Chronic lymphocytic leukemia (CLL) is the most common form of lymphoid malignancy in Western countries1. Recent multi-stage genome-wide association studies (GWAS) have shown that part of the eight-fold increased risk of CLL seen in first-degree relatives of patients can be ascribed to the co-inheritance of multiple low-risk variants.2, 3, 4, 5, 6
Current projections for the number of independent regions harbouring common variants that are associated with CLL suggest that additional risk loci conferring modest effects should be identified by the expansion of discovery GWAS data sets.2
In this study, we have made use of a meta-analysis of GWAS data, followed by validation in multiple independent case–control series, to identify a novel susceptibility locus for CLL at 12q24.13.
The discovery phase comprised two previously described GWAS conducted in the United Kingdom2, 5 (see Supplementary Methods). UK-GWAS-1; 517 CLL cases (155 enriched for genetic susceptibility by virtue of family history) genotyped using Illumina HumanCNV370-Duo BeadChips5 and 2698 controls from the Wellcome Trust Case Control Consortium 2 (WTCCC2) 1958 Birth cohort, typed using Hap1.2M-Duo Custom array.7 UK-GWAS-2; 1271 CLL cases genotyped using the Illumina Omni Express BeadChip and 2501 UK Blood Service Donor controls typed using Hap1.2M-Duo Custom arrays.2 To harmonise GWAS data sets we recovered untyped genotypes by imputation using IMPUTEv2 with 1000genomes as a reference (phase 1 integrated variant set (b37) from March 2012) (Supplementary Methods). Genomic control lambda values for UK-GWAS1 and UK-GWAS2 were 1.04 and 1.05, respectively, thereby excluding significant differential genotyping or cryptic population substructure.2
Post quality control the two GWAS provided data on 1739 cases and 5199 controls. In a meta-analysis we identified 156 common SNPs (minor allele frequency>0.01), typed in either UK GWAS-1 or 2, that showed good evidence of an association (ie P<1.5 × 10−4) and did not map to any of the 30 loci that have previously been associated with CLL risk.2
Seven SNPs chosen on the basis of strength of association and/or biological plausibility of the annotated gene (that is, a role in B-cell or cancer biology) were genotyped in the UK replication series (Supplementary Table 1), which comprised 1195 CLL cases ascertained from an ongoing national study being conducted by the Institute of Cancer Research and 2568 controls ascertained through the National Study of Colorectal Cancer8 (Supplementary Methods).
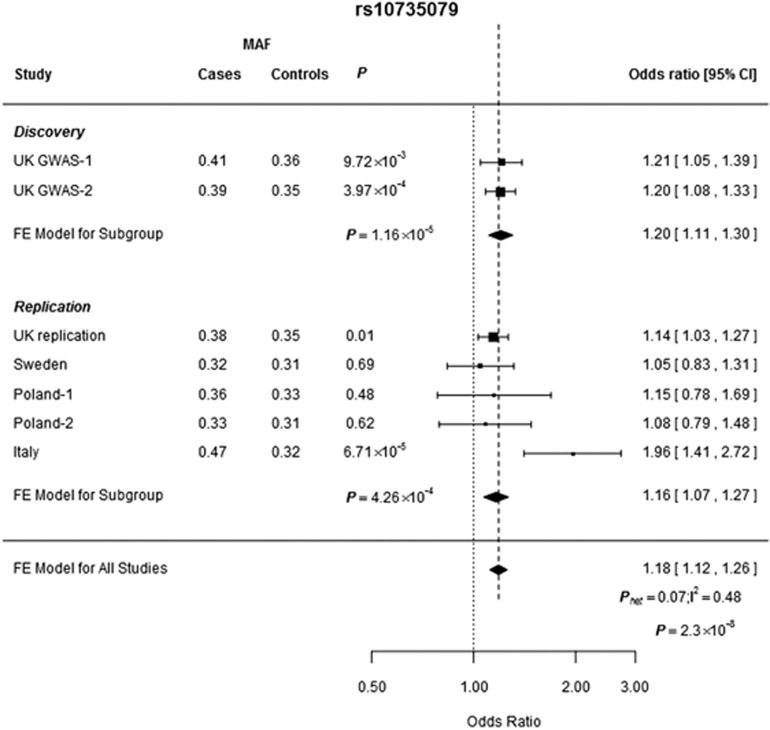
Two SNPs, rs10735079 and rs17512800, provided further evidence for an association with CLL risk (ie P<0.05) and these two SNPs were taken forward for genotyping in a further replication series from Sweden, which comprised 347 CLL cases and 342 controls (Supplementary Table 1). This case control analysis provided additional evidence for an association between rs10735079 and CLL risk. Subsequently we genotyped rs10735079 in three further case–control series, Poland-1 (105 cases, 101 controls), Poland-2 (176 cases, 209 controls) and Italy (186 cases, 155 controls) (Supplementary Methods). In the combined analysis of all series the association between rs10735079 and CLL attained genome-wide significance (combined OR per allele=1.18, 95% CI:1.12–1.26, P=2.34 × 10−8) (Figure 1). The association was not restricted to IGHV mutation and showed no relationship with either sex or age (Supplementary Table 2).
