Abstract
Loss of neurons that express the neuropeptide hypocretin (Hcrt) has been implicated in narcolepsy, a debilitating disorder characterized by excessive daytime sleepiness and cataplexy. Cell replacement therapy, using Hcrt-expressing neurons generated in vitro, is a potentially useful therapeutic approach, but factors sufficient to specify Hcrt neurons are unknown. Using zebrafish as a high-throughput system to screen for factors that can specify Hcrt neurons in vivo, we identified the LIM homeobox transcription factor Lhx9 as necessary and sufficient to specify Hcrt neurons. We found that Lhx9 can directly induce hcrt expression and we identified two potential Lhx9 binding sites in the zebrafish hcrt promoter. Akin to its function in zebrafish, we found that Lhx9 is sufficient to specify Hcrt-expressing neurons in the developing mouse hypothalamus. Our results elucidate an evolutionarily conserved role for Lhx9 in Hcrt neuron specification that improves our understanding of Hcrt neuron development.
Keywords: Hypocretin, Lhx9, Zebrafish
Summary: The transcription factor Lhx9 is sufficient to drive the specification of zebrafish and mouse hypocretin-expressing neurons, the neuron type that is affected in narcolepsy.
INTRODUCTION
The neuropeptide hypocretin (Hcrt) is conserved among vertebrates and plays key roles in regulating sleep, metabolism, feeding, anxiety, reward and addiction (Bonnavion and de Lecea, 2010; Tsujino and Sakurai, 2009). Hcrt is particularly important in promoting arousal, as loss of Hcrt neurons is thought to cause narcolepsy (Peyron et al., 2000; Thannickal et al., 2000), a disorder characterized by daytime sleepiness, fragmented sleep-wake states and cataplexy. Narcolepsy affects approximately 1 in 2000 individuals, but treatments are limited to symptom management (Dauvilliers et al., 2007). Despite the importance of the Hcrt system, little is known about the developmental processes that give rise to Hcrt neurons. A recent study found that mice lacking the LIM domain homeobox transcription factor Lhx9 had fewer Hcrt neurons (Dalal et al., 2013), suggesting that Lhx9 is required to specify a subset of Hcrt neurons. However, overexpression of Lhx9 in adult mice or in a mouse neuroblastoma cell line had no effect on Hcrt cell number or expression. Therefore, the role of Lhx9 in Hcrt neuron specification remains unclear and the set of factors sufficient to specify Hcrt neurons remains unknown. Identifying these factors would help elucidate how a key neural circuit that governs sleep is established, and could lead to novel therapies for narcolepsy.
The zebrafish Danio rerio is a powerful genetic model of vertebrate development that provides several advantages for studying Hcrt neuron specification. First, the hypothalamus is remarkably conserved (Blackshaw et al., 2010; Machluf et al., 2011; Tessmar-Raible et al., 2007), suggesting that developmental mechanisms identified in zebrafish are likely to be relevant to mammals. Several studies have shown that the mammalian Hcrt system is functionally and anatomically conserved in zebrafish (Chiu and Prober, 2013; Elbaz et al., 2013). Whereas the rodent hypothalamus contains thousands of Hcrt neurons, larval and adult zebrafish contain only approximately 10 and 40 Hcrt neurons, respectively (Faraco et al., 2006; Kaslin et al., 2004), making zebrafish a more tractable system to study Hcrt neuron development. Second, the external development and transparency of zebrafish embryos facilitate the observation of developing Hcrt neurons. Third, high-throughput genetic gain- and loss-of-function assays facilitate efficient screens to identify developmental regulators. We exploited these features of zebrafish to identify genes that regulate Hcrt neuron specification.
RESULTS
Microarray analysis identifies transcripts enriched in Hcrt neurons
Previous studies showed that the number of Hcrt neurons in zebrafish and rodents increases as animals develop and mature to adulthood (Faraco et al., 2006; Kaslin et al., 2004; Sawai et al., 2010). We reasoned that cell-autonomous factors required to specify Hcrt neurons might still be expressed in Hcrt neurons shortly after they are specified. To identify these factors, we generated transgenic zebrafish that express monomeric red fluorescent protein (mRFP) in Hcrt neurons and enhanced green fluorescent protein (EGFP) in neurons that express the hypothalamic neuropeptide QRFP (supplementary material Fig. S1). QRFP has been implicated in regulating locomotor activity (Takayasu et al., 2006), feeding (Chartrel et al., 2003; Takayasu et al., 2006) and nociception (Yamamoto et al., 2009) in rodents, and sleep/wake behaviors in zebrafish (C.N.C., A. Chen and D.A.P., unpublished).
Expression of hcrt and qrfp (si:ch211-185o22.2, incorrectly annotated as lincRNA) is first detected in zebrafish embryos at 24 h post-fertilization (hpf) in bilateral hypothalamic nuclei of 4-6 cells (Fig. 1A,B), which expand to 10-15 cells by 120 hpf (Fig. 1C,D). Hcrt and QRFP are expressed in neighboring neurons throughout development, but are never co-expressed within the same cells (Fig. 1B,D). To identify genes with enriched expression in Hcrt neurons, we dissociated pools of 100-300 Tg(hcrt:mRFP, qrfp:EGFP) embryos at 26 hpf into single cells and isolated EGFP- and mRFP-expressing neurons by fluorescence-activated cell sorting (FACS) (Fig. 1E; supplementary material Fig. S2). FACS gates for EGFP and mRFP populations were set using wild-type embryos (0/10,000 EGFP+ or mRFP+ events). In a representative experiment, we obtained 250 EGFP+ cells and 528 mRFP+ cells from 150 Tg(hcrt:mRFP, qrfp:EGFP) double-heterozygous embryos. To verify the fidelity of FACS, we visually screened for fluorescence in sorted cells (Fig. 1F). In the sorted qrfp:EGFP population, we observed EGFP in 99/117 cells (85%) but no mRFP (0/117). In the sorted hcrt:mRFP population, we observed mRFP in 110/146 cells (75%) but no EGFP (0/146). These values are likely to underestimate the purity of the sorted cells because FACS is more sensitive than visual inspection.
Fig. 1.
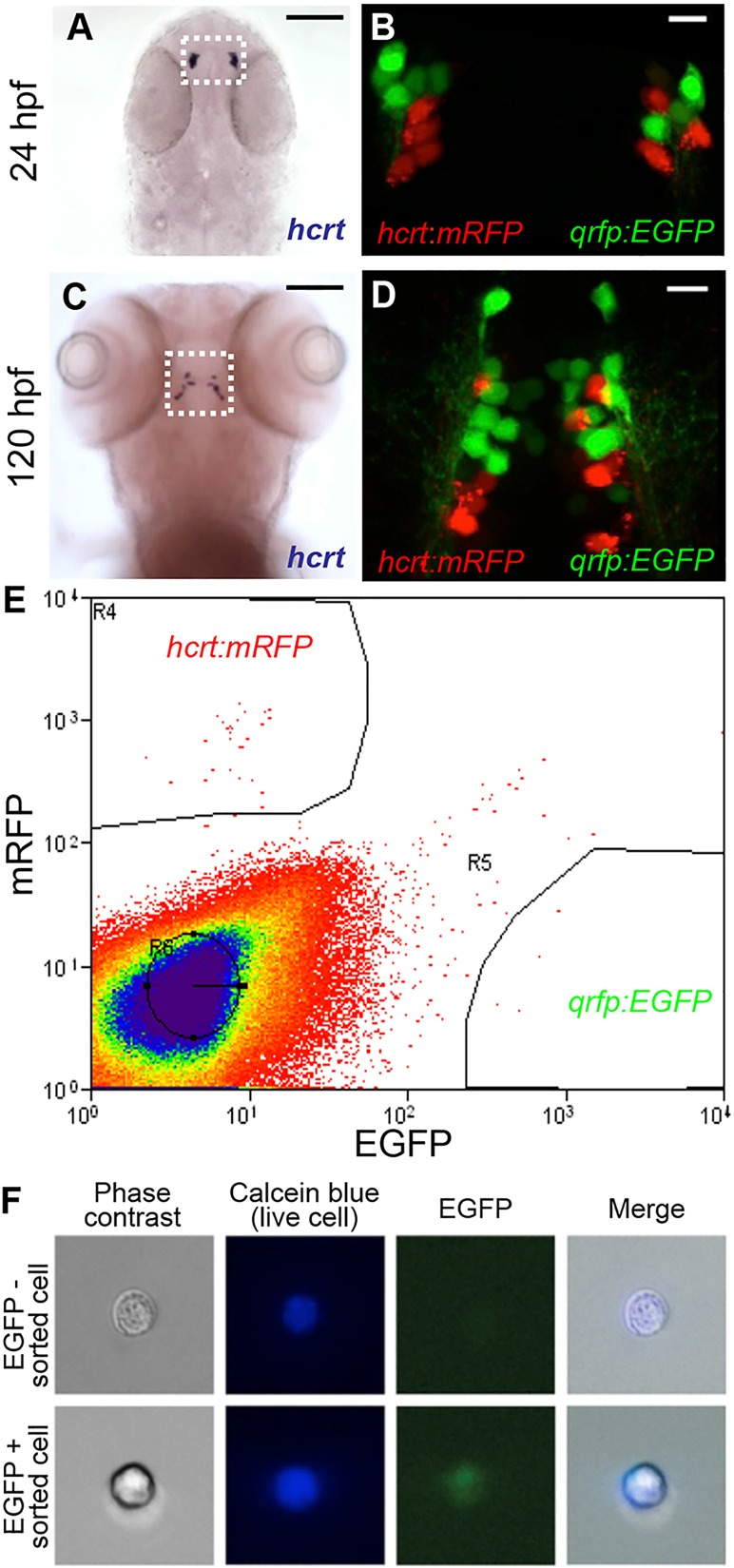
Isolation of hcrt- and qrfp-expressing neurons from zebrafish embryos. (A-D) hcrt is expressed in bilateral populations of 4-6 neurons at 24 hpf (A) and 10-15 neurons at 120 hpf (C). Fluorescence in Tg(hcrt:mRFP, qrfp:EGFP) embryos is first observed at ∼24 hpf. mRFP- and EGFP-labeled neurons are intermingled, but the markers are never co-expressed in the same cell (B,D). Boxed regions in A,C are shown at higher magnification in B,D. Scale bars: 100 μm in A,C; 10 μm in B,D. (E) Tg(hcrt:mRFP, qrfp:EGFP) embryos were dissociated into single cells at 26 hpf and mRFP- and EGFP-expressing cells were isolated by FACS. The proportion of mRFP-positive and EGFP-positive cells was consistent with the number of Hcrt and QRFP neurons, respectively, in a 26 hpf embryo. (F) EGFP is observed in a sorted qrfp:EGFP+ cell.
