Abstract
Background:
Major depression is a severe mental illness that causes heavy social and economic burdens worldwide. A number of studies have shown that interaction between individual genetic vulnerability and environmental risk factors, such as stress, is crucial in psychiatric pathophysiology. In particular, the experience of stressful events in childhood, such as neglect, abuse, or parental loss, was found to increase the risk for development of depression in adult life. Here, to reproduce the gene x environment interaction, we employed an animal model that combines genetic vulnerability with early-life stress.
Methods:
The Flinders Sensitive Line rats (FSL), a validated genetic animal model of depression, and the Flinders Resistant Line (FRL) rats, their controls, were subjected to a standard protocol of maternal separation (MS) from postnatal days 2 to 14. A basal comparison between the two lines for the outcome of the environmental manipulation was performed at postnatal day 73, when the rats were into adulthood. We carried out a global proteomic analysis of purified synaptic terminals (synaptosomes), in order to study a subcellular compartment enriched in proteins involved in synaptic function. Two-dimensional gel electrophoresis (2-DE), mass spectrometry, and bioinformatic analysis were used to analyze proteins and related functional networks that were modulated by genetic susceptibility (FSL vs. FRL) or by exposure to early-life stress (FRL + MS vs. FRL and FSL + MS vs. FSL).
Results:
We found that, at a synaptic level, mainly proteins and molecular pathways related to energy metabolism and cellular remodeling were dysregulated.
Conclusions:
The present results, in line with previous works, suggest that dysfunction of energy metabolism and cytoskeleton dynamics at a synaptic level could be features of stress-related pathologies, in particular major depression.
Keywords: early-life stress, frontal cortex, hippocampus, prefrontal cortex, proteomics, synaptosome
Introduction
Major depression (MD) is a severe mental illness that causes heavy social and economic burdens worldwide (World Health Organization, 2008). It has been estimated that more than 30 million people suffer from MD in Europe, with a prevalence of 6.9% each year (Wittchen et al., 2011). Stress-related psychiatric disorders such as depression are complex diseases and, although the pathophysiology of MD is still essentially unknown, several lines of evidence show that the interplay between individual genetic vulnerability and environmental risk factors, such as stress, may precipitate pathology (Caspi et al., 2002; Caspi and Moffitt, 2006; Krishnan and Nestler, 2008). Indeed, stressful adverse events in childhood (early-life stress) have been found to increase the likelihood of developing the pathology in adult life (Heim and Nemeroff, 2001; Caspi et al., 2003; Nugent et al., 2011).
In this study, to reproduce the gene x environment (GxE) interaction we employed an animal model that combines genetic vulnerability with early-life stress (Wörtwein et al., 2006; Husum et al., 2008; Mallei et al., 2008; Musazzi et al., 2010; Piubelli et al., 2011; Wegener et al., 2012). Therefore, we subjected a genetic animal model of depression, the Flinders Sensitive Line (FSL) rats, to a standard protocol of maternal separation (MS; Plotsky and Meaney, 1993). Indeed, MS has been shown to induce hypothalamic-pituitary-adrenal axis alteration, decrease saccharin intake, and increase anxiety-like behavior (Vazquez et al., 2000; Gardner et al., 2005; Mallei et al., 2008). The FSL rats, a well-validated animal model of depression, were genetically selected from Sprague-Dawley rats for their sensitivity to cholinergic agents (Overstreet and Russell, 1982). They address the validation criteria of construct, face, and predictive validity for a good animal model. In fact, the FSL rats display many features of depressed individuals, such as reduced general activity, decreased appetite and body weight, decreased libido, alteration of REM sleep, anhedonia (after stress), and biochemical and behavioral abnormalities (Overstreet et al., 2005; Jiménez-Vasquez et al., 2007; Neumann et al., 2011; Overstreet and Wegener, 2013). However, it is worth mentioning that, like other selectively-bred animal models of depression, FSL rats exhibit some similarities to depressed humans, but cannot reproduce all features of human pathology. For instance, FSL rats in non-stressed conditions do not show anhedonia, a key symptom of depression in humans. Therefore, all conclusions of the present study must be taken with the caveat that FSL, like other animal models, cannot explain all aspects of pathology.
Proteomics allows the simultaneous analysis of hundreds of proteins and is a powerful approach to gain insight into the molecular mechanisms underlying vulnerability to psychiatric disorders and response to stress (Rohlff and Hollis, 2003; Fountoulakis, 2004; Vercauteren et al., 2007; Mallei et al., 2008). Thus, proteomic analysis allows us to move beyond the single gene/protein/pathway and to explore multiple biological pathways and functions related to the pathophysiology of MD.
In this study, we carried out a global proteomics analysis of purified synaptic terminals (synaptosomes) from the prefrontal and frontal cortices (PFC/FC) and hippocampi (HPC) of FSL rats and their controls, the Flinders Resistant Line (FRL) rats, in order to study a subcellular compartment enriched in proteins involved in synaptic function. Thus, two-dimensional gel electrophoresis (2-DE), mass spectrometry, and bioinformatic analysis were used to analyze proteins and related functional networks that are modulated by genetic susceptibility (FSL vs. FRL) or by exposure to early-life stress (FRL + MS vs. FRL and FSL + MS vs. FSL).
We found that, at a synaptic level, proteins and molecular pathways related to energy metabolism and cellular remodeling were mainly dysregulated.
Methods
Animals
FSL and FRL from the rat colonies maintained at the Karolinska Institutet were housed in standard cages (26 x 42 x 15cm) on a 12h light/dark cycle (lights on at 07:00 hours) with controlled room temperature (22±1°C) and relative humidity (45–55%). Food (Lactamin R36) and tap water were available ad libitum. Stockholm’s Ethical Committee for the Protection of Animals approved the study, and all animal handling and procedures were conducted in conformity with the Karolinska Institutet’s Guidelines for the Care and Use of Laboratory Animals, in accordance with the European Community Council Directive 86/609/EEC. All efforts were made to minimize animal distress and to reduce the numbers of animals used in this study.
Maternal Separation
A standard MS procedure was carried out as described before (Plotsky and Meaney, 1993; El Khoury et al., 2006). Briefly, the day of delivery was designated as postnatal day (PND) 0. Litters from each rat strain were randomly assigned to the MS group and were separated from the dam for 180min/day from PND 2 to PND 14, beginning at 10:00 hours. Dams were removed from the home cages and placed in a new individual cage, then litters were removed from the nest and placed in clean plastic chambers in a incubator at 30–33°C. At the end of the separation period, first the litters and then the dams were returned to the home cages. Control litters were left undisturbed and not handled at any time except for during changes of bedding twice a week. After weaning at PND 22, rats were housed in groups of three to five per cage. Only male rats were used in this study (Figure 1).
Figure 1.
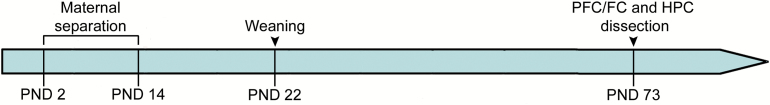
Schematic representation of the experimental design. Flinders Resistant Line (FRL) and Flinders Sensitive Line (FSL) rats were separated from the dam for 180min/day from postnatal day (PND) 2 to PND 14. Control FSL and FRL rats were not separated. All rats were weaned at PND 22. On PND 73 animals were sacrificed and prefrontal and frontal cortices (PFC/FC) and hippocampi (HPC) were removed.
Purification of Synaptosomes
Synaptosomes were prepared from FSL/FRL rats at the Karolinska Institutet, and synaptosome pellets were frozen and transferred for processing. Rats (16–20 per experimental group) were sacrificed on PND 73, the HPC and the whole frontal lobe (PFC/FC) were dissected on ice according to brain atlas coordinates (for PFC/FC plates 4–11; Paxinos and Watson, 1998), and synaptosomes were prepared by the Percoll gradient procedure according to Dunkley and colleagues (1986), with minor modifications (Mallei et al., 2008; Musazzi et al., 2010), from fresh brain tissue. Briefly, HPC or PFC/FC were homogenized in 10vol of homogenization buffer (0.28M sucrose, 10mM HEPES pH 7.4, 0.1mM EGTA, 20mM NaF, 5mM Na2PO4, 1mM Na3VO4, and 2 µl/ml protease inhibitor cocktail [Sigma Aldrich]) using a glass/teflon tissue grinder with clearance of 0.25mm. The homogenate was centrifuged 5min at 1 000 g, the supernatant was stratified on a discontinuous Percoll gradient (6, 10, and 20% v/v in Tris-buffered sucrose) and centrifuged at 33 500 g for 5min. The layer between 10 and 20% Percoll (synaptosomes) was collected and washed by centrifugation, and the resulting pellet was stored at -80°C.
2-DE and Proteome Analysis
2-DE and Imaging
2-DE was carried out as previously described (Mallei et al., 2008 2011). Synaptosome pellets were dissolved in isoelectric focusing (IEF) buffer (7M urea, 2M thiourea, 40mM Tris, 3mM tributylphosphine, 2% CHAPS, 1% carrier ampholytes [GE Healthcare], and protease inhibitors [Roche Diagnostic]). An aliquot of each pellet was dialyzed in 1% sodium dodecyl sulfate in distilled water to measure protein concentration by bicinchoninic acid assay (Pierce Chemical). Next, 115 µg of synaptosomes were dissolved in 125 µl of IEF buffer containing 10mM iodoacetamide as an alkylating agent and a trace of bromophenol blue, and separated by IEF in 7cm pH 3–10 non-linear immobilized pH gradient (IPG) strips (Bio-Rad). IEF was performed at 15°C at a maximum of 4000V for a total of 28 000 Vh using Protean IEF Cell (Bio-Rad).
Prior to the second dimension, the IPG strips were equilibrated in a solution containing 6M urea, 2% SDS, 375mM Tris pH 8.8, and 4mM tributylphosphine. After equilibration, the IPG strips were placed on top of 8–18% T-gradient polyacrylamide gels, and sealed with 0.5% agarose in running buffer. The 2-DE gels were then fixed and stained with SYPRO Ruby (Bio-Rad). The 2-DE gel images were digitally acquired by VersaDoc imaging system (Bio-Rad). Image and statistical analysis were carried out by PDQuest software (Bio-Rad), to compare replicate groups and identify sets of protein spots that show a statistically significant difference with a confidence level of 0.05.
Mass Fingerprinting and Protein Identification
Differently expressed spots were cut from gel with a spot cutter (Bio-Rad), digested with trypsin, and identified by peptide mass fingerprinting at the Proteomics Core Facility of the University of Geneva (Scherl et al., 2002). Mascot (Matrix Science Ltd.; Perkins et al., 1999) and Profound software (PROWL; http://prowl.rockefeller.edu/prowl-cgi/profound.exe) and Aldente tools (http://au.expasy.org/cgi-bin/aldente/form.cgi) were used to analyze spectra. The research was conducted against SWISS-PROT, TrEMBL, and NCBInr databases.
Western Blot Analysis
Western blotting was carried out as previously described (Musazzi et al., 2010). Briefly, synaptosomal proteins were separated on 12% polyacrylamide gels and blotted on polyvinylidene fluoride membranes (GE Healthcare). Blocking was performed for 1 hour at room temperature in 5% nonfat dry milk in Tris-buffered saline containing 0.1% Tween 20 (TBST). Membranes were then incubated overnight in 5% nonfat dry milk in TBST with primary antibodies for aconitate hydratase (1:2000, a generous gift from Professor Szweda, Oklahoma Medical Research Foundation), N-ethylmaleimide sensitive factor (NSF, 1:1000, Cell Signalling Technology Inc.), syntaxin-binding protein 1 (1:3000, BD Biosciences Italy), adenosine triphosphate synthase alpha (1:3000, Life Technologies Italia), synaptosomal-associated protein 25 (SNAP-25, 1:2000, Synaptic Systems GmbH), dihydropyrimidinase-related protein 2 (DRP-2, 1:2000, Sigma-Aldrich), and β-actin (1:10000, Sigma-Aldrich). Following incubation with peroxidase-coupled secondary antibodies, protein bands were visualized with StoS Protein Detection System (GeneSpin) on Hyperfilm ECL films (GE Healthcare). All protein bands used were within linear range, and normalized for β-actin levels in the same membrane. Quantity One software (Bio-Rad) was used for standardization and quantitation.
