Abstract
Regeneration requires that the identities of new cells are properly specified to replace missing tissues. The Wnt signaling pathway serves a central role in specifying posterior cell fates during planarian regeneration. We identified a gene encoding a homolog of the Teashirt family of zinc-finger proteins in the planarian Schmidtea mediterranea to be a target of Wnt signaling in intact animals and at posterior-facing wounds. Inhibition of Smed-teashirt (teashirt) by RNA interference (RNAi) resulted in the regeneration of heads in place of tails, a phenotype previously observed with RNAi of the Wnt pathway genes β-catenin-1, wnt1, Dvl-1/2 or wntless. teashirt was required for β-catenin-1-dependent activation of posterior genes during regeneration. These findings identify teashirt as a transcriptional target of Wnt signaling required for Wnt-mediated specification of posterior blastemas.
Keywords: Wnt, Neoblasts, Patterning, Planarians, Regeneration, Teashirt
Summary: The zinc finger protein teashirt is a transcriptional target of Wnt signaling and is required for Wnt-mediated specification of cell fate during regeneration in planarians.
INTRODUCTION
A central issue in animal regeneration is how positional information is maintained and regenerated for proper specification of cell types, organs and tissue patterns (Wolpert, 1969; French et al., 1976; Reddien, 2011). The planarian Schmidtea mediterranea is a flatworm with nearly unlimited capacity to regenerate, and is an emerging system for the study of positional information in regeneration. Planarian regeneration requires a population of proliferative cells called neoblasts, which includes pluripotent stem cells (Wagner et al., 2011). Neoblasts can specialize to produce lineage-specific progenitors for the regeneration of missing target tissues (Lapan and Reddien, 2011; Scimone et al., 2011, 2014a; Wenemoser et al., 2012; Cowles et al., 2013; Currie and Pearson, 2013; Reddien, 2013; van Wolfswinkel et al., 2014), and must receive appropriate instructions during regeneration in order to replace missing tissues correctly. The choice of whether to regenerate a head or a tail at transverse amputation planes, sometimes called regeneration polarity, is a classic problem in regeneration and a paradigm for molecular dissection of the positional information that guides regeneration (Morgan, 1898; Reddien, 2011). In a fragment lacking both a head and a tail, anterior cells must receive instructions to produce head tissues, whereas posterior cells must be specified to form tail tissues. This head-versus-tail regeneration decision occurs at transverse wounds at essentially any position along the head-to-tail axis (Randolph, 1897; Morgan, 1898).
Wnt signaling regulates tissue identity in a wide array of processes across many investigated metazoan phyla (Peifer et al., 1991; Harland and Gerhart, 1997; Hobmayer et al., 2000; Eisenmann, 2005; Stoick-Cooper et al., 2007; Petersen and Reddien, 2009a). Wnts are expressed in the posterior during early development and have a central role in patterning the primary axis in many organisms (Petersen and Reddien, 2009a; Niehrs, 2010). In planarian regeneration, Wnt signaling inhibits head and promotes tail regeneration at posterior-facing wounds. RNAi of the Wnt pathway components Smed-β-catenin-1 (β-catenin-1), Smed-wnt1 (wnt1, formerly wntP-1) and Smed-wntless (wntless) results in the regeneration of heads at posterior-facing wounds (Gurley et al., 2008; Iglesias et al., 2008; Petersen and Reddien, 2008, 2009b; Adell et al., 2009). wnt1 expression in intact animals is restricted to a set of cells near the tip of the tail referred to as the posterior pole (Petersen and Reddien, 2008). Following injury, wnt1 transcription is generically induced, such that wnt1 is expressed at essentially all wound sites (Petersen and Reddien, 2009b). RNAi of the gene Smed-notum (notum) results in the regeneration of tails in place of heads, a phenotype opposite to that observed in wnt1 or β-catenin-1 RNAi animals (Petersen and Reddien, 2011). notum is expressed in a cluster of cells at head tips referred to as the anterior pole (Petersen and Reddien, 2011). Following injury, notum expression is generically induced, but its expression is more robust at anterior-facing wounds than at posterior-facing wounds (Petersen and Reddien, 2011). For example, a flank incision that is allowed to seal will induce notum transcription, but preferentially on the posterior side of the incision (anterior facing). notum is a negative regulator of Wnt signaling, inhibiting the pathway at anterior-facing wounds and allowing head regeneration to occur at suitable wounds (Petersen and Reddien, 2011). Whereas these early steps of the planarian head-tail regeneration choice have been identified, little is known about how the output of the Wnt pathway is modulated or what downstream genes are targeted to mediate regeneration polarity.
Teashirt proteins are found across the Bilateria and can interact with Wnt signaling in several developmental contexts. teashirt was first identified in Drosophila and collaborates with homeotic genes to control prothoracic segment specification (Fasano et al., 1991; Roder et al., 1992; Andrew et al., 1994; de Zulueta et al., 1994; McCormick et al., 1995). Drosophila teashirt mutants can have segment polarity defects similar to those caused by disrupted Wnt signaling, and these defects can be suppressed by ectopic stabilized β-catenin (Fasano et al., 1991; Peifer et al., 1991; Gallet et al., 1998). In Xenopus, a Teashirt homolog (XTsh3) is expressed at the presumptive dorsal embryo side near the site of gastrulation. XTsh3 inhibition causes ventralization, a phenotype also seen following perturbation of β-catenin (Onai et al., 2007). XTsh3 can enhance Wnt signaling activity in a β-catenin-dependent manner (Onai et al., 2007). In both flies and frogs, Teashirt can localize to the nucleus and bind β-catenin, but its exact molecular function remains unclear (Gallet et al., 1999; Onai et al., 2007). In other vertebrates, Teashirt proteins have been implicated in myogenesis and in the development of the nervous system, particularly the specification of hindbrain structures; whether Teashirt functions with the Wnt pathway is unknown in several of these contexts (Koebernick et al., 2006; Caubit et al., 2010; Erickson et al., 2011).
Here, we present the characterization of a teashirt gene in planarians. teashirt was previously unstudied in regeneration and in non-Ecdysozoan protostomes. We present evidence that planarian Smed-teashirt (teashirt) is an early target of Wnt signaling and is necessary for the output of Wnt signaling in planarian regeneration, promoting tail and inhibiting head regeneration.
RESULTS
Planarian Teashirt
The planarian Schmidtea mediterranea genome encodes one Teashirt-family zinc-finger protein. Smed-teashirt encodes a predicted 1107 amino acid protein, with 23.9% sequence identity to Drosophila Teashirt and 23.5% sequence identity to Xenopus XTsh3. The sequence contains two Cx2Cx12HMx4H-type zinc-finger motifs, sharing unique domain structure with other Teashirt family proteins (supplementary material Fig. S1). The presence of multiple copies of this atypical zinc-finger motif is a hallmark of Teashirt family proteins (Manfroid et al., 2004). Smed-Teashirt lacks the basic motif, the homeodomain and the second C-terminal zinc finger found in vertebrate orthologs, which are also lacking in Drosophila Teashirt and the Drosophila Teashirt paralog Tiptop (Onai et al., 2007). The three mouse teashirt genes can rescue the Drosophila teashirt loss-of-function phenotype, suggesting that Teashirt proteins have similar biochemical activities in highly divergent animals (Manfroid et al., 2004). We also identified an 11 amino acid domain (NPLSXLQSVM) in planarian Teashirt that is present in all known vertebrate orthologs (82% identity) but absent from Ecdysozoan orthologs (supplementary material Fig. S1).
Teashirt and regeneration polarity
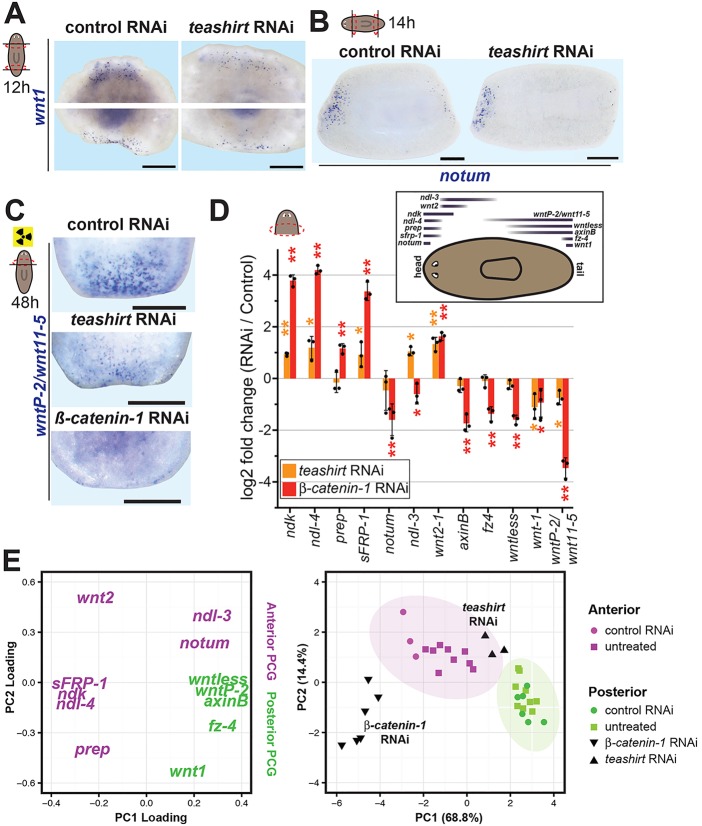
RNAi of teashirt resulted in a highly penetrant polarity phenotype with heads being regenerated at posterior-facing wounds (Fig. 1A; supplementary material Fig. S2; n=36/49 trunks). teashirt RNAi with head fragments (see Materials and Methods, 4-day RNAi protocol) also yielded a two-headed phenotype (19/47 two-headed and 18/47 abnormal tail blastemas; 38/38 controls were normal). In situ hybridization to detect the neuronal marker choline acetyltransferase (Smed-chat) showed that teashirt(RNAi) animals regenerated a symmetrical nervous system with brains at both ends of the primary axis, similar to the β-catenin-1(RNAi) phenotype (Fig. 1B; supplementary material Fig. S3). The ectopic presence of anterior marker expression (Smed-sFRP-1) and the absence of posterior marker expression (Smed-wntP-2, also known as wnt11-5) in teashirt(RNAi) posterior blastemas confirmed that animals regenerated heads in place of tails (Fig. 1C,D). wntP-2/wnt11-5 orthology is difficult to assign, making it unclear whether this gene encodes a Wnt11-family member (Gurley et al., 2010); we refer to the gene here as wntP-2 or wntP-2/wnt11-5. Less stringent RNAi resulted in additional posterior defects, including lack of a blastema, a misshapen blastema or partial head regeneration (supplementary material Fig. S2). Similar posterior blastema defects have also been described for wnt1(RNAi) animals (Adell et al., 2009; Petersen and Reddien, 2009b). teashirt RNAi combined with weak RNAi of wnt1 (under conditions that do not cause a two-headed regeneration phenotype for RNAi of either gene alone) caused enhancement of the phenotype (two-headed animals were observed), demonstrating partial Wnt signaling perturbation can cause synthetic defects when combined with partial teashirt inhibition (Fig. 1E).