We extracted total mRNA from each cellular fraction and used cDNA microarrays to compare gene expression in Hcrt and QRFP neurons. We also compared gene expression in Hcrt neurons with expression in neurons labeled by a pan-neuronal marker, Tg(elavl3:EGFP), and with expression in subtypes of sensory neurons: Tg(trpa1b:EGFP), Tg(isl1:Gal4VP16, 14xUAS:EGFP) and Tg(p2rx3b:EGFP). elavl3 encodes an RNA-binding protein that is expressed in most postmitotic neurons (Park et al., 2000); trpa1b encodes a transient receptor potential (TRP) channel that is activated by chemical irritants (Prober et al., 2008); p2rx3b encodes an ATP-gated ion channel in non-peptidergic nociceptors (Kucenas et al., 2006); and islet1 (isl1) encodes a LIM homeobox transcription factor that is expressed in sensory neurons and motoneurons (Higashijima et al., 2000). We used an isl1 enhancer that drives expression in a subset of sensory neurons (Sagasti et al., 2005). The Tg(trpa1b:EGFP) and Tg(isl1:Gal4VP16, 14xUAS:EGFP) lines express EGFP in largely non-overlapping subsets of trigeminal and Rohon-Beard sensory neurons (Pan et al., 2012). p2rx3b is expressed in all cells labeled in Tg(trpa1b:EGFP) embryos and in a quarter of cells labeled in Tg(isl1:Gal4VP16, 14xUAS:EGFP) embryos.
These samples allowed five pairwise comparisons of Hcrt neurons with different purified neuron populations (supplementary material Fig. S2 and Table S1). This analysis provided greater statistical power than previous studies that compared Hcrt neurons with a single outgroup (Cvetkovic-Lopes et al., 2010; Dalal et al., 2013). An additional study examined changes in gene expression across multiple brain regions after the onset of narcolepsy (Honda et al., 2009); however, this approach is unlikely to identify developmentally relevant transcripts. We focused on 19 highly ranked genes that encode transcription factors or secreted proteins (Tables 1 and 2), as both classes of proteins have well-established roles in neural development (Blackshaw et al., 2010; Wilson and Houart, 2004); as expected, hcrt was the most highly enriched gene in Hcrt neurons (Table 2). The complete microarray dataset is available through ArrayExpress, accession number E-MTAB-3317.
Table 1.
Transcription factors enriched in Hcrt neurons
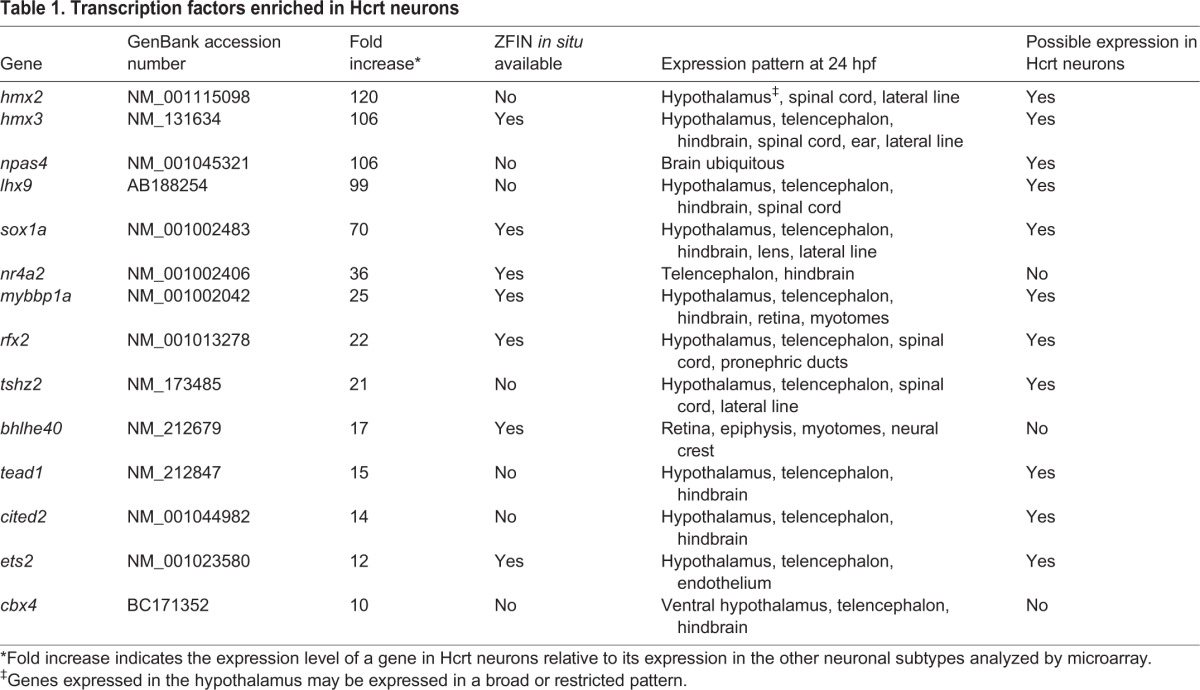
Table 2.
Secreted proteins enriched in Hcrt neurons
Expression patterns of candidate genes validate the microarray results
High quality in situ hybridization (ISH) images were available in the ZFIN database for seven candidate genes (supplementary material Fig. S3). We determined the expression patterns of the remaining 12 genes using ISH on 24 hpf embryos (Fig. 2; supplementary material Fig. S3 and Movie 1). We found that 11 of 14 genes encoding transcription factors and five of five genes encoding secreted proteins are expressed in a similar or overlapping domain to Hcrt neurons. Some genes, such as the transcription factor lhx9, are expressed in all Hcrt cells throughout early development (supplementary material Fig. S4). The microarray therefore accurately reported the expression of most candidate genes.
Fig. 2.
Examples of expression patterns of Hcrt neuron-enriched genes. (A,C,E) ISH performed on 24 hpf zebrafish embryos using probes specific for hcrt, hmx2 and lhx9. Dashed boxes indicate the hypothalamic regions shown in B,D,F. (B,D,F) A 1.25-μm confocal section of fluorescent ISH (red) and hcrt:EGFP immunofluorescence (green). All hcrt:EGFP neurons express hmx2 and lhx9. hmx2 expression extends ventrally (C,D) and lhx9 is expressed in broader domains of the telencephalon and diencephalon (E,F), as well as the hindbrain and spinal cord (not shown). Supplementary material Movie 1 contains the complete confocal image stack for F. (G) Mean±s.e.m. number of cells in the boxed regions in A,C,E. n, number of brains. Scale bars: 10 μm.
Lhx9 is sufficient to specify Hcrt neurons
To determine whether any candidate gene is sufficient to induce specification of Hcrt neurons, we cloned each gene downstream of a heat shock-inducible promoter in a vector containing Tol2 transposase sites, and injected the plasmid together with tol2 transposase mRNA into zebrafish embryos at the 1-cell stage. As a result, the plasmid inserts into the genome of a random subset of cells. We transiently overexpressed each gene by performing a heat shock at 24 hpf (Fig. 3A). This approach provides an efficient, inducible method for overexpressing different candidate genes in 10-20% of cells (Fig. 3C).
Fig. 3.
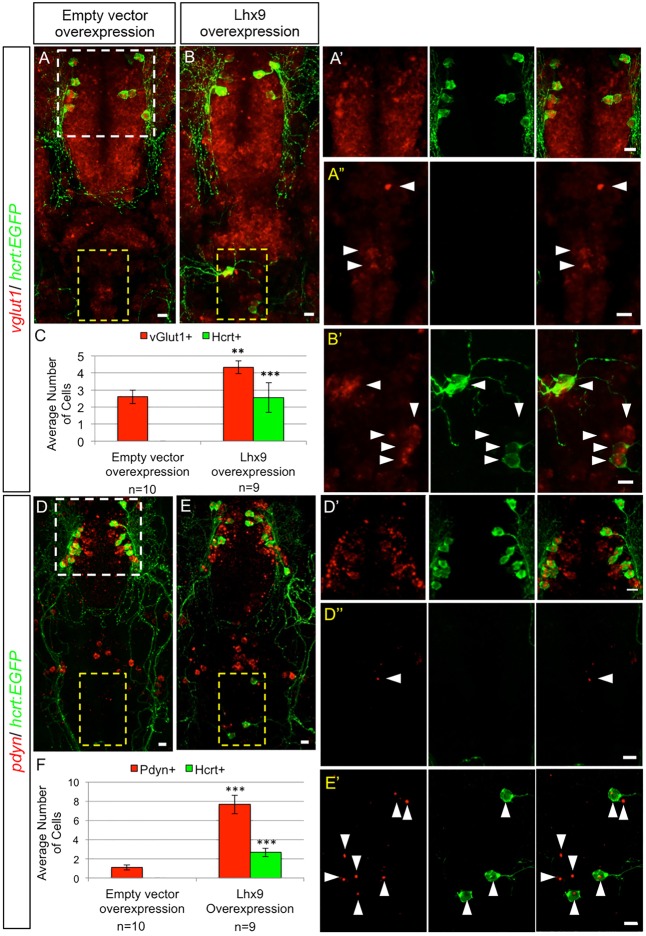
Transient lhx9 overexpression induces ectopic Hcrt neurons. (A) Zebrafish embryos were injected with tol2 transposase mRNA and a plasmid in which a heat shock-inducible promoter regulates the expression of a candidate gene (hs:gene x). (B,C) Anterior views of 26 hpf embryos after ISH showing endogenous hcrt expression (B) and mosaic expression of gene x 1 h after heat shock (C). Approximately 10-20% of cells overexpress gene x. White box in B,C indicates the hypothalamus. (D) Control embryos injected with an hs:EGFP plasmid exhibit normal hcrt expression at 120 hpf. (E) Embryos overexpressing lhx9 contain ectopic Hcrt cells that are dorsal and caudal relative to endogenous Hcrt neurons. (F) Ectopic Hcrt cells exhibit neuronal morphology, as visualized using Tg(hcrt:EGFP) larvae. White and yellow boxes in D-F indicate endogenous and ectopic Hcrt cells, respectively.