Bioinformatic Analysis
Functional, canonical pathways and networks analyses were generated using Ingenuity Pathways Analysis (IPA, Ingenuity Systems, http://www.ingenuity.com). All proteins identified by mass spectrometry were considered for the analyses. The software identified the biological functions/canonical pathways in the Ingenuity Pathways Knowledge Base that were most significant to the data set. Fischer’s exact test was used to calculate a p-value determining the probability that each biological function assigned to that data set is due to chance alone. For network generation, a data set containing protein identifiers and relative fold change values was uploaded into the application and mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base. Networks of these focus genes were then algorithmically generated based on their connectivity. Fischer’s exact test was used to calculate a p-value determining the probability that each biological function assigned to that network is due to chance alone. The p-value is then expressed as a score (i.e. -log10 p-value); a score of 8 or higher is considered extremely significant.
Statistical Analysis
In the proteomic study, protein spot intensities were first log transformed to fit the normal distribution curve and then analyzed with student’s t-tests and Partial Least Squares tests using PDQuest software. Unpaired two-tailed non-parametric Mann-Whitney tests were used to analyze Western blotting data using GraphPad Prism 4 (GraphPad Software Inc.). In both proteomic and Western blotting analyses, significance was assumed at p < 0.05.
Results
In this study we employed 2-DE to analyze basal differences in synaptosomal protein expression patterns in FSL and FRL rats. Moreover, the long-term effect of early-life stress on the synaptosomal protein expression pattern was assessed in both rat lines. In addition, IPA was used to identify the biological functions and cellular processes most relevant to the differently-expressed proteins, and to explore relative pathways/networks of the proteins involved.
Synaptosomal Proteome Maps
PFC/FC and HPC synaptosomes were prepared from FSL and FRL rats, naïve or subjected to MS. Proteins were separated by 2-DE gels and only proteomic maps belonging to the same electrophoretic run were analyzed in the same match-set in order to reduce experimental variability. Examples of rat PFC/FC and HPC 2-DE maps are reported in Figure 2.
Figure 2.
Representative two-dimensional gel electrophoresis gel images of Sypro-Ruby-stained synaptosomal proteins from (A) prefrontal and frontal cortices or (B) hippocampi in FSL rats. Circles and numbers indicate differently-regulated proteins spots (see also Tables 2 and 3).
The map containing the largest number of spots was chosen as the reference master map and the corresponding spots in all gels were matched. The average spot number (± standard deviation [SD]) detected in a gel was 470±53 in PFC/FC, with an average percentage of matched spots across gels of 87%. In HPC, the average number of spots (± SD) detected was 370±15, with an average percentage of matched spots across gels of 96.5%.
The following comparisons between experimental groups were carried out in both brain areas: FSL vs. FRL, to evaluate basal (genetic) differences between rat lines; FRL + MS vs. FRL; and FSL + MS vs. FSL to evaluate the effects of GxE interaction.
The number of spots differently modulated in the different comparisons are reported in Table 1. Spots with statistically significant different levels were excised from gels and identified by peptide mass fingerprinting analysis (Table 2 and Table 3).
Table 1.
Comparisons of Experimental Groups
| Comparison | Spots per gel (mean ± SD) | % of matched spots (mean) | Modulated spots | t-test | PLS | Common to t-test and PLS |
|---|---|---|---|---|---|---|
| PFC/FC | ||||||
| FSL vs. FRL | 497±62 | 88 | 27 | 25 | 11 | 9 |
| FRL+MS vs. FRL | 436±28 | 86 | 19 | 14 | 9 | 4 |
| FSL+MS vs. FSL | 504±51 | 88 | 43 | 34 | 20 | 11 |
| HPC | ||||||
| FSL vs. FRL | 376±14 | 96 | 17 | 15 | 10 | 8 |
| FRL+MS vs. FRL | 372±6 | 96 | 19 | 17 | 7 | 5 |
| FSL+MS vs. FSL | 368±21 | 97 | 19 | 14 | 9 | 4 |
The average number of spots per gel with standard deviation (SD) is reported in column 2 for the considered comparison. The average percentage of gel spots matched with the standard map for each comparison set is reported in column 3. Two statistical tests were performed: student’s t-test and Partial Least Squares analysis (PLS). The total number of spots showing a statistically-significant modulation in student’s t-test (t-test, p < 0.05, column 5) or in PLS analysis (95% significance level, column 5) and the number of shared spots between the two tests are indicated (column 6). HPC, hippocampus; MS, maternal separation; PFC/FC, prefrontal and frontal cortices.
Table 2.
Proteins Identified by Mass-Spectrometry in PFC/FC
| Spot No.a | Protein name | Acc. No.b | No. of matched peptides | Coverage (%)c | Mascot score | Fold change | T-test | PLSd |
|---|---|---|---|---|---|---|---|---|
| Spot differently modulated in FSL vs. FRL | ||||||||
| 308 | ATPase H+ transporting, V0 subunit D isoform 1 | Q5M7T6 | 6 | 15 | 56 | 1,4 | Y | N |
| 1209 | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | Q6AY95 | 7 | 22 | 50 | 2,5 | Y | N |
| 1706 | NADH-ubiquinone oxidoreductase 75kDa subunit, mitochondrial | Q66HF1 | 12 | 16 | 68 | -1,8 | Y | N |
| Heat shock cognate 71 kDA protein | P63018 | 16 | 27 | 113 | ||||
| 2605 | Histamine N-methyltransferase | Q01984 | 6 | 17 | 38 | 1,6 | Y | N |
| 3201 | Peroxiredoxin 6 | O35244 | 4 | 17 | 28 | 1,5 | Y | N |
| 3404 | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial precursor | Q99NA5 | 7 | 21 | 42 | 1,1 | Y | N |
| 5404 | Fructose bisphosphate aldolase C | P09117 | 5 | 19 | 31 | 1,5 | Y | N |
| 5607 | Syntaxin-binding protein 1 | P61765 | 11 | 17 | 57 | -3,0 | Y | N |
| 5807 | Dynamin-1 | P21575 | 24 | 23 | 174 | -1,7 | Y | N |
| 7601 | Transketolase | P50137 | 7 | 13 | 45 | 3,4 | N | Y |
| Spot differently modulated in FRL+MS vs. FRL | ||||||||
| 1209 | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | Q6AY95 | 7 | 22 | 50 | 1,8 | N | Y |
| 1308 | Guanine nucleotide binding protein G(o) alpha subunit 2 | P30033 | 9 | 25 | 63 | -2,1 | Y | Y |
| Calcium binding protein 39-like | Q5XIJ7 | 7 | 13 | 40 | ||||
| 1706 | NADH-ubiquinone oxidoreductase 75kDa subunit, mitochondrial | Q66HF1 | 12 | 16 | 68 | 1,2 | Y | N |
| Heat shock cognate 71 kDA protein | P63018 | 16 | 27 | 113 | ||||
| 1707 | NADH-ubiquinone oxidoreductase 75kDa subunit, mitochondrial | Q66HF1 | 16 | 26 | 115 | -1,6 | Y | Y |
| Heat shock cognate 71 kDA protein | P63018 | 8 | 15 | 38 | ||||
| 2601 | 60kDa heat shock protein, mitochondrial precursor | P63039 | 15 | 30 | 108 | -1,1 | Y | N |
| 3201 | Peroxiredoxin-6 | O35244 | 4 | 17 | 28 | -1,3 | Y | N |
| 3404 | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial precursor | Q99NA5 | 7 | 21 | 42 | -1,9 | Y | Y |
| 5404 | Fructose-biphosphate aldolase C | P09117 | 5 | 19 | 31 | -1,3 | Y | N |
| 8306 | Malate dehydrogenase, mitochondrial | P04636 | 10 | 34 | 75 | -1,4 | Y | N |
| Glyceraldehyde-3-phosphate dehydrogenase | P04797 | 6 | 25 | 43 | ||||
| Spot differently modulated in FSL+MS vs. FSL | ||||||||
| 207 | 14-3-3 protein zeta/delta | P63102 | 13 | 40 | 89 | 1,5 | Y | N |
| 14-3-3 protein eta | P68511 | 9 | 29 | 58 | ||||
| 14-3-3 protein beta/alpha | P35213 | 9 | 27 | 51 | ||||
| 14-3-3 protein gamma | P61983 | 8 | 27 | 44 | ||||
| 306 | Clathrin light chain B | P08082 | 6 | 17 | 60 | -1,8 | Y | Y |
| 308 | ATPase H+ trasporting V0 subunit D, isoform 1 | Q5M7T6 | 6 | 15 | 56 | -1,6 | Y | N |
| 1106 | NADH-ubiquinone oxidoreductase 23kDa subunit, mitochondrial precursor (mouse) | Q8K3J1 | 4 | 19 | 31 | -1,4 | Y | N |
| 2202 | Prohibitin | P67779 | 7 | 31 | 57 | -1,6 | Y | N |
| 2203 | NADH-ubiquinone oxidoreductase 30 kDA subunit, mitochondrial precursor (mouse) | Q9DCT2 | 8 | 25 | 71 | -2,4 | Y | Y |
| 2207 | NADH-ubiquinone oxidoreductase 30 kDA subunit, mitochondrial precursor (mouse) | Q9DCT2 | 7 | 27 | 57 | -1,7 | Y | N |
| 2209 | Prohibitin | P67779 | 7 | 31 | 68 | -1,8 | Y | Y |
| 2302 | Guanine nucleotide binding protein G(o) alpha subunit 2 | P30033 | 6 | 19 | 35 | -1,5 | Y | N |
| 3201 | Peroxiredoxin 6 | O35244 | 4 | 17 | 28 | -1,6 | Y | N |
| 3309 | Cytosolic malate dehydrogenase | O88989 | 9 | 28 | 60 | -1,4 | Y | N |
| 5103 | Cytochrome C1 protein fragment (mouse) | Q63ZW4 | 5 | 18 | 31 | -1,5 | Y | N |
| 5306 | Voltage-dependent anion selective channel protein 1 | Q9Z2L0 | 5 | 23 | 45 | -1,2 | Y | N |
| 5607 | Syntaxin-binding protein 1 | P61765 | 11 | 17 | 57 | 1,9 | Y | N |
| 5704 | N-ethylmaleimide sensitive factor | Q9QUL6 | 8 | 13 | 36 | -1,2 | Y | N |
| 6610 | Pyruvate kinase isozymes M1/M2 | P11980 | 12 | 19 | 69 | -1,8 | N | Y |
| 6710 | Aconitate hydratase, mitochondrial precursor | Q9ER34 | 20 | 25 | 147 | -1,3 | Y | N |
| 7407 | Creatine kinase mitochondrial 1, ubiquitous | Q5BJT9 | 13 | 30 | 97 | 1,8 | N | Y |
| 7507 | ATP synthase alpha chain, mitochondrial precursor | P15999 | 11 | 23 | 68 | -2,3 | N | Y |
| 7601 | Transketolase | P50137 | 7 | 13 | 45 | -3,1 | Y | N |
| 8502 | ATP synthase alpha chain, mitochondrial precursor | P15999 | 14 | 26 | 88 | -1,8 | N | Y |
aProtein spot number as indicated in Figure 2A
bSwissProt or TrEMBL database accession number
cPercentage of identified sequence of the known protein
dPartial Least Square
ATP, adenosine triphosphate; FRL, Flinders Resistant Line; FSL, Flinders Sensitive Line; MS, maternal separation; PFC/FC, prefrontal and frontal cortices.
