Information) were collected and analyzed in mass spectrometry (Table 3). These analyses confirmed that the products formed were kynurenine and N-formyl-kynurenine derivatives (Scheme 3). Accurate mass determination was performed on N-fluoroethyl-N-formyl-kynurenine (19) (FT-MS (positive): m/z = 283.108862 (calculated), 283.108707 (measured) [M + H]) and N-fluoroethyl-kynurenine (20) (FT-MS (positive): m/z = 255.113947 (calculated), 255.113825 (measured) [M + H]) confirming the nature of these compounds.
Table 3. HPLC Analyses of the Enzymatic Assays of l-Tryptophan and Derivatives with rhIDO (Retention Times and MS).
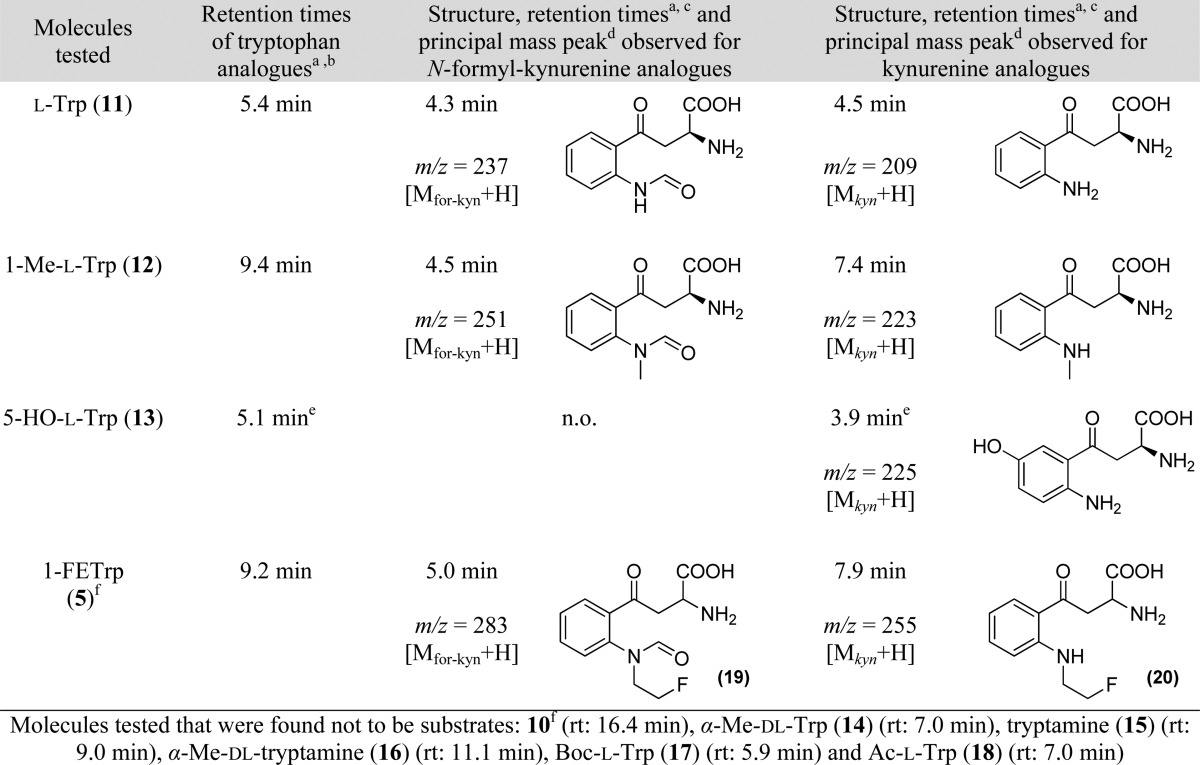
Retention times determined by HPLC analyses (or UPLC analyses; see Supporting Information). Elution conditions: CH3CN/ammonium acetate (10 mM), 10/90 (v/v), flow 0.7 mL/min.
UV detection (method A) at 221 and 284 nm.
UV detection (method A) at 321 or 360 nm.
Mass spectra (see Supporting Information).
Retention times determined by HPLC analyses (or UPLC analyses; see Supporting Information). Elution conditions: CH3CN/ammonium acetate (10 mM), 5/95 (v/v), flow 0.7 mL/min.
Racemate. n.o.: not observed because hydrolysis of the formyl substituent was too fast in these conditions.
Scheme 3. General Enzymatic Oxidative Indole Ring Opening of rhIDO Substrates and Hydrolysis in Perchloric Acid.
