Abstract
Establishing a transcriptomic profile of human hepatocellular liver cancer (HCC) progression is a complex undertaking. A rat model of HCC was employed to develop a transcriptomic profile. Using three interventions, preneoplastic lesions appeared after 30 days and they progressed to HCC by 9 months. Preneoplastic and cancer lesions were characterized for transcriptomic analysis, and RNA from total liver homogenates was obtained at 1, 7, 11 and 16 days after the initiation treatment. RNA from dissected persistent preneoplastic lesions, adjacent tissue or cancer tissue was used for 30 days, and 5, 9, 12 and 18 months. The GeneChip® Rat Exon 1.0 ST arrays, Partek software and an Affymetrix console were employed for these analyses. LGALS3BP was differentially expressed at each time point, from the initial period, through the preneoplastic evolution period and until the end of cancer progression period. Twelve differentially expressed genes common to the preneoplastic evolution and to the cancer progression period were detected, which included ABCC3. Validation of the microarrays was confirmed by reverse transcription-quantitative polymerase chain reaction of six genes, including LGALS3BP and ABCC3. Of note, the proteins of these two genes are associated with the multidrug response complex, and evasion of immune surveillance and negative regulation of T cell proliferation. This model is useful for identifying candidate genes, and to validate them with regards to determining their relevance in rat HCC progression.
Keywords: liver cancer, animal model, gene expression profile, cancer markers, therapeutic targets
Introduction
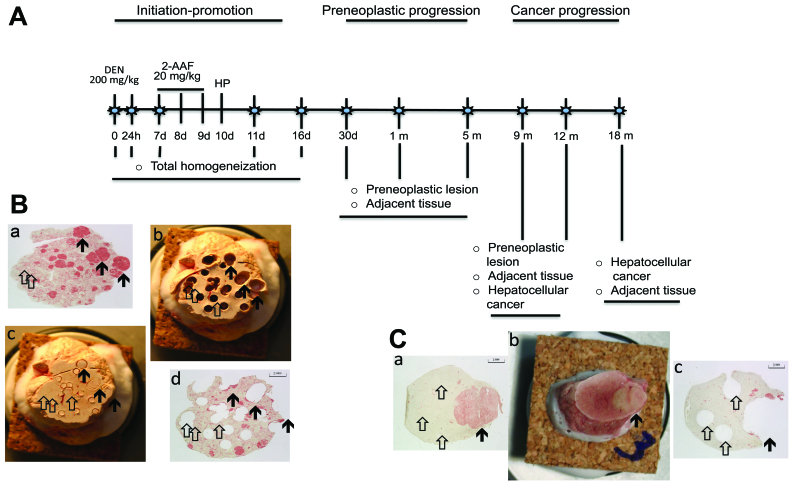
The global molecular changes during the development of liver cancer are not well characterized despite the availability of large-scale screening techniques capable of detecting these alterations. Cancer progression stages involve genetic and epigenetic events that transform a normal cell into a malignant cell. Underlying these changes are genome instability, inflammation, a reprogramming of energy metabolism and evasion of the immune system giving rise to a cell with progressive autonomy that has a sustained proliferation, with insensitivity to inhibitory growth, resistance to death, replicative immortality, an increment in angiogenesis and activation of invasion and metastasis (1). In humans, a liver cancer progression analysis is uncertain; it is difficult to establish a demonstrative transcriptomic profile of the time-points representative of the common stages of cancer progression. An issue that increases the complexity of investigating cancer in humans is the numerous etiological factors involved in its occurrence (2). Despite these limitations, important achievements have been made through microarray studies using human tumors to obtain the characteristic gene expression profiles (GEP) that are referred to as ‘signatures’. In mammary cancers, GEP predict the result of treatment (3), and in liver cancer, GEP forecast an early recurrence following treatment (4). GEP has been used to identify the origins of metastatic tumors (5). Additionally, there are examples of successful studies in experimental animal liver cancer models. In a microarray study in rodents, the HSP70 protein was proposed to be an early marker of hepatocellular cancer (6). In another animal model, epigenetic modulation of protein expression was detected through gene expression profiling by selecting differentially expressed genes and studying their non-coding, regulatory regions (7). One potential method of studying types of human cancer is to use experimental animal models (8). The advantage of this methodology is the well-defined and reproducible stages of tumor evolution (9), which allows for a sound global molecular study of cancer progression through GEP that can then be extrapolated to humans. The aim of the present study was to characterize the liver GEP associated with hepatocellular liver cancer (HCC) progression in a rat model of hepatocarcinogenesis. The resistant hepatocyte model, as modified in laboratory 50 (Department of Cell Biology, Center for Research and Advanced Studies, Mexico) is suitable for this purpose (10) as it reproducibly exhibits the initiation, promotion, preneoplastic and tumor progression stages (Fig. 1A).