Table 3.
Proteins Identified by Mass-Spectrometry in HPC
| Spot No.a | Protein name | Acc. No.b | No. of matched peptides | Coverage (%)c | Mascot score | Fold change | T-test | PLSd |
|---|---|---|---|---|---|---|---|---|
| Spot differently modulated in FSL vs. FRL | ||||||||
| 601 | Beta actin, cytoplasmic 1 | P60711 | 7 | 22 | 18 | -3,9 | N | Y |
| 903 | Neurofilament light polypeptide | P19527 | 14 | 25 | 81 | -2,0 | N | Y |
| 1602 | Beta actin, cytoplasmic 1 | P60711 | 13 | 31 | 82 | 2,8 | Y | Y |
| 2203 | NADH-dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial precursor (mouse) | Q9DCT2 | 6 | 22 | 35 | 1,1 | Y | N |
| 2403 | Beta actin, cytoplasmic 1 | P60711 | 11 | 29 | 73 | 1,8 | Y | Y |
| Gamma actin, cytoplasmic 2 | P63259 | 11 | 29 | 73 | ||||
| Guanine nucleotide binding prot. G(o) subunit alpha 2 | P30033 | 6 | 19 | 27 | ||||
| 3404 | Beta actin, cytoplasmic 1 | P60711 | 10 | 29 | 78 | 4,4 | Y | N |
| Gamma actin, cytoplasmic 2 | P63259 | 10 | 29 | 78 | ||||
| Guanine nucleotide binding prot. G(o) subunit alpha 2 | P30033 | 6 | 19 | 34 | ||||
| 3602 | Beta actin, cytoplasmic 1 | P60711 | 10 | 34 | 90 | 1,3 | Y | N |
| 4503 | Succinyl-CoA ligase [ADP-forming] beta chain, mitochondrial precursor (mouse) | Q9Z2I9 | 8 | 12 | 44 | -2,1 | Y | Y |
| 4902 | Dihydropyrimidinase-related protein 2 | P47942 | 8 | 15 | 56 | 1,8 | Y | Y |
| 5902 | N-ethylmaleimide sensitive factor | Q9QUL6 | 14 | 18 | 65 | -1,1 | Y | N |
| 9501 | Aspartate aminotransferase, mitochondrial precursor | P00507 | 15 | 31 | 145 | 1,4 | Y | N |
| Spot differently modulated in FRL+MS vs. FRL | ||||||||
| 101 | Synaptosomal-associated protein 25 | P60881 | 10 | 54 | 79 | 1,3 | Y | Y |
| 604 | NSFL1 cofactor p47 | O35987 | 12 | 37 | 108 | -7,2 | Y | N |
| Beta actin, cytoplasmic 1 | P60711 | 9 | 26 | 67 | ||||
| Gamma enolase | P07323 | 7 | 19 | 44 | ||||
| 1103 | NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial precursor | P19234 | 9 | 45 | 77 | -1,4 | Y | N |
| Rho GDP-dissociation inhibitor 1 (mouse) | Q99PT1 | 7 | 37 | 53 | ||||
| 1903 | Heat shock cognate 71kDa protein | P63018 | 20 | 34 | 111 | -3,6 | Y | Y |
| Heat shock-related 70kDa protein 2 | P14659 | 10 | 16 | 31 | ||||
| 1907 | Heat shock cognate 71kDa protein | P63018 | 21 | 37 | 201 | -7,5 | Y | N |
| Heat shock-related 70kDa protein 2 | P14659 | 9 | 15 | 52 | ||||
| 2704 | Vacuolar ATP synthase subunit B, brain isoform | P62815 | 8 | 17 | 54 | -1,8 | N | Y |
| 2902 | Heat shock cognate 71kDa protein | P63018 | 18 | 32 | 156 | -1,4 | Y | N |
| 3002 | Superoxide dismutase [Cu-Zn] | Q6LDS4 | 5 | 26 | 49 | 1,2 | Y | N |
| 3201 | Prohibitin | P67779 | 9 | 30 | 59 | -1,2 | Y | N |
| 3402 | Beta actin, cytoplasmic 1 | P60711 | 10 | 34 | 80 | -1,8 | Y | Y |
| Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial precursor | Q99NA5 | 6 | 20 | 35 | ||||
| 5201 | Vacuolar ATP synthase subunit E1 | Q6PCU2 | 9 | 30 | 62 | 1,2 | Y | N |
| 5805 | Dihydropyrimidinase-related protein 2 | P47942 | 5 | 10 | 17 | -100,0 | Y | N |
| 7701 | Glutamate dehydrogenase 1, mitochondrial precursor | P10860 | 18 | 28 | 70 | 1,9 | Y | Y |
| ATP synthase subunit alpha, mitochondrial precursor | P15999 | 14 | 26 | 61 | ||||
| Vimentin | P31000 | 14 | 24 | 56 | ||||
| 7705 | ATP synthase subunit alpha, mitochondrial precursor | P15999 | 12 | 23 | 93 | -1,9 | Y | N |
| 8101 | Superoxide dismutase [Mn], mitochondrial precursor | P07895 | 5 | 23 | 45 | -1,1 | Y | N |
| 8506 | Isocitrate dehydrogenase [NAD] subunit beta, mitochondrial precursor | Q68FX0 | 12 | 25 | 77 | -2,2 | Y | Y |
| Aspartate aminotransferase, cytoplasmic | P13221 | 8 | 16 | 47 | ||||
| Spot differently modulated in FSL+MS vs. FSL | ||||||||
| 201 | 14-3-3 protein zeta/delta | P63102 | 15 | 45 | 77 | 1,3 | Y | N |
| 14-3-3 protein theta | P68255 | 10 | 30 | 42 | ||||
| 14-3-3 protein gamma | P61983 | 9 | 31 | 37 | ||||
| 14-3-3 protein alpha/beta | P35213 | 9 | 26 | 36 | ||||
| 14-3-3 protein eta | P68511 | 8 | 26 | 26 | ||||
| 903 | Neurofilament light polypeptide | P19527 | 14 | 25 | 81 | 2,0 | N | Y |
| 1804 | Alpha-internexin | P23565 | 17 | 29 | 64 | -1,8 | N | Y |
| 60kDa heat shock protein, mitochondrial precursor | P63039 | 9 | 18 | 35 | ||||
| 1805 | Alpha-internexin | P23565 | 22 | 31 | 199 | 2,8 | N | Y |
| Neurofilament light polypeptide | P19527 | 9 | 12 | 48 | ||||
| 1905 | Heat shock cognate 71kDa protein | P63018 | 15 | 25 | 59 | 1,5 | Y | N |
| NADH-ubiquinone oxidoreductase 75kDa subunit, mitochondrial precursor | Q66HF1 | 10 | 14 | 32 | ||||
| 1911 | Mitochondrial inner membrane protein (mouse) | Q8CAQ8 | 12 | 15 | 57 | 1,7 | N | Y |
| 2901 | Heat shock cognate 71kDa protein | P63018 | 23 | 38 | 61 | 1,3 | Y | N |
| Heat shock-related 70kDa protein 2 | P14659 | 16 | 24 | 24 | ||||
| 3701 | Vacuolar ATP synthase subunit B, brain isoform | P62815 | 13 | 27 | 113 | 1,1 | Y | N |
| 4905 | Dihydropyrimidinase-related protein 2 | P47942 | 9 | 18 | 62 | -2,1 | N | Y |
| 5603 | Elongation factor Tu, mitochondrial precursor (mouse) | Q8BFR5 | 11 | 28 | 95 | -1,4 | Y | N |
| 6602 | Calcium/calmodulin-dependent protein kinase type II alpha chain | P11275 | 8 | 18 | 53 | 4,5 | Y | N |
| 6702 | Glutamate dehydrogenase 1, mitochondrial precursor | P10860 | 16 | 29 | 83 | -1,4 | Y | N |
| ATP synthase subunit alpha, mitochondrial precursor | P15999 | 11 | 20 | 38 | ||||
| 8504 | Phosphoglycerate kinase 1 | P16617 | 8 | 18 | 44 | -1,9 | Y | Y |
| 8703 | ATP synthase subunit alpha, mitochondrial precursor | P15999 | 22 | 38 | 135 | -3,6 | Y | Y |
aProtein spot number as indicated in Figure 2B
bSwissProt or TrEMBL database accession number
cPercentage of identified sequence of the known protein
dPartial Least Square
ATP, adenosine triphosphate; FSL, Flinders Sensitive Line; FRL, Flinders Resistant Line; HPC, hippocampus; MS, maternal separation.
Prefrontal and Frontal Cortices Synaptoproteomics
Basal Differences Between FSL-FRL in the Prefrontal and Frontal Cortices
2-DE proteomic analysis revealed 27 differently-expressed protein spots between FSL and FRL rats. Among them, 10 were successfully identified by mass spectrometry, representing 11 distinct proteins (one spot containing a mixture of two proteins; Table 2). IPA functional analysis tool identified the biological functions relevant to the upregulated or downregulated proteins in basal comparison (Figure 3A). The top three molecular and cellular functions identified were nucleic acid metabolism, small molecule biochemistry, and energy production (Table 4). Moreover, IPA was used to explore canonical pathways statistically relevant in the same comparison. The top three canonical pathways were pentose phosphate pathway, acetyl-CoA biosynthesis, and sucrose degradation (Figure 3B and Table 5).
Figure 3.
Bioinformatic analysis in the prefrontal and frontal cortices. Major biological functions and cellular processes most relevant to the proteins differently expressed in: (A) Flinders Sensitive Line (FSL) vs. Flinders Resistant Line (FRL), (C) FRL after maternal separation (MS), and (E) FSL after MS, as calculated by Ingenuity Pathways Analysis software. Canonical pathways most relevant to the proteins differently expressed in: (B) FSL vs. FRL, (D) FRL after MS, and (F) FSL after MS. Biological functions and canonical pathways are listed based on higher statistical significance [-log(p value)], with a threshold set at 1.3.
Table 4.