Similar in vitro enzymatic assays with recombinant human tryptophan 2,3-dioxygenase (rhTDO; Scheme 1) were performed to determine the specificity of the substrates (11, 12, 13, and 5) for rhIDO.36,37 The solutions obtained after incubation and the addition of perchloric acid were analyzed by HPLC with fluorescence detection. As expected, l-tryptophan was found to be an excellent substrate of rhTDO, while 5-HO-l-Trp was a poor substrate. 1-FETrp and 1-Me-l-Trp, however, were found not to be substrates (Table 4). For the highest enzyme concentration tested, consumption of only 2 or 3% of 1-FETrp and 1-Me-l-Trp was observed, but these uptakes were too low to consider these molecules as substrates of rhTDO. This specificity of 5 (and a fortiori for 12) could be explained by a broader substrate-selectivity of rhIDO due to a higher flexibility of its heme as compared to that of rhTDO.8
Table 4. In vitro Enzymatic Assays Performed with rhTDO (37 °C, pH 7.5).
| percentage of substrate
consumed after 1 h of incubation |
||
|---|---|---|
| substrates tested | rhTDO 1 μM | rhTDO 10 μM |
| l-Trp (11) | 67%a | 100%a |
| 1-Me-l-Trp (12) | 0%a | 3%a |
| 5-HO-l-Trp (13) | 0%a | 14%a |
| 1-FETrp (5)b | 0%a | 2%a |
Substrate concentration: 100 μM.
Racemate.
In summary, this letter describes the synthesis of two tryptophan derivatives, 5 and 10, both modified on the nitrogen of the indole ring. These compounds were tested as potential substrates of rhIDO. In vitro enzymatic assays with 1-FETrp show that this molecule is a good and specific substrate of rhIDO but that it is not a substrate of rhTDO. Unfortunately, 10 is found not to be a substrate of rhIDO.
As 5 shows encouraging results in the in vitro enzymatic assays, a simplified n.c.a. radiosynthesis of 1-(2-[18F]-fluoroethyl)-tryptophan (1-[18F]FETrp) could be designed. This radioactive isotopomer, currently being developed in our laboratory, could be a valuable tool for the preclinical and the clinical validation of new potential inhibitors. Furthermore, 1-[18F]FETrp could also facilitate a more specific detection of hIDO expressing cells in cancer imaging.
Glossary
Abbreviations
- AMT
α-methyl-l-tryptophan
- 1-FETrp
1-(2-fluoroethyl)-tryptophan
- 3-HAA
3-hydroxyanthranilic acid
- hIDO
human indoleamine 2,3-dioxygenase
- IFN-γ
interferon-γ
- KYN
kynurenine
- KYNA
kynurenic acid
- NAD+
nicotinamide adenine dinucleotide
- hTDO
human tryptophan 2,3-dioxygenase
- Trp
l-tryptophan
Supporting Information Available
Experimental sections, enzymatic equations, enzymatic curves obtained with 5, NMR analyses of some compounds (4, 5, 9, and 10), UPLC chromatograms, and mass spectra of the rhIDO substrates. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors gratefully acknowledge the Region Wallonne (Keymarker Project and Cantol Project (BioWin)) for their financial support. Alain Plenevaux is a senior research associate from FRS-FNRS Belgium.
The authors declare no competing financial interest.
Supplementary Material
References
- Vecsei L.; Szalardy L.; Fulop F.; Toldi J. Kynurenines in the CNS: recent advances and new questions. Nat. Rev. Drug Discovery 2013, 12, 64–82. [DOI] [PubMed] [Google Scholar]
- Platten M.; Wick W.; Van den Eynde B. J. Tryptophan catabolism in cancer: beyond IDO and tryptophan depletion. Cancer Res. 2012, 72, 5435–5440. [DOI] [PubMed] [Google Scholar]
- Takikawa O. Biochemical and medical aspects of the indoleamine 2,3-dioxygenase-initiated l-tryptophan metabolism. Biochem. Biophys. Res. Commun. 2005, 338, 12–19. [DOI] [PubMed] [Google Scholar]
- Adams S.; Braidy N.; Bessesde A.; Brew B. J.; Grant R.; Teo C.; Guillemin G. J. The kynurenine pathway in brain tumor pathogenesis. Cancer Res. 2012, 72, 5649–5657. [DOI] [PubMed] [Google Scholar]
- Uyttenhove C.; Pilotte L.; Theate I.; Stroobant V.; Colau D.; Parmentier N.; Boon T.; Van den Eynde B. J. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003, 9, 1269–1274. [DOI] [PubMed] [Google Scholar]