Fig. 1.
teashirt RNAi causes posterior blastema polarity reversal. (A) Trunk fragments 12 days after amputation. Control RNAi trunks regenerated posterior tails (n=56/56), whereas teashirt(RNAi) trunks regenerated posterior heads with eyes (white arrowheads) (n=36/49). (B) teashirt(RNAi) animals regenerated cephalic ganglia in anterior and posterior blastemas, shown by in situ hybridization with chat. (C,D) teashirt(RNAi) animals expressed the anterior marker sFRP-1 at both ends (C) and did not express the posterior marker wntP-2 (D) (100% of double-headed animals; n>4, each marker). (A-D) Animals viewed dorsally; anterior is leftwards; dotted lines indicate approximate blastema/old tissue boundary. (E) Weak individual wnt1 and teashirt RNAi using dsRNA diluted to one-third of the working concentration with control (C. elegans unc-22) dsRNA failed to cause regeneration of two-headed planarians (n=8, each). By contrast, combining the diluted dsRNA for each gene (one-third wnt1, one-third teashirt, one-third unc-22) caused regeneration of two-headed animals (n=4/8 with another animal displaying failed posterior regeneration). Posterior is downwards. (F,G) Fluorescence in situ hybridization for cintillo (Oviedo et al., 2003) showed a decrease in putative mechanosensory cells in teashirt(RNAi) and β-catenin-1(RNAi) 7-day anterior blastemas (Hoechst marks nuclei in white). (H) Fluorescence in situ hybridization for the pigment cup marker tph showed mis-shaped eyes in both teashirt (n=5/14) and β-catenin-1 (n=4/6) RNAi 7-day anterior blastemas. Scale bars: 200 µm in A-D; 500 µm in E; 50 µm in F,H. Error bars indicate s.d.
Previously, inhibition of genes encoding core components of the Wnt secretion and signal transduction pathway (β-catenin-1, wnt1, Dvl-1/2 and wntless) was shown to result in the regeneration of posterior heads (Gurley et al., 2008; Petersen and Reddien, 2008, 2009b; Adell et al., 2009). The similarity of the teashirt(RNAi) phenotype to the phenotype caused by inhibition of these core Wnt pathway components therefore raises the possibility that teashirt has an essential role in the outcome of Wnt signaling in planarian regeneration. In Xenopus, a Teashirt protein can act with β-catenin to establish the dorsoventral (DV) axis, and Drosophila teashirt mutants mis-specify certain tissues along the anteroposterior (AP) axis (Fasano et al., 1991; Onai et al., 2007); however, teashirt genes were not previously known to be required for establishment of the polarity of an AP axis, and this raises the possibility that, like Wnt signaling (Petersen and Reddien, 2009a), Teashirt proteins will be involved in AP axis polarization in many metazoans.
Teashirt and other Wnt-dependent processes
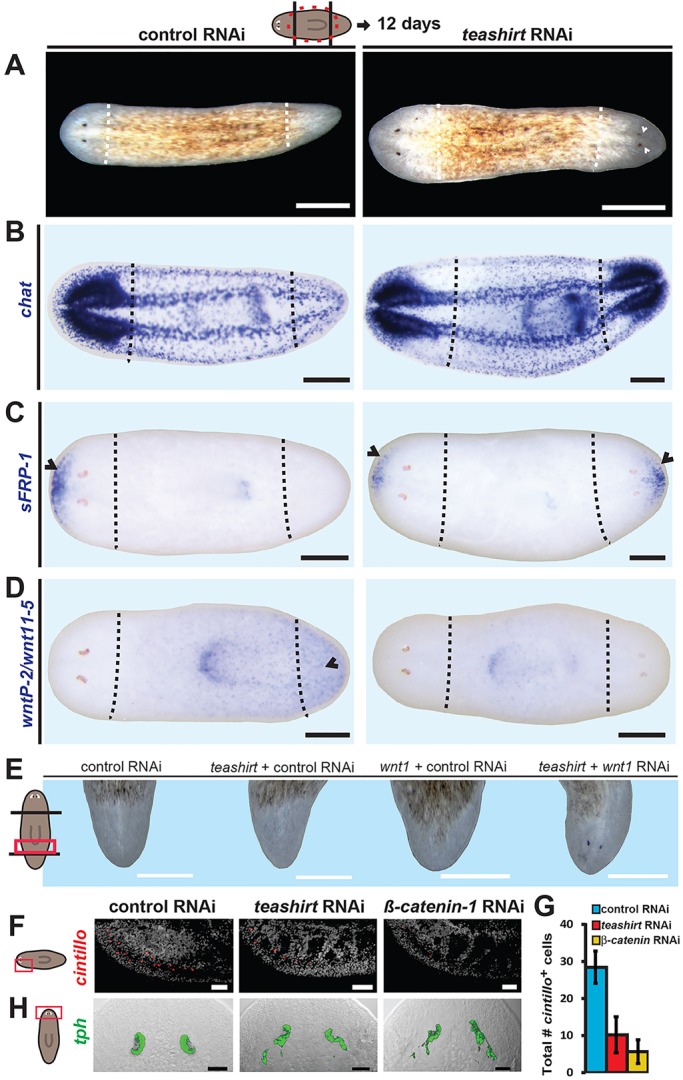
Teashirt proteins can have roles in many developmental contexts, including Drosophila eye, wing disk and gut development, as well as hindbrain patterning in zebrafish and myogenesis in mouse (Pan and Rubin, 1998; Erkner et al., 1999; Waltzer et al., 2001; Singh et al., 2002; Wu and Cohen, 2002; Erickson et al., 2011; Faralli et al., 2011). We therefore examined tissue markers in regenerating teashirt(RNAi) animals for additional defects. Both teashirt and β-catenin-1 RNAi animals showed significant decreases in the number of head rim candidate mechanosensory cells (cintillo+) (Fig. 1F,G). In addition, both teashirt and β-catenin-1 RNAi resulted in minor morphology defects in regenerated eyes (Fig. 1H). Drosophila teashirt, together with wingless, dpp, eyeless and homothorax, is known to control eye morphogenesis (Singh et al., 2002; Bessa and Casares, 2005). Overall brain morphology and the presence of other neuronal lineages (e.g. cholinergic and serotonergic), however, were largely unaffected by teashirt or β-catenin-1 RNAi (supplementary material Fig. S3). These data indicate that in addition to polarity phenotypes, RNAi of Smed-teashirt and β-catenin-1 can also result in similar nervous system defects.
teashirt affects the expression of anterior and posterior position control genes during tail regeneration
The teashirt(RNAi) polarity defects suggest Teashirt might regulate expression of genes at posterior-facing wounds that are involved in the specification of head and tail tissue identity. There are numerous genes expressed regionally and constitutively in adult planarians that are candidates to specify positional information to tissues and are referred to as position control genes (PCGs) (Reddien, 2011; Witchley et al., 2013). PCGs display regional expression and are implicated in patterning, either because PCG RNAi causes a patterning phenotype or because the PCG encodes a predicted component of a pathway associated with planarian patterning (Reddien, 2011; Witchley et al., 2013). PCGs are expressed in planarian body wall muscle cells and define a candidate ‘GPS’-like coordinate system of positional information (Witchley et al., 2013). We examined the expression of PCGs that normally display anterior- or posterior-restricted expression during regeneration in teashirt(RNAi) animals.
wnt1 is wound induced as the first known step in posterior regeneration (Petersen and Reddien, 2009b). notum is expressed preferentially at anterior-facing wounds by 6 h of regeneration and is the earliest known anterior regeneration-specific step (Petersen and Reddien, 2011). Polarized notum expression is β-catenin dependent (Petersen and Reddien, 2011). Wound-induced wnt1 and notum expression was similar in teashirt and control RNAi trunk fragments, suggesting that the teashirt(RNAi) phenotype is not explained by a defect in regulation of this phase of the head-versus-tail regeneration decision (Fig. 2A,B). Both wnt1 and wntP-2 promote posterior fate in regeneration, and wnt1 promotes wntP-2 expression (Petersen and Reddien, 2009b). teashirt RNAi resulted in decreased wntP-2 expression at posterior-facing wounds 48 h after amputation (Fig. 2C). Overall, we did not detect a requirement for teashirt in the β-catenin-1-dependent expression of notum early at anterior-facing wounds but did detect a requirement for teashirt in early expression of a posterior-specific PCG during regeneration (Fig. 2C).