Figure 1.
Forest plot of the ORs for the association between CLL and rs10735079. Studies were weighted according to the inverse of the variance of the log of the OR calculated by unconditional logistic regression. Horizontal lines: 95% CI. Box: OR point estimate; box area is proportional to the weight of the study. Diamond (and broken line): overall summary estimate, with CI given by its width. Unbroken vertical line: null value (OR=1.0). FE, fixed effects; MAF, minor allele frequency.
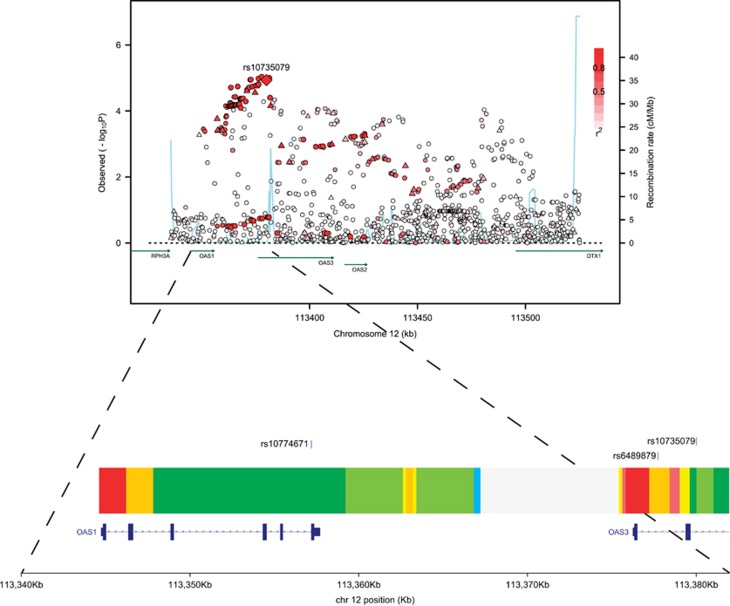
rs10735079 maps to intron 2 of the 2′-5′-oligoadenylate synthetase 3 (OAS3) gene, one of three OAS genes clustering at 12q24.13 (Figure 2), and is in LD (r2=0.87) with the splice acceptor variant of OAS1, rs10774671, which mediates alternative splicing of OAS1 transcription and affects enzymatic activity.9 Although attractive as the basis of the 12q24.13 association the association with CLL is stronger for rs10735079 than rs10774671 (P=1.16 × 10−5 and 1.74 × 10−4, respectively; Supplementary Table 3).
Figure 2.
Regional plot of association results, recombination rates and chromatin state segmentation track for 12q24.13 susceptibility locus. Association results of both genotyped (triangles) and imputed (circles) SNPs in the GWAS samples and recombination rates. −log10 P-values (y axis) of the SNPs are shown according to their chromosomal positions (x axis). rs10735079 is shown as a large diamond and is labelled by its rsID. Colour intensity of each symbol reflects the extent of LD with the top genotyped SNP; white (r2=0) through to dark red (r2=1.0) Genetic recombination rates, estimated using HapMap Utah residents of Western and Northern European ancestry (CEU) samples, are shown with a light blue line. Physical positions are based on NCBI Build 37 of the human genome. Also shown are the relative positions of genes and transcripts mapping to the region of association. Genes have been redrawn to show the relative positions; therefore, maps are not to physical scale. The lower panel shows the chromatin state segmentation track (ChromHMM) for LCL data derived from the ENCODE project and the positions of SNPs of interest (produced using visPIG-Visual Plotting Interface for Genetics).