- Macchiarulo A.; Camaioni E.; Nuti R.; Pellicciari R. Highlights at the gate of tryptophan catabolism: a review on the mechanisms of activation and regulation of indoleamine 2,3-dioxygenase (IDO), a novel target in cancer disease. Amino Acids 2009, 37, 219–229. [DOI] [PubMed] [Google Scholar]
- Capece L.; Lewis-Ballester A.; Yeh S.-R.; Estrin D. A.; Marti M. A. The complete reaction mechanism of indoleamine 2,3-dioxygenase as revealed by QM/MM simulations. J. Phys. Chem. B 2012, 116, 1401–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis-Ballester A.; Batabyal D.; Egawa T.; Lu C.; Lin Y.; Marti M. A.; Capece L.; Estrin D. A.; Yeh S.-R. Evidence for a ferryl intermediate in a heme-based dioxygenase. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 17371–17376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myint A. M.; Kim Y. K. Cytokine–serotonin interaction through IDO: a neurodegeneration hypothesis of depression. Med. Hypotheses 2003, 61, 519–525. [DOI] [PubMed] [Google Scholar]
- Maes M.; Leonard B. E.; Myint A. M.; Kubera M.; Verkerk R. The new ‘5-HT’ hypothesis of depression: Cell-mediated immune activation induces indoleamine 2,3-dioxygenase, which leads to lower plasma tryptophan and an increased synthesis of detrimental tryptophan catabolites (TRYCATs), both of which contribute to the onset of depression. Prog. Neuropsychopharmacol. Biol. Psychiatry 2011, 35, 702–721. [DOI] [PubMed] [Google Scholar]
- Wainwright D. A.; Balyasnikova I. V.; Chang A. L.; Ahmed A. U.; Moon K.-S.; Auffinger B.; Tobias A. L.; Han Y.; Lesniak M. S. IDO expression in brain tumors increases the recruitment of regulatory T cells and negatively impacts survival. Clin. Cancer Res. 2012, 18, 6110–6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn D. H.; Mellor A. L. Indoleamine 2,3-dioxygenase and metabolic control of immune responses. Trends Immunol. 2013, 34, 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwidzinski E.; Bechmann I. IDO expression in the brain: a double-edged sword. J. Mol. Med. 2007, 85, 1351–1359. [DOI] [PubMed] [Google Scholar]
- Muller A. J.; Prendergast G. C. Indoleamine 2,3-dioxygenase in immune suppression and cancer. Curr. Cancer Drug Targets 2007, 7, 31–40. [DOI] [PubMed] [Google Scholar]
- Juhász C.; Nahleh Z.; Zitron I.; Chugani D. C.; Janabi M. Z.; Bandyopadhyay S.; Ali-Fehmi R.; Mangner T. J.; Chakraborty P. K.; Mittal S.; Muzik O. Tryptophan metabolism in breast cancers: molecular imaging and immunohistochemistry studies. Nucl. Med. Biol. 2012, 39, 926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor A. L.; Munn D. H. Tryptophan catabolism and T-cell tolerance: immunosuppression by starvation?. Immunol. Today 1999, 20, 469–473. [DOI] [PubMed] [Google Scholar]
- Munn D. H.; Mellor A. L. IDO and tolerance to tumors. Trends Mol. Med. 2004, 10, 15–18. [DOI] [PubMed] [Google Scholar]
- Dolušić E.; Frédérick R. Indoleamine 2,3-dioxygenase inhibitors: a patent review (2008 – 2012). Expert Opin. Ther. Pat. 2013, 23, 1367–1381. [DOI] [PubMed] [Google Scholar]
- Washburn L. C.; Sun T. T.; Byrd B. L.; Hayes R. L.; Butler T. A. dl-[Carboxyl-11C]tryptophan, a potential agent for pancreatic imaging; production and preclinical investigations. J. Nucl. Med. 1979, 20 (8), 857–864. [PubMed] [Google Scholar]
- Hartvig P.; Lindner K. J.; Tedroff J.; Andersson Y.; Bjurling P.; Långström B. Brain kinetics of 11C-labelled l-tryptophan and 5-hydroxy-l-tryptophan in the Rhesus monkey. A study using positron emission tomography. J. Neural Transm. 1992, 88 (1), 1–10. [DOI] [PubMed] [Google Scholar]
- Lundquist P.; Hartvig P.; Blomquist G.; Hammarlund-Udenaes M.; Långström B. 5-Hydroxy-l-[β-11C]tryptophan versus α-[11C]Methyl-l-tryptophan for positron emission tomography imaging of serotonin synthesis capacity in the rhesus monkey brain. J. Cereb. Blood Flow Metab. 2006, 27 (4), 821–830. [DOI] [PubMed] [Google Scholar]
