Abstract
Alternative splicing of precursor mRNA is an essential mechanism to increase the complexity of gene expression, and it plays an important role in cellular differentiation and organism development. Regulation of alternative splicing is a complicated process in which numerous interacting components are at work, including cis-acting elements and trans-acting factors, and is further guided by the functional coupling between transcription and splicing. Additional molecular features, such as chromatin structure, RNA structure and alternative transcription initiation or alternative transcription termination, collaborate with these basic components to generate the protein diversity due to alternative splicing. All these factors contributing to this one fundamental biological process add up to a mechanism that is critical to the proper functioning of cells. Any corruption of the process may lead to disruption of normal cellular function and the eventuality of disease. Cancer is one of those diseases, where alternative splicing may be the basis for the identification of novel diagnostic and prognostic biomarkers, as well as new strategies for therapy. Thus, an in-depth understanding of alternative splicing regulation has the potential not only to elucidate fundamental biological principles, but to provide solutions for various diseases.
Keywords: alternative splicing, regulation, precursor mRNA, mechanism, disease
1. Introduction
The discovery of the phenomenon that viral sequences are removed from a pre-mRNA and the remaining sequences are joined together led to a fundamental principle governing biology, known as RNA splicing. The identification stimulated theories for protein diversity, such as alternative splicing, which over time have been realized repeatedly through experiments. Gilbert (1) first proposed the concept of alternative splicing in 1978, which is currently the mechanism that accounts for the discrepancy between the number of protein-coding genes (~25,000) in humans and the >90,000 different proteins that are actually generated (2, 3). The notion of ‘one gene-one RNA-one protein’ is no longer relevant. More than 95% of human genes have been found to undergo splicing in a developmental, tissue-specific or signal transduction-dependent manner (4).
Constitutive splicing is the process of intron removal and exon ligation of the majority of the exons in the order in which they appear in a gene. Alternative splicing is a deviation from this preferred sequence where certain exons are skipped resulting in various forms of mature mRNA. Weaker splicing signals at alternative splice sites, shorter exon length or higher sequence conservation surrounding orthologous alternative exons influence the exons that are ultimately included in the mature mRNA (5). This process is mediated by a dynamic and flexible macromolecular machine, the spliceosome, which works in a synergistic and antistatic manner (as explained below) (6, 7). Three possible mechanisms, exon shuffling, exonization of transposable elements and constitutively spliced exons, have been proposed for the origin of alternative splicing (8).
Numerous studies have reiterated the critical and fundamental role of alternative splicing across biological systems (9). The species of higher eukaryotes have been discovered to exhibit a higher proportion of alternatively spliced genes, which is an underlying indication of a prominent role for the mechanism in evolution. Alternative splicing mediates diverse biological processes over the entire life span of organisms, from before birth to death (10, 11). Conserved splicing to species-specific splice variants play a significant functional role in species differentiation and genome evolution (12, 13), as well as in the development of functionally simple to complex tissues with diverse cell types, such as the brain, testis and the immune system. Alternative splicing even participates in RNA processing itself, from pre- to post-transcriptional events.
Thus, alternative splicing has a role in almost every aspect of protein function, including binding between proteins and ligands, nucleic acids or membranes, localization and enzymatic properties. Taken together, alternative splicing is a central element in gene expression (14).
2. Molecular mechanisms of alternative spicing
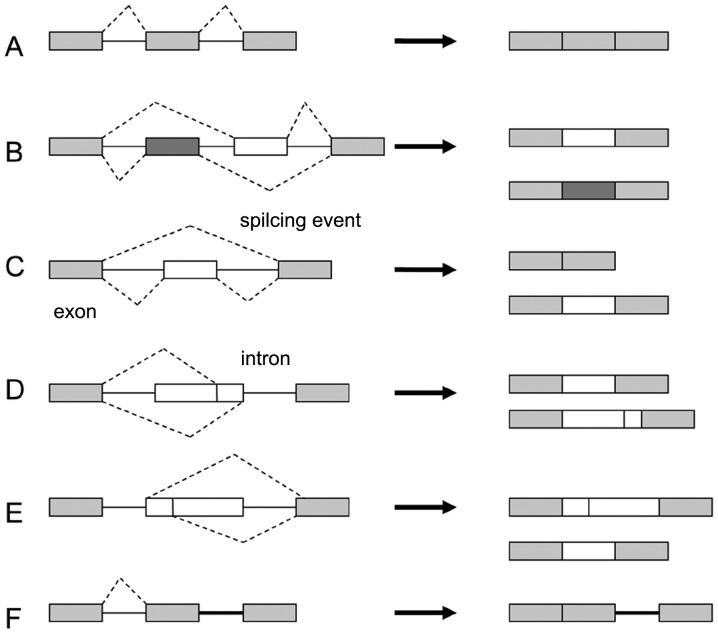
Systematic analyses of ESTs and microarray data have so far revealed seven main types of alternative splicing (12) (Fig. 1). The most prevalent pattern (~30%) is the cassette-type alternative exon (exon skipping) in vertebrates and invertebrates (Fig. 1C), while in lower metazoans, it is intron retention (Fig. 1F) (15). Intron retention in human transcripts is positioned primarily in the untranslated regions (UTRs) (16) and has been associated with weaker splice sites, short intron length and the regulation of cis-regulatory elements (17).