Molecular and Cellular Functions Identified by IPA in PFC/FC and HPC
| Molecular and Cellular Functions | p-value | Proteinsa |
|---|---|---|
| FSL vs. FRL (PFC/FC) | ||
| Nucleic Acid Metabolism | 2,49E-06 | HSPA8, NDUFS1, TKT, ATP6V0D1, IDH3A, PDHB |
| Small Molecule Biochemistry | 2,49E-06 | HSPA8, STXBP1, NDUFS1, HNMT, TKT, ATP6V0D1, IDH3A, PDHB, PRDX6 |
| Energy Production | 4,91E-05 | HSPA8, NDUFS1, ATP6V0D1 |
| Cellular Assembly and Organization | 9,52E-05 | DNM1, HSPA8, NDUFS1, STXBP1 |
| Cellular Function and Maintenance | 9,52E-05 | HSPA8, DNM1, NDUFS1, STXBP1 |
| FRL+MS vs. FRL (PFC/FC) | ||
| Nucleic Acid Metabolism | 1,59E-06 | HSPA8, NDUFS1, GNAO1, GAPDH, IDH3A, HSPD1, MDH2, PDHB |
| Small Molecule Biochemistry | 1,59E-06 | HSPA8, NDUFS1, GNAO1, GAPDH, IDH3A, HSPD1, MDH2, PDHB, PRDX6 |
| Post-Translational Modification | 3,39E-05 | HSPA8, HSPD1, PRDX6, ALDOC |
| Protein Folding | 3,39E-05 | HSPA8, HSPD1 |
| Cell Death and Survival | 1,36E-04 | HSPA8, NDUFS1, GNAO1, GAPDH, HSPD1, PRDX6, ALDOC |
| FSL+MS vs. FSL (PFC/FC) | ||
| Nucleic Acid Metabolism | 4,53E-06 | NSF, ATP5A1, TKT, GNAO1, ATP6V0D1, PKM, CKMT1A/CKMT1B, MDH1, VDAC1 |
| Small Molecule Biochemistry | 4,53E-06 | STXBP1, YWHAH, TKT, ATP5A1, PKM, ACO2, CKMT1A/CKMT1B, YWHAZ, MDH1, PRDX6, NSF, ATP6V0D1, GNAO1, VDAC1 |
| Molecular Transport | 7,63E-06 | STXBP1, YWHAH, YWHAB, ATP5A1, PKM, YWHAZ, CKMT1A/CKMT1B, MDH1, PRDX6, NSF, GNAO1, ATP6V0D1, VDAC1 |
| DNA Replication, Recombination, and Repair | 2,81E-04 | NSF, ATP5A1, ATP6V0D1, GNAO1, CKMT1A/CKMT1B |
| Energy Production | 2,81E-04 | NSF, ATP5A1, ATP6V0D1, PKM, MDH1, VDAC1 |
| FSL vs. FRL (HPC) | ||
| Cellular Assembly and Organization | 3,84E-05 | DPYSL2, NSF, NEFL, ACTB, GNAO1, ACTG1 |
| Small Molecule Biochemistry | 8,91E-05 | DPYSL2, NSF, SUCLA2, GNAO1, GOT2 |
| Cell Morphology | 1,76E-04 | DPYSL2, NSF, NEFL, ACTB, GNAO1 |
| Cellular Development | 1,76E-04- | DPYSL2, NEFL, ACTB, GNAO1 |
| Cellular Function and Maintenance | 1,76E-04 | DPYSL2, NSF, NEFL, ACTB, GNAO1, ACTG1 |
| FRL+MS vs. FRL (HPC) | ||
| Free Radical Scavenging | 1,58E-06 | SOD2, SOD1, ACTB, ARHGDIA |
| Small Molecule Biochemistry | 9,45E-06 | DPYSL2, SOD1, ATP5A1, GLUD1, VIM, SNAP25, ATP6V1E1, HSPA8, SOD2, GOT1, IDH3A, IDH3B, ATP6V1B2 |
| Carbohydrate Metabolism | 1,57E-05 | SOD1, SOD2, GOT1, IDH3A, IDH3B |
| Amino Acid Metabolism | 3,3E-05 | DPYSL2, SOD1, GLUD1, GOT1 |
| Nucleic Acid Metabolism | 3,42E-05 | ATP6V1E1, HSPA8, DPYSL2, SOD2, SOD1, ATP5A1, IDH3A, IDH3B |
| FSL+MS vs. FSL (HPC) | ||
| Protein Trafficking | 8,23E-06 | YWHAQ, YWHAG, YWHAB, YWHAZ |
| Cellular Assembly and Organization | 5,09E-05 | HSPA8, DPYSL2, NDUFS1, YWHAG, CAMK2A, IMMT, YWHAH, NEFL, YWHAZ, INA |
| Cellular Function and Maintenance | 5,09E-05 | HSPA8, DPYSL2, NDUFS1, YWHAG, CAMK2A, YWHAH, IMMT, NEFL, INA, HSPD1, HSPA2, ATP6V1B2 |
| Molecular Transport | 7,05E-05 | HSPA8, DPYSL2, NDUFS1, CAMK2A, YWHAH, YWHAB, ATP5A1, YWHAZ, ATP6V1B2 |
| Nucleic Acid Metabolism | 7,3E-05 | PGK1, HSPA8, DPYSL2, NDUFS1, ATP5A1, HSPD1 |
aProteins are indicated with gene name
FSL, Flinders Sensitive Line; FRL, Flinders Resistant Line; HPC, hippocampus; IPA, Ingenuity Pathways Analysis; MS, maternal separation; PFC/FC, prefrontal and frontal cortices prefrontal cortex.
Table 5.
Canonical Pathways identified by IPA in PFC/FC and HPC.
| Ingenuity Canonical pathways | p-value | Proteinsa |
|---|---|---|
| FSL vs. FRL (PFC/FC) | ||
| Pentose Phosphate Pathway (Non-oxidative Branch) | 4,25E-03 | TKT |
| Acetyl-CoA Biosynthesis I (Pyruvate Dehydrogenase Complex) | 4,25E-03 | PDHB |
| Sucrose Degradation V (Mammalian) | 5,66E-03 | ALDOC |
| Pentose Phosphate Pathway | 7,07E-03 | TKT |
| Glutathione Redox Reactions I | 1,12E-02 | PRDX6 |
| FRL+MS vs. FRL (PFC/FC) | ||
| Gluconeogenesis I | 4,4E-07 | GAPDH, MDH2, ALDOC |
| TCA Cycle II (Eukaryotic) | 9,39E-05 | IDH3A, MDH2 |
| Glycolysis I | 1,2E-04 | GAPDH, ALDOC |
| NADH Repair | 1,93E-03 | GAPDH |
| Synaptic Long Term Depression | 3,42E-03 | GNAO1, PRDX6 |
| FSL+MS vs. FSL (PFC/FC) | ||
| Cell Cycle: G2/M DNA Damage Checkpoint Regulation | 2,95E-07 | YWHAG, YWHAH, YWHAB, YWHAZ |
| Myc Mediated Apoptosis Signaling | 1E-06 | YWHAG, YWHAH, YWHAB, YWHAZ |
| ERK5 Signaling | 1,4E-06 | YWHAG, YWHAH, YWHAB, YWHAZ |
| IGF-1 Signaling | 7,91E-06 | YWHAG, YWHAH, YWHAB, YWHAZ |
| 14-3-3-mediated Signaling | 1,66E-05 | YWHAG, YWHAH, YWHAB, YWHAZ |
| FSL vs. FRL (HPC) | ||
| RhoGDI Signaling | 1,07E-04 | ACTB, GNAO1, ACTG1 |
| Signaling by Rho Family GTPases | 2,49E-04 | ACTB, GNAO1, ACTG1 |
| Remodeling of Epithelial Adherent Junctions | 7,08E-04 | ACTB, ACTG1 |
| FAK Signaling | 1,12E-3 | ACTB, ACTG1 |
| VEGF Signaling | 1,25E-3 | ACTB, ACTG1 |
| FRL+MS vs. FRL (HPC) | ||
| Superoxide Radicals Degradation | 2,36E-05 | SOD1, SOD2 |
| TCA Cycle II (Eukaryotic) | 3,93E-04 | IDH3A, IDH3B |
| Mitochondrial Dysfunction | 6,73E-04 | SOD2, NDUFV2, ATP5A1 |
| NRF2-mediated Oxidative Stress Response | 1,51E-03 | SOD1, SOD2, ACTB |
| Glutamate Biosynthesis II | 2,58E-03 | GLUD1 |
| FSL+MS vs. FSL (HPC) | ||
| Cell Cycle: G2/M DNA Damage Checkpoint Regulation | 1,45E-09 | YWHAQ, YWHAG, YWHAH, YWHAB, YWHAZ |
| Myc Mediated Apoptosis Signaling | 6,83E-09 | YWHAQ, YWHAG, YWHAH, YWHAB, YWHAZ |
| ERK5 Signaling | 1,04E-08 | YWHAQ, YWHAG, YWHAH, YWHAB, YWHAZ |
| IGF-1 Signaling | 9,32E-08 | YWHAQ, YWHAG, YWHAH, YWHAB, YWHAZ |
| 14-3-3-mediated Signaling | 2,39E-07 | YWHAQ, YWHAG, YWHAH, YWHAB, YWHAZ |
aProteins are indicated with gene name
FSL, Flinders Sensitive Line; FRL, Flinders Resistant Line; HPC, hippocampus; IPA, Ingenuity Pathways Analysis; MS, maternal separation; PFC/FC, prefrontal and frontal cortices.
Effect of Early-Life Stress in FRL Prefrontal and Frontal Cortices
In FRL rats, MS differently regulated 19 spots and mass spectrometry identified 9 spots representing 13 distinct proteins/protein isoforms (four spots containing a mixture of two proteins; Table 2). The three top molecular and cellular functions revealed by IPA analysis were nucleic acid metabolism, small molecule biochemistry, and post-translational modification (Figure 3C and Table 4). The top three canonical pathways were gluconeogenesis, TCA cycle, and glycolysis (Figure 3D and Table 5).
Effects of Early-Life Stress in FSL Prefrontal and Frontal Cortices
MS differently modulated 43 spots in FSL and mass spectrometry identified 21 spots representing 24 distinct proteins/protein isoforms (one spot containing a mixture of four proteins; Table 2). The top three molecular and cellular functions, revealed by bioinformatic analysis with IPA, were nucleic acid metabolism, small molecule biochemistry, and molecular transport (Figure 3E and Table 4). The top three canonical pathways were cell cycle (G2/M DNA damage checkpoint regulation), Myc mediated apoptosis signaling, and ERK5 signaling (Figure 3F and Table 5).
It is worth mentioning that in the PFC/FC the bioionformatic analysis evidenced biological functions and canonical pathways mostly related to cellular energy metabolism, including the pentose phosphate pathway, acetyl-CoA biosynthesis, sucrose degradation/glycolysis/gluconeogenesis, and TCA cycle (Table 5). Moreover, early-life stress modulated twice the number of spots in FSL compared to FRL.
Hippocampus Synaptoproteomics
Basal Differences Between FSL-FRL in Hippocampus
In HPC, 17 protein spots were differently expressed between FSL and FRL rats. Among them, 11 were successfully identified by mass spectrometry, representing 15 distinct proteins/protein isoforms (two spots containing a mixture of three proteins; Table 3). The top three biological functions identified by IPA were cellular assembly and organization, small molecule biochemistry, and cell morphology (Figure 4A and Table 4). The top three pathways were RhoGDI signaling, signaling by Rho family GTPases, and remodeling of epithelial adherents junctions (Figure 4B and Table 5).
Figure 4.
Bioinformatic analysis in the hippocampus. Major biological functions and cellular processes most relevant to the proteins differently expressed in: (A) Flinders Sensitive Line (FSL) vs. Flinders Resistant Line (FRL), (C) FRL after maternal separation (MS), and (E) FSL after MS, as calculated by Ingenuity Pathways Analysis software. Canonical pathways most relevant to the proteins differently expressed in: (B) FSL vs. FRL, (D) FRL after MS, and (F) FSL after MS. Biological functions and canonical pathways are listed based on higher statistical significance [-log(p value)], with a threshold set at 1.3.
Effect of Early-Life Stress in FRL Hippocampus
In FRL rats, MS differently regulated 19 spots and mass spectrometry identified 16 spots representing 25 distinct proteins/protein isoforms (five spots containing a mixture of two proteins and two spots containing three proteins; Table 3). The three top molecular and cellular functions identified by IPA analysis were free radical scavenging, small molecule biochemistry, and carbohydrate metabolism (Figure 4C and Table 4). The top three pathways were superoxide radicals degradation, TCA cycle, and mitochondrial dysfunction (Figure 4D and Table 5).
Effects of Early-Life Stress in FSL Hippocampus
MS differently modulated 19 spots in FSL and mass spectrometry identified 14 spots representing 23 distinct proteins/protein isoforms (five spots containing a mixture of two proteins, and one spot containing a mixture of five proteins; Table 3). The top three molecular and cellular functions, revealed by bioinformatic analysis with IPA, were protein trafficking, cellular assembly and organization, and cellular function and maintenance (Figure 4E and Table 4). The top three canonical pathways were cell cycle (G2/M DNA damage checkpoint regulation), Myc-mediated apoptosis signaling, and ERK5 signaling (Figure 4F and Table 5).