- Juhász C.; Muzik O.; Chugani D. C.; Chugani H. T.; Sood S.; Chakraborty P. K.; Barger G. R.; Mittal S. Differential kinetics of α-[11C]methyl-l-tryptophan on PET in low-grade brain tumors. J. Neuro-Oncol. 2011, 102, 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharib A.; Balende C.; Sarda N.; Weissmann D.; Plenevaux A.; Luxen A.; Bobillier P.; Pujol J.-F. Biochemical and autoradiographic measurements of brain serotonin synthesis rate in the freely moving rat: a reexamination of the α-methyl-l-tryptophan method. J. Neurochem. 1999, 72, 2593–2600. [DOI] [PubMed] [Google Scholar]
- Sun T.; Tang G.; Tian H.; Wang X.; Chen X.; Chen Z.; Wang S. Radiosynthesis of 1-[18F]fluoroethyl-l-tryptophan as a novel potential amino acid PET tracer. Appl. Radiat. Isot. 2012, 70, 676–680. [DOI] [PubMed] [Google Scholar]
- Schultz A. W.; Lewis C. A.; Luzung M. R.; Baran P. S.; Moore B. S. Functional characterization of the cyclomarin/cyclomarazine prenyltransferase CymD directs the biosynthesis of unnatural cyclic peptides. J. Nat. Prod. 2010, 73, 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard C.; Önen E.; Aufort M.; Beauvière S.; Samson E.; Herscovici J. Reusable polymer-supported catalyst for the [3 + 2] Huisgen cycloaddition in automation protocols. Org. Lett. 2006, 8, 1689–1692. [DOI] [PubMed] [Google Scholar]
- Bock V. D.; Hiemstra H.; van Maarseveen J. H. CuI-catalyzed alkyne-azide ″click″ cycloadditions from a mechanistic and synthetic perspective. Eur. J. Org. Chem. 2006, 2006, 51–68. [Google Scholar]
- Crumpton J. B.; Santos W. L. Site-specific incorporation of diamondoids on DNA using click chemistry. Chem. Commun. 2012, 48, 2018–2020. [DOI] [PubMed] [Google Scholar]
- Basran J.; Rafice S. A.; Chauhan N.; Efimov I.; Cheesman M. R.; Ghamsari L.; Raven E. L. A kinetic, spectroscopic, and redox study of human tryptophan 2,3-dioxygenase. Biochemistry 2008, 47, 4752–4760. [DOI] [PubMed] [Google Scholar]
- Lu C.; Lin Y.; Yeh S.-R. Inhibitory substrate binding site of human indoleamine 2,3-dioxygenase. J. Am. Chem. Soc. 2009, 131, 12866–12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan N.; Thackray S. J.; Rafice S. A.; Eaton G.; Lee M.; Efimov I.; Basran J.; Jenkins P. R.; Mowat C. G.; Chapman S. K.; Raven E. L. Reassessment of the reaction mechanism in the heme dioxygenases. J. Am. Chem. Soc. 2009, 131, 4186–4187. [DOI] [PubMed] [Google Scholar]
- Basran J.; Efimov I.; Chauhan N.; Thackray S. J.; Krupa J. L.; Eaton G.; Griffith G. A.; Mowat C. G.; Handa S.; Raven E. L. The mechanism of formation of N-formylkynurenine by heme dioxygenases. J. Am. Chem. Soc. 2011, 133, 16251–16257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujigaki H.; Saito K.; Lin F.; Fujigaki S.; Takahashi K.; Martin B. M.; Chen C. Y.; Masuda J.; Kowalak J.; Takikawa O.; Seishima M.; Markey S. P. Nitration and inactivation of IDO by peroxynitrite. J. Immunol. 2006, 176, 372–379. [DOI] [PubMed] [Google Scholar]
- Meisel R.; Zibert A.; Laryea M.; Göbel U.; Däubener W.; Dilloo D. Human bone marrow stromal cells inhibit allogeneic T-cell responses by indoleamine 2,3-dioxygenase-mediated tryptophan degradation. Blood 2004, 103, 4619–4621. [DOI] [PubMed] [Google Scholar]
- Zitron I. M.; Kamson D. O.; Kiousis S.; Juhász C.; Mittal S. In vivo metabolism of tryptophan in meningiomas is mediated by indoleamine 2,3-dioxygenase 1. Cancer Biol. Ther. 2013, 14, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolušić E.; Larrieu P.; Moineaux L.; Stroobant V.; Pilotte L.; Colau D.; Pochet L.; Van den Eynde B. t.; Masereel B.; Wouters J.; Frédérick R. Tryptophan 2,3-Dioxygenase (TDO) inhibitors. 3-(2-(pyridyl)ethenyl)indoles as potential anticancer immunomodulators. J. Med. Chem. 2011, 54, 5320–5334. [DOI] [PubMed] [Google Scholar]
- Dolušić E.; Larrieu P.; Blanc S.; Sapunaric F.; Pouyez J.; Moineaux L.; Colette D.; Stroobant V.; Pilotte L.; Colau D.; Ferain T.; Fraser G.; Galleni M.; Frère J.-M.; Masereel B.; Van den Eynde B.; Wouters J.; Frédérick R. Discovery and preliminary SARs of keto-indoles as novel indoleamine 2,3-dioxygenase (IDO) inhibitors. Eur. J. Med. Chem. 2011, 46, 3058–3065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.