Figure 1.
Five main types of alternative splicing events are depicted. (A) Constitutive splicing; (B) mutually exclusive exons; (C) cassette alternative exon; (D) alternative 3′ splice site; (E) alternative 5′ splice site; and (F) intron retention.
Alternative selection of 5′ or 3′ splice sites within exon sequences (~25%) may lead to subtle changes in the coding sequence (Fig. 1D and E), and an additional layer of complexity arises with mutually exclusive alternative exons (Fig. 1B). One example of a transcript that undergoes alternative splicing, which generates variation in the protein, is FGFR2. Differences in the splicing machinery in different cell types and unique cis-acting elements in the FGF-R2 pre-mRNA lead to altered tissue specific choices that create either FGF-R2IIIb or FGF-R2IIIc mature transcripts (18).
The protein expression is further regulated by alternative polyadenylation of mRNA, which influences the coding potential or the 3′UTR length by modifying the binding availability of microRNA or RNA (19). Of note, it has been demonstrated that each type of alternative splicing can operate in a stochastic manner, and different splice-site identification and processing mechanisms do not necessarily occur at the same frequencies among all biological kingdoms (20).
The mechanisms outlined above are just one indication of the complexity, as numerous molecules are involved in alternative splicing in a coordinated manner. Even the basic nucleotide components and the essential molecules that recognize them can introduce diversity in the synthesis of mature transcripts.
Two major steps constitute the basic process of splicing: Assembly of the spliceosome followed by the actual splicing of pre-mRNA. The spliceosome is mainly composed of U1, U2 small nuclear ribonucleic proteins (snRNPs) and the U4/U6.U5 tri-snRNP, and configure in identify a core set of splicing signals: The 5′ splice site, the branch point sequence and the 3′ splice site (Fig. 2). Specific spliceosomal complexes (E, A, B and others) and eight evolutionarily conserved DExD/H-type RNA-dependent ATPases/helicases assemble in a proposed stepwise manner and execute multiple splicing steps that result in exon ligation and intron excision. Numerous steps in the pathway are reversible (21).
Figure 2.
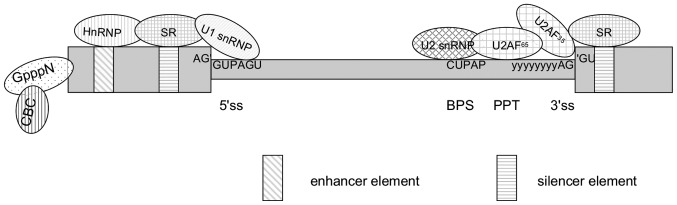
Schematic representation of the sequence elements and proteins at 5′ and 3′ exon-intron boundaries in an RNA transcript. The diagram illustrates the appropriate relative distributions of the molecules and core splicing signals with its consensus sequence in regulation of the alternative splicing. The enhancer elements [(exonic splicing enhancers (ESEs) and intronic splicing enhancers (ISEs)] are recognized by activator proteins (the SR protein family), and the silencer elements [exonic splicing silencers (ESSs) and intronic splicing silencers (ISSs)] are bound by repressor proteins [the heterogeneous nuclear ribonucleoproteins (hnRNP) protein family]. These two protein families are engaged to promote or inhibit spliceosome assembly at weak splice sites, respectively.