The biological functions and canonical pathways most relevant to the differently-expressed proteins in HPC were related to cellular remodeling, including cellular assembly and organization, cellular function and maintenance, cell morphology, and Rho GTPase signaling.
Network Analysis
IPA analysis was used to determine networks of proteins significantly enriched in the various comparisons. IPA identified four networks in PFC/FC and four in HPC. The networks identified are shown in Figures 5 and 6, respectively. The proteins involved and the network scores and biofunctions associated are shown in Tables 6 and 7, respectively.
Figure 5.
Significant pathway networks based on the proteins differently expressed between (A) Flinders Sensitive Line (FSL) vs. Flinders Resistant Line (FRL), (B) FRL after maternal separation (MS), and (C and D) FSL after MS, in prefrontal and frontal cortices. Protein nodes with a colored background correspond to the identified proteins in the various comparisons, while other proteins were added from the Ingenuity database. The intensity of node colors indicates the degree of upregulation (red) or downregulation (green). Fischer’s exact test was used to calculate a p-value determining the probability that each biological function assigned to that network is due to chance alone.
Figure 6.
Significant pathway networks based on the differently expressed protein between (A) Flinders Sensitive Line (FSL) vs. Flinders Resistant Line (FRL), (B and C) FRL after maternal separation (MS), and (D) FSL after MS, in the hippocampus. Protein nodes with a colored background correspond to the identified proteins in the various comparisons, while other proteins were added from the Ingenuity database. The intensity of node colors indicates the degree of upregulation (red) or downregulation (green). Fischer’s exact test was used to calculate a p-value determining the probability that each biological function assigned to that network is due to chance alone.
Table 6.
Networks of Proteins Identified by IPA in PFC/FC.
| Proteins in the networka | Scoreb | No. of focus proteins | High-level functions |
|---|---|---|---|
| FSL vs. FRL | |||
| ALDOC, ATP6V0D1, DLAT, DLD, DNAJA4, DNAJB12, DNM1, FBXO11, FBXO45, FH, HNMT, HSPA8, HTT, IDH3A, INSR, MAPK3, NDUFS1, PDHB, PDHX, PRDX6, PRNP, SNAP25, Snare, SNPH, STX2, STX3, STXBP1, STXBP5, SYN3, SYTL3, TGFB1, TKT, TMEM132A, UBC, ZFAND2A | 32 | 11 | Cellular Function and Maintenance, Molecular Transport, Lipid Metabolism |
| FRL+MS vs. FRL | |||
| Akt, ALDOC, ASB9, CD3, DNAJB12, DNAJC6, DNAJC13, ETFB, FAM103A1, FBXO45, GAPDH, GNAO1, GPR158, HIF1A, HSPA8, HSPD1, IDH3A, IDH3B, IDH3G, IKBKG, MDH2, MPST, MYC, NCALD, NDUFS1, NGB, p85 (pik3r), PDHB, Pgk, PRDX6, RGS17, TMEM132A, TST, UBC, ZFAND2A | 28 | 10 | Nucleic Acid Metabolism, Small Molecule Biochemistry, Carbohydrate Metabolism |
| FSL+MS vs. FSL | |||
| 14-3-3, 14-3-3 (β,ε,ζ), 14-3-3 (β,γ,θ,η,ζ), 14-3-3 (η,θ,ζ), AANAT, Akt, ARRB2, ATP5A1, CBY1, CKMT1A/CKMT1B, CLTB, DENND4A, FAM13B, FAM53C, HECTD4, Histone h3, KIAA0101, LRFN1, MLXIP, Phb, PKM, PRDX6, RALGPS2, REM1, REM2, SAMD4A, SAMD4B, SH3BP5L, SYNPO2, TKT, TNF, YWHAB, YWHAG, YWHAH, YWHAZ | 26 | 11 | Cell Morphology, Embryonic Development, Organ Development |
| ACO2, ATP6V0D1, CSNK1G3, CYC1, GNAO1, GPR158, MDH1, NAPG, ND2, ND3, ND4L, NDUFA6, NDUFA8, NDUFA9, NDUFAF1, NDUFAF3, NDUFAF4, NDUFB6, NDUFS3, NDUFS5, NDUFS6, NDUFS7, NDUFS8, NDUFV2, NGB, NOA1, NSF, PANK2, RGS17, SLC25A24, STXBP1, SYTL3, UBC, UQCR10, VDAC1 | 23 | 10 | Hereditary Disorder, Metabolic Disease, Cardiovascular Disease |
aProteins are indicated with gene name
bScores >3 were considered significant (p<0.001)
FSL, Flinders Sensitive Line; FRL, Flinders Resistant Line; IPA, Ingenuity Pathways Analysis; MS, maternal separation; PFC/FC, prefrontal and frontal cortices.
Table 7.
Networks of Proteins identified by IPA in HPC.
| Proteins in the networka | Scoreb | No. of focus proteins | High-level functions |
|---|---|---|---|
| FSL vs. FRL | |||
| ACTB, ACTG1, Actin, CAP2, CFL2, COTL1, CPNE2, Dmd, DPYSL2, EPS8L1, EPS8L2, GAS8, GNAO1, GOT2, GPR158, HIP1R, KCNQ2, KLHL17, MAF1, NCALD, NDUFS3, NEFL, NSF, PCYT1B, PHACTR1, PLS1, RAB8B, RGS17, SPTBN2, SSH1, SUCLA2, SULT2B1, TRPM7, UBC, XPO6 | 26 | 9 | Cellular Assembly and Organization, Cancer, Connective Tissue Disorders |
| FRL+MS vs. FRL | |||
| 26s Proteasome, ACTB, Actin, Akt, ATP5A1, Beta Tubulin, BLK, BMP, DNAJC6, DPYSL2, ENO2, FSH, GLUD1, GOT1, Hsp70, Hsp90, HSP, HSPA2, HSPA8, Insulin, MT3, NFkB (complex), NMDA Receptor, NTF3, p85 (pik3r), PACRG, RTN4R, SEMA3A, SNAP25, SOD1, SOD2, Sod, Spectrin, Tubulin, VIM | 30 | 12 | Free Radical Scavenging, Cell Morphology, Cellular Assembly and Organization |
| AP2A1, ARHGDIA, ASPSCR1, ATP6V1A, ATP6V1B1, ATP6V1B2, ATP6V1E1, ATP6V1G2, AXIN1, CRMP1, DBI, GDI1, GLMN, IDH3A, IDH3B, IDH3G, IPO9, MECP2, MYC, NDUFS7, NDUFV2, NGLY1, NSFL1C, PHAX, Phb, ROCK2, RPL22, RTN4, SRP14, TUBA1A, UBC, UBXN6, UBXN2A, UBXN2B, VAV1 | 18 | 8 | Carbohydrate Metabolism, Nucleic Acid Metabolism, Small Molecule Biochemistry |
| FSL+MS vs. FSL | |||
| 14-3-3, 14-3-3 (β,γ,θ,η,ζ), 14-3-3 (η,θ,ζ), 26s Proteasome, Actin, Akt, ATP5A1, Calmodulin, CAMK2A, CaMKII, CD3, DPYSL2, ERK, Histone h3, HSP, HSPA2, HSPA8, HSPD1, IMMT, INA, ITPKA, NDUFS1, NEFL, p85 (pik3r), PGK1, PI3K (complex), REM1, REM2, TCR, TUFM, YWHAB, YWHAG, YWHAH, YWHAQ, YWHAZ | 48 | 17 | Protein Trafficking, Cellular Assembly and Organization, Cellular Function and Maintenance |
aProteins are indicated with gene name
bScores >3 were considered significant (p<0.001)
FSL, Flinders Sensitive Line; FRL, Flinders Resistant Line; HPC, hippocampus; IPA, Ingenuity Pathways Analysis; MS, maternal separation.
Western Blot Analysis
To validate our proteomic results, we used Western analysis to measure the expression levels of six proteins of interest (NSF, ATP synthase alpha, aconitate hydratase, syntaxin-binding protein 1, DRP-2, and SNAP-25; Figures 7 and Table 8). Consistent with our 2-DE results, Western analysis confirmed significant decreases for NSF (Figure 7A), ATP synthase alpha (Figure 7B), and aconitate hydratase (Figure 7C) levels in FSL+MS as compared with FSL+MS in PFC/FC. Western analysis also confirmed significantly increased levels of DRP-2 (Figure 7E) in FSL compared to FRL, while SNAP-25 (Figure 7F) showed a trend toward increases in FSL+MS compared to FSL+MS in the HPC (Table 8). Conversely, Western analysis for syntaxin-binding protein 1 (Figure 7D) levels in FSL+MS compared to FSL in PFC/FC did not confirm the 2-DE results, showing decreased rather than increased level.
Figure 7.
Validation of two-dimensional gel electrophoresis results. (A) N-ethylmaleimide sensitive factor (NSF), (B) ATP synthase α, (C) aconitate hydratase, and (D) Syntaxin-binding protein 1 protein levels were analyzed in prefrontal and frontal cortices synaptosomes of Flinders Sensitive Line (FSL) and Flinders Resistant Line (FRL) after maternal separation (MS) by Western blot. Protein levels were normalized to β-actin and expressed as % relative to FSL rats. Data are presented as mean ± standard error of the mean (SEM; n = 6–8). Mann-Whitney test, *p < 0.01; **p < 0.001. (E) Dihydropyrimidinase-related protein 2 (DRP-2) protein levels were analyzed in hippocampus (HPC) synaptosomes of FRL and FSL by Western blot. Protein levels were normalized to β-actin and expressed as % relative to FRL rats. Data are presented as mean ± SEM (n = 9–12). Mann-Whitney test, **p < 0.001. (F) Synaptosomal-associated protein 25 (SNAP-25) protein levels were analyzed in HPC synaptosomes of FRL and FRL after MS by Western blot. Protein levels were normalized to β-actin and expressed as % relative to FRL rats. Data are presented as mean ± SEM (n = 10–11). Mann-Whitney test.
Table 8.
Proteins Selected for Western Blotting Validations.
| Proteins | Fold change WB | p-value WBa | Fold change 2-DE spots |
|---|---|---|---|
| FSL+MS vs. FSL (PFC/FC) | |||
| NSF | -1.23 | p = 0,0019 | -1.2 |
| ATP synthase subunit alpha(b) | -1.25 | p = 0,0080 | -2.3; -1.8 |
| Aconitate hydratase | -1.31 | p = 0,0002 | -1.3 |
| Syntaxin-binding protein 1 | -1.25 | p = 0,0047 | 1.9 |
| FSL vs. FRL (HPC) | |||
| DRP-2 | 1.45 | p = 0,0003 | 1.8 |
| FRL+MS vs. FRL (HPC) | |||
| SNAP-25 | 1.12 | p = 0,0726 | 1.3 |
aUnpaired two-tailed Mann-Whitney test p-value
bMultiple spots of this protein were dysreguated in 2-DE
2-DE, two-dimensional gel electrophoresis; ATP, adenosine triphosphate; DRP-2, dihydropyrimidinase-related protein 2; FSL, Flinders Sensitive Line; FRL, Flinders Resistant Line; HPC, hippocampus; MS, maternal separation; NSF, N-ethylmaleimide sensitive factor; PFC/FC, prefrontal and frontal cortices; SNAP-25, synaptosomal-associated protein 25; WB, Western blot analysis.