The significant dose relationship between rs10735079 genotype and OAS3 expression in blood, with the risk allele being associated with reduced levels of mRNA (P=5.4 × 10−29; Supplementary Table 4), supports a role for rs10735079 genotype mediating its effect on CLL through differential OAS3 expression rather than impacting on OAS1.
Although rs10735079 is not predicted to lie in an active promoter or strong enhancer element, the correlated SNP rs6489879 (r2=0.99) that maps to intron 1 of OAS3 resides in a region predicted to be a strong enhancer in lymphoblastoid GM12878 cells and to be involved in binding of a number of transcription factors including IRF4 (interferon regulatory factor-4), a lymphocyte-specific transcription factor (Figure 2; Supplementary Table 3).
OAS is induced by interferon in response to viral infection activating 2-5A-dependent RNase L degradation of viral RNA10 and variation in OAS genes has been reported to be a determinant of viral susceptibility.9, 11, 12, 13 Given the possible role of viral response in the pathogenesis of CLL, although speculative, it is therefore possible that genetic variation in OAS3 influences risk of developing CLL through differing response to antigenic challenge. Moreover, OAS3 is a B-cell receptor (BCR) signature gene.14 Intriguingly as variation in the BCR genes IRF4 (ref. 5), BCL2 (ref. 3) and HLA-DQA1 (ref. 15) has previously been implicated by GWAS as determinants of CLL risk this suggests a common aetiological pathway through differential BCR-activation.
Although further functional studies are required to fully elucidate the biological basis of the 12q24.13 association, our finding brings the total number of risk loci identified for CLL thus far to 31 and provides additional support for the role of inherited genetic factors in the aetiology of CLL.
URLS
Blood eQTL browser: http://genenetwork.nl/bloodeqtlbrowser/
PLINK: http://pngu.mgh.harvard.edu/~purcell/plink/
Illumina: http://www.illumina.com/
Kaspar: http://www.lgcgenomics.com/genotyping/kasp-genotyping-chemistry/
SNAP: http://www.broadinstitute.org/
Haploreg: http://www.broadinstitute.org/mammals/haploreg/haploreg.php
visPIG-Visual Plotting Interface for Genetics: http://vispig.icr.ac.uk/
Acknowledgments
Leukaemia and Lymphoma Research provided principal funding for the study (LRF05001, LRF06002 and LRF13044). We acknowledge support from Cancer Research UK (C1298/A8362 supported by the Bobby Moore Fund) and the Arbib Fund. GS is in receipt of a PhD studentship from the Institute of Cancer Research. We also acknowledge National Health Service funding to the Royal Marsden/Institute of Cancer Research; National Institute for Health Research Biomedical Research Centre. The study made use of genotyping data on the 1958 Birth Cohort; a full list of the investigators who contributed to the generation of these data is available at http://www.wtccc.org.uk/. We thank L. Padyukov (Karolinska Institutet) and the Epidemiological Investigation of Rheumatoid Arthritis (EIRA) group for providing control samples from the Swedish population for the Swedish replication study. This study makes use of data generated by the Wellcome Trust Case-Control Consortium 1 and 2. A full list of the investigators who contributed to the generation of the data is available at http://www.wtccc.org.uk/. We are grateful to all investigators and all the patients and individuals for their participation. We also thank the clinicians, other hospital staff and study staff that contributed to the blood sample and data collection for this study.