The exons that end up in the mature mRNA during the process of alternative splicing is entirely defined by the interaction between cis-acting elements and trans-acting factors. Cis-acting elements include exonic splicing enhancers (ESEs) and intronic splicing enhancers (ISE) that are bound by positive trans-acting factors, such as SR proteins (serine/arginine-rich family of nuclear phosphoproteins), whereas exonic splicing silencers (ESSs) and intronic splicing silencers are bound by negative acting factors, such as heterogeneous nuclear ribonucleoproteins (hnRNPs). The collaboration between these elements results in the promotion or inhibition of splicesome assembly of the weak splice sites, respectively (Fig. 2) (22, 23). In general, the cis-acting elements function additively. The enhancing elements tend to play dominant roles in constitutive splicing, while the silencers are relatively more important in the control of alternative splicing (22). Enhancer activity has been shown to be abolished by a stable stem-loop structure as short as 7 base pairs in an RNA transcript owing to the mechanisms of physical competition, long-range RNA pairing, a structure splice code and co-transcription splicing (24, 25). Furthermore, the specificity of cis-acting enhancer elements for introns or exons has been investigated. In these experiments, an ESE was found to act as an ISE depending on its location in an exon or intron (26).
HnRNPs are highly conserved from nematodes to mammals and have several critical roles in pre-mRNA maturation. Their function is to bind to the ESS to the exclusion of SR proteins. A looping out pre-mRNA leads to exonic sequestration from the rest of pre-mRNA transcript (27). HnRNPs A/B are a family of RNA-binding proteins, its diversification roles in the modulation of alternative splicing have evolved based on differing affinities for their cognate nucleic acids (28). HnRNP H and F serve to alter the proteolipid protein (PLP/DM20) ratio via the variation in the recruitment of U1 snRNP (29). Similarly, the antagonistic role of hnRNP M to the splicing factor Nova-1 generates alternatively spliced dopamine receptor pre-mRNAs, which create isoforms associated with diverse key physical functions, such as control, reward, learning and memory (30). In addition, hnRNP L and phosphorylation of ser513 have been recently shown to be involved in the regulation of alternative splicing through dynamic membrane depolarization and Ca2+/calmodulin-dependent protein kinase IV activation (31, 32).
In addition to the coupling of SR proteins to enhancer elements, SR proteins interact with U1 snRNP and the 35 kDa subunit of the heterodimeric factor, U2AF. The second subunit of U2AF, U2AF65, binds SF1 and the pyrimidine tract simultaneously, on the basis of the arginine/serine (RS)-rich domain, which results in recognition and stability of the branch point, as well as polypyrimidine tract sequences. Approximately 10–12 serines in the N-terminal region of the RS domain are rapidly phosphorylated by the binding of SR-specific protein kinase to serine/arginine-rich splicing factor 1 with an unusually high affinity. This continuous phosphorylation/dephosphorylation cycle of SR proteins facilitates the shuttling of SR proteins between the nucleus and the cytoplasm, and is critically required for the regulation of alternative splicing by growth signals transduced to the nucleus (33, 34). SR proteins have also been proposed to participate in post-splicing activities, such as mRNA nuclear export, nonsense-mediated decay (NMD) and mRNA translation (35).
In general, positive or negative splice-site recognition is regulated through various mechanisms, such as the local concentration or activity of splicing regulatory factors, under diverse physiological or pathological conditions. How these elements function together to precisely select a regulated splice site is, however, only partially explained by these results (36).
3. Coupling of alternative splicing to transcription
Since the first significant observation of co-transcriptional spliceosome assembly from electron micrographs of Drosophila melanogaster embryonic transcription units (37), increasing evidence supports the idea that transcription and splicing are physically and functionally coupled, and has also uncovered the intricate association between mRNA splicing, RNA polymerase II (Pol II) and chromatin structure (38, 39).
A large number of components associated with the physical interaction between splicing and transcription have been purified, with particular attention on the carboxyl terminal domain (CTD) of the large subunit of RNAPII (40). The CTD consists of 52 tandem repeats of the heptapeptide YSPTSPS in mammals (26 tandem repeats in yeast) (41), which act as a special platform to recruit different factors to the nascent transcripts via dynamic phosphorylation of serine residues. Kinases that phosphorylate specific CTD serine residues have been identified and are components of the protein apparatus driving the specific function. For example, ser5 phosphorylation is associated with transcription initiation through cyclin-dependent kinase 7 (CDK7) of the general transcription factor IIH (TFIIH), whereas ser2 phosphorylation is preferentially linked with CTD activity at the 3′-end of genes through CDK9 of the positive transcription elongation factor b (42). In addition, phosphorylation of ser7 has been found to facilitate elongation and splicing (43). Thus, phosphorylation is a mechanism that clearly demonstrates that functional coupling exists between transcription and alternative splicing.