Fig. 2.
teashirt is required for normal PCG expression during tail regeneration. (A) teashirt(RNAi) trunk fragments properly expressed wnt1 12 h after amputation (teashirt RNAi: 6/6 anterior- and posterior-facing wounds; control: 7/7 anterior-facing and 6/7 posterior-facing wounds). Anterior is upwards. Scale bars: 200 µm. (B) Irradiated teashirt(RNAi) trunks properly expressed notum 14 h after amputation (n=11/12). Anterior is leftwards. Scale bars: 200 µm. (C) teashirt RNAi resulted in decreased wntP-2 expression at posterior wounds in irradiated head fragments after 48 h of regeneration (n=16/31, three experiments). Anterior is upwards. Scale bars: 200 µm. (D) Quantitative RT-PCR (qRT-PCR) of position control genes (PCG) following teashirt or β-catenin-1 RNAi. Cartoon displays PCG expression domains. RNA was harvested from dissected posterior-facing wound sites of dsRNA-injected head fragments 72 h post-amputation. Biological triplicates consisted of pooled wound sites from six to nine individual animals. Data depict mean log2-fold changes in transcript levels between teashirt or β-catenin-1 RNAi specimens and control RNAi specimens. Error bars indicate s.d. Individual replicates are indicated by dots. Asterisks indicate significance determined by t-tests between sets of biological triplicate delta-Ct values (*P<0.05) and with standard Bonferroni adjustment (**P<0.0042). (E) Principal component analysis (PCA) of PCG transcriptional signatures in 72 h post-amputation wound sites. Left: PCA loadings indicate relative contributions of each gene to principal components 1 and 2. Right: PCA plot. Each dot represents a pooled wound site sample. Colors and shapes indicate anterior versus posterior wound orientation or RNAi treatment. Percentages of total dataset variance explained by each principal component are shown. Plot regions corresponding to anterior or posterior-specific transcriptional states are indicated by shaded ellipses.
We next used qRT-PCR to examine the expression of multiple PCGs during regeneration. nou darake (ndk), ndl-4, sFRP-1, notum and prep were selected as anterior PCGs (Cebrià et al., 2002; Gurley et al., 2008; Petersen and Reddien, 2008, 2011; Rink et al., 2009; Felix and Aboobaker, 2010); wnt2 and ndl-3 were selected as prepharyngeal PCGs (Petersen and Reddien, 2008; Rink et al., 2009); and wntless, wntP-2, frizzled-4 (fz-4), axinB and wnt1 were selected as posterior PCGs (Gurley et al., 2008; Petersen and Reddien, 2008; Adell et al., 2009; Iglesias et al., 2011). We used a highly parallel microfluidic qRT-PCR platform (Fluidigm) to assess gene expression in wild-type, control, β-catenin-1 and teashirt RNAi animals (supplementary material Fig. S4). At 72 h of regeneration, a timepoint that allowed robust detection of regenerative PCG expression by qRT-PCR, teashirt and β-catenin-1 RNAi animals deviated from the control in a similar manner (Fig. 2D). Specifically, in head fragments regenerating a tail, many anterior PCGs were expressed more highly in teashirt and β-catenin-1 RNAi animals than in the control, whereas some posterior PCGs were expressed at lower levels in teashirt and β-catenin-1 RNAi animals than in the control (Fig. 2D). In general, the effect of β-catenin-1 RNAi on PCG gene expression during regeneration was more robust than teashirt RNAi, which is expected, given the incomplete penetrance of the teashirt RNAi phenotype. Whereas most genes behaved similarly in teashirt and β-catenin-1 RNAi animals, the effects of RNAi of these two genes on ndl-3 expression were opposite. ndl-3 expression differs from the other PCGs in that its expression is largely prepharyngeal and extends from the anterior into the midbody (Rink et al., 2009); these results indicate that regulation of this gene might differ from the other PCGs. Possibly because of its low levels of expression at posterior wounds, notum levels were variable between biological replicates. However, notum behaved similarly to posterior (rather than anterior) PCGs. Whereas notum expression during regeneration is preferentially at anterior-facing wounds at early (6-18 h) timepoints, it is also expressed at low levels during posterior regeneration (Petersen and Reddien, 2011). These results suggest that expression of notum during posterior regeneration is Wnt signaling dependent.
We next used principal component analysis (PCA) to assess underlying transcriptional signatures in teashirt(RNAi) regenerating tissues. PCA was performed using transcript levels for 12 PCGs and compared 72 h anterior- or posterior-facing wound sites from wild-type, control RNAi, β-catenin-1 RNAi and teashirt RNAi animals. Contributions of each of the 12 PCGs to the first two principal components are illustrated in a loadings plot (Fig. 2E, left). With the exception of ndl-3, which is expressed prepharyngeally, and notum, for which expression is β-catenin-1 dependent, anterior and posterior-specific PCGs are oppositely weighted in the first principal component (PC1). PC1 explains the majority (>65%) of the total variance in the dataset, and distinguishes anterior- from posterior-facing wounds for wild-type and control RNAi specimens (Fig. 2E, right panel). Posterior-facing wounds from teashirt and β-catenin-1 RNAi animals did not cluster with posterior-facing wounds in control animals. Instead, these samples more closely resembled the expression signature of anterior-facing wounds (Fig. 2E). We conclude that teashirt and β-catenin-1 are similarly required for the differential expression of anterior and posterior PCGs during tail regeneration.
teashirt is expressed in the posterior
In uninjured adults, teashirt mRNA was present in a broad gradient along the AP axis with the signal strongest in the tail (Fig. 3A). teashirt was expressed more broadly than previously known PCGs, including in the central nervous system (cephalic ganglia and nerve cords), in the pharynx (and strongly just anterior to the pharynx), in the dorsal head rim and in the epidermis (Fig. 3A,B). Posterior teashirt expression was present in subepidermal cells, which included but was not restricted to muscle cells marked by collagen expression (Fig. 3C). The posterior-to-anterior graded expression and strong expression at the anterior end of the pharynx is similar to the wntP-2-expression pattern (Petersen and Reddien, 2008) (Fig. 1D), and these two genes were co-expressed in cells at the anterior end of the pharynx compartment (Fig. 3D).
Fig. 3.
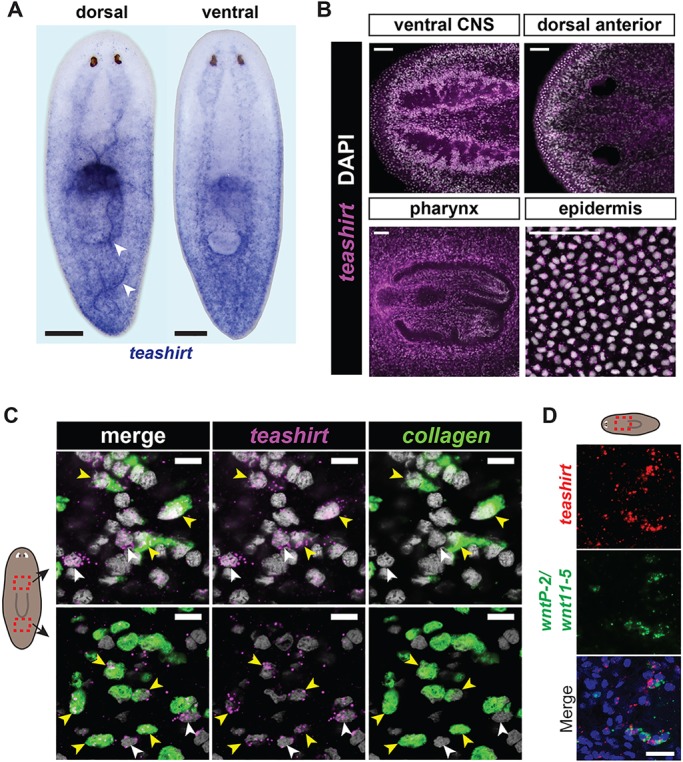
teashirt is expressed in the planarian posterior. (A) Whole-mount in situ hybridization for teashirt showed graded expression highest at the posterior end. teashirt was also expressed anterior to the pharynx, and in the brain and nerve cords (ventral view). Arrowheads indicate the signal in folds of epidermis. Anterior is upwards. Scale bars: 200 µm. (B) Single confocal planes following fluorescence in situ hybridization show teashirt expression in the brain, dorsal head rim cells, anterior to and in the pharynx, and in the epidermis. Anterior is leftwards. Scale bars: 50 µm. (C) Double fluorescence in situ hybridization showed teashirt and collagen co-expression in some (yellow arrowheads), but not all (white arrowheads), teashirt+ subepidermal cells (single confocal planes). DAPI-labeled nuclei are in gray. Scale bars: 10 µm. (D) Double fluorescence in situ hybridization showed that teashirt and wntP-2 were co-expressed in cells anterior to the pharynx. Scale bar: 20 µm.