Figure 1.
Experimental protocol for the sample selection. (A) Model and time-points selected. Initial progression period; initiation stage over 24 h and 7 days and promotion stage over 11 and 16 days. The preneoplastic progression period includes the evolution of persistent preneoplastic lesions over 30 days, and 5 and 9 months. The cancer progression period includes the growth of hepatocellular carcinomas over 9, 12 and 18 months. (B) Sample analysis. For tissue gene expression analysis, whole homogenates were used of the sample at 24 h and 7, 11 and 16 days. Representative samples of (B) 30 days and (C) 9 months after treatment are presented to illustrate the enrichment of samples. White arrows, adjacent tissue; black arrows, (B) preneoplastic lesions or (C) tumor lesion.
Ten different time-points from day 1 through 18 months after the initiation of treatment were selected to assess the GEP in whole tissue, dysplastic nodules, tumors and tissue surrounding neoplastic alterations, corresponding to the initiation, promotion, preneoplastic lesion evolution and cancer progression stages. As the genes identified in the present study and possibly those that are directly involved in cancer progression, ABCC3 and LGALS3BP genes are discussed as candidates for further analysis to establish their protein expression profiles associated with liver cancer progression, and consequently, to validate them as early detection markers or therapeutic targets. In the present model, based only in the comparative gene profile expression during progression of preneoplastic lesions and tumors, it is possible to identify candidate genes and their respective proteins to validate them in determining their relevance in rat HCC progression.
Materials and methods
Animals
All the experiments were performed in accordance with and approval by the Internal Committee for the Care and Use of Laboratory Animals of the Center for Research and Advanced Studies of the National Polytechnic Institute (CINVESTAV-IPN) under the protocol no. 0001-02. Male Fischer 344 rats (180–200 g) were obtained from the Unit for Production of Experimental Laboratory Animals (UPEAL Cinvestav, Mexico City, Mexico). The animals had free access to food (PMI Feeds Inc., Laboratory Diet, Richmond, IN, USA) and water. The animals were maintained in a holding room under controlled conditions with 12-h light/dark cycles, 50% relative humidity and a temperature of 21 ˚ C. Animal care followed the institutional guidelines for the use of laboratory animals.
Experimental protocol
The animals were administered 200 mg/kg diethylnitrosamine (Sigma-Aldrich, St. Louis, MO, USA) (11). Subsequently, they received 3 daily dosages of 20 mg/kg 2-acetylaminofluoeren (Sigma-Aldrich) on days 7, 8 and 9, and a 75% partial hepatectomy was performed on day 10. Three groups of non-treated animals (n=4/group) were sacrificed by exsanguination at 0 h, and 9 and 12 months. Treated animals (n=4/group) were sacrificed at 1, 7, 11, 16 and 30 days and 5, 9, 12 and 18 months (Fig. 1A). Their livers were excised, washed in physiological saline solution, frozen in 2-methyl butane with liquid nitrogen (Sigma-Aldrich) and stored at −80 ˚ C. Frozen liver sections were used for the microarray assays (n=4/group). Paraffin-fixed liver samples were also prepared for histochemical and immunohistochemical examination.
Histochemical analysis and tissue selection
Histological analysis of preneoplastic and neoplastic lesions was performed using hematoxylin and eosin staining and γ-glutamil transpeptidase (GGT) histochemical (12) (Fig. 1B). Images of GGT-positive lesions were captured with a digital camera (Color view 12) and quantified with AnalySIS software (AnalySIS) (Soft Imaging System GmbH, Muenster, Germany). Total liver homogenates were used to analyze the specimens obtained at 0 h and 1, 7, 11 and 16 days. Based on the GGT histochemical analysis, tissues from preneoplastic lesions corresponding to the persistent nodules, tumors and the adjacent tissue were selected to be analyzed at 30 days and 5, 9, 12 and 18 months (Fig. 1B).