Quantification of hypothalamic Hcrt neurons using ISH at 120 hpf showed no significant differences between larvae overexpressing candidate genes and controls (supplementary material Fig. S5). However, 14% of larvae overexpressing lhx9 contained additional, ectopic Hcrt expression in the medial hindbrain (7/50 larvae; Fig. 3E,F). These larvae had 2.7 ectopic Hcrt cells on average, a 17% increase in the total number of Hcrt neurons. No other candidate gene was sufficient to specify ectopic Hcrt neurons. Like endogenous hypothalamic Hcrt neurons, the ectopic Hcrt neurons expressed vesicular glutamate transporter 1 (vglut1; slc17a7a) and prodynorphin (pdyn) (Fig. 4). vglut1 is widely expressed in the hypothalamus and hindbrain, but its expression is particularly strong in Hcrt neurons (Fig. 4A,B; supplementary material Movie 2). Hcrt neurons also exhibit intense, punctate pdyn labeling (Fig. 4D,E; supplementary material Movie 3) that is likely to indicate sites of transcription (Hanisch et al., 2012; Kosman et al., 2004). We did not detect significant expression of vglut2a (slc17a6b) or vglut2b (slc17a6a) in Hcrt neurons (supplementary material Figs S8 and S9).
Fig. 4.
Hypothalamic and ectopic Hcrt neurons share biomarkers. Confocal projections of 120 hpf Tg(hcrt:EGFP) zebrafish larval brains containing endogenous and ectopic Hcrt neurons labeled with an anti-EGFP antibody and fluorescent ISH probes specific for vglut1 (A,B) and pdyn (D,E). White and yellow boxes in A,D indicate endogenous Hcrt neurons in the hypothalamus and endogenous vglut1 and pdyn expression in the hindbrain, respectively, shown enlarged in A′,D′ and in A″,D″. Yellow boxes in B,E indicate ectopic Hcrt neurons, as enlarged in B′,E′. All hypothalamic and ectopic Hcrt neurons express vglut1 and pdyn. Larvae injected with hs:lhx9 and heat-shocked at 24 hpf contain ectopic Hcrt neurons in the hindbrain and more cells with strong vglut1 (B,B′) and punctate pdyn (E,E′) expression, compared with controls injected with an empty heat shock vector (A,A″,D,D″). Arrowheads indicate cells with strong vglut1 or pdyn expression. (C,F) Mean±s.e.m. number of cells in the yellow boxed regions. n, number of brains quantified. **P<0.01, ***P<0.001 compared with empty heat shock vector (Student's t-test). Scale bars: 10 μm.
lhx9 overexpression generated more cells in the medial hindbrain with strong vglut1 expression or punctate pdyn expression than ectopic Hcrt cells (Fig. 4C,F), suggesting that Lhx9 can specify multiple cell types. Indeed, most lhx9-overexpressing neurons also expressed qrfp at 1 h post-heat shock (data not shown), and we observed an average of three ectopic QRFP neurons in the medial hindbrain 96 h after Lhx9 overexpression (8/40 larvae; supplementary material Fig. S6; note that lhx9 was enriched in hcrt neurons compared with elavl3, isl1, trpa1b and p2rx3b neurons, but not compared with qrfp neurons, in the microarray analysis). Similar to Hcrt neurons, both hypothalamic and ectopic QRFP neurons express Lhx9 (supplementary material Figs S6 and S7). Ectopic QRFP neurons did not co-express hcrt (0/24 QRFP neurons), suggesting that they remain developmentally distinct populations.
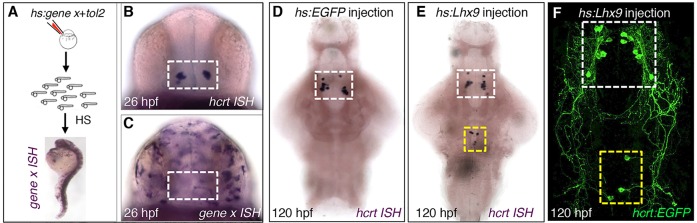
Both mammalian and zebrafish Hcrt neurons project to several brain regions, including the noradrenergic locus coeruleus (LC) (Horvath et al., 1999; Prober et al., 2006). To determine whether ectopic Hcrt neurons project to this endogenous Hcrt neuron target, we overexpressed lhx9 in transgenic zebrafish that expressed the photoconvertible fluorescent protein Kaede in Hcrt neurons and EGFP in dopamine beta-hydroxylase (dbh)-expressing LC neurons (supplementary material Fig. S1). We used a 405 nm laser to photoconvert Hcrt neurons in the hindbrain, but not in the hypothalamus, from green to red fluorescence. We observed that all red fluorescent ectopic Hcrt neurons project to the LC (15/15 neurons) (Fig. 5A,C). By contrast, stochastic labeling of neurons in the medial hindbrain with an elavl3:Kaede transgene indicates that only ∼20% of cells in this region target the LC (9/50 neurons) (Fig. 5B,C). These experiments indicate that Lhx9 is sufficient to specify Hcrt neurons in vivo that are genetically and anatomically similar to endogenous Hcrt neurons.
Fig. 5.
Ectopic Hcrt neurons project to the locus coeruleus. (A) Confocal projection of a 120 hpf Tg(hcrt:Kaede, dbh:EGFP) larva injected with an hs:lhx9 plasmid shows that ectopic photoconverted Hcrt cells in the medial hindbrain (red) project to the locus coeruleus (LC). (B) Confocal projection of a 120 hpf Tg(dbh:EGFP) larva injected with an elavl3:Kaede plasmid to stochastically label neurons shows that some neurons in the medial hindbrain project to the LC (arrowhead) whereas others do not (arrow; axon projects orthogonally to the image). (C) All ectopic Hcrt neurons project to the LC, but only 20% of elavl3:Kaede-labeled cells in the same brain region project to the LC. n, number of neurons analyzed. ***P<0.001 compared with elavl3:Kaede (Fisher's exact test). Yellow boxes in A,B indicate the regions used for quantification. (D) Schematic of hcrt, pdyn and vglut1 expression in the ventral zebrafish brain at 120 hpf. The locations of ectopic Hcrt neurons, dbh-expressing LC neurons and the photoconverted region are shown. Scale bars: 10 μm.
Lhx9 is necessary for Hcrt neuron specification
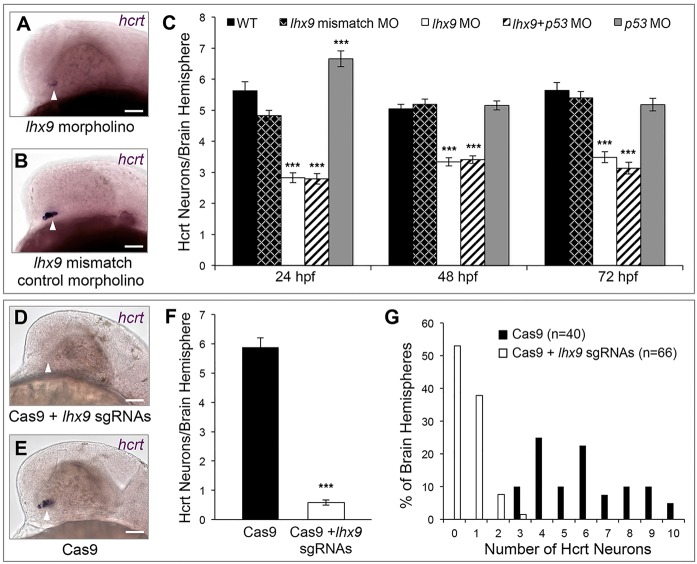
To test whether Lhx9 is necessary for Hcrt neuron specification, we used a splice-blocking morpholino to knock down lhx9 expression. RT-PCR revealed robust inhibition of lhx9 mRNA splicing, and thus of functional Lhx9 protein production, up to 72 hpf (supplementary material Fig. S10). Morphants had ∼40% fewer Hcrt neurons when assessed by ISH (Fig. 6C), and the remaining Hcrt neurons had reduced hcrt expression compared with embryos injected with a 5 bp mismatch control morpholino (Fig. 6A,B). qRT-PCR revealed that lhx9 morphants express 62% less hcrt transcript than control morphants (s.e.m.=5%, n=3 replicates), confirming that Hcrt neurons that persist in lhx9 morphants contain less hcrt transcript than controls.
Fig. 6.
Lhx9 is required for Hcrt neuron specification. (A-C) hcrt ISH at 24 hpf shows that morpholino-mediated knockdown of lhx9 reduces the number of Hcrt neurons and the level of hcrt expression (A) compared with embryos injected with a control morpholino (B). (C) Quantification of Hcrt neurons per brain hemisphere at 24, 48 and 72 hpf revealed that lhx9 morphants have ∼40% fewer Hcrt neurons. Co-injecting a p53 morpholino did not affect this phenotype. Mean±s.e.m. is shown. At least 22 embryos were quantified for each condition. ***P<0.001 compared with control morpholino (one-way ANOVA followed by Bonferroni's correction for multiple comparisons). (D-G) hcrt ISH at 24 hpf shows that co-injection of Cas9 protein and ten lhx9 sgRNAs eliminates hcrt expression (D) compared with embryos injected with Cas9 alone (E). (F) Quantification of Hcrt neurons per brain hemisphere at 24 hpf. Mean±s.e.m. is shown. ***P<0.001 compared with Cas9 alone (one-way ANOVA). (G) The percentage of brain hemispheres containing the indicated number of Hcrt neurons. n, number of brain hemispheres analyzed in F,G. Arrowheads indicate endogenous Hcrt neuron region. Scale bars: 50 μm.