CTD participates in gene expression-related functions ranging from 5′ capping, splicing, poly-adenylation and chromatin remodeling (44). Of note, mutation and deletion analysis of CTD has revealed multiple defects in mRNA processing (45), therefore, CTD and additional components of the two machineries have emerged as a central element in governing the interactions between transcription and splicing. Taken together, functional coupling appears to maintain an important role in alternative splicing in driving determinative physiological changes, and fine-tune gene expression in mathematical modeling approaches (46).
Two models have been suggested to explain the co-transcription process of how transcription coupled repair influences alternative splicing. The mechanism of the recruitment model may mainly depend on specific features of CTD (as mentioned above), whereas the kinetic model is based on the different elongation rates of Pol II, which in turn determine the timing of the presentation of splices sites (47, 48).
Fundamentally, the aforementioned mechanism influences patterns of alternative splicing via the variations in Pol II elongation and recruitment of splicing factors by specific histone marks (49). Thus, alternative splicing is highly influenced not only by transcription, but also by the chromatin structure, which underscores chromatin as another layer in the regulation of alternative splicing. The resultant mature mRNA is thus a reflection of numerous DNA modifications, such as patterns of histone methylation at exons, modulation of histone modifications and increased DNA methylation at exons (50, 51). Conversely, a previous study indicated that splicing may mediate chromatin remodeling via deposition of histone marks on DNA or numerous associations between splicing factors and elongation proteins (38).
Adding additional complexity to the regulation network is alternative transcription initiation (ATI) and alternative transcription termination (ATT) sites. ATI and ATT significantly contribute to the diversity of the human and mouse transcriptomes to a degree that may exceed alternative splicing, when considering the number of possibilities available through alternative nucleotides, isoforms and introns (52, 53). In contrast to the prevalence of alternative splicing that occurs within coding sequences (CDSs), the dominant class of alternative events, which includes ATI and ATT, occur in UTRs. This discovery reflects the preferential regulation of large distinct groups of genes with different mechanisms, such as strong coupling with alternative splicing in 5′ and 3′UTRs (54).
Despite the strong correlation between alternative splicing and transcription, alternative transcription mainly results in variations of the transcript number or the 5′/3′ terminal protein variants due to differential transcriptional start or terminal sites. By contrast, alternative splicing associated alterations mostly lie within the protein sequence, potentially affecting almost all areas of protein function (14, 55).
4. Alternative splicing and nonsense-mediated decay
NMD is an extensive and complicated mechanism, ranging from yeast to human, exploited to achieve another level of robustness in post-transcriptional gene expression control. Studies have revealed that up to one-third of human alternative splicing events contain premature termination codons (PTC), which are recognized and lead to the degradation of transcripts containing NMD cis-elements in their 3′ UTRs (56, 57). In vertebrates, it has been proposed that the coupling of the exon junction complex (EJC) to mRNA transcripts, followed by binding of 3′UTRs to EJCs, triggers vertebrate specific NMD (58). The sensitivity of mRNA transcripts to NMD is modulated by alternative splicing events in the 5′ or 3′UTRs and aids with the wide range of protein biosynthesis (59). Furthermore, analysis of quantitative alternative splicing microarray profiling has demonstrated that individual knockdown of NMD factors [Up-Frameshift (UPF)] strongly affects PTC-introducing alternative splicing events, indicating a role for different UPF factor requirements in alternative splicing regulation (60). In a second example, regulation of intron retention by alternative splicing-NMD in a specific differentiation event has been recently observed (61).
5. Trans-splicing
Trans-splicing is a common phenomenon in trypanosomes, nematodes, Drosophila and even humans, and refers to the novel and unusual splicing of exons from independent pre-mRNAs (62, 63). The phenomenon has been explored as a therapeutic option for a variety of genetic diseases, particularly in the treatment of cancer (64). The carcinoembryonic antigen (CEA), for example, is associated with a variety of neoplastic processes and was exploited as a target for trans-splicing. A CEA RNA-targeting trans-splicing ribozyme was designed to perform RNA replacement through a trans-splicing reaction specifically in CEA expressing cells (65). The activity of the ribozyme simultaneously reduced CEA expression and introduced the thymidine kinase gene, which rendered the cells sensitive to ganciclovir treatment. RNA trans-splicing has also been utilized for the potential treatment of neurodegenerative diseases through a novel technology, spliceosome mediated trans-splicing (SMaRT). SMaRT was successfully used in vivo to re-engineer tau mRNA transcripts to include E10, and therefore, offers the opportunity potential to correct tau mis-splicing and treat the underlying disease (66).