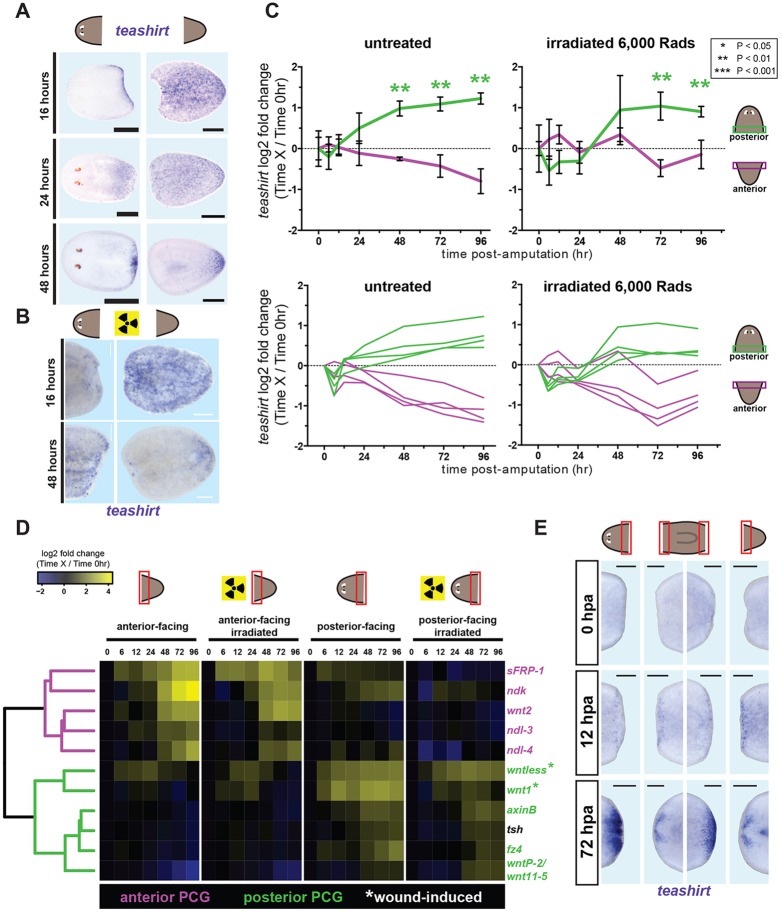
We next assessed teashirt expression during regeneration. Consistent with its elevated posterior expression in uninjured animals, teashirt levels became elevated at posterior-facing wounds by 16-24 h post-amputation (Fig. 4A). In amputated tail fragments, in which teashirt is initially broadly expressed, the posterior teashirt expression domain retracted towards the posterior pole over time (Fig. 4A). This process of gene expression retraction to the posterior, and away from anterior-facing wounds, likely reflects the rescaling of existing tissues to a smaller total body size in a regenerative process called morphallaxis (Reddien and Sánchez Alvarado, 2004). Irradiation can ablate all proliferating cells (neoblasts) in adult planarians and blocks regeneration (Dubois, 1949; Reddien et al., 2005). Dynamic expression of several PCGs still occurs in amputated animals exposed to irradiation (Petersen and Reddien, 2009b; Gurley et al., 2010; Witchley et al., 2013), demonstrating that some PCG expression domain changes can occur in existing differentiated tissue after injury. Dynamic teashirt expression features during regeneration (at posterior-facing wounds and rescaling in tail fragments) were also observed in amputated irradiated fragments (Fig. 4B,C).
Fig. 4.
teashirt is expressed in the posterior during regeneration. (A) In regenerating fragments viewed dorsally, new teashirt expression occurred at posterior wounds by 16 h (n=21/27). By 48 h, teashirt expression in tail fragments retracted towards the posterior. (B) teashirt expression at posterior-facing wounds also occurred in irradiated animals (n=45/56), and expression also retracted towards the tail tip at 48 h. (C) Top, log2-fold changes in teashirt levels for each timepoint relative to time zero. Error bars indicate s.d. Asterisks denote significance determined by t-tests between each timepoint and time zero using sets of biological triplicate delta-Ct values. Colors indicate wound orientation. Bottom, four independent primer sets for teashirt display similar trends. (D) Heatmap of gene expression changes during anterior or posterior regeneration, determined by qRT-PCR. Total RNA was harvested from wound sites at specified times post-amputation. Some specimens were γ-irradiated (6000 rads) 4 days prior to amputation, as indicated. Heatmap displays mean log2-fold changes in transcript levels between each timepoint and time zero. Dendrogram reflects hierarchical clustering of genes based on Euclidean distance. Anterior and posterior-specific PCGs are indicated in magenta and green, respectively. (E) Whole-mount in situ hybridization showing teashirt wound-site expression at 0, 12 and 72 h post-amputation (hpa). Short development times were used to allow detection of wound-site expression without other expression locations obscuring signal. Background level was determined with a teashirt ‘sense’ probe (see supplementary material Fig. S5B). Above-background teashirt expression at wounds was evident at 12 and 72 hpa. Anterior expression of teashirt at 72 hpa was confined to presumptive brain regions. Scale bars: 200 μm.
We next used qRT-PCR to compare the expression of teashirt during regeneration with the expression of several anterior and posterior-specific PCGs during regeneration. Time course analysis of PCG expression at anterior- and posterior-facing wounds confirmed anterior regeneration-biased elevation of sFRP-1, ndk, wnt2, ndl-3 and ndl-4, and posterior-biased elevation of wntless, wnt1, axinB, fz4, wntP-2 and teashirt (Fig. 4D). Consistent with previous observations (Adell et al., 2009; Petersen and Reddien, 2009b), transient early (by 6 h post-injury) wound-induced expression of wntless and wnt1 at anterior-facing wounds was also apparent. Most expression change trends were similar in irradiated animals, confirming that dynamic expression features of many PCGs at wounds are largely independent of new tissue formation. Hierarchical clustering revealed that teashirt expression patterns were highly similar to those of other posterior PCGs (Fig. 4D). Posterior teashirt expression occurred after the wound-induced phase of wnt1 expression (∼6-12 h), but before wnt1 expression becomes polarized to be present only in posterior-facing blastemas (∼48 h) in regenerating posterior poles (Petersen and Reddien, 2009b). Posterior teashirt expression also occurred at a similar time or before the detected expression of other known posterior genes (Fig. 4D; supplementary material Fig. S5A), such as wntP-2 (Petersen and Reddien, 2009b). During head regeneration, two small symmetrical teashirt expression domains appeared in anterior-facing blastemas, probably representing expression in the developing brain (Fig. 4E).
By qRT-PCR teashirt showed a robust expression increase at posterior- but not anterior-facing wounds. However, by in situ hybridization some teashirt expression was apparent at 12 h or 24 h at all wound sites, including at anterior-facing wounds and side incisions (Fig. 4E; supplementary material Fig. S5B,C). This expression was not strong enough to be detected above the basal overall level of teashirt expression in all tissues of wound-site fragments used in qRT-PCR experiments. These data indicate that teashirt expression is also moderately wound induced. Over time, after this wound-induced phase teashirt expression became strong at posterior-facing wounds and was largely restricted to clusters of apparent regenerating cephalic ganglia at anterior-facing wounds (Fig. 4E).
The teashirt expression pattern has three key characteristics that are consistent with its role as a factor involved in positional identity specification during regeneration: regionalized expression in uninjured animals, polarized expression during regeneration of heads and tails, and expression during early regeneration that is independent of new cell production. Few genes (including notum, wnt1, wntP-2, wntless and bmp4) are known to possess these expression characteristics and an axial tissue-identity RNAi phenotype in regeneration (Molina et al., 2007; Orii and Watanabe, 2007; Reddien et al., 2007; Adell et al., 2009; Petersen and Reddien, 2009b, 2011; Gurley et al., 2010). Sharing a relatively rare set of expression characteristics with this exclusive group points to teashirt as an important regulator of early tissue identity specification in regeneration.
Posterior teashirt expression requires Wnt signaling
Constitutive expression of tissue patterning factors could provide a system for maintaining proper regional tissue identity during cell turnover and for maintaining body proportions during growth and degrowth (Reddien, 2011). Consistent with this hypothesis, β-catenin-1 RNAi causes gradual loss of AP polarity in uninjured animals, eventually resulting in hypercephalized animals (Gurley et al., 2008; Iglesias et al., 2008; Petersen and Reddien, 2008). This suggests that Wnt signaling maintains positional information along the AP axis during homeostatic tissue turnover. The posterior teashirt expression domain was substantially reduced 4 days after RNAi of β-catenin-1 in uninjured animals (Fig. 5A). This rapid change occurred prior to the loss of posterior cell types [assessed by continued expression of Smed-fibrillin (fibrillin) or Smed-mag-1 (mag-1)], and before detectable anteriorization occurred (assessed with sFRP-1 expression) (Fig. 5A). Other posterior PCG expression (wnt11-1 and wnt1) was not robustly decreased at this early time point (4 days) following β-catenin-1 RNAi (supplementary material Fig. S6). Together, these data indicate that the loss of posterior teashirt expression in β-catenin-1(RNAi) animals is the consequence of reduction in Wnt signaling, rather than an indirect consequence of a gradual loss of posterior tissue. teashirt inhibition in uninjured animals resulted in an enlarged pharynx, occurring at low penetrance (n=4/52) (supplementary material Fig. S7), and we did not detect expression abnormalities for posterior markers (wntP-2, frizzled-4 and plox4) in uninjured teashirt(RNAi) animals (supplementary material Fig. S7). However, it is possible that the lack of effect in tissue turnover is the consequence of incomplete inhibition by RNAi.
Fig. 5.