To determine whether the missing Hcrt neurons in lhx9 morphants lack hcrt expression or were absent, we examined the effect of lhx9 knockdown on vglut1 and pdyn (supplementary material Fig. S11). lhx9 morphants had no gross defects in vglut1 expression compared with controls. However, the number of cells with intense punctate pdyn expression, which includes all Hcrt cells, was reduced by ∼40%. The loss of Hcrt cells was not caused by nonspecific morpholino-induced apoptosis, as embryos co-injected with an apoptosis-suppressing p53 (tp53) morpholino (Robu et al., 2007) and the lhx9 morpholino showed the same phenotype (Fig. 6C). Furthermore, lhx9 morpholino-injected embryos stained with Acridine Orange, which labels apoptotic cells, showed no increase in apoptosis (supplementary material Fig. S12A-E). The morpholino phenotype was partially rescued by co-injecting the lhx9 morpholino with the hs:lhx9 plasmid and performing a heat shock at 24 hpf (supplementary material Fig. S12F,G), indicating that the reduction in Hcrt cells was a specific effect of lhx9 knockdown. hcrt expression was weaker in rescued cells than in endogenous Hcrt neurons, presumably because rescued cells only received a pulse of lhx9 whereas endogenous Hcrt neurons continuously express lhx9 (supplementary material Fig. S4). We also tested morpholinos against hmx2 or hmx3, which were more highly enriched in Hcrt neurons than lhx9 in our microarray analysis. Although RT-PCR confirmed that these morpholinos were effective, they had no effect on Hcrt neuron specification (data not shown).
Some Hcrt neurons persisted in lhx9 morphants, possibly owing to incomplete lhx9 knockdown (supplementary material Fig. S10) or because Lhx9 is only necessary to specify a subset of Hcrt neurons, as in mice (Dalal et al., 2013). To distinguish between these possibilities, we used the CRISPR/Cas9 system (Jao et al., 2013; Hwang et al., 2013) to introduce mutations into lhx9. We co-injected Cas9 protein (Gagnon et al., 2014) with a set of short guide RNAs (sgRNAs) that target lhx9 into embryos at the 1-cell stage to generate biallelic mutations and a loss-of-function phenotype in the injected animals (Jao et al., 2013). Embryos injected with Cas9+lhx9 sgRNAs had 90% fewer Hcrt cells than embryos injected with Cas9 alone (Fig. 6D-F). Furthermore, over half of brain hemispheres of embryos injected with Cas9+lhx9 sgRNAs completely lacked Hcrt cells, whereas embryos injected with Cas9 alone had at least three Hcrt cells in each brain hemisphere (Fig. 6G). This phenotype is unlikely to be due to off-target effects of particular sgRNAs, as we observed a similar, albeit weaker, phenotype in embryos injected with Cas9 and independent subsets of lhx9 sgRNAs (supplementary material Fig. S13). We conclude that lhx9 is necessary to specify all Hcrt neurons in zebrafish embryos.
Because lhx9 overexpression was sufficient to specify QRFP neurons, we asked whether lhx9 is also necessary for QRFP neuron specification. We observed 63% fewer QRFP neurons in embryos injected with Cas9+lhx9 sgRNAs compared with Cas9 alone, and over 20% of brain hemispheres lacked QRFP neurons (supplementary material Fig. S13). Injection of embryos with Cas9 and subsets of lhx9 sgRNAs produced a similar, albeit weaker, phenotype. We conclude that lhx9 is necessary to specify all QRFP neurons.
Lhx9 directly promotes hcrt expression
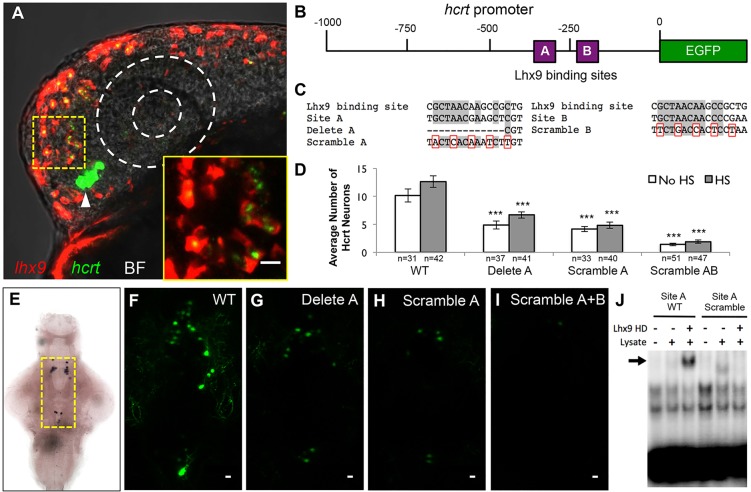
As Lhx9 is a transcription factor, we hypothesized that it might promote hcrt expression directly. A previous study tested this hypothesis in vitro by co-expressing Lhx9 and a mouse Hcrt promoter-luciferase reporter in a neuroblastoma cell line, and in vivo by lentiviral transduction of Lhx9 into the hypothalamus of adult mice, but observed no effect on Hcrt expression (Dalal et al., 2013). However, when we performed double-fluorescent ISH against lhx9 and hcrt on zebrafish embryos fixed 1 h after heat shock, we observed hcrt expression in almost all lhx9-overexpressing cells (Fig. 7A; supplementary material Fig. S14 and Movie 4). The number of ectopic lhx9- and hcrt-expressing cells is reduced at 8 h after heat shock (supplementary material Fig. S14D), and few ectopic cells are observed 24 h after heat shock (supplementary material Fig. S14F), similar to the number observed at 120 hpf (supplementary material Fig. S14H). Importantly, ectopic hcrt-expressing neurons also express ectopic lhx9 at all time points. These findings support the hypothesis that Lhx9 can directly regulate hcrt expression.
Fig. 7.
Lhx9 can directly induce hcrt expression. (A) Ectopic lhx9-expressing neurons also express hcrt in embryos injected with an hs:lhx9 plasmid, fixed 1 h after heat shock at 24 hpf, and analyzed using double fluorescent ISH with hcrt-specific and lhx9-specific probes. Arrowhead indicates endogenous Hcrt neurons. A single 1.5 μm confocal section is shown. Supplementary material Movie 4 contains the complete confocal image stack. The bright field (BF) overlay and dashed white circles show the position of the eye. The boxed region is shown at higher magnification in the inset. (B) Schematic of the zebrafish 1 kb hcrt promoter, including putative Lhx9 binding sites A and B. (C) Sequence of a previously characterized mammalian Lhx9 binding site compared with sites A and B in the zebrafish hcrt promoter and in mutated constructs. Gray shading, dashes and red boxes indicate conserved nucleotides, deleted nucleotides and mutated nucleotides, respectively. (D-I) Embryos were injected with a plasmid containing both hcrt:EGFP and hs:lhx9, and some injected embryos were heat-shocked at 24 hpf. Deletion or scrambling of putative Lhx9 binding sites reduced the number (D,F-I) and intensity (F-I) of EGFP-expressing cells. Cell counts indicate Hcrt cells per brain with (endogenous and ectopic Hcrt cells) and without (endogenous Hcrt cells only) heat shock. Mean±s.e.m. is shown. n, number of brains analyzed. ***P<0.001 compared with the wild-type promoter (one-way ANOVA followed by Bonferroni's correction for multiple comparisons). (E) Yellow box indicates area shown in F-I. (J) EMSA showing that the Lhx9 homeodomain (HD) binds to the wild-type (WT) site A (arrow), but not the scrambled site A, in vitro. Scale bars: 10 μm.
A previous study identified an Lhx9 binding site upstream of the wilms tumor 1 gene (Wilhelm and Englert, 2002), and we observed two similar sites in the zebrafish hcrt promoter (sites A and B) (Fig. 7B,C). Notably, site A corresponds to a region previously shown to be important for hcrt expression in zebrafish (Faraco et al., 2006). We tested whether these sites are necessary for endogenous (i.e. hypothalamic) and Lhx9-induced ectopic hcrt expression by injecting wild-type embryos with plasmids containing the hcrt promoter, in which one or both putative Lhx9 binding sites were mutated, placed upstream of EGFP. Each plasmid also contained a heat shock-inducible lhx9 transgene downstream of the EGFP reporter. Thus, any cell that contains the plasmid will have both the hcrt:EGFP reporter and the hs:lhx9 transgene. Injected embryos were heat-shocked at 24 hpf and analyzed at 120 hpf for hypothalamic and ectopic hcrt:EGFP-expressing neurons (Fig. 7D, HS samples). Some injected embryos were not heat-shocked (Fig. 7D, no HS samples) to determine whether the putative Lhx9 binding sites are required for EGFP expression in the endogenous Hcrt domain alone. Mutating site A, either by deletion or by scrambling every third nucleotide, reduced the number and intensity of endogenous and ectopic cells labeled with EGFP as compared with the wild-type hcrt promoter (Fig. 7C,D,F-H). Scrambling the sequences of both sites A and B virtually abolished EGFP expression (Fig. 7D,I). Notably, ectopic hcrt cells were never observed for the double-mutant reporter. These experiments indicate that sites A and B are crucial for hcrt expression in vivo.