Discussion
Early-life stress has been recognized as a risk factor for depression (Heim and Nemeroff, 2001). In the present study we employed a standard MS protocol to analyze the impact of early-life adverse events on the proteomic profile of purified synaptic terminals in the FSL/FRL genetic animal model of depression. We have previously explored the outcome of the interaction of early-life stress with the background of vulnerability of FSL rats. We found marked alterations in key regulators of presynaptic release/neurotransmission in the basal FSL rats as a result of early-life stress, such as blunted responses to the stress of synaptic Erk-MAP kinases. These findings suggested the occurrence of synaptic dysfunction in the GxE model. This was accompanied by remodeling of N-methyl-D-aspartate receptor-dependent hippocampal synaptic plasticity (Ryan et al., 2009; Musazzi et al., 2010). At the peripheral level (serum), proteomic analysis evidenced changes in pathways related with inflammation and regulation of metabolism (Carboni et al., 2010).
In the present work, for the first time, we carried out a global proteomic analysis of the FSL/FRL model at the synaptic level.
Here we used a bioinformatic approach to analyze the functional relevance of the numerous proteins found dysregulated in FSL versus FRL following early-life stress. Interestingly, we found a brain region–specific pattern of pathway enrichment. Indeed, we found that expression of the proteins involved in energy metabolism pathways was mostly changed in the PFC/FC area, suggesting that in this area energy metabolism is particularly affected by genetic vulnerability and early-life stress. This is not surprising since the brain is a high energy-demanding structure, especially the cortical areas, where glutamatergic synapses and neurotransmission are predominant (Bélanger et al., 2011). On the contrary, we found that expression of the proteins related to cellular remodeling pathways was mainly changed in the HPC. Indeed, numerous studies have highlighted the notion that changes in brain morphology represent a key factor in the response to stress, pathophysiology of depression, and antidepressant action. Indeed, volumetric changes in the PFC/FC and HPC have been reported both in depressed patients and animal models of depression. Furthermore, stress is able to induce dendritic retraction and reduction of spine number, thus affecting synaptic transmission (McEwen, 2005; Krishnan and Nestler, 2008; Gorman and Docherty, 2010; Sanacora et al., 2012; Sousa and Almeida, 2012).
Basal Differences Between FSL and FRL in Prefrontal and Frontal Cortices
In the present work, we found a number of proteins involved in energy metabolism pathways differently expressed in the basal comparison of FSL versus FRL in PFC/FC synaptosomes. Indeed, we found the energy metabolism proteins isocitrate dehydrogenase subunit alpha, fructose bisphosphate aldolase C and pyruvate dehydrogenase subunit beta were upregulated in FSL rats versus their control FRL rats. These modifications are in line with previous clinical and preclinical studies. Fructose bisphosphate aldolase C was found to be upregulated in the frontal cortices of patients with MD in two proteomic studies (Johnston-Wilson et al., 2000; Beasley et al., 2006), while pyruvate dehydrogenase (subunit alpha) was found to increase in total homogenate of PFC/FC of FSL in a previous proteomic study (Piubelli et al., 2011). Moreover, mitochondrial disorders (including mutation in the gene coding for pyruvate dehydrogenase subunit alpha) have been diagnosed in depressed adolescents (Koene et al., 2009).
In addition, we found that several proteins involved in synaptic function were dysregulated as a consequence of genetic vulnerability. In basal FSL versus FRL PFC/FC synaptosomes, proteins related to synaptic vesicle fusion and recycling were downregulated. Indeed, we found lower levels of syntaxin-binding protein 1 (also known as Munc-18), a protein involved in synaptic vesicle exocytosis (Jahn and Fasshauer, 2012); dynamin 1, a GTPase involved in clathrin-coated synaptic vesicles budding; and heat shock cognate 71 (also known as heat shock cognate 70, HSPA8), an ATPase involved in chaperoning SNAP-25 during synaptic function and in the clathrin uncoating of recycled vesicles (Sharma et al., 2011; McMahon and Boucrot, 2011). Interestingly, a recent hypothesis suggested that clathrin-dependent membrane and protein trafficking might be core processes involved in the pathophysiology of schizophrenia and bipolar disorder (Schubert et al., 2012). Furthermore, a proteomic study that analyzed the postmortem dorsolateral PFC from depressed patients, found reduced phosphorylation of dynamin-1 at different phosphorylation sites (Martins-de-Souza et al., 2012).
Effect of Early-Life Stress in FRL Prefrontal and Frontal Cortices
Early-life stress had an important effect on brain metabolic pathways in PFC/FC FRL rats. Firstly, we found that the genetic vulnerability in FSL and early-life stress in FRL result in a similar effect on pyruvate dehydrogenase beta (i.e., increased expression), again consistent with the previous proteomic study of FSL/FRL (Piubelli et al., 2011). Secondly, we found that early-life stress in FRL synaptosomes reduced the expression of several proteins involved in glycolysis, tricarboxilic acid cycle and oxidative phosphorylation (glyceraldehyde-3-phosphate dehydrogenase, fructose-bisphosphate aldolase C, isocitrate dehydrogenase, malate dehydrogenase, NADH-ubiquinone oxidoreductase 75kDa), suggesting a reduction of energy production. Indeed, a previous proteomic work on maternally-separated rats reported a downregulation of isocitrate dehydrogenase and glyceraldehyde-3-phosphate dehydrogenase similar to what we found here for FRL rats subjected to the same protocol of stress (Marais et al., 2009). Interestingly, a previous proteomic study from our group showed a similar downregulation of several mitochondrial and energy metabolism proteins in PFC/FC synaptosomes following induction of learned helplessness in rats (Mallei et al., 2011).
Effect of Early-Life Stress in FSL Prefrontal and Frontal Cortices
Early MS had a deeper impact on the synaptic proteome of FSL compared with FRL rats, with twice the number of proteins found dysregulated. In FSL, we found a decrease in proteins related to energy metabolism, such as components of oxidative phosphorylation complexes I (NADH-ubiquinone oxidoreductases), III (cytochrome C1), and V (ATP synthase alpha), malate dehydrogenase, and aconitate hydratase. Notably, neuroimaging studies have shown reduced levels of ATP in the brains of patients with mood disorders compared to controls, thus indicating lower metabolic brain activity in depressed patients (Moretti et al., 2003). Indeed, we found reduced levels of ATP synthase alpha in FSL rats subjected to MS. On the contrary, we found increased levels of creatine kinase, the enzyme that catalyzes the conversion of creatine to its high energy phosphorylated form phosphocreatine with use of ATP (Béard and Braissant, 2010). This upregulation of creatine kinase in response to early-life stress could be explained by a compensatory mechanism for the possible reduction of ATP, due to lower ATP synthase alpha level.
Early-life stress also reduced expression of two proteins involved in synaptic transmission: NSF, an ATPase involved in the disassembly of SNARE complexes and synaptic vesicle recycling (Südhof and Rizo, 2011) and clathrin light chain B. This piece of evidence further supports the involvement of clathrin-mediated endocythosis/trafficking in the pathophysiology of psychiatric diseases (Schubert et al., 2012).
Basal Differences Between FSL and FRL in Hippocampus
In the present study we report modifications in the expression of several proteins involved in cellular remodeling processes; it is interesting to note that these modifications were found mainly in HPC. Actin is a key cytoskeletal protein involved in axon guidance, synapse development, and synaptic plasticity (Yao et al., 2006; Wong et al., 2013; Chia et al., 2013). Here we found a dysregulation of several actin isoforms in basal FSL versus FRL rats. This result is in agreement with a previous proteomic work in the same GxE animal model, that analyzed the total homogenate of PFC/FC and HPC (Piubelli et al., 2011). Moreover, in the present study, neurofilament light neuropeptide (NEFL), an intermediate neurofilament protein involved in axonal and dendritic growth (Yuan et al., 2006), was found to be modulated by genetic vulnerability (FSL vs. FRL).
Effect of Early-Life Stress in FRL Hippocampus
In FRL, early-life stress modulated levels of actin isoforms in HPC synaptosomes; similar modifications have been found previously in the HPC total homogenate of FRL rats subjected to the same protocol of MS (Piubelli et al., 2011). In addition, we found a downregulation of DRP-2 in both FRL and FSL (see below). Notably, we found an increased expression of the SNARE-complex protein SNAP-25 and downregulation of three distinct protein isoforms of HSPA8, a component of the chaperone complex that prevents misfolding of SNAP-25 (Sharma et al., 2011; Südhof, 2013).
Effect of Early-Life Stress in FSL Hippocampus
NEFL and alpha-internexin, other intermediate neurofilament proteins involved in axonal and dendritic growth (Yuan et al., 2006), were found to be modulated by early-life stress in FSL HPC. In line with the present findings, modifications of these two proteins were also found in other animal models of depression, such as the psychosocial stress (Carboni, Piubelli, et al., 2006) and the learned helplessness model (Reinés et al., 2004), or after long-term treatment with the stress hormone corticosterone (Zhao et al., 2009). Notably, enriched environment, physical exercise, chronic electroconvulsive shock, or treatment with valproic acid (but not fluoxetine) modulate NEFL levels in rodents (Vaidya et al., 2000; Ding et al., 2006; Ferrero et al., 2007; Sifonios et al., 2009).
In the present study, we found additional evidence involving cytoskeleton-remodeling pathways as an outcome of early-life stress. The 14-3-3 proteins are adaptor proteins involved in intracellular signaling, cell growth, apoptosis, ion channel function, and neurotransmission (Berg et al., 2003). Moreover, 14-3-3 proteins interact with NEFL and have a role in NEFL disassembly (Miao et al., 2013). The increased levels of 14-3-3 proteins, found here in both PFC/FC and HPC synaptosomes following MS in FSL rats, suggest increased disassembly of neurofilaments and possibly altered dynamics of neurofilaments at the synaptic level. Increased levels of 14-3-3 have also been found in the chronic unpredictable stress model of depression (Mu et al., 2007). Interestingly, 14-3-3 proteins have been found to be altered in schizophrenia and bipolar disorder (Altar et al., 2009) after treatment with the antidepressant fluoxetine, or the mood stabilizers valproate and lithium (Altar et al., 2009; Nanavati et al., 2011).
DRP-2 (also known as CRMP2) is a cytosolic protein involved in axonal guidance and growth, cell migration, signal transduction, and neuronal differentiation (Charrier et al., 2003; Mallei et al., 2011; Ip et al., 2014). DRP-2 interacts with the cytoskeleton proteins tubulin, actin, and vimentin, and is able to regulate microtubule dynamics, based on its phosphorylated state, by stabilizing/destabilizing tubulin heterodimer (Khanna et al., 2012). We found DRP-2 downregulated following MS in both FRL and FSL rats. Two proteomic studies of post-mortem brain of depressed patients found DRP-2 downregulated as in the present study (Johnston-Wilson et al., 2000; Martins-de-Souza et al., 2012), while a third found DRP-2 upregulated (Beasley et al., 2006). Involvement of DRP-2 in the pathophysiology of depression is confirmed also by several preclinical studies (Khawaja et al., 2004; Carboni, Vighini, et al., 2006; Mallei et al., 2011).
In the present work, as already mentioned, the changes in proteins involved in energy metabolism were found mainly in PFC/FC; however, in HPC we also found a reduction of ATP synthase alpha following early-life stress in FSL, similar to what we observed in PFC/FC.