Author Contributions
RSH obtained financial support, designed the project and provided overall project management; GPS and RSH drafted the manuscript; GPS and HES performed project management and supervised genotyping; JMA obtained financial support and supervised genotyping; GPS and MCDB performed bioinformatic and statistical analyses; GPS and AH performed genotyping; YW and MCDB performed imputation analysis; MJSD, NJS, HM, LM, SD, LK, IF, DW, KJ, GJ, KES, SJ, AM, CD, AGH, TM, GHJ, GS, RJH, ARP, DJA, JRB, GP, CP, CF, RR, DC and JMA acquired samples for the replication cohorts.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Sellick GS, Catovsky D, Houlston RS. Familial chronic lymphocytic leukemia. Seminars Oncol. 2006;33:195–201. doi: 10.1053/j.seminoncol.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Speedy HE, Di Bernardo MC, Sava GP, Dyer MJ, Holroyd A, Wang Y, et al. A genome-wide association study identifies multiple susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2014;46:56–60. doi: 10.1038/ng.2843. [DOI] [PubMed] [Google Scholar]
- Berndt SI, Skibola CF, Joseph V, Camp NJ, Nieters A, Wang Z, et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat Genet. 2013;45:868–876. doi: 10.1038/ng.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slager SL, Skibola CF, Di Bernardo MC, Conde L, Broderick P, McDonnell SK, et al. Common variation at 6p21.31 (BAK1) influences the risk of chronic lymphocytic leukemia. Blood. 2012;120:843–846. doi: 10.1182/blood-2012-03-413591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bernardo MC, Crowther-Swanepoel D, Broderick P, Webb E, Sellick G, Wild R, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008;40:1204–1210. doi: 10.1038/ng.219. [DOI] [PubMed] [Google Scholar]
- Crowther-Swanepoel D, Broderick P, Di Bernardo MC, Dobbins SE, Torres M, Mansouri M, et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat Genet. 2010;42:132–136. doi: 10.1038/ng.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power C, Elliott J. Cohort profile: 1958 British birth cohort (National Child Development Study) Int J Epidemiol. 2006;35:34–41. doi: 10.1093/ije/dyi183. [DOI] [PubMed] [Google Scholar]
- Penegar S, Wood W, Lubbe S, Chandler I, Broderick P, Papaemmanuil E, et al. National study of colorectal cancer genetics. Br J Cancer. 2007;97:9. doi: 10.1038/sj.bjc.6603997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi S, Hamano E, Matsushita I, Hijikata M, Ito H, Nagase T, et al. Differential effects of a common splice site polymorphism on the generation of OAS1 variants in human bronchial epithelial cells. Hum Immunol. 2013;74:395–401. doi: 10.1016/j.humimm.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S, Paranjape JM, Silverman RH. RNase L dimerization in a mammalian two-hybrid system in response to 2′,5′-oligoadenylates. Nucleic Acids Res. 1998;26:1522–1527. doi: 10.1093/nar/26.6.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Kang H, Ji Y, Chen X. Evaluate the relationship between polymorphisms of OAS1 gene and susceptibility to chronic hepatitis C with high resolution melting analysis. Clin Exp Med. 2013;13:171–176. doi: 10.1007/s10238-012-0193-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JK, Lisco A, McDermott DH, Huynh L, Ward JM, Johnson B, et al. Genetic Variation in OAS1 Is a Risk Factor for Initial Infection with West Nile Virus in Man. PLoS Pathog. 2009;5:e1000321. doi: 10.1371/journal.ppat.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnevie-Nielsen V, Leigh Field L, Lu S, Zheng D-J, Li M, Martensen PM, et al. Variation in antiviral 2′,5′-oligoadenylate synthetase (2′5′AS) enzyme activity is controlled by a single-nucleotide polymorphism at a splice-acceptor site in the OAS1 Gene. Am J Hum Genet. 2005;76:623–633. doi: 10.1086/429391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herishanu Y, Perez-Galan P, Liu D, Biancotto A, Pittaluga S, Vire B, et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-kappaB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood. 2011;117:563–574. doi: 10.1182/blood-2010-05-284984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slager SL, Rabe KG, Achenbach SJ, Vachon CM, Goldin LR, Strom SS, et al. Genome-wide association study identifies a novel susceptibility locus at 6p21.3 among familial CLL. Blood. 2011;117:1911–191. doi: 10.1182/blood-2010-09-308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.