To test whether Lhx9 can interact with these sites, we performed an EMSA using the zebrafish Lhx9 homeodomain (Lhx9 HD) and radiolabeled oligonucleotides that include the wild-type or scrambled sequences for sites A and B. We found that Lhx9 HD binds to the wild-type site A probe, but not to the scrambled site A probe (Fig. 7J). We failed to observe an interaction between Lhx9 HD and the site B probe (data not shown), possibly owing to non-optimal in vitro binding or electrophoresis conditions. This result indicates that Lhx9 can bind to site A in vitro and suggests that Lhx9 directly regulates hcrt expression in vivo via binding to this site.
Lhx9 overexpression in mouse embryos induces Hcrt neuron specification
A previous study that overexpressed Lhx9 in the hypothalamus of adult mice observed no effect on Hcrt neuron specification (Dalal et al., 2013). To determine whether Lhx9 can promote Hcrt neuron specification earlier in mammalian development, we used micro in utero electroporation (Matsui et al., 2011) to focally overexpress EYFP and the murine Lhx9 ortholog, or EYFP alone, in the developing murine diencephalon at embryonic day (E) 10.5 and assayed Hcrt expression at postnatal day (P) 6. Embryos overexpressing Lhx9 had significantly more Hcrt-expressing neurons in the lateral hypothalamus than embryos overexpressing EYFP alone (Fig. 8; supplementary material Figs S15 and S16). This effect appears to be specific, since Lhx9 overexpression had no effect on the expression of other hypothalamic markers, including Cartpt, Foxp2 and Gal (data not shown). Lhx9 overexpression in the subthalamic zona incerta (supplementary material Fig. S17) or cerebral cortex (data not shown) did not induce ectopic Hcrt expression. We conclude that Lhx9 is capable of promoting Hcrt neuron specification exclusively in its endogenous domain during mouse embryogenesis.
Fig. 8.
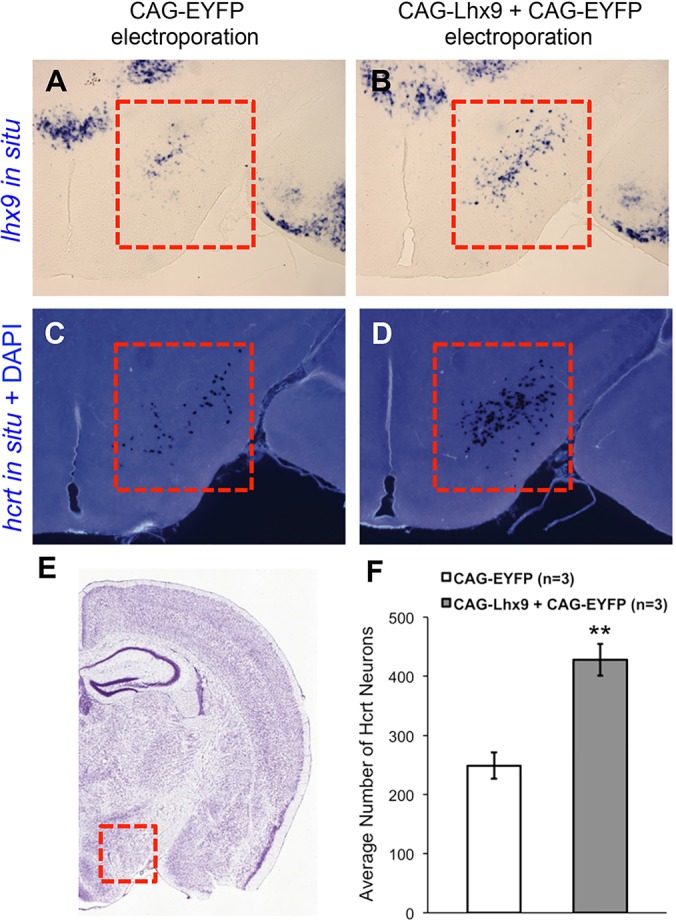
Lhx9 overexpression in mouse embryos promotes Hcrt neuron specification. (A-D) E10.5 mouse embryos were electroporated in utero into the right brain hemisphere lateral hypothalamus with either CAG-EYFP or with CAG-Lhx9+CAG-EYFP and analyzed at P6 for expression of Lhx9 and Hcrt by ISH in serial coronal sections. Mice electroporated with CAG-Lhx9+CAG-EYFP have increased Lhx9 expression (B) and more Hcrt cells (D) in the lateral hypothalamus (red boxes) than controls electroporated with CAG-EYFP alone (A,C). Coronal sections (A-D) are approximately Bregma −1.6 mm. (E) The entire right hemisphere of a comparable Nissl-stained section is shown for reference, with the red box indicating the lateral hypothalamus. Image adapted from the Allen Mouse Brain Atlas (Lein et al., 2006). (F) Quantification of Hcrt cell number in the lateral hypothalamus of the right brain hemisphere following electroporation with CAG-Lhx9+CAG-EYFP compared with CAG-EYFP alone. Mean±s.e.m. is shown for three experimental and three control brain hemispheres (see supplementary material Fig. S15 for quantification details). **P<0.01 (Student's t-test).
DISCUSSION
Using microarray gene expression analysis and high-throughput gene overexpression assays in zebrafish, we found that the LIM homeobox transcription factor Lhx9 is both necessary and sufficient to specify Hcrt neurons in zebrafish and is sufficient to specify Hcrt neurons in mouse embryos. We found that Lhx9 is also necessary and sufficient to specify QRFP neurons in zebrafish, which are located adjacent to Hcrt neurons in the hypothalamus. To our knowledge, this is the first study to identify a factor that is capable of inducing the specification of these neurons, or of any terminal neural subtype in the lateral hypothalamus.
lhx9 was identified as enriched in Hcrt neurons by both our analysis of zebrafish embryos and by a previous study that used adult mice (Dalal et al., 2013). We analyzed zebrafish neurons just after the initiation of hcrt expression, enabling us to screen for transcripts that are likely to play a role in the specification of Hcrt neurons. We isolated purified cell populations by FACS, analyzed their gene expression patterns by microarray, and performed multiple pairwise comparisons of purified Hcrt neurons with a closely related cell type (QRFP-expressing neuron) and to more distantly related neurons (pan-neuronal or sensory neurons). By contrast, Dalal and colleagues used a translational profiling approach in which a tagged ribosomal subunit is expressed in Hcrt neurons. Biochemical purification of this subunit from a whole brain homogenate isolates transcripts that are actively translated in Hcrt neurons. Although this approach allowed profiling of the rare Hcrt cell population, its statistical power was diminished by contamination with nonspecific transcripts, as indicated by the presence of glial transcripts. In addition to lhx9, the 112 genes most enriched in Hcrt neurons in our study and the 188 most enriched genes identified by Dalal and colleagues include the definitive Hcrt neuron markers hcrt and pdyn, the transcription factors hmx2 and rfx4, and scg2, agrp, glipr1 and fam46a. The absence of more overlapping genes can be attributed in part to ambiguity in the gene assignment of microarray probes and the imperfect annotation of the zebrafish genome. Furthermore, the stringent criteria for significance used by both studies are likely to underestimate the true complement of Hcrt enriched genes shared between zebrafish embryos and adult mice.
Lhx9 belongs to the LIM homeobox family of transcription factors, which is conserved from invertebrates to mammals. These proteins have essential roles in tissue patterning and differentiation, particularly in the brain (Hobert and Westphal, 2000). In mice, several LIM homeobox proteins are expressed dynamically to demarcate regions of the developing hypothalamus (Shimogori et al., 2010). However, the developmental roles of specific LIM homeobox genes have been difficult to distinguish; loss-of-function phenotypes are subtle and similar LIM homeobox family members often exhibit redundancy. For instance, double knockdown of lhx9 and lhx2 dramatically altered thalamus and forebrain patterning in zebrafish, but knockdown of either gene alone had no gross effects (Peukert et al., 2011). Similarly, Lhx9 knockout mice survive to adulthood without gross brain defects (Birk et al., 2000).
Although we tested morpholinos against several candidate genes, only the lhx9 morpholino decreased the number of Hcrt neurons, with an average decrease of 40%. The remaining Hcrt neurons, which expressed hcrt at reduced levels, are likely to result from incomplete lhx9 knockdown. Indeed, co-injection of Cas9 protein with sgRNAs targeting lhx9 completely abolished hcrt expression in over half of brain hemispheres analyzed, indicating that lhx9 is required for the specification of all Hcrt neurons. This result contrasts with Lhx9 knockout mice, in which Hcrt neurons are only reduced by 39% (Dalal et al., 2013). This discrepancy is likely to be due to the expression of lhx9 in all Hcrt neurons in zebrafish but in only a subset in mice (Shimogori et al., 2010).
In zebrafish, lhx9 overexpression at 24 hpf was sufficient to produce ectopic Hcrt neurons in the medial hindbrain, but the number of Hcrt neurons in the hypothalamus remained unchanged. We characterized these ectopic Hcrt neurons at 120 hpf, when the Hcrt neuronal circuit is functional (Prober et al., 2006; Elbaz et al., 2012), and confirmed that all zebrafish Hcrt neurons in the hypothalamus and hindbrain express pdyn and vglut1, two markers of mammalian Hcrt neurons (Chou et al., 2001; Rosin et al., 2003). Unlike previous studies (Appelbaum et al., 2009; Rosin et al., 2003), we did not observe expression of vglut2a or vglut2b in hypothalamic or hindbrain Hcrt neurons, indicating that Hcrt neurons in larval zebrafish express different VGLUT family genes than adult zebrafish or rats. We also observed that all ectopic Hcrt neurons project to the LC, a target of Hcrt neurons in zebrafish and mammals (Horvath et al., 1999; Prober et al., 2006). Thus, despite their location in the hindbrain, Lhx9-induced ectopic Hcrt neurons express the same genetic markers and project to the same target as endogenous Hcrt neurons.