RNA extraction and microarray hybridization
RNA was extracted using Tripure Isolation Reagent (Roche, Indianapolis, IN, USA). The microarray analysis was performed using GeneChip® Rat Exon 1.0 ST arrays, which are whole-genome arrays containing over 1 million probe sets, with <4 perfect match (PM) probes each, spread across the exons of all the known genes, plus a number of additional regions based on other annotation sources, including GenScan predictions and ESTs from dbEST. Microarray hybridizations were performed in four replicates for each analysis point according to the GeneChip Whole Transcript (WT) Sense Target Labeling Assay user manual (www.affymetrix.com). The data were collected using Affymetrix GCOS software, and the quality of the results was analyzed with the Affymetrix Gene Expression Console (Affymetrix, Inc., Santa Clare, CA, USA).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Certain gene products that were differentially expressed using the microarray gene analysis were validated by RT-qPCR. Total RNA was extracted from the tissue samples using TRIzol reagent and was reverse transcribed into cDNA using SuperScrit II RT and oligo(dTs) were used according to the manufacturer's instructions (Invitrogen, Carlsbad, CA, USA). qPCR assays of the transcripts were performed with gene-specific fluorescent labeled probes on a 7000 Sequence Detector (Applied Biosystems, Foster City, CA, USA). The specificity of S100A10, SPINT1, TXNRD1, LGALS3BP, ABCC3 and LYST primers was designed using Primer Express software (Applied Biosystems). The reference gene, 18S, was used to normalize the mRNA data. The PCR reaction mixture contained 1 µ1 cDNA, 7.5 µ1 1X TaqMan Universal PCR Master Mix and 1 µ1 of the primers and probe. The following cycling protocol was employed: 1 cycle at 50 ˚ C for 2 min, 1 cycle at 95 ˚ C for 10 min and 40 cycles at 95 ˚ C and 60 ˚ C for 15 sec and 1 min, respectively. The results were evaluated according to the comparative Ct method.
Principal component analysis, Venn diagram and pathway analysis
Microarray data was performed (Affymetrix, Inc.) and analyzed (Affymetrix gene expression console and Partek software) determining the transcriptional changes during liver cancer progression.
Partek® software, version 6.5 (Partek, Inc., St. Louis, MO, USA) was used to perform the principal component analysis. After statistical analysis using Partek® software we realized the Venn diagram in each stage. The ingenuity pathway analysis (IPA) software program was used to assign genes to specific biological functions and canonical and toxicological pathways.
Data analysis
Data analysis was performed using Partek Genomics Suite software (Partek, Inc.). Normalization and probe summarization were performed using the Robust Multi-array average algorithm, and differential gene expression was evaluated using a one-way analysis of variance with Tukey's post hoc test. GEPs were selected based on a fold-change ≤ 1 and a P-value <0.05, which was considered to indicate a statistically significant difference.
Results
GEP analysis
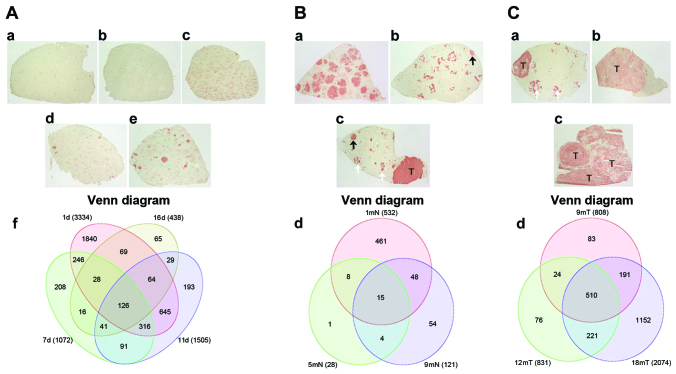
GEP from the liver of non-treated rats and in 9 different times after the corresponding treatment interventions were obtained (Fig. 1A) and analyzed using the Affymetrix gene expression console and Partek software. As shown in Fig. 1, to obtain tissue from preneoplastic lesion (Fig. 1Ba-d) sections were stained with GGT+ to localize preneoplastic lesion and with a cork borer were extracted. The same operation was performed to obtain tumor tissue (Fig. 1 Ca-c). In the progression period, from 1, 7, 11 days until 16 days after the initiation of treatment, the expression of 3,334, 1,072, 1,505 and 438 transcripts were differentially expressed, respectively (Fig. 2A-f). Of the 3,334 altered-expression genes of day 1, ~60% were overexpressed. There is a clear tendency for the number of differentially expressed genes to decrease that coincides with the minimum number of nodular lesions being observed at 5 months (data not shown). At this time, significant changes in the histology of GGT+ tissue were observed, mainly in the persistence of nodules with highly modified phenotypes, which was in contrast to the detection of the fewest gene expression changes, with only 28 differentially expressed genes in the nodular regions and eight in the surrounding tissue (Fig. 2B-d). When a cancer was exhibited and progressed, a continuous increase in the number of differentially expressed genes was observed. Tumors exhibited 808, 831 and 1,465 differentially expressed genes at 9, 12 and 18 months (Fig. 2C-d). Of note, from 9 months onward, in contrast to the early period from 24 h to 16 days, the number of underexpressed genes was greater than the number of overexpressed genes.