6. Alternative splicing and non-coding RNA
Non-coding RNAs (ncRNAs), including microRNA and small interfering RNA, have recently emerged as novel regulators in alternative splicing, generally through the modulation of the expression of key splicing factors during development and differentiation (67).
7. Alternative splicing and disease
Stringent regulation of alternative splicing is necessary for the functional requirements of complex tissues under normal conditions, whereas aberrant splicing appears to an underlying cause for an extremely high fraction of dysfunction and disease (68). Aberrant splicing has been suggested to root in alterations of the cellular concentration, composition, localization and activity of regulatory splicing factors, as well as mutations in components of core splicing machinery (69). A changed efficiency of splice site recognition is the immediate consequence, while irregularities in protein isoforms in different systems ultimately establish the disease state. Any of these alterations affecting alternative splicing can facilitate the appearance of characteristics in cancer cells, including the inappropriate proliferation, migration, methylation changes and resistance to apoptosis and chemotherapy (70). Alternative splicing has been implicated in nearly all aspects of cancer development, and therefore, is a main participant in the disease.
Understanding the basic mechanisms and patterns of splicing in tumor progress will shed light on the biology of cancer and lay the foundation for diagnostic, prognostic and therapeutic tools with minimum treatment toxicity in cancer (71). Extensive research efforts have already committed to developing drugs that target specific cancer protein isoforms. Several examples are genes associated with apoptosis [BCL2L1 (BCL-X), FAS, BIRC5 (survivin) and MDM2], immortality (human telomerase reverse transcriptase), and angiogenesis (vascular endothelial growth factor-A) (72, 73).
However, limited success has been achieved by simply activating or inhibiting cancer-associated genes, possibly due to the expression of target genes in normal and cancers cells, such as angiogenic and anti-angiogenic isoforms (74). The lack of specificity of numerous molecular targets for cancer cells favors the development of isoform-specific diagnostic markers as therapeutic targets (75). Therefore, the key task for cancer treatment in the future should be to detect and target the expression of a gene at the gene level.
8. Conclusion
The combination of an alternative splicing database, tandem mass spectrometry, and even the latest synthetic alternative splicing database may aid with the identification, analysis and characterization of potential alternative splicing isoforms. Over two-thirds of human genes and 40% of Drosophila genes contain one or more alternative exons, and >90% of the protein-coding genes associated with alternative splicing events according to the >60,000 studies since the discovery of splicing (76). Alternative splicing appears to be prevalent in almost all multi-exon genes. However, what limits our insight into a more complete and accurate usage of alternative splicing are factors such as biased coverage of ESTs toward the 5′- and 3′-ends of transcripts, insufficient widespread analyses, subtle alternative splicing associated changes and advanced alternative splicing networks involved in various mechanisms and numbers of regulatory proteins. All these deficiencies lead to an incomplete understanding of the alternative splicing mechanism and may prevent the correct prediction of splice events in other species, such as the chimpanzee or plant (77, 78). Distinguishing alternative splicing from other regulatory mechanisms in the gene regulation is also difficult. Alternative splicing, alternative trans-splicing, NMD, transcriptional efficiency, exon duplication and RNA editing (79) all contribute to an extensive mechanism for generating protein diversity. In addition, the difference between artificial experimental systems and real-life scenarios makes it challenging to transfer functional studies from cells to whole organisms. Numerous questions remain regarding the global impact of alternative splicing on cellular and organismal homeostasis, as well as its underlying molecular mechanisms. Finally, with regards to cancer-associated alternative splicing, whether a particular splice site selection causes the observed effect or is merely the result of the cancerous transformation is hard to distinguish. The data collected regarding alternative splicing is likely to represent only the tip of the iceberg, with further information yet to be revealed in future studies.
Acknowledgements
The present study was supported by grants from the National Science Foundation of China (No. 81271912) and the Education Department of Jiangxi province, China (No. GJJ13041).