Although it might be surprising that we did not detect an effect of lhx9 overexpression on the number of hypothalamic Hcrt neurons, there are several possible explanations for this result. First, because lhx9 induces few ectopic Hcrt neurons in our assay and the number of endogenous Hcrt neurons is variable, it is possible that lhx9 induced additional Hcrt neurons in the hypothalamus but the difference was not large enough to detect. Second, endogenous lhx9 is likely to act in concert with other factors to specify Hcrt neurons because only a subset of endogenous lhx9-expressing cells expresses hcrt. If these co-factors are only present in hypothalamic neurons that express endogenous hcrt, lhx9 overexpression will have no effect on the number of Hcrt neurons in the hypothalamus. Other genes identified by our microarray analysis might encode these co-factors. However, overexpressing two or more candidate genes using our assay was not feasible due to DNA toxicity. Reducing the concentration of each injected plasmid to offset this toxicity also reduces the extent of gene overexpression. We were thus unable to observe ectopic Hcrt neurons when the hs:lhx9 plasmid was co-injected with a second plasmid, and the number of hypothalamic Hcrt neurons was unaffected (data not shown).
We detected widespread co-expression of lhx9 and hcrt 1 h after heat shock-induced lhx9 overexpression. Because ectopic hcrt expression was observed so soon after heat shock, it seemed likely that Lhx9 was directly inducing hcrt expression. Consistent with this hypothesis, we identified two putative Lhx9 binding sites in the zebrafish hcrt promoter and found that they are necessary for both endogenous and ectopic hcrt expression in vivo. We also found that one binding site can form a complex with the Lhx9 homeodomain in vitro. However, this hypothesis is complicated by the fact that lhx9 is widely expressed in the embryonic zebrafish brain, whereas hcrt is normally expressed exclusively in the hypothalamus. Because the extent of both lhx9 overexpression and of ectopic hcrt expression is dramatically reduced by 24 h after heat shock (supplementary material Fig. S14), we propose that the ability of Lhx9 to drive hcrt expression may depend on Lhx9 levels. High Lhx9 levels might directly induce hcrt expression, whereas endogenous Lhx9 levels require one or more co-factors in the hypothalamus and medial hindbrain to promote and maintain hcrt expression.
Since the expression patterns of hcrt and lhx9 are conserved between zebrafish and mammals (Peukert et al., 2011; Shimogori et al., 2010), we tested whether Lhx9 overexpression could induce Hcrt neuron specification in mice. Indeed, Lhx9 overexpression by in utero electroporation was sufficient to specify additional Hcrt neurons, although they were only observed in the endogenous Hcrt neuron domain. This result suggests that the zone of cells competent to specify Hcrt neurons is more spatially restricted in mice than in zebrafish. Our findings differ from a previous study that saw no change in Hcrt expression after viral transduction of an Lhx9-overexpression construct in the hypothalamus of adult mice (Dalal et al., 2013). These results suggest that Lhx9 can induce Hcrt neuron specification in the embryonic, but not adult, mouse hypothalamus, possibly because cells competent to specify Hcrt neurons are fully differentiated in adults.
Our study demonstrates the utility of zebrafish to identify and test genes that regulate vertebrate development. Furthermore, the ability of Lhx9 to induce Hcrt neuron specification suggests a therapeutic approach to compensate for the loss of Hcrt neurons that is thought to cause narcolepsy. This strategy would use Lhx9 to generate HCRT-expressing neurons from human pluripotent stem cells in vitro, followed by screening and selection of Hcrt neurons to be transplanted into the hypothalamus. The promise of this approach is highlighted by the recent demonstration that narcoleptic-like sleep induced by the lesion of Hcrt neurons in rats is diminished by the transplantation of Hcrt neurons into the lateral hypothalamus (Arias-Carrión and Murillo-Rodríguez, 2014).
MATERIALS AND METHODS
Ethics statement
Zebrafish experiments followed standard protocols (Westerfield, 1993) in accordance with Caltech Institutional Animal Care and Use Committee guidelines. Mouse procedures were approved by the RIKEN Institutional Animal Care Committee.
Transgenic zebrafish
A 1-kb fragment of zebrafish genomic DNA upstream of qrfp was cloned upstream of EGFP using primers 5′-CTGACTCTCCCATCAGTCCT-3′ and 5′-CTGAAATTTAAGGAATAATTTAAAGTTG-3′. A 1-kb zebrafish genomic fragment upstream of hcrt (Faraco et al., 2006) was subcloned upstream of mRFP and Kaede using primers 5′-ATAATAAATAAAT-CTGATGGGGTTTT-3′ and 5′-GAGTTTAGCTTCTGTCCCCTG-3′. The dbh:EGFP transgene was generated by cloning a 1.1-kb fragment upstream of zebrafish dbh using primers 5′-ACTTGAACCAGCGACCTTCT-3′ and 5′-GGTTTGAAGGCCTTTCTAAGTTTTT-3′. Transgenes were co-injected with tol2 transposase mRNA to generate stable transgenic lines. The Tg(hcrt:EGFP), Tg(elavl3:EGFP), Tg(trpa1b:EGFP), Tg(isl1:Gal4VP16, UAS:EGFP), Tg(p2rx3b:EGFP) and Tg(vglut2a:RFP) lines have been described (Prober et al., 2006; Park et al., 2000; Pan et al., 2012; Sagasti et al., 2005; Kucenas et al., 2006; Koyama et al., 2011). Hindbrain neurons were stochastically labeled by injecting a Tg(elavl3:Kaede) transgene (Sato et al., 2006).
Microarray analysis
We analyzed embryos co-expressing qrfp:EGFP and hcrt:mRFP, as well as separate transgenic lines expressing EGFP in all neurons [Tg(elavl3:EGFP)] or in subsets of sensory neurons [Tg(trpa1b:EGFP), Tg(isl1:Gal4VP16, 14xUAS:EGFP), Tg(p2rx3b:EGFP)]. Dechorionated embryos were anesthetized with tricaine at 26 hpf, incubated at room temperature in 0.25% trypsin-EDTA (Life Technologies) and manually dissociated. The cells were passed through a 40 μm strainer, pelleted and resuspended in cell culture medium for FACS (Manoli and Driever, 2012). Cells were incubated with Calcein Blue-AM (Life Technologies) to ensure the sorting of live cells. Sorting gates were set using wild-type embryos. Cells expressing EGFP or mRFP formed clear populations that were visually confirmed. Cells were sorted into a final volume of 100 μl PicoPure XB lysis buffer (Arcturus) and incubated at 42°C for 30 min. At least two independent sorting experiments were performed using each transgenic line. RNA was isolated (Arcturus PicoPure RNA, Life Technologies), amplified (MessageAmp II aRNA, Ambion) and quantified by bioanalyzer. cDNA libraries were prepared in duplicate or triplicate and labeled with Cy3 or Cy5 for hybridization to NimbleGen zv7 microarrays. Fluorophores were switched between replicates to minimize labeling bias. After scanning, data were normalized and pairwise comparisons were performed in R between Hcrt neurons and the five other sorted neuron populations. For each comparison, we generated a list of the most significantly Hcrt-enriched genes by intersecting the subset of probes that were at least fourfold upregulated in Hcrt neurons with the subset of probes with the most statistically significant differential expression using Bayesian statistics. The five pairwise comparisons were then intersected to identify probe sets consistently enriched in Hcrt neurons.
Zebrafish ISH and immunohistochemistry
Samples were fixed in 4% paraformaldehyde for 12-16 h at room temperature. ISH was performed using digoxigenin (DIG)-labeled antisense riboprobes (Thisse and Thisse, 2008). Images were acquired using a Zeiss Axio ImagerM1 microscope. Fluorescent ISH used DIG- and 2,4-dinitrophenol (DNP)-labeled antisense riboprobes with the TSA Plus DNP System (PerkinElmer). Rabbit polyclonal anti-GFP (1:1000; MBL International, #598), rabbit polyclonal anti-orexin-A (Hcrt) (1:1000; Millipore, #AB3704) and goat anti-rabbit Alexa488 (1:500; Invitrogen, #A-11008) antibodies were used. Images were acquired using a Zeiss LSM 780 confocal microscope and analyzed using Fiji (Schindelin et al., 2012).
Candidate gene overexpression
The coding sequence of each candidate gene was amplified from 24 hpf zebrafish cDNA and cloned into pENTR-D-TOPO (Invitrogen). Gateway recombination (Invitrogen) was used to clone each gene downstream of a heat shock-inducible promoter (Halloran et al., 2000), and the entire cassette was flanked by Tol2 transposase sites. We co-injected individual overexpression plasmids with tol2 transposase mRNA into zebrafish embryos at the 1-cell stage. Gene overexpression was induced by incubating embryos in a 37°C water bath for 1 h.
Morpholino-mediated knockdown
Morpholinos (GeneTools) were injected into wild-type embryos at the 1-cell stage. We used a splice-blocking morpholino to knock down Lhx9 (5′-AGCCTCAAAGTTAATGCTTACCTGT-3′). A morpholino with a 5 bp mismatch to the target was injected as a negative control (5′-AGCG-TGAAACTTAATCCTTACCTCT-3′). Potential apoptosis was suppressed by co-injecting a p53 morpholino (5′-GCGCCATTGCTTTGCAAGA-ATTG-3′). To verify knockdown efficacy, we isolated RNA from pools of five injected embryos and used RT-PCR (Superscript III First-Strand Synthesis System, Invitrogen) to amplify a fragment of the lhx9 transcript that spans exon 2. To detect apoptosis, 24-hpf embryos were bathed in 1 μg/ml Acridine Orange for 1 h at room temperature, followed by three 10 min washes with E3 medium. Splice-blocking morpholinos were designed for hmx2 (5′-TGGGAACGTCACTCACCGAGACAGA-3′) and hmx3 (5′-TGCTGCTACAGTAATAGAGGCCAAA-3′) to retain the first intron of each gene, resulting in an early stop codon.