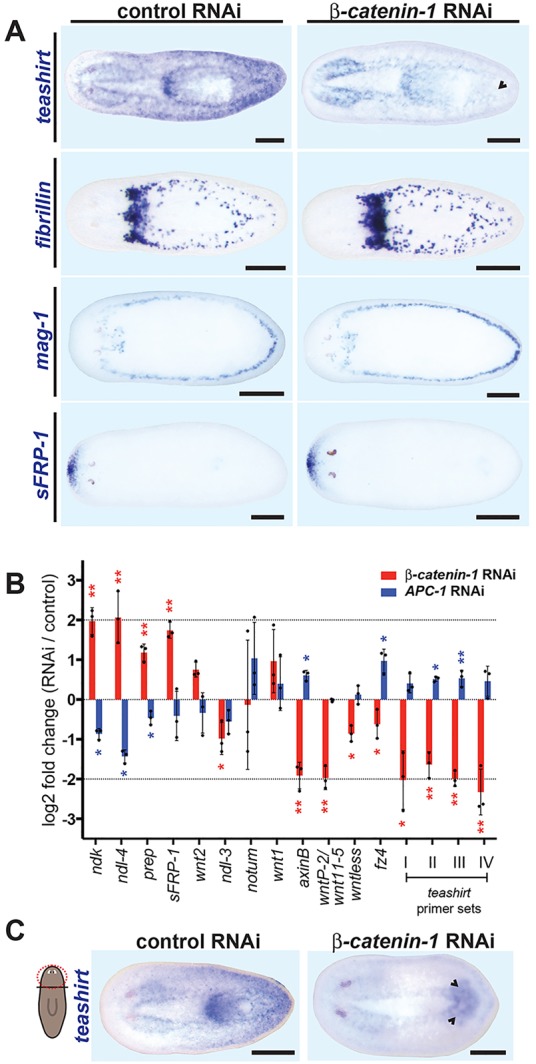
Posterior teashirt expression is dependent on Wnt signaling. (A) Posterior teashirt expression was reduced by 4 days after RNAi of β-catenin-1, whereas brain expression persisted (n=12/15). This occurred before loss of posterior cell types, indicated by in situ hybridization for fibrillin (n=10/10) and mag-1 (n=12/12). Anterior-restricted sFRP-1 expression was not altered, indicating that major polarity defects have not yet occurred (n=7/7). (B) qRT-PCR analysis of position control gene (PCG) transcriptional changes following β-catenin-1 or APC RNAi in intact uninjured animals. RNA was harvested from uninjured animals after 10 days of RNAi. Biological triplicates consisted of four individual animals each. teashirt transcriptional changes were measured by four independent primer sets (I-IV). Error bars indicate s.d. Individual replicates are indicated by dots. Asterisks indicate significance determined by t-tests between sets of biological triplicate delta-Ct values (*P<0.05) and with standard Bonferroni adjustment (**P<0.0031). (C) teashirt was expressed in both anterior and posterior brains of head fragments subjected to β-catenin-1 RNAi (n=8/9). (A,C) Anterior is leftwards. Scale bars: 200 µm.
APC is a negative regulator of β-catenin protein stability in the Wnt signaling pathway (Stamos and Weis, 2013). Inhibition of β-catenin-1 and APC cause opposite phenotypes in planarians, with APC RNAi causing regeneration of tails in place of heads (Gurley et al., 2008). Inhibition of β-catenin-1 and APC in uninjured animals resulted in opposite changes in many PCGs by 10 days of RNAi (Fig. 5B). β-catenin-1 RNAi resulted in general upregulation of anterior PCGs and downregulation of posterior PCGs, whereas APC(RNAi) caused the opposite effect. teashirt was downregulated in β-catenin-1(RNAi) animals similar to the case for other posterior PCGs (Fig. 5B). In APC(RNAi) animals, four different primer pairs for teashirt consistently showed moderate upregulation of teashirt but the effect was modest and not significant for all primer sets (Fig. 5B). In general, APC RNAi caused weaker PCG expression changes than did β-catenin-1 RNAi in these experiments in uninjured animals.
Whereas the posterior-specific teashirt expression domain was dependent upon β-catenin-1, expression of teashirt in the nervous system did not decrease following β-catenin-1 RNAi (Fig. 5A). In β-catenin-1(RNAi) head fragments, teashirt was expressed in newly regenerated posterior brains and maintained in pre-existing anterior brains, suggesting that Wnt-independent regulation of teashirt transcription in the nervous system exists (Fig. 5C). In Drosophila, teashirt expression in the trunk is initially Wnt dependent, but becomes Wnt independent as development progresses, and transcription factors such as Antennapedia and Eyeless can positively regulate its expression (Roder et al., 1992; Gallet et al., 1998; Pan and Rubin, 1998).
Wnt signaling promotes teashirt expression during tail regeneration
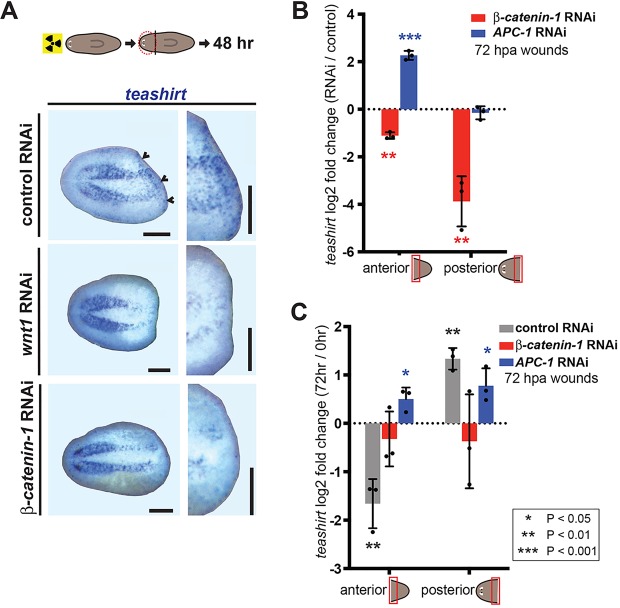
We next tested whether teashirt expression during regeneration was dependent upon Wnt signaling. First, we assessed teashirt expression in irradiated head fragments 48 h after amputation. Both β-catenin-1(RNAi) and wnt1(RNAi) head fragments showed decreased or absent teashirt expression (Fig. 6A). We next assessed teashirt expression at anterior-facing and posterior-facing wounds (from tail and head fragments, respectively) by qRT-PCR. At 72 h after injury, teashirt displayed lower expression in β-catenin-1(RNAi) animals than in the control, with the effect most pronounced at posterior-facing wounds (Fig. 6B). teashirt RNAi did not affect β-catenin-1 transcript levels (supplementary material Fig. S8). Conversely, RNAi of APC caused increased teashirt expression at anterior-facing wounds compared with the control (Fig. 6B). We next assessed the effects of β-catenin-1 and APC RNAi on both the posterior induction and the anterior rescaling of teashirt by directly comparing transcript levels at 72 h with those at 0 h post-amputation. teashirt levels failed to change from its already low levels during regeneration in β-catenin-1(RNAi) animals, and were elevated with time at both wound types in APC(RNAi) animals (Fig. 6C; supplementary material Fig. S8). Together, these findings demonstrate that teashirt transcriptional regulation at wounds in planarian regeneration is positively influenced by Wnt signaling.
Fig. 6.
Wnt signaling regulates teashirt expression during regeneration. (A) teashirt was expressed at posterior wounds in irradiated head fragments at 48 h. Less expression (see individual blue dots, which represent cells) was observed at head fragment posterior-facing wounds following RNAi of wnt1 (n=16/32) and β-catenin-1 (n=16/16). Animals viewed ventrally; anterior is leftwards. Scale bars: 200 µm. (B,C) qRT-PCR analysis of teashirt levels in β-catenin-1 and APC RNAi animals. RNA was harvested from anterior- or posterior-facing wound sites (0 h or 72 h post-amputation) cut from animals after 10 days of β-catenin-1, APC or control RNAi. Biological triplicates consisted of five to eight wound sites per sample. (B) Data are mean log2-fold changes in teashirt levels at 72 h wound sites between β-catenin-1 or APC RNAi and control RNAi samples. (C) Data are mean log2-fold changes in teashirt levels between 72 h and 0 h wound sites for each RNAi condition. Error bars indicate s.d. Individual replicates are indicated by dots. Asterisks denote significance determined by t-tests between sets of biological triplicate delta-Ct values. See also supplementary material Fig. S8.
DISCUSSION
teashirt and the head-versus-tail regeneration decision
The choice made at transverse amputation planes to regenerate a head or a tail in planarians has emerged as a paradigm for studying the molecular logic of how wounds initiate suitable regeneration programs (Reddien, 2011). This process requires Wnt signaling in planarians, with homeotic phenotypes (replacing heads with tails or vice versa) caused by activating or inhibiting the Wnt pathway. Here, we describe the identification of an additional gene, teashirt, that is required for this head-tail regeneration decision. The teashirt RNAi phenotype is highly similar to that caused by Wnt pathway inhibition in planarians at the molecular and regeneration outcome levels. Furthermore, teashirt transcriptional activation during posterior regeneration requires Wnt signaling. The preferential expression of teashirt during posterior regeneration occurred after initial wound-induced activation of wnt1. teashirt was required for the subsequent activation of posterior-specific position control genes, such as wntP-2. Therefore, teashirt is regulated by Wnt signaling and is required for the output of Wnt signaling in regeneration (Fig. 7). We propose a model for the initial phases of planarian head-versus-tail regeneration in which teashirt is required for proper PCG expression (Fig. 7). In this model, wounding initiates wnt1 expression 3-6 h following injury. Notum inhibits Wnt signaling at anterior-facing wounds to promote head regeneration. At posterior-facing wounds that will regenerate a tail, Wnt signaling activates a program of gene expression involving posterior PCGs, rather than anterior PCGs. teashirt is required for this Wnt-dependent gene expression program.