References
- 1.Gilbert W. Why genes in pieces? Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 2.C. elegans Sequencing Consortium, corp-author. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 3.Human Genome Sequencing Consortium I International Human Genome Sequencing C, corp-author. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 4.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng CL, Fu XD, Gribskov M. Characteristics and regulatory elements defining constitutive splicing and different modes of alternative splicing in human and mouse. RNA. 2005;11:1777–1787. doi: 10.1261/rna.2660805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Effenberger KA, Perriman RJ, Bray WM, et al. A high-throughput splicing assay identifies new classes of inhibitors of human and yeast spliceosomes. J Biomol Screen. 2013;18:1110–1120. doi: 10.1177/1087057113493117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Kim E, Goren A, Ast G. Alternative splicing: current perspectives. Bioessays. 2008;30:38–47. doi: 10.1002/bies.20692. [DOI] [PubMed] [Google Scholar]
- 9.Irimia M, Penny D, Roy SW. Coevolution of genomic intron number and splice sites. Trends Genet. 2007;23:321–325. doi: 10.1016/j.tig.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Fu G, Condon KC, Epton MJ, et al. Female-specific insect lethality engineered using alternative splicing. Nat Biotechnol. 2007;25:353–357. doi: 10.1038/nbt1283. [DOI] [PubMed] [Google Scholar]
- 11.Hurt KJ, Sezen SF, Champion HC, et al. Alternatively spliced neuronal nitric oxide synthase mediates penile erection. Proc Natl Acad Sci USA. 2006;103:3440–3443. doi: 10.1073/pnas.0511326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 13.Nygard AB, Cirera S, Gilchrist MJ, et al. A study of alternative splicing in the pig. BMC Res Notes. 2010;3:123. doi: 10.1186/1756-0500-3-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelemen O, Convertini P, Zhang Z, et al. Function of alternative splicing. Gene. 2013;514:1–30. doi: 10.1016/j.gene.2012.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim E, Magen A, Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007;35:125–131. doi: 10.1093/nar/gkl924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galante PA, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ. Detection and evaluation of intron retention events in the human transcriptome. RNA. 2004;10:757–765. doi: 10.1261/rna.5123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakabe NJ, de Souza SJ. Sequence features responsible for intron retention in human. BMC Genomics. 2007;8:59. doi: 10.1186/1471-2164-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldstrohm AC, Greenleaf AL, Garcia-Blanco MA. Co-transcriptional splicing of pre-messenger RNAs: considerations for the mechanism of alternative splicing. Gene. 2001;277:31–47. doi: 10.1016/S0378-1119(01)00695-3. [DOI] [PubMed] [Google Scholar]
- 19.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mastrangelo AM, Marone D, Laido G, et al. Alternative splicing: enhancing ability to cope with stress via transcriptome plasticity. Plant Sci. 2012;185–186:40–49. doi: 10.1016/j.plantsci.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Hoskins AA, Moore MJ. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem Sci. 2012;37:179–188. doi: 10.1016/j.tibs.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Xiao X, Van Nostrand E, Burge CB. General and specific functions of exonic splicing silencers in splicing control. Mol Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin Y, Yang Y, Zhang P. New insights into RNA secondary structure in the alternative splicing of pre-mRNAs. RNA Biol. 2011;8:450–457. doi: 10.4161/rna.8.3.15388. [DOI] [PubMed] [Google Scholar]
- 25.Liu W, Zhou Y, Hu Z, et al. Regulation of splicing enhancer activities by RNA secondary structures. FEBS Lett. 2010;584:4401–4407. doi: 10.1016/j.febslet.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McManus CJ, Graveley BR. RNA structure and the mechanisms of alternative splicing. Curr Opin Genet Dev. 2011;21:373–379. doi: 10.1016/j.gde.