Quantitative reverse-transcription PCR (qRT-PCR)
We isolated total RNA from three biological replicates (25 embryos each) of lhx9 morpholino-injected and control morpholino-injected embryos. TURBO DNase I was used to remove genomic DNA (TURBO DNA-free Kit, Invitrogen). We then generated cDNA (Superscript III First-Strand Synthesis System, Invitrogen) and amplified hcrt transcripts with primers 5′-GAGCATCAAGACTTTTCGATACA-3′ and 5′-ATGAAGACGAGCA-CCTGGAG-3′. Transcripts of the rpl13a reference gene were amplified with primers 5′-TCTGGAGGACTGTAAGAGGTATGC-3′ and 5′-AGA-CGCACAATCTTGAGAGCAG-3′. Each qRT-PCR reaction was run in triplicate on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Relative fold-change in expression was calculated using the 2−ΔΔCt method (Schmittgen and Livak, 2008).
CRISPR/Cas9
Ten target sites within the lhx9 open reading frame were chosen using CHOPCHOP (Montague et al., 2014) (supplementary material Table S2). Cas9 protein was mixed with all ten sgRNAs and injected into embryos at the 1-cell stage (Gagnon et al., 2014). At 24 hpf, deformed embryos were removed and the remainder were fixed in 4% paraformaldehyde for ISH. To control for potential sgRNA off-target effects, we co-injected Cas9 protein with independent subsets of the sgRNAs (subset 1 comprised sgRNAs 1, 3, 5, 7, 9; subset 2 comprised sgRNAs 2, 4, 6, 8, 10) (supplementary material Table S2).
hcrt enhancer constructs
We mutated one or both putative Lhx9 binding sites in the zebrafish hcrt promoter by PCR and Gibson assembly (Gibson et al., 2009). Enhancer fragments were placed upstream of the EGFP coding sequence. An hs:lhx9 cassette was placed downstream of hcrt:EGFP in a vector containing Tol2 transposase sites.
Electrophoretic mobility shift assay (EMSA)
The following oligonucleotide probes (5′-3′) were used: site A wild-type probe, GTTGGTATTTGCTAACGAAGCTCGTCCTCCTGTCCA; site A scramble probe, GTTGGTATTTACTCACAAATCTTGTCCTCCTGTCCA; site B wild-type probe, TGACAAAGATGCTAACAACCCCGAAAAAT-CCTTTGT; site B scramble probe, TGACAAAGATTCTGACCACTCCT-AAAAATCCTTTGT. Probes were radiolabeled using [γ-32P]ATP and T4 polynucleotide kinase for 1 h at 37°C and column-purified (Illustra Microspin G-50, GE Healthcare). EMSAs were performed using a truncated form of zebrafish Lhx9 (Lhx9 HD) that contains the DNA-binding homeodomain but lacks both LIM domains (amino acids 224-396 of the 396 amino acid protein), as described (Wilhelm and Englert, 2002). Lhx9 HD was synthesized in vitro (TNT SP6 Quick Coupled Transcription/Translation System, Promega). 1 μl normalized radiolabeled oligo and 4.5 μl TNT lysate were added to a final volume of 30 μl binding buffer, containing 100 mM KCl, 1 mM MgCl2, 10 μM ZnSO4, 10 mM Tris pH 7.5, 4% glycerol, 1 mg/ml BSA, 200 ng poly(dIdC) and 0.5 mM DTT. After 1 h at room temperature, DNA-protein complexes were resolved by electrophoresis at 4°C on a 6% polyacrylamide DNA retardation gel (Invitrogen) at 100 V for 90 min in 0.5× TBE buffer. The gel was dried at 60°C for 2 h using a gel dryer (Bio-Rad) and analyzed by phosphorimaging (GE Healthcare).
Mouse experiments
Outbred ICR (CD-1) timed-pregnant mice were obtained from Japan SLC. Midday of the day of vaginal plug discovery was considered embryonic (E) day 0.5. Early postnatal mice were anesthetized with a lethal dose of pentobarbitone (100 mg/kg) and, after three failed attempts to elicit a foot withdrawal reflex, the animals were transcardially perfused with 4% paraformaldehyde in PBS. For ISH, brains were fixed overnight in 30% sucrose/4% paraformaldehyde and sectioned in the coronal plane on a Leica sledge microtome at 28 μm. Sections were mounted on slides and processed for non-radioactive ISH as described (Grove et al., 1998). DNA for riboprobes and electroporation were obtained from FANTOM clones (Carninci et al., 2005). Mouse Lhx9 (GenBank NM_001025565) was subcloned into a CAG vector as described (Onishi et al., 2010) to generate an overexpression plasmid. In utero electroporation was performed as described (Matsui et al., 2011).
Supplementary Material
Acknowledgements
We thank Farhad Imam for assistance with microarray data analysis, Anthony Kirilushna for EMSA assistance and Daniel Lee for comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
D.A.P. and A.F.S. conceived and directed the project; J.L., F.T.M., J.A.G., T.S. and D.A.P. performed experiments; A.V.G. and C.N.C. generated and maintained reagents; I.G.W. provided microarray data; J.L., F.T.M., T.S., A.F.S. and D.A.P. wrote the paper.
Funding
This work was supported by grants from the National Institutes of Health [F31NS77844 to J.L.; 1R21NS071598, 5K99NS083713 to F.T.M.; R01HL109525, R21NS071598 to A.F.S.; R00NS060996, R01NS070911, R01DA031367 to D.A.P.], the American Cancer Society [to J.A.G. and I.G.W.], the Jane Coffin Childs Memorial Fund for Medical Research [to F.T.M.], the RIKEN Brain Institute [to T.S.], the Mallinckrodt Foundation, the Rita Allen Foundation, and the Brain and Behavior Research Foundation [to D.A.P.]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.117424/-/DC1
References
- Appelbaum L., Wang G. X., Maro G. S., Mori R., Tovin A., Marin W., Yokogawa T., Kawakami K., Smith S. J., Gothilf Y. et al. (2009). Sleep-wake regulation and hypocretin-melatonin interaction in zebrafish. Proc. Natl. Acad. Sci. USA 106, 21942-21947 10.1073/pnas.906637106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias-Carrión O. and Murillo-Rodríguez E. (2014). Effects of hypocretin/orexin cell transplantation on narcoleptic-like sleep behavior in rats. PLoS ONE 9, e95342 10.1371/journal.pone.0095342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birk O. S., Casiano D. E., Wassif C. A., Cogliati T., Zhao L., Zhao Y., Grinberg A., Huang S., Kreidberg J. A., Parker K. L. et al. (2000). The LIM homeobox gene Lhx9 is essential for mouse gonad formation. Nature 403, 909-913 10.1038/35002622 [DOI] [PubMed] [Google Scholar]
- Blackshaw S., Scholpp S., Placzek M., Ingraham H., Simerly R. and Shimogori T. (2010). Molecular pathways controlling development of thalamus and hypothalamus: from neural specification to circuit formation. J. Neurosci. 30, 14925-14930 10.1523/JNEUROSCI.4499-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnavion P. and de Lecea L. (2010). Hypocretins in the control of sleep and wakefulness. Curr. Neurol. Neurosci. Rep. 10, 174-179 10.1007/s11910-010-0101-y [DOI] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M. C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C. et al. (2005). The transcriptional landscape of the mammalian genome. Science 309, 1559-1563 10.1126/science.1112014 [DOI] [PubMed] [Google Scholar]
- Chartrel N., Dujardin C., Anouar Y., Leprince J., Decker A., Clerens S., Do-Régo J.-C., Vandesande F., Llorens-Cortes C., Costentin J. et al. (2003). Identification of 26RFa, a hypothalamic neuropeptide of the RFamide peptide family with orexigenic activity. Proc. Natl. Acad. Sci. USA 100, 15247-15252 10.1073/pnas.2434676100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C. N. and Prober D. A. (2013). Regulation of zebrafish sleep and arousal states: current and prospective approaches. Front. Neural Circuits 7, 58 10.3389/fncir.2013.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T. C., Lee C. E., Lu J., Elmquist J. K., Hara J., Willie J. T., Beuckmann C. T., Chemelli R. M., Sakurai T., Yanagisawa M. et al. (2001). Orexin (hypocretin) neurons contain dynorphin. J. Neurosci. 21, RC168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovic-Lopes V., Bayer L., Dorsaz S., Maret S., Pradervand S., Dauvilliers Y., Lecendreux M., Lammers G.-J., Donjacour C. E. H. M., Du Pasquier R. A. et al. (2010). Elevated Tribbles homolog 2-specific antibody levels in narcolepsy patients. J. Clin. Invest. 120, 713-719 10.1172/JCI41366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal J., Roh J. H., Maloney S. E., Akuffo A., Shah S., Yuan H., Wamsley B., Jones W. B., Strong C. d. G., Gray P. A. et al. (2013). Translational profiling of hypocretin neurons identifies candidate molecules for sleep regulation. Genes Dev. 27, 565-578 10.1101/gad.207654.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvilliers Y., Arnulf I. and Mignot E. (2007). Narcolepsy with cataplexy. Lancet 369, 499-511 10.1016/S0140-6736(07)60237-2 [DOI] [PubMed] [Google Scholar]
- Elbaz I., Yelin-Bekerman L., Nicenboim J., Vatine G. and Appelbaum L. (2012). Genetic ablation of hypocretin neurons alters behavioral state transitions in zebrafish. J. Neurosci. 32, 12961-12972 10.1523/JNEUROSCI.1284-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz I., Foulkes N. S., Gothilf Y. and Appelbaum L. (2013). Circadian clocks, rhythmic synaptic plasticity and the sleep-wake cycle in zebrafish. Front. Neural Circuits 7, 9 10.3389/fncir.2013.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraco J. H., Appelbaum L., Marin W., Gaus S. E., Mourrain P. and Mignot E. (2006). Regulation of hypocretin (orexin) expression in embryonic zebrafish. J. Biol. Chem. 281, 29753-29761 10.1074/jbc.M605811200 [DOI] [PubMed] [Google Scholar]
- Gagnon J. A., Valen E., Thyme S. B., Huang P., Ahkmetova L., Pauli A., Montague T. G., Zimmerman S., Richter C. and Schier A. F. (2014). Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE 9, e98186 10.1371/journal.pone.0098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A. and Smith H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343-345 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Grove E. A., Tole S., Limon J., Yip L. and Ragsdale C. W. (1998). The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development 125, 2315-2325. [DOI] [PubMed] [Google Scholar]