Network Analysis
In genetically complex diseases such as depression, multiple genetic factors of small effect interact among themselves and with environmental factors to precipitate the pathology (Wong and Licinio, 2001). In this context it is important not only to identify the many genes and proteins involved but also to unveil the molecular pathways associated. It is interesting to note that in the networks related to basal comparison in both PFC/FC (Figure 5A) and HPC (Figure 6A), a major hub is formed by ubiquitin C (Ubc). Ubc, in combination with the 26S proteasome, has a central role in proteolytic degradation of substrates in multiple processes but is involved also in non-proteolytic regulatory mechanisms, including membrane protein endocytosis and intracellular trafficking (Hochstrasser, 2009). Moreover, the ubiquitin-proteasome pathway has been also involved in synaptic plasticity (Hegde, 2010). In the basal PFC/FC network (Figure 5A), Ubc is connected to a cluster formed by proteins involved in synaptic function, such as dynamin 1, syntaxin-binding protein 1, and synaptosomal associated protein of 25kDa. Moreover, Ubc is also linked through multiple connections to a cluster of proteins that form the pyruvate dehydrogenase complex, a key mitochondrial enzyme that links the glycolysis to the tricarboxylic acid cycle (Patel and Korotchkina, 2006). Instead, in the network related to the basal comparison in HPC (Figure 6A), Ubc is connected to a large cluster of proteins that interact with the cytoskeletal protein actin. Therefore, network analysis of the present results shows cross-interaction of pathways for energy metabolism, cytoskeleton remodeling, and synaptic transmission.
Validation by Western Blot Analysis
Validation of dysregulated proteins by independent methods is an important step of a proteomic study. In this work, we used Western analysis to confirm the differential expression of six proteins found dysregulated in the present study. These proteins were selected based on expression levels observed in 2-DE maps, fold-changes, and availability of commercial antibodies. The six proteins were NSF, ATP synthase alpha, aconitate hydratase, syntaxin-binding protein 1, DRP-2, and SNAP-25. Our results show a strong agreement with the 2-DE expression changes, with the exception of syntaxin-binding protein 1, which showed change in the opposite direction (decrease). This discrepancy could be explained by the fact that 2-DE is able to detect the differential expression of single isoforms or post-translational modifications of a protein, while Western analysis can measure only the total amount of that protein. Indeed, syntaxin-binding protein 1 has two alternatively-spliced isoforms and several phosphorylation sites (Latham and Meunier, 2007). It is possible that 2-DE experiment highlighted a change in protein expression of a low-abundant post-translational modified isoform. Furthermore, in a previous work we found a similar reduction of syntaxin-binding protein 1 in HPC synaptosomes from FSL subjected to MS by using Western blotting analysis (Musazzi et al., 2010). The case for syntaxin-binding protein 1 in this study highlights the importance of protein post-translational modifications. This is exemplified by the finding that some proteins appear on gels in multiple spots. For instance, HSPA8 protein appears on the gel in Figure 2B in at least 4 spots (1903, 1907, 2901, 2902) with same molecular weight, and 1 spot (1905) with higher molecular weight. The first 4 spots are likely to be different post-translational modifications of the same protein. Therefore, changes in post-translational modifications can be at least as important as expression changes in regulating protein function, and in turn the phenotype of FSL.
Conclusion
Taken together, our results indicate an overall dysregulation of proteins related to energy metabolism in PFC/FC of FSL/FRL as a consequence of genetic vulnerability and early-life stress. Indeed, mitochondrial dysfunction and mitochondrial regulation of energy metabolism could be central in the pathophysiology of stress-related pathologies such as depression (Morava and Kozicz, 2013; Picard et al., 2014). In addition, the dysregulation of structural proteins found here in HPC suggests that genetic vulnerability and early-life stress may cause perturbation of cytoskeleton dynamics at the synaptic level, which is now being recognized as a cellular/molecular feature of depression (Nakatani et al., 2007; Wong et al., 2013).
Interestingly, it was recently found that the depressed-like phenotype of FSL rats could be linked to hyperfunctioning of a G protein–coupled, inward rectifying potassium channel (GIRK). Indeed, GIRK knockout mice showed reduced hypothermic responses to virtually all drugs that typically induce greater hypothermic response in FSL rats. Possible hyperfunctioning of GIRK could explain the increased sensitivity to multiple G protein–coupled receptors in FSL (Overstreet and Wegener, 2013). We wonder whether the marked proteomic changes we found in FSL could be linked in some way to GIRK abnormalities in this rat line. Additional experiments are required to explore this interesting hypothesis.
Statement of Interest
None.
Acknowledgments
This work was supported by a European Union (6th Framework Program) grant for project GENDEP to Drs Popoli and Mathé (grant number LSHB-CT-2003–503428); the Swedish Medical Research Council grant to Dr Mathé (grant number 10414); and the Karolinska Institutet.
References
- Altar CA, Vawter MP, Ginsberg SD. (2009). Target identification for CNS diseases by transcriptional profiling. Neuropsychopharmacology 34:18–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béard E, Braissant O. (2010). Synthesis and transport of creatine in the CNS: importance for cerebral functions. J Neurochem 115:297–313. [DOI] [PubMed] [Google Scholar]
- Beasley CL, Pennington K, Behan A, Wait R, Dunn MJ, Cotter D. (2006). Proteomic analysis of the anterior cingulate cortex in the major psychiatric disorders: Evidence for disease-associated changes. Proteomics 6(11):3414–3425. [DOI] [PubMed] [Google Scholar]
- Bélanger M, Allaman I, Magistretti PJ. (2011). Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab 14:724–738. [DOI] [PubMed] [Google Scholar]
- Berg D, Holzmann C, Riess O. (2003). 14-3-3 proteins in the nervous system. Nat Rev Neurosci 4:752–762. [DOI] [PubMed] [Google Scholar]
- Carboni L, Piubelli C, Pozzato C, Astner H, Arban R, Righetti PG, Hamdan M, Domenici E. (2006). Proteomic analysis of rat hippocampus after repeated psychosocial stress. Neuroscience 137:1237–1246. [DOI] [PubMed] [Google Scholar]
- Carboni L, Vighini M, Piubelli C, Castelletti L, Milli A, Domenici E. (2006). Proteomic analysis of rat hippocampus and frontal cortex after chronic treatment with fluoxetine or putative novel antidepressants: CRF1 and NK1 receptor antagonists. Eur Neuropsychopharmacol 16:521–537. [DOI] [PubMed] [Google Scholar]
- Carboni L, Becchi S, Piubelli C, Mallei A, Giambelli R, Razzoli M, Mathé AA, Popoli M, Domenici E. (2010). Early-life stress and antidepressants modulate peripheral biomarkers in a gene-environment rat model of depression. Prog Neuropsychopharmacol Biol Psychiatry 34:1037–1048. [DOI] [PubMed] [Google Scholar]
- Caspi A, Moffitt TE. (2006). Gene-environment interactions in psychiatry: joining forces with neuroscience. Nat Rev Neurosci 7:583–590. [DOI] [PubMed] [Google Scholar]
- Caspi A, McClay J, Moffitt TE, Mill J, Martin J, Craig IW, Taylor A, Poulton R. (2002). Role of genotype in the cycle of violence in maltreated children. Science 297:851–854. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R. (2003). Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science 301:386–389. [DOI] [PubMed] [Google Scholar]
- Chia PH, Li P, Shen K. (2013). Cellular and molecular mechanisms underlying presynapse formation. J Cell Biol 203:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier E, Reibel S, Rogemond V, Aguera M, Thomasset N, Honnorat J. (2003). Collapsin response mediator proteins (CRMPs): involvement in nervous system development and adult neurodegenerative disorders. Mol Neurobiol 28:51–64. [DOI] [PubMed] [Google Scholar]
- Ding Q, Vaynman S, Souda P, Whitelegge JP, Gomez-Pinilla F. (2006). Exercise affects energy metabolism and neural plasticity-related proteins in the hippocampus as revealed by proteomic analysis. Eur J Neurosci 24:1265–1276. [DOI] [PubMed] [Google Scholar]
- Dunkley PR, Jarvie PE, Heath JW, Kidd GJ, Rostas JA. (1986). A rapid method for isolation of synaptosomes on Percoll gradients. Brain Res 372:115–129. [DOI] [PubMed] [Google Scholar]
- El Khoury A, Gruber SH, Mørk A, Mathé AA. (2006). Adult life behavioral consequences of early maternal separation are alleviated by escitalopram treatment in a rat model of depression. Prog Neuropsychopharmacol Biol Psychiatry 30:535–540. [DOI] [PubMed] [Google Scholar]
- Ferrero AJ, Cereseto M, Sifonios LL, Reinés A, Peixoto E, Rubio MC, Wikinski S. (2007). Cytoskeleton of hippocampal neurons as a target for valproic acid in an experimental model of depression. Prog Neuropsychopharmacol Biol Psychiatry 31:1419–1428. [DOI] [PubMed] [Google Scholar]
- Fountoulakis M. (2004). Application of proteomics technologies in the investigation of the brain. Mass Spectrom Rev 23:231–258. [DOI] [PubMed] [Google Scholar]
- Gardner KL, Thrivikraman KV, Lightman SL, Plotsky PM, Lowry CA. (2005). Early life experience alters behavior during social defeat: focus on serotonergic systems. Neuroscience 136:181–191. [DOI] [PubMed] [Google Scholar]
- Gorman JM, Docherty JP. (2010). A hypothesized role for dendritic remodeling in the etiology of mood and anxiety disorders. J Neuropsychiatry Clin Neurosci 22:256–264. [DOI] [PubMed] [Google Scholar]
- Heim C, Nemeroff CB. (2001). The role of childhood trauma in the neurobiology of mood and anxiety disorders: preclinical and clinical studies. Biol Psychiatry 49:1023–1039. [DOI] [PubMed] [Google Scholar]
- Hegde AN. (2010). The ubiquitin-proteasome pathway and synaptic plasticity. Learn Mem 17:314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. (2009). Origin and function of ubiquitin-like proteins. Nature 458:422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husum H, Wörtwein G, Andersson W, Bolwig TG, Mathé AA. (2008). Gene-environment interaction affects substance P and neurokinin A in the entorhinal cortex and periaqueductal grey in a genetic animal model of depression: implications for the pathophysiology of depression. Int J Neuropsychop 11:93–101. [DOI] [PubMed] [Google Scholar]
- Ip JP, Fu AK, Ip NY. (2014). CRMP2: functional roles in neural development and therapeutic potential in neurological diseases. Neuroscientist. Advance online publication. Retrieved 10 Feb 2014. 10.1177/1073858413514278. [DOI] [PubMed] [Google Scholar]
- Jahn R, Fasshauer D. (2012). Molecular machines governing exocytosis of synaptic vesicles. Nature 490:201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Vasquez PA, Diaz-Cabiale Z, Caberlotto L, Bellido I, Overstreet D, Fuxe K, Mathé AA. (2007). Electroconvulsive stimuli selectively affect behavior and neuropeptide Y (NPY) and NPY Y(1) receptor gene expressions in hippocampus and hypothalamus of Flinders Sensitive Line rat model of depression. Eur Neuropsychopharmacol 17:298–308. [DOI] [PubMed] [Google Scholar]
- Johnston-Wilson NL, Sims CD, Hofmann JP, Anderson L, Shore AD, Torrey EF, Yolken RH. (2000). Disease-specific alterations in frontal cortex brain proteins in schizophrenia, bipolar disorder, and major depressive disorder. The Stanley Neuropathology Consortium. Mol Psychiatry 5:142–149. [DOI] [PubMed] [Google Scholar]