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nasim FU, Hutchison S, Cordeau M, Chabot B. High-affinity hnRNP A1 binding sites and duplex-forming inverted repeats have similar effects on 5′ splice site selection in support of a common looping out and repression mechanism. RNA. 2002;8:1078–1089. doi: 10.1017/S1355838202024056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han SP, Kassahn KS, Skarshewski A, Ragan MA, Rothnagel JA, Smith R. Functional implications of the emergence of alternative splicing in hnRNP A/B transcripts. RNA. 2010;16:1760–1768. doi: 10.1261/rna.2142810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang E, Cambi F. Heterogeneous nuclear ribonucleoproteins H and F regulate the proteolipid protein/DM20 ratio by recruiting U1 small nuclear ribonucleoprotein through a complex array of G runs. J Biol Chem. 2009;284:11194–11204. doi: 10.1074/jbc.M809373200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park E, Iaccarino C, Lee J, et al. Regulatory roles of heterogeneous nuclear ribonucleoprotein M and Nova-1 protein in alternative splicing of dopamine D2 receptor pre-mRNA. J Biol Chem. 2011;286:25301–25308. doi: 10.1074/jbc.M110.206540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu G, Razanau A, Hai Y, et al. A conserved serine of heterogeneous nuclear ribonucleoprotein L (hnRNP L) mediates depolarization-regulated alternative splicing of potassium channels. J Biol Chem. 2012;287:22709–22716. doi: 10.1074/jbc.M112.357343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J, Hai Y, Liu G, Fang T, Kung SK, Xie J. The heterogeneous nuclear ribonucleoprotein L is an essential component in the Ca2+/calmodulin-dependent protein kinase IV-regulated alternative splicing through cytidine-adenosine repeats. J Biol Chem. 2009;284:1505–1513. doi: 10.1074/jbc.M805113200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghosh G, Adams JA. Phosphorylation mechanism and structure of serine-arginine protein kinases. FEBS J. 2011;278:587–597. doi: 10.1111/j.1742-4658.2010.07992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Twyffels L, Gueydan C, Kruys V. Shuttling SR proteins: more than splicing factors. FEBS J. 2011;278:3246–3255. doi: 10.1111/j.1742-4658.2011.08274.x. [DOI] [PubMed] [Google Scholar]
- 35.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 36.Kalnina Z, Zayakin P, Silina K, Linē A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer. 2005;42:342–357. doi: 10.1002/gcc.20156. [DOI] [PubMed] [Google Scholar]
- 37.Beyer AL, Bouton AH, Miller OL., Jr Correlation of hnRNP structure and nascent transcript cleavage. Cell. 1981;26:155–165. doi: 10.1016/0092-8674(81)90299-3. [DOI] [PubMed] [Google Scholar]
- 38.Kornblihtt AR, de la Mata M, Fededa JP, et al. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montes M, Becerra S, Sanchez-Alvarez M, Sune C. Functional coupling of transcription and splicing. Gene. 2012;501:104–117. doi: 10.1016/j.gene.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 40.Ryman K, Fong N, Bratt E, Bentley DL, Ohman M. The C-terminal domain of RNA Pol II helps ensure that editing precedes splicing of the GluR-B transcript. RNA. 2007;13:1071–1078. doi: 10.1261/rna.404407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Bartkowiak B, Liu P, Phatnani HP, et al. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24:2303–2316. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim H, Erickson B, Luo W, et al. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol. 2010;17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 45.Rosonina E, Blencowe BJ. Analysis of the requirement for RNA polymerase II CTD heptapeptide repeats in pre-mRNA splicing and 3′-end cleavage. RNA. 2004;10:581–589. doi: 10.1261/rna.5207204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schor IE, Gómez Acuña LI, Kornblihtt AR. Coupling between transcription and alternative splicing. Cancer Treat Res. 2013;158:1–24. doi: 10.1007/978-3-642-31659-3_1. [DOI] [PubMed] [Google Scholar]
- 48.Dujardin G, Lafaille C, Petrillo E, et al. Transcriptional elongation and alternative splicing. Biochim Biophys Acta. 2013;1829:134–140. doi: 10.1016/j.bbagrm.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 49.Gómez Acuña LI, Fiszbein A, Alló M, et al. Connections between chromatin signatures and splicing. Wiley Interdiscip Rev RNA. 2013;4:77–91. doi: 10.1002/wrna.1142. [DOI] [PubMed] [Google Scholar]
- 50.Iannone C, Valcárcel J. Chromatin's thread to alternative splicing regulation. Chromosoma. 2013;122:465–474. doi: 10.1007/s00412-013-0425-x. [DOI] [PubMed] [Google Scholar]
- 51.Shukla S, Oberdoerffer S. Co-transcriptional regulation of alternative pre-mRNA splicing. Biochim Biophys Acta. 2012;1819:673–683. doi: 10.1016/j.bbagrm.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma X, Li-Ling J, Huang Q, et al. Systematic analysis of alternative promoters correlated with alternative splicing in human genes. Genomics. 2009;93:420–425. doi: 10.1016/j.ygeno.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 53.Xin D, Hu L, Kong X. Alternative promoters influence alternative splicing at the genomic level. PLoS One. 2008;3:e2377. doi: 10.1371/journal.pone.0002377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shabalina SA, Spiridonov AN, Spiridonov NA, Koonin EV. Connections between alternative transcription and alternative splicing in mammals. Genome Biol Evol. 2010;2:791–799. doi: 10.1093/gbe/evq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roy B, Haupt LM, Griffiths LR. Review: Alternative splicing (AS) of genes as an approach for generating protein complexity. Curr Genomics. 2013;14:182–194. doi: 10.2174/1389202911314030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lareau LF, Brooks AN, Soergel DA, Meng Q, Brenner SE. The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv Exp Med Biol. 2007;623:190–211. doi: 10.1007/978-0-387-77374-2_12. [DOI] [PubMed] [Google Scholar]