Fig. 7.
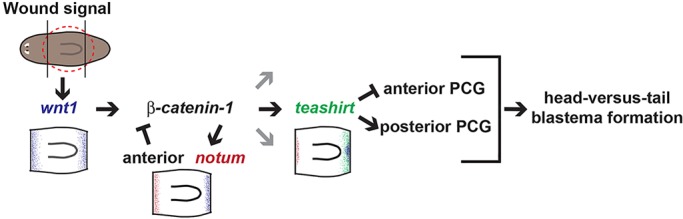
Model for the role of teashirt in planarian head-tail regeneration. A developmental pathway for the initiation of head versus tail regeneration. wnt1 is activated by wounding and, through Wnt signaling, activates teashirt. During anterior regeneration, notum inhibits Wnt signaling (later notum roles are not the focus of this model). During initiation of posterior regeneration, teashirt helps promote the expression of posterior position control genes (PCGs) (such as wntP-2) and inhibit the expression of anterior PCGs to give a tail regeneration signature. It is unknown whether these effects (depicted by black arrows) reflect direct action. Gray arrows indicate that it is unknown whether all action of β-catenin-1 will involve teashirt.
Anterior and posterior poles are small clusters of cells at the head and tail tip, respectively, and are required for normal planarian head and tail regeneration and PCG expression (Felix and Aboobaker, 2010; Hayashi et al., 2011; Blassberg et al., 2013; Chen et al., 2013; Scimone et al., 2014b; Vásquez-Doorman and Petersen, 2014; Vogg et al., 2014). However, in contrast to teashirt RNAi, inhibition of either anterior or posterior pole formation does not cause a reversal of the head-tail regeneration choice. Unlike inhibition of pole formation, inhibition of β-catenin-1 or teashirt causes a gene expression signature switch: posterior-facing wounds not only fail to express tail PCGs but inappropriately express head PCGs. These findings suggest that Wnt signaling and teashirt regulate a broad program of gene expression events – giving wounds anterior or posterior character – and that pole formation is just one component of this program.
The Teashirt family and Wnt signaling
teashirt encodes a broadly conserved zinc-finger protein, with orthologs found throughout the animal kingdom. In other organisms, it has been suggested that Teashirt can function to increase the relative strength of the cellular response to Wnt (Gallet et al., 1998; Onai et al., 2007). We found that teashirt in planarians is required for the outcome of β-catenin-dependent processes during regeneration. Planarians are members of the superphylum the Lophotrochozoa. Xenopus is a chordate and part of the deuterostomes, and Drosophila is an arthropod and a representative from the ecdysozoans. Thus, whereas teashirt is still understudied, connections between Teashirt proteins and the outcome of Wnt signaling now exist for all three major branches of the Bilateria. This raises the possibility that Teashirt proteins will be involved in many Wnt-dependent processes throughout the Metazoa, a possibility that can be explored by study of Wnt signaling and Teashirt in distinct tissue and organism contexts.
In the case of Xenopus, Xtsh3 can affect DV axis development through its role in β-catenin activity for Spemann's organizer formation, which coordinates early embryonic patterning (Onai et al., 2007). After gastrulation in Xenopus, Wnt signaling promotes AP axis polarization; whether a Teashirt protein has a role in this AP axis polarization process remains unknown. Wnt signaling promotes AP axis polarization in many animals throughout the animal kingdom (Petersen and Reddien, 2009a). Data from planarians therefore raise the possibility that Teashirt proteins will have a broad role, together with Wnt signaling, in AP axis development in the Metazoa. Wnt signaling controls a large array of important processes in addition to AP axis development, including stem cell proliferation, wound healing, polarization of the animal-vegetal axis of early embryos in many species and posterior growth. Because of this diversity of roles, Wnt signaling components have been the subject of intensive investigation, and have been targeted for the development of chemical therapeutic approaches. An important direction will therefore be to determine the diversity of contexts in which Teashirt activity is important for Wnt-signaling outcomes, such as in the context of human diseases.
We conclude that Teashirt is a key factor in regulating the response to Wnt signaling in the polarization of the planarian primary body axis during regeneration. These findings in planarians predict that Teashirt proteins will prove to be involved in the development of the primary axis in many other animals. Furthermore, these findings support the possibility that Teashirt will prove to be an important but presently understudied determinant of the outcome of Wnt signaling activity in many of the diverse and important biological processes controlled by Wnt signaling throughout the Metazoa.
MATERIALS AND METHODS
Animal culture and radiation treatment
Asexual S. mediterranea (strain CIW4) animals were starved 7-14 days prior to experiments. Animals were exposed to 6000 rads using a dual Gammacell-40 cesium-137 source and amputated 4-6 days after irradiation.
Smed-teashirt and RNAi
The teashirt sequence was determined using RACE (Ambion). teashirt was amplified (5′-CAACGAAATCCGGAGGAAA-3′ and 5′-TTTGGACTCTTGGACGGTTC-3′) from cDNA. β-catenin-1, wnt1, wntP-2, APC, notum and control (C. elegans unc-22) constructs have been previously described (Petersen and Reddien, 2008, 2009b, 2011). RNAi experiments used dsRNA-expressing bacteria mixed with liver as food or dsRNA injection (Petersen and Reddien, 2009b). All RNAi involved matched control RNAi animals that had undergone the same procedure. Injection protocol: animals underwent amputation and injection on day 0; animals were then injected on day 1; wound sites were removed and animals were injected on day 3; animals were then injected on day 4; wound sites were removed and animals were injected on day 6; animals were then injected on day 7. Wound site removal removed ∼200 μm of pre-existing tissue next to the injury. This protocol was performed with trunk fragments (lacking heads and tails; Fig. 1A-H and Fig. 2A,B) or head fragments (lacking trunk and tail; Fig. 2C-E and Fig. 6A). Final wound site removal (experimental amputation, day 6) in head fragments used for gene expression analyses amputated tissue that would include most wound-induced gene expression (≥250 μm). A short ‘4-day RNAi protocol’ with head fragments (day 0, amputate and inject; day 1, inject; day 3, amputate and inject; day 4, inject) also generated a teashirt phenotype. For Fig. 5A, β-catenin-1 and control dsRNA-expressing bacteria were fed and animals were fixed 4 days later. For Figs 5 and 6, β-catenin-1, APC-1 and control dsRNA-expressing bacteria were fed to animals on day 0, 2, 4 and 7; on day 10, RNA was collected for Fig. 5B and amputation occurred for Fig. 6B,C. For long-term teashirt inhibition experiments (supplementary material Fig. S7), animals were fed dsRNA-containing food every 4 days for 8 weeks.
In situ hybridizations
Whole-mount in situ hybridization and fluorescence in situ hybridization were performed as described previously (Pearson et al., 2009).
Quantitative reverse-transcriptase PCR (qRT-PCR)
Samples were processed as in supplementary material Fig. S4 and analyzed on the Fluidigm Biomark platform using 96.96 Dynamic Array IFC chips. Dissected wound sites or whole animals were collected and dissociated in 0.75 ml Trizol (Life Technologies) by gentle trituration with a P1000 tip (for ∼1-2 min, room temperature) and then stored (−80°C). The RNA from the samples was purified according to manufacturer guidelines and resuspended in dH2O. RNA concentrations were determined by Qubit using the RNA HS Assay kit (Life Technologies) and normalized to 100 ng/μl in dH2O. Each sample (500 ng) was dispensed into a 96-well plate and treated with 1 U of amplification-grade DNAse I (Life Technologies) for 15 min (at room temperature). DNAse was heat-inactivated for 10 min (at 65°C) in the presence of ∼2.5 mM EDTA. Samples were diluted 1:100 in dH2O and stored (−80°C). Multiplex reverse-transcription (50°C for 20 min) followed by 15 PCR amplification cycles were performed using pooled (outer) primers at 50 nM each for 28 target transcripts (see supplementary material Table S1) and a Superscript III/Platinum Taq enzyme mix (Life Technologies). Outer primers were digested with ExoI (15 U/well, New England Biolabs) at 37°C (30 min), followed by enzyme inactivation (80°C for 15 min). Samples were diluted 1:5 in TE and 1 μl of each was used to evaluate g6pd levels by conventional qRT-PCR (7500 Fast PCR System, Applied Biosystems). Samples were loaded onto a 96.96 Dynamic Array IFC chip and transcript levels assessed using a separate inner primer set (see supplementary material Table S1) for each transcript. Amplified cDNA (3.3 μl/sample) was mixed with 2.7 μl of EvaGreen Taq Supermix (Bio-Rad) supplemented with 20× DNA-Binding Mix (Fluidigm). For each inner primer pair, 10 μM of forward and reverse primer mixes were diluted 1:1 with 2× Assay Loading Reagent (Fluidigm). Six technical replicates for each of four housekeeping transcripts (g6pd, clathrin, luc7 and ubiquilin) and three technical replicates for the remaining 24 transcripts were prepared for each chip. Primer mixes and samples were loaded onto the left and right sides of the IFC chip, respectively; priming and loading of the IFCs and operation of the Biomark instrument were performed according to the manufacturer's instructions. Temperature cycling used a thermal mixing step, 44 fast PCR cycles (96°C for 5 s; 60°C for 20 s) and melting (60-95°C). Data were analyzed using Fluidigm Real-Time PCR Analysis software using the Linear (Derivative) Baseline Correction Method and the Auto (Global) Ct Threshold Method. For each target transcript, melt curve analysis was used to eliminate Ct measurements associated with abnormally high or low Tm values.