- Halloran M. C., Sato-Maeda M., Warren J. T., Su F., Lele Z., Krone P. H., Kuwada J. Y. and Shoji W. (2000). Laser-induced gene expression in specific cells of transgenic zebrafish. Development 127, 1953-1960. [DOI] [PubMed] [Google Scholar]
- Hanisch A., Holder M. V., Choorapoikayil S., Gajewski M., Ozbudak E. M. and Lewis J. (2012). The elongation rate of RNA polymerase II in zebrafish and its significance in the somite segmentation clock. Development 140, 444-453 10.1242/dev.077230 [DOI] [PubMed] [Google Scholar]
- Higashijima S., Hotta Y. and Okamoto H. (2000). Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J. Neurosci. 20, 206-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. and Westphal H. (2000). Functions of LIM-homeobox genes. Trends Genet. 16, 75-83 10.1016/S0168-9525(99)01883-1 [DOI] [PubMed] [Google Scholar]
- Honda M., Eriksson K. S., Zhang S., Tanaka S., Lin L., Salehi A., Hesla P. E., Maehlen J., Gaus S. E., Yanagisawa M. et al. (2009). IGFBP3 colocalizes with and regulates hypocretin (orexin). PLoS ONE 4, e4254 10.1371/journal.pone.0004254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath T. L., Peyron C., Diano S., Ivanov A., Aston-Jones G., Kilduff T. S. and van den Pol A. N. (1999). Hypocretin (orexin) activation and synaptic innervation of the locus coeruleus noradrenergic system. J. Comp. Neurol. 415, 145-159 [DOI] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., Sander J. D., Peterson R. T., Yeh J.-R. J. and Joung J. K. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227-229 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao L.-E., Wente S. R. and Chen W. (2013). Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 110, 13904-13909 10.1073/pnas.1308335110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaslin J., Nystedt J. M., Östergård M., Peitsaro N. and Panula P. (2004). The orexin/hypocretin system in zebrafish is connected to the aminergic and cholinergic systems. J. Neurosci. 24, 2678-2689 10.1523/JNEUROSCI.4908-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D., Mizutani C. M., Lemons D., Cox W. G., McGinnis W. and Bier E. (2004). Multiplex detection of RNA expression in Drosophila embryos. Science 305, 846 10.1126/science.1099247 [DOI] [PubMed] [Google Scholar]
- Koyama M., Kinkhabwala A., Satou C., Higashijima S.-i. and Fetcho J. (2011). Mapping a sensory-motor network onto a structural and functional ground plan in the hindbrain. Proc. Natl. Acad. Sci. USA 108, 1170-1175 10.1073/pnas.1012189108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucenas S., Soto F., Cox J. A. and Voigt M. M. (2006). Selective labeling of central and peripheral sensory neurons in the developing zebrafish using P2X3 receptor subunit transgenes. Neuroscience 138, 641-652 10.1016/j.neuroscience.2005.11.058 [DOI] [PubMed] [Google Scholar]
- Lein E. S., Hawrylycz M. J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A. F., Boguski M. S., Brockway K. S., Byrnes E. J. et al. (2006). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168-176 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Machluf Y., Gutnick A. and Levkowitz G. (2011). Development of the zebrafish hypothalamus. Ann. N. Y. Acad. Sci. 1220, 93-105 10.1111/j.1749-6632.2010.05945.x [DOI] [PubMed] [Google Scholar]
- Manoli M. and Driever W. (2012). Fluorescence-activated cell sorting (FACS) of fluorescently tagged cells from zebrafish larvae for RNA isolation. Cold Spring Harb. Protoc. 2012, ppii: pdb.prot069633 10.1101/pdb.prot069633 [DOI] [PubMed] [Google Scholar]
- Matsui A., Yoshida A. C., Kubota M., Ogawa M. and Shimogori T. (2011). Mouse in utero electroporation: controlled spatiotemporal gene transfection. J. Vis. Exp. e3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague T. G., Cruz J. M., Gagnon J. A., Church G. M. and Valen E. (2014). CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, W401-W407 10.1093/nar/gku410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi A., Peng G.-H., Poth E. M., Lee D. A., Chen J., Alexis U., de Melo J., Chen S. and Blackshaw S. (2010). The orphan nuclear hormone receptor ERRβ controls rod photoreceptor survival. Proc. Natl. Acad. Sci. USA 107, 11579-11584 10.1073/pnas.1000102107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y. A., Choy M., Prober D. A. and Schier A. F. (2012). Robo2 determines subtype-specific axonal projections of trigeminal sensory neurons. Development 139, 591-600 10.1242/dev.076588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.-C., Kim C.-H., Bae Y.-K., Yeo S.-Y., Kim S.-H., Hong S.-K., Shin J., Yoo K.-W., Hibi M., Hirano T. et al. (2000). Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev. Biol. 227, 279-293 10.1006/dbio.2000.9898 [DOI] [PubMed] [Google Scholar]
- Peukert D., Weber S., Lumsden A. and Scholpp S. (2011). Lhx2 and Lhx9 determine neuronal differentiation and compartition in the caudal forebrain by regulating Wnt signaling. PLoS Biol. 9, e1001218 10.1371/journal.pbio.1001218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyron C., Faraco J., Rogers W., Ripley B., Overeem S., Charnay Y., Nevsimalova S., Aldrich M., Reynolds D., Albin R. et al. (2000). A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat. Med. 6, 991-997 10.1038/79690 [DOI] [PubMed] [Google Scholar]
- Prober D. A., Rihel J., Onah A. A., Sung R.-J. and Schier A. F. (2006). Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J. Neurosci. 26, 13400-13410 10.1523/JNEUROSCI.4332-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prober D. A., Zimmerman S., Myers B. R., McDermott B. M., Kim S.-H., Caron S., Rihel J., Solnica-Krezel L., Julius D., Hudspeth A. J. et al. (2008). Zebrafish TRPA1 channels are required for chemosensation but not for thermosensation or mechanosensory hair cell function. J. Neurosci. 28, 10102-10110 10.1523/JNEUROSCI.2740-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu M. E., Larson J. D., Nasevicius A., Beiraghi S., Brenner C., Farber S. A. and Ekker S. C. (2007). p53 activation by knockdown technologies. PLoS Genet. 3, e78 10.1371/journal.pgen.0030078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin D. L., Weston M. C., Sevigny C. P., Stornetta R. L. and Guyenet P. G. (2003). Hypothalamic orexin (hypocretin) neurons express vesicular glutamate transporters VGLUT1 or VGLUT2. J. Comp. Neurol. 465, 593-603 10.1002/cne.10860 [DOI] [PubMed] [Google Scholar]
- Sagasti A., Guido M. R., Raible D. W. and Schier A. F. (2005). Repulsive interactions shape the morphologies and functional arrangement of zebrafish peripheral sensory arbors. Curr. Biol. 15, 804-814 10.1016/j.cub.2005.03.048 [DOI] [PubMed] [Google Scholar]
- Sato T., Takahoko M. and Okamoto H. (2006). HuC:Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis 44, 136-142 10.1002/gene.20196 [DOI] [PubMed] [Google Scholar]
- Sawai N., Ueta Y., Nakazato M. and Ozawa H. (2010). Developmental and aging change of orexin-A and -B immunoreactive neurons in the male rat hypothalamus. Neurosci. Lett. 468, 51-55 10.1016/j.neulet.2009.10.061 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D. and Livak K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101-1108 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Shimogori T., Lee D. A., Miranda-Angulo A., Yang Y., Wang H., Jiang L., Yoshida A. C., Kataoka A., Mashiko H., Avetisyan M. et al. (2010). A genomic atlas of mouse hypothalamic development. Nat. Neurosci. 13, 767-775 10.1038/nn.2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayasu S., Sakurai T., Iwasaki S., Teranishi H., Yamanaka A., Williams S. C., Iguchi H., Kawasawa Y. I., Ikeda Y., Sakakibara I. et al. (2006). A neuropeptide ligand of the G protein-coupled receptor GPR103 regulates feeding, behavioral arousal, and blood pressure in mice. Proc. Natl. Acad. Sci. USA 103, 7438-7443 10.1073/pnas.0602371103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessmar-Raible K., Raible F., Christodoulou F., Guy K., Rembold M., Hausen H. and Arendt D. (2007). Conserved sensory-neurosecretory cell types in annelid and fish forebrain: insights into hypothalamus evolution. Cell 129, 1389-1400 10.1016/j.cell.2007.04.041 [DOI] [PubMed] [Google Scholar]
- Thannickal T. C., Moore R. Y., Nienhuis R., Ramanathan L., Gulyani S., Aldrich M., Cornford M. and Siegel J. M. (2000). Reduced number of hypocretin neurons in human narcolepsy. Neuron 27, 469-474 10.1016/S0896-6273(00)00058-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C. and Thisse B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59-69 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- Tsujino N. and Sakurai T. (2009). Orexin/hypocretin: a neuropeptide at the interface of sleep, energy homeostasis, and reward system. Pharmacol. Rev. 61, 162-176 10.1124/pr.109.001321 [DOI] [PubMed] [Google Scholar]
- Westerfield M. (1993). The Zebrafish Book. A Guide for The Laboratory Use of Zebrafish (Danio rerio), 4th edn Eugene, OR: University of Oregon Press. [Google Scholar]
- Wilhelm D. and Englert C. (2002). The Wilms tumor suppressor WT1 regulates early gonad development by activation of Sf1. Genes Dev. 16, 1839-1851 10.1101/gad.220102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S. W. and Houart C. (2004). Early steps in the development of the forebrain. Dev. Cell 6, 167-181 10.1016/S1534-5807(04)00027-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Miyazaki R. and Yamada T. (2009). Intracerebroventricular administration of 26RFa produces an analgesic effect in the rat formalin test. Peptides 30, 1683-1688 10.1016/j.peptides.2009.05.027 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.