- 57.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nyiko T, Kerenyi F, Szabadkai L, et al. Plant nonsense-mediated mRNA decay is controlled by different autoregulatory circuits and can be induced by an EJC-like complex. Nucleic Acids Res. 2013;41:6715–6728. doi: 10.1093/nar/gkt366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kalyna M, Simpson CG, Syed NH, et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012;40:2454–2469. doi: 10.1093/nar/gkr932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saltzman AL, Kim YK, Pan Q, et al. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ge Y, Porse BT. The functional consequences of intron retention: alternative splicing coupled to NMD as a regulator of gene expression. Bioessays. 2014;36:236–243. doi: 10.1002/bies.201300156. [DOI] [PubMed] [Google Scholar]
- 62.Herai RH, Yamagishi ME. Detection of human interchromosomal trans-splicing in sequence databanks. Brief Bioinform. 2010;11:198–209. doi: 10.1093/bib/bbp041. [DOI] [PubMed] [Google Scholar]
- 63.Horiuchi T, Aigaki T. Alternative trans-splicing: a novel mode of pre-mRNA processing. Biol Cell. 2006;98:135–140. doi: 10.1042/BC20050002. [DOI] [PubMed] [Google Scholar]
- 64.Lee SW, Jeong JS. Use of tumor-targeting trans-splicing ribozyme for cancer treatment. Methods Mol Biol. 2014;1103:83–95. doi: 10.1007/978-1-62703-730-3_7. [DOI] [PubMed] [Google Scholar]
- 65.Jung HS, Lee SW. Ribozyme-mediated selective killing of cancer cells expressing carcinoembryonic antigen RNA by targeted trans-splicing. Biochem Biophys Res Commun. 2006;349:556–563. doi: 10.1016/j.bbrc.2006.08.073. [DOI] [PubMed] [Google Scholar]
- 66.Avale ME, Rodriguez-Martin T, Gallo JM. Trans-splicing correction of tau isoform imbalance in a mouse model of tau mis-splicing. Hum Mol Genet. 2013;22:2603–2611. doi: 10.1093/hmg/ddt108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luco RF, Misteli T. More than a splicing code: integrating the role of RNA, chromatin and non-coding RNA in alternative splicing regulation. Curr Opin Genet Dev. 2011;21:366–372. doi: 10.1016/j.gde.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang J, Manley JL. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 2013;3:1228–1237. doi: 10.1158/2159-8290.CD-13-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shkreta L, Bell B, Revil T, et al. Cancer-associated perturbations in alternative pre-messenger RNA splicing. Cancer Treat Res. 2013;158:41–94. doi: 10.1007/978-3-642-31659-3_3. [DOI] [PubMed] [Google Scholar]
- 71.Kim YJ, Kim HS. Alternative splicing and its impact as a cancer diagnostic marker. Genomics Inform. 2012;10:74–80. doi: 10.5808/GI.2012.10.2.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lapuk AV, Volik SV, Wang Y, Collins CC. The role of mRNA splicing in prostate cancer. Asian J Androl. 2014;16:515–521. doi: 10.4103/1008-682X.127825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pajares MJ, Ezponda T, Catena R, et al. Alternative splicing: an emerging topic in molecular and clinical oncology. Lancet Oncol. 2007;8:349–357. doi: 10.1016/S1470-2045(07)70104-3. [DOI] [PubMed] [Google Scholar]
- 74.Sitohy B, Nagy JA, Dvorak HF. Anti-VEGF/VEGFR therapy for cancer: reassessing the target. Cancer Res. 2012;72:1909–1914. doi: 10.1158/0008-5472.CAN-11-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pal S, Gupta R, Davuluri RV. Alternative transcription and alternative splicing in cancer. Pharmacol Ther. 2012;136:283–294. doi: 10.1016/j.pharmthera.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 76.Pan Q, Shai O, Lee LJ, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 77.Calarco JA, Xing Y, Caceres M, et al. Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev. 2007;21:2963–2975. doi: 10.1101/gad.1606907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reddy AS, Rogers MF, Richardson DN, Hamilton M, Ben-Hur A. Deciphering the plant splicing code: experimental and computational approaches for predicting alternative splicing and splicing regulatory elements. Front Plant Sci. 2012;3:18. doi: 10.3389/fpls.2012.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tang W, Fei Y, Page M. Biological significance of RNA editing in cells. Mol Biotechnol. 2012;52:91–100. doi: 10.1007/s12033-012-9498-7. [DOI] [PubMed] [Google Scholar]