Normalization and analysis of qRT-PCR Data
Processing and normalization of qRT-PCR data was performed using the R statistical computing environment. For each chip, technical replicates were averaged and fold changes calculated using the ‘Delta Delta Ct’ (ΔΔCt) method. For each of three biological replicates, samples were first normalized to the average Ct value of four housekeeping transcripts: g6pd, clathrin, luc7 and ubiquilin (ΔCt). ΔCt values were then subtracted between samples and a reference (e.g. control RNAi or time zero) to generate ΔΔCt values reflecting log2-fold changes. Significance was determined by t-tests (unpaired, equal variance) between sets of biological triplicate ΔCt values. Bar plots of qRT-PCR data represent mean log2-fold changes (±s.d.). Dots represent individual ΔΔCt values. Heatmaps were generated using the heatmap.2 function in R, and dendrograms reflect hierarchical clustering based on Euclidean distance. For principal component analysis (PCA), each sample was normalized to average housekeeping levels as above. Transcript levels were then converted to z-scores, and PCA was performed on scaled, centered data using the prcomp function. PCA plots and ‘PC loading’ plots were generated using ggplot2.
Note added in proof
Analysis of the role of teashirt in planarians has also been described by Reuter et al. (2015), published during final publication preparations of this manuscript.
Supplementary Material
Acknowledgements
We thank Sylvain Lapan for the cintillo probe, and Lucila Scimone, Katrina Longenecker, Irving Wang and the Reddien lab for manuscript comments.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
J.H.O., D.E.W., C.-C.C., C.P.P. and P.W.R. designed the experiments. D.E.W. and C.P.P. identified teashirt as expressed at posterior wounds. J.H.O. identified and characterized the teashirt RNAi phenotype. Experiments leading to: Fig. 1 were carried out by J.H.O. and C.-C.C.; Fig. 2, D.E.W., J.H.O., C.-C.C. and P.W.R.; Fig. 3, D.E.W. and J.H.O.; Fig. 4, D.E.W., J.H.O. and C.-C.C.; Fig. 5, J.H.O. and D.E.W.; Fig. 6, D.E.W. and J.H.O.; supplementary material Fig. S1, J.H.O.; supplementary material Fig. S2 J.H.O. and C.-C.C.; supplementary material Fig. S3, C.-C.C.; supplementary material Fig. S4, D.E.W.; supplementary material Fig. S5, D.E.W. and C.-C.C.; supplementary material Fig. S6, C.-C.C., supplementary material Fig. S7, J.H.O.; and supplementary material Fig. S8, D.E.W. and P.W.R.
Funding
P.W.R. is an Investigator of the Howard Hughes Medical Institute and an associate member of the Broad Institute of Harvard and MIT. We acknowledge support from the National Institutes of Health (NIH) [R01GM080639] and from the Keck Foundation. Deposited in PMC for release after 6 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.119685/-/DC1
References
- Adell T., Salo E., Boutros M. and Bartscherer K. (2009). Smed-Evi/Wntless is required for beta-catenin-dependent and -independent processes during planarian regeneration. Development 136, 905-910 10.1242/dev.033761 [DOI] [PubMed] [Google Scholar]
- Andrew D. J., Horner M. A., Petitt M. G., Smolik S. M. and Scott M. P. (1994). Setting limits on homeotic gene function: restraint of Sex combs reduced activity by teashirt and other homeotic genes. EMBO J. 13, 1132-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa J. and Casares F. (2005). Restricted teashirt expression confers eye-specific responsiveness to Dpp and Wg signals during eye specification in Drosophila. Development 132, 5011-5020 10.1242/dev.02082 [DOI] [PubMed] [Google Scholar]
- Blassberg R. A., Felix D. A., Tejada-Romero B. and Aboobaker A. A. (2013). PBX/extradenticle is required to re-establish axial structures and polarity during planarian regeneration. Development 140, 730-739 10.1242/dev.082982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caubit X., Thoby-Brisson M., Voituron N., Filippi P., Bevengut M., Faralli H., Zanella S., Fortin G., Hilaire G. and Fasano L. (2010). Teashirt 3 regulates development of neurons involved in both respiratory rhythm and airflow control. J. Neurosci. 30, 9465-9476 10.1523/JNEUROSCI.1765-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrià F., Kobayashi C., Umesono Y., Nakazawa M., Mineta K., Ikeo K., Gojobori T., Itoh M., Taira M., Sánchez Alvarado A. et al. (2002). FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature 419, 620-624 10.1038/nature01042 [DOI] [PubMed] [Google Scholar]
- Chen C.-C. G., Wang I. E. and Reddien P. W. (2013). pbx is required for pole and eye regeneration in planarians. Development 140, 719-729 10.1242/dev.083741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles M. W., Brown D. D. R., Nisperos S. V., Stanley B. N., Pearson B. J. and Zayas R. M. (2013). Genome-wide analysis of the bHLH gene family in planarians identifies factors required for adult neurogenesis and neuronal regeneration. Development 140, 4691-4702 10.1242/dev.098616 [DOI] [PubMed] [Google Scholar]
- Currie K. W. and Pearson B. J. (2013). Transcription factors lhx1/5-1 and pitx are required for the maintenance and regeneration of serotonergic neurons in planarians. Development 140, 3577-3588 10.1242/dev.098590 [DOI] [PubMed] [Google Scholar]
- de Zulueta P., Alexandre E., Jacq B. and Kerridge S. (1994). Homeotic complex and teashirt genes co-operate to establish trunk segmental identities in Drosophila. Development 120, 2287-2296. [DOI] [PubMed] [Google Scholar]
- Dubois F. (1949). Contribution a l'étude de la migration des cellules de régénération chez les Planaires Dulcicoles. Bull. Biol. Fr. Belg. 83, 213-283. [Google Scholar]
- Eisenmann D. M. (2005). Wnt signaling. In WormBook (ed. The C. elegans Research Community), http://www.wormbook.org/pidoi/10.1895/wormbook.1.7.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson T., Pillay L. M. and Waskiewicz A. J. (2011). Zebrafish Tshz3b negatively regulates Hox function in the developing hindbrain. Genesis 49, 725-742 10.1002/dvg.20781 [DOI] [PubMed] [Google Scholar]
- Erkner A., Gallet A., Angelats C., Fasano L. and Kerridge S. (1999). The role of Teashirt in proximal leg development in Drosophila: ectopic Teashirt expression reveals different cell behaviours in ventral and dorsal domains. Dev. Biol. 215, 221-232 10.1006/dbio.1999.9452 [DOI] [PubMed] [Google Scholar]
- Faralli H., Martin E., Core N., Liu Q.-C., Filippi P., Dilworth F. J., Caubit X. and Fasano L. (2011). Teashirt-3, a novel regulator of muscle differentiation, associates with BRG1-associated factor 57 (BAF57) to inhibit myogenin gene expression. J. Biol. Chem. 286, 23498-23510 10.1074/jbc.M110.206003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano L., Röder L., Coré N., Alexandre E., Vola C., Jacq B. and Kerridge S. (1991). The gene teashirt is required for the development of Drosophila embryonic trunk segments and encodes a protein with widely spaced zinc finger motifs. Cell 64, 63-79 10.1016/0092-8674(91)90209-H [DOI] [PubMed] [Google Scholar]
- Felix D. A. and Aboobaker A. A. (2010). The TALE class homeobox gene Smed-prep defines the anterior compartment for head regeneration. PLoS Genet. 6, e1000915 10.1371/journal.pgen.1000915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French V., Bryant P. J. and Bryant S. V. (1976). Pattern regulation in epimorphic fields. Science 193, 969-981 10.1126/science.948762 [DOI] [PubMed] [Google Scholar]
- Gallet A., Erkner A., Charroux B., Fasano L. and Kerridge S. (1998). Trunk-specific modulation of wingless signalling in Drosophila by teashirt binding to armadillo. Curr. Biol. 8, 893-902 10.1016/S0960-9822(07)00369-7 [DOI] [PubMed] [Google Scholar]
- Gallet A., Angelats C., Erkner A., Charroux B., Fasano L. and Kerridge S. (1999). The C-terminal domain of armadillo binds to hypophosphorylated teashirt to modulate wingless signalling in Drosophila. EMBO J. 18, 2208-2217 10.1093/emboj/18.8.2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley K. A., Rink J. C. and Sánchez Alvarado A. (2008). Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319, 323-327 10.1126/science.1150029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley K. A., Elliott S. A., Simakov O., Schmidt H. A., Holstein T. W. and Sánchez Alvarado A. (2010). Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev. Biol. 347, 24-39 10.1016/j.ydbio.2010.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland R. and Gerhart J. (1997). Formation and function of Spemann's organizer. Annu. Rev. Cell Dev. Biol. 13, 611-667 10.1146/annurev.cellbio.13.1.611 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Motoishi M., Yazawa S., Itomi K., Tanegashima C., Nishimura O., Agata K. and Tarui H. (2011). A LIM-homeobox gene is required for differentiation of Wnt-expressing cells at the posterior end of the planarian body. Development 138, 3679-3688 10.1242/dev.060194 [DOI] [PubMed] [Google Scholar]
- Hobmayer B., Rentzsch F., Kuhn K., Happel C. M., von Laue C. C., Snyder P., Rothbächer U. and Holstein T. W. (2000). WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature 407, 186-189 10.1038/35025063 [DOI] [PubMed] [Google Scholar]
- Iglesias M., Gomez-Skarmeta J. L., Salo E. and Adell T. (2008). Silencing of Smed-betacatenin1 generates radial-like hypercephalized planarians. Development 135, 1215-1221 10.1242/dev.020289 [DOI] [PubMed] [Google Scholar]