- Khanna R, Wilson SM, Brittain JM, Weimer J, Sultana R, Butterfield A, Hensley K. (2012). Opening Pandora’s jar: a primer on the putative roles of CRMP2 in a panoply of neurodegenerative, sensory and motor neuron, and central disorders. Future Neurol 7:749–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khawaja X, Xu J, Liang JJ, Barrett JE. (2004). Proteomic analysis of protein changes developing in rat hippocampus after chronic antidepressant treatment: implications for depressive disorders and future therapies. J Neurosci Res 75:451–460. [DOI] [PubMed] [Google Scholar]
- Koene S, Kozicz TL, Rodenburg RJ, Verhaak CM, de Vries MC, Wortmann S, van de Heuvel L, Smeitink JA, Morava E. (2009). Major depression in adolescent children consecutively diagnosed with mitochondrial disorder. J Affect Disord 114:327–332. [DOI] [PubMed] [Google Scholar]
- Krishnan V, Nestler EJ. (2008). The molecular neurobiology of depression. Nature 455:894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham CF, Meunier FA. (2007). Munc18a: Munc-y business in mediating exocytosis. Int J Biochem Cell Biol 39:1576–1581. [DOI] [PubMed] [Google Scholar]
- Mallei A, Giambelli R, el Khoury A, Gruber SHM, Musazzi L, Barbiero VS, Tardito D, Vollmayr B, Gass P, Mathé AA, Racagni G, Popoli M. (2008). Synaptoproteomics of existing and new animal models of depression. In: Biomarkers for psychiatric disorders (Turk CW, ed), pp185–202. New York: Springer. [Google Scholar]
- Mallei A, Giambelli R, Gass P, Racagni G, Mathé AA, Vollmayr B, Popoli M. (2011). Synaptoproteomics of learned helpless rats involve energy metabolism and cellular remodeling pathways in depressive-like behavior and antidepressant response. Neuropharmacology 60:1243–1253. [DOI] [PubMed] [Google Scholar]
- Marais L, Hattingh SM, Stein DJ, Daniels WM. (2009). A proteomic analysis of the ventral hippocampus of rats subjected to maternal separation and escitalopram treatment. Metab Brain Dis 24:569–586. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Guest PC, Vanattou-Saifoudine N, Rahmoune H, Bahn S. (2012). Phosphoproteomic differences in major depressive disorder postmortem brains indicate effects on synaptic function. Eur Arch Psychiatry Clin Neurosci 262:657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS. (2005). Glucocorticoids, depression, and mood disorders: structural remodeling in the brain. Metabolism 54:20–23. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. (2011). Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 12:517–533. [DOI] [PubMed] [Google Scholar]
- Miao L, Teng J, Lin J, Liao X, Chen J. (2013). 14-3-3 proteins interact with neurofilament protein-L and regulate dynamic assembly of neurofilaments. J Cell Sci 126:427–436. [DOI] [PubMed] [Google Scholar]
- Moretti A, Gorini A, Villa RF. (2003). Affective disorders, antidepressant drugs and brain metabolism. Mol Psychiatry 8:773–785. [DOI] [PubMed] [Google Scholar]
- Morava E, Kozicz T. (2013). Mitochondria and the economy of stress (mal)adaptation. Neurosci Biobehav Rev 37:668–680. [DOI] [PubMed] [Google Scholar]
- Mu J, Xie P, Yang ZS, Yang DL, Lv FJ, Luo TY, Li Y. (2007). Neurogenesis and major depression: implications from proteomic analyses of hippocampal proteins in a rat depression model. Neurosci Lett 416:252–256. [DOI] [PubMed] [Google Scholar]
- Musazzi L, Mallei A, Tardito D, Gruber SH, El Khoury A, Racagni G, Mathé AA, Popoli M. (2010). Early-life stress and antidepressant treatment involve synaptic signaling and Erk kinases in a gene-environment model of depression. J Psychiatr Res 44:511–520. [DOI] [PubMed] [Google Scholar]
- Nakatani N, Ohnishi T, Iwamoto K, Watanabe A, Iwayama Y, Yamashita S, Ishitsuka Y, Moriyama K, Nakajima M, Tatebayashi Y, Akiyama H, Higuchi T, Kato T, Yoshikawa T. (2007). Expression analysis of actin-related genes as an underlying mechanism for mood disorders. Biochem Biophys Res Commun 352:780–786. [DOI] [PubMed] [Google Scholar]
- Nanavati D, Austin DR, Catapano LA, Luckenbaugh DA, Dosemeci A, Manji HK, Chen G, Markey SP. (2011). The effects of chronic treatment with mood stabilizers on the rat hippocampal post-synaptic density proteome. J Neurochem 119:617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann ID, Wegener G, Homberg JR, Cohen H, Slattery DA, Zohar J, Olivier JD, Mathé AA. (2011). Animal models of depression and anxiety: What do they tell us about human condition? Prog Neuropsychopharmacol Biol Psychiatry 35(6):1357–1375. [DOI] [PubMed] [Google Scholar]
- Nugent NR, Tyrka AR, Carpenter LL, Price LH. (2011). Gene-environment interactions: early life stress and risk for depressive and anxiety disorders. Psychopharmacology (Berl) 214:175–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overstreet DH, Friedman E, Mathé AA, Yadid G. (2005). The Flinders Sensitive Line rat: a selectively bred putative animal model of depression. Neurosci Biobehav Rev 29:739–759. [DOI] [PubMed] [Google Scholar]
- Overstreet DH, Russell RW. (1982). Selective breeding for diisopropyl fuorophosphate-sensitivity: behavioural effects of cholinergic agonists and antagonists. Psychopharmacology (Berl) 78:150–155. [DOI] [PubMed] [Google Scholar]
- Overstreet DH, Wegener G. (2013). The flinders sensitive line rat model of depression–25 years and still producing. Pharmacol Rev 65:143–55. [DOI] [PubMed] [Google Scholar]
- Patel MS, Korotchkina LG. (2006). Regulation of the pyruvate dehydrogenase complex. Biochem Soc Trans 34:217–222. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. (1998). The rat brain in stereotaxic coordinates, fourth edition. New York: Academic Press. [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567. [DOI] [PubMed] [Google Scholar]
- Picard M, Juster RP, McEwen BS. (2014). Mitochondrial allostatic load puts the ‘gluc’ back in glucocorticoids. Nat Rev Endocrinol 10:303–310. [DOI] [PubMed] [Google Scholar]
- Piubelli C, Vighini M, Mathé AA, Domenici E, Carboni L. (2011). Escitalopram modulates neuron-remodelling proteins in a rat gene-environment interaction model of depression as revealed by proteomics. Part I: genetic background. Int J Neuropsychop 14:796–833. [DOI] [PubMed] [Google Scholar]
- Plotsky PM, Meaney MJ. (1993). Early, postnatal experience alters hypothalamic corticotropin-releasing factor (CRF) mRNA, median eminence CRF content and stress-induced release in adult rats. Brain Res Mol Brain Res 18:195–200. [DOI] [PubMed] [Google Scholar]
- Reinés A, Cereseto M, Ferrero A, Bonavita C, Wikinski S. (2004). Neuronal cytoskeletal alterations in an experimental model of depression. Neuroscience 129:529–38. [DOI] [PubMed] [Google Scholar]
- Rohlff C, Hollis K. (2003). Modern proteomic strategies in the study of complex neuropsychiatric disorders. Biol Psychiatry 53:847–853. [DOI] [PubMed] [Google Scholar]
- Ryan B, Musazzi L, Mallei A, Tardito D, Gruber SH, El Khoury A, Anwyl R, Racagni G, Mathé AA, Rowan MJ, Popoli M. (2009). Remodelling by early-life stress of NMDA receptor-dependent synaptic plasticity in a gene-environment rat model of depression. Int J Neuropsychop 12:553–559. [DOI] [PubMed] [Google Scholar]
- Sanacora G, Treccani G, Popoli M. (2012). Towards a glutamate hypothesis of depression: an emerging frontier of neuropsychopharmacology for mood disorders. Neuropharmacology 62:63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherl A, Couté Y, Déon C, Callé A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ. (2002). Functional proteomic analysis of human nucleolus. Mol Biol Cell 13:4100–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert KO, Föcking M, Prehn JH, Cotter DR. (2012). Hypothesis review: are clathrin-mediated endocytosis and clathrin-dependent membrane and protein trafficking core pathophysiological processes in schizophrenia and bipolar disorder? Mol Psychiatry 17:669–681. [DOI] [PubMed] [Google Scholar]
- Sharma M, Burré J, Südhof TC. (2011). CSPα promotes SNARE-complex assembly by chaperoning SNAP-25 during synaptic activity. Nat Cell Biol 13:30–39. [DOI] [PubMed] [Google Scholar]
- Sifonios L, Trinchero M, Cereseto M, Ferrero A, Cladouchos ML, Macedo GF, Reinés A, Wikinski S. (2009). An enriched environment restores normal behavior while providing cytoskeletal restoration and synaptic changes in the hippocampus of rats exposed to an experimental model of depression. Neuroscience 164:929–940. [DOI] [PubMed] [Google Scholar]
- Sousa N, Almeida OF. (2012). Disconnection and reconnection: the morphological basis of (mal)adaptation to stress. Trends Neurosci 35:742–751. [DOI] [PubMed] [Google Scholar]
- Südhof TC. (2013). Neurotransmitter release: the last millisecond in the life of a synaptic vesicle. Neuron 80:675–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC, Rizo J. (2011). Synaptic vesicle exocytosis. Cold Spring Harb Perspect Biol 3:a005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya VA, Terwilliger RZ, Duman RS. (2000). Alterations in heavy and light neurofilament proteins in hippocampus following chronic ECS administration. Synapse 35:137–143. [DOI] [PubMed] [Google Scholar]
- Vazquez DM, Lopez JF, Van Hoers H, Watson SJ, Levine S. (2000). Maternal deprivation regulates serotonin 1A and 2A receptors in the infant rat. Brain Res 855:76–82. [DOI] [PubMed] [Google Scholar]
- Vercauteren FG, Arckens L, Quirion R. (2007). Applications and current challenges of proteomic approaches, focusing on two-dimensional electrophoresis. Amino Acids 33:405–414. [DOI] [PubMed] [Google Scholar]
- Wegener G, Mathé AA, Neumann ID. (2012). Selectively bred rodents as models of depression and anxiety. Curr Top Behav Neurosci 12:139–187. [DOI] [PubMed] [Google Scholar]
- Wittchen HU, Jacobi F, Rehm J, Gustavsson A, Svensson M, Jönsson B, Olesen J, Allgulander C, Alonso J, Faravelli C, Fratiglioni L, Jennum P, Lieb R, Maercker A, van Os J, Preisig M, Salvador-Carulla L, Simon R, Steinhausen HC. (2011). The size and burden of mental disorders and other disorders of the brain in Europe 2010. Eur Neuropsychopharmacol 21:655–679. [DOI] [PubMed] [Google Scholar]
- Wong GT, Chang RC, Law AC. (2013). A breach in the scaffold: the possible role of cytoskeleton dysfunction in the pathogenesis of major depression. Ageing Res Rev 12:67–75. [DOI] [PubMed] [Google Scholar]
- Wong ML, Licinio J. (2001). Research and treatment approaches to depression. Nat Rev Neurosci 2:343–351. [DOI] [PubMed] [Google Scholar]
- World Health Organization (2008). The Global Burden of Disease: 2004 Update. Geneva, Switzerland: World Health Organization. [Google Scholar]
- Wörtwein G, Husum H, Andersson W, Bolwig TG, Mathé AA. (2006). Effects of maternal separation on neuropeptide Y and calcitonin gene-related peptide in “depressed” Flinders Sensitive Line rats: a study of gene-environment interactions. Prog Neuropsychopharmacol Biol Psychiatry 30:684–693. [DOI] [PubMed] [Google Scholar]
- Yao J, Qi J, Chen G. (2006). Actin-dependent activation of presynaptic silent synapses contributes to long-term synaptic plasticity in developing hippocampal neurons. J Neurosci 26:8137–8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan A, Rao MV, Sasaki T, Chen Y, Kumar A, Veeranna, Liem RK, Eyer J, Peterson AC, Julien JP, Nixon RA. (2006). Alpha-internexin is structurally and functionally associated with the neurofilaments triplet proteins in the mature CNS. J Neurosci 26:10006–10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xie W, Dai J, Wang Z, Huang Y. (2009). The varying effects of short-term and long-term corticosterone injections on depression-like behavior in mice. Brain Res 1261:82–90. [DOI] [PubMed] [Google Scholar]