- Iglesias M., Almuedo-Castillo M., Aboobaker A. A. and Saló E. (2011). Early planarian brain regeneration is independent of blastema polarity mediated by the Wnt/beta-catenin pathway. Dev. Biol. 358, 68-78 10.1016/j.ydbio.2011.07.013 [DOI] [PubMed] [Google Scholar]
- Koebernick K., Kashef J., Pieler T. and Wedlich D. (2006). Xenopus Teashirt1 regulates posterior identity in brain and cranial neural crest. Dev. Biol. 298, 312-326 10.1016/j.ydbio.2006.06.041 [DOI] [PubMed] [Google Scholar]
- Lapan S. W. and Reddien P. W. (2011). dlx and sp6–9 control optic cup regeneration in a prototypic eye. PLoS Genet. 7, e1002226 10.1371/journal.pgen.1002226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfroid I., Caubit X., Kerridge S. and Fasano L. (2004). Three putative murine Teashirt orthologues specify trunk structures in Drosophila in the same way as the Drosophila teashirt gene. Development 131, 1065-1073 10.1242/dev.00977 [DOI] [PubMed] [Google Scholar]
- McCormick A., Core N., Kerridge S. and Scott M. P. (1995). Homeotic response elements are tightly linked to tissue-specific elements in a transcriptional enhancer of the teashirt gene. Development 121, 2799-2812. [DOI] [PubMed] [Google Scholar]
- Molina M. D., Saló E. and Cebrià F. (2007). The BMP pathway is essential for re-specification and maintenance of the dorsoventral axis in regenerating and intact planarians. Dev. Biol. 311, 79-94 10.1016/j.ydbio.2007.08.019 [DOI] [PubMed] [Google Scholar]
- Morgan T. H. (1898). Experimental studies of the regeneration of Planaria maculata. Arch. Entw. Mech. Org. 7, 364-397. [Google Scholar]
- Niehrs C. (2010). On growth and form: a Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development 137, 845-857 10.1242/dev.039651 [DOI] [PubMed] [Google Scholar]
- Onai T., Matsuo-Takasaki M., Inomata H., Aramaki T., Matsumura M., Yakura R., Sasai N. and Sasai Y. (2007). XTsh3 is an essential enhancing factor of canonical Wnt signaling in Xenopus axial determination. EMBO J. 26, 2350-2360 10.1038/sj.emboj.7601684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orii H. and Watanabe K. (2007). Bone morphogenetic protein is required for dorso-ventral patterning in the planarian Dugesia japonica. Dev. Growth Differ. 49, 345-349 10.1111/j.1440-169X.2007.00931.x [DOI] [PubMed] [Google Scholar]
- Oviedo N. J., Newmark P. A. and Sánchez Alvarado A. (2003). Allometric scaling and proportion regulation in the freshwater planarian Schmidtea mediterranea. Dev. Dyn. 226, 326-333 10.1002/dvdy.10228 [DOI] [PubMed] [Google Scholar]
- Pan D. and Rubin G. M. (1998). Targeted expression of teashirt induces ectopic eyes in Drosophila. Proc. Natl. Acad. Sci. USA 95, 15508-15512 10.1073/pnas.95.26.15508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson B. J., Eisenhoffer G. T., Gurley K. A., Rink J. C., Miller D. E. and Sánchez Alvarado A. (2009). Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev. Dyn. 238, 443-450 10.1002/dvdy.21849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M., Rauskolb C., Williams M., Riggleman B. and Wieschaus E. (1991). The segment polarity gene armadillo interacts with the wingless signaling pathway in both embryonic and adult pattern formation. Development 111, 1029-1043. [DOI] [PubMed] [Google Scholar]
- Petersen C. P. and Reddien P. W. (2008). Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science 319, 327-330 10.1126/science.1149943 [DOI] [PubMed] [Google Scholar]
- Petersen C. P. and Reddien P. W. (2009a). Wnt signaling and the polarity of the primary body axis. Cell 139, 1056-1068 10.1016/j.cell.2009.11.035 [DOI] [PubMed] [Google Scholar]
- Petersen C. P. and Reddien P. W. (2009b). A wound-induced Wnt expression program controls planarian regeneration polarity. Proc. Natl. Acad. Sci. USA 106, 17061-17066 10.1073/pnas.0906823106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C. P. and Reddien P. W. (2011). Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science 332, 852-855 10.1126/science.1202143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph H. (1897). Observations and experiments on regeneration in planarians. Arch. Entw. Mech. Org. 5, 352-372. [Google Scholar]
- Reddien P. W. (2011). Constitutive gene expression and the specification of tissue identity in adult planarian biology. Trends Genet. 27, 277-285 10.1016/j.tig.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W. (2013). Specialized progenitors and regeneration. Development 140, 951-957 10.1242/dev.080499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W. and Sánchez Alvarado A. (2004). Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 20, 725-757 10.1146/annurev.cellbio.20.010403.095114 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Oviedo N. J., Jennings J. R., Jenkin J. C. and Sánchez Alvarado A. (2005). SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327-1330 10.1126/science.1116110 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Bermange A. L., Kicza A. M. and Sánchez Alvarado A. (2007). BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development 134, 4043-4051 10.1242/dev.007138 [DOI] [PubMed] [Google Scholar]
- Reuter H., März M., Vogg M. C., Eccles D., Grífol-Boldú L., Wehner D., Owlarn S., Adell T., Weidinger G. and Bartscherer K. (2015). β-catenin-dependent control of positional information along the AP body axis in planarians involves a Teashirt family member. Cell Rep. 13, 253-265 10.1016/j.celrep.2014.12.018 [DOI] [PubMed] [Google Scholar]
- Rink J. C., Gurley K. A., Elliott S. A. and Sánchez Alvarado A. (2009). Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science 326, 1406-1410 10.1126/science.1178712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roder L., Vola C. and Kerridge S. (1992). The role of the teashirt gene in trunk segmental identity in Drosophila. Development 115, 1017-1033. [DOI] [PubMed] [Google Scholar]
- Scimone M. L., Srivastava M., Bell G. W. and Reddien P. W. (2011). A regulatory program for excretory system regeneration in planarians. Development 138, 4387-4398 10.1242/dev.068098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone M. L., Kravarik K. M., Lapan S. W. and Reddien P. W. (2014a). Neoblast specialization in regeneration of the planarian Schmidtea mediterranea. Stem Cell Rep. 3, 339-352 10.1016/j.stemcr.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone M. L., Lapan S. W. and Reddien P. W. (2014b). A forkhead transcription factor is wound-induced at the planarian midline and required for anterior pole regeneration. PLoS Genet. 10, e1003999 10.1371/journal.pgen.1003999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Kango-Singh M. and Sun Y. H. (2002). Eye suppression, a novel function of teashirt, requires Wingless signaling. Development 129, 4271-4280. [DOI] [PubMed] [Google Scholar]
- Stamos J. L. and Weis W. I. (2013). The beta-catenin destruction complex. Cold Spring Harb. Perspect. Biol. 5, a007898 10.1101/cshperspect.a007898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoick-Cooper C. L., Weidinger G., Riehle K. J., Hubbert C., Major M. B., Fausto N. and Moon R. T. (2007). Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development 134, 479-489 10.1242/dev.001123 [DOI] [PubMed] [Google Scholar]
- van Wolfswinkel J. C., Wagner D. E. and Reddien P. W. (2014). Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell 15, 326-339 10.1016/j.stem.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vásquez-Doorman C. and Petersen C. P. (2014). zic-1 Expression in planarian neoblasts after injury controls anterior pole regeneration. PLoS Genet. 10, e1004452 10.1371/journal.pgen.1004452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogg M. C., Owlarn S., Pérez Rico Y. A., Xie J., Suzuki Y., Gentile L., Wu W. and Bartscherer K. (2014). Stem cell-dependent formation of a functional anterior regeneration pole in planarians requires Zic and Forkhead transcription factors. Dev. Biol. 390, 136-148 10.1016/j.ydbio.2014.03.016 [DOI] [PubMed] [Google Scholar]
- Wagner D. E., Wang I. E. and Reddien P. W. (2011). Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811-816 10.1126/science.1203983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltzer L., Vandel L. and Bienz M. (2001). Teashirt is required for transcriptional repression mediated by high Wingless levels. EMBO J. 20, 137-145 10.1093/emboj/20.1.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D., Lapan S. W., Wilkinson A. W., Bell G. W. and Reddien P. W. (2012). A molecular wound response program associated with regeneration initiation in planarians. Genes Dev. 26, 988-1002 10.1101/gad.187377.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witchley J. N., Mayer M., Wagner D. E., Owen J. H. and Reddien P. W. (2013). Muscle cells provide instructions for planarian regeneration. Cell Rep. 4, 633-641 10.1016/j.celrep.2013.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert L. (1969). Positional information and the spatial pattern of cellular differentiation. J. Theor. Biol. 25, 1-47 10.1016/S0022-5193(69)80016-0 [DOI] [PubMed] [Google Scholar]
- Wu J. and Cohen S. M. (2002). Repression of Teashirt marks the initiation of wing development. Development 129, 2411-2418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.