Figure 2.
Histochemistry analysis of the rat tissue samples from modified reistant hepatocyte model, and the Venn diagrams of the differentially expressed genes. (A) Initiation-promotion period. γ-glutamil transpeptidase (GGT) histochemistry stain of representative tissue samples from (a) non-treated rats and (b) 1, (c) 7, (d) 11 and (e) 16 days after treatment. A few scattered small preneoplastic lesion are present in (d) and several are present in (e). (f) Venn diagram shows that of the 3,334 genes differentially expressed after 24 h of experiment initiation, only 126 are common to the four time-points. (B) Preneoplastic progression period. Samples from rats sacrificed after (a) 30 days in which a maximum of preneoplastic nodule lesions are present; (b) 5 months in which the majority of preneoplastic nodule lesions become remodeled (white arrows) and a few persistent preneoplastic remain (black arrows) and (c) 9 months in which persistent nodule lesions (black arrow), remodeled preneoplastic (white arrow) lesions and tumor lesions (T) coexist. (d) Venn diagram where the amount of differentially expressed genes decreased to a minimum at 5 months and only 15 differentially expressed genes are in common. (C) Hepatocellular liver cancer (HCC) progression period. Samples from rats sacrificed at (a) 9 months in which remodeled preneoplastic nodule lesions (white arrows) coexist with HCC (T); rats sacrificed at (b) 12 months in which HCC becomes consolidated and at (c) 18 months in which the majority of liver tissue is occupied by tumor (T). (d) Venn diagram where the increment of differentially expressed genes occurs and 510 genes are common to these three time-points.
IPA
Pathway and global functional analyses were performed using IPA 6.0 (www. ingenuity.com). Analysis of the top four biological functions determined by IPA was cancer with 1,335 genes altered, neurological disease with 1,310 genes altered, cellular growth and proliferation with 1,271 genes altered and cell death with 1,260 genes altered. The top genes from those determined as altered for cancer by the number of frequency were LGALS3BP, GADD45B, ABCC3 and EPH1. The other genes of the remaining pathways that were of note for the number of frequency were CREM, ANNXA2, GNAI2, BAK1, ARNT and CCN1. This data provide information to find new markers for the early detection of HCC or target genes for chemoprevention.
Common GEP in the modified resistant hepatocyte model
A comparative analysis using Venn diagrams allows for the detection of the number of genes exclusive to each time-point in the three progression periods. Between 1, 7, 11 and 16 days, there were 126 genes in common with altered expression; this GEP could be considered a characteristic of the initial progression period of cancer development (Fig. 2A). In the preneoplastic evolution period of persistent nodular lesions, which was measured at 1, 5 and 9 months, there were 15 commonly altered genes (Fig. 2B), of which 11 were overexpressed and only four were underexpressed. Several genes are highly associated with detoxification processes, lipid metabolism, redox reactions and energy production. In tumor tissue samples from 9, 12 and 18 months (Fig.2C), there were 510 modified genes in common, which represent a group of genes that may be considered as markers of cancer progression (Fig. 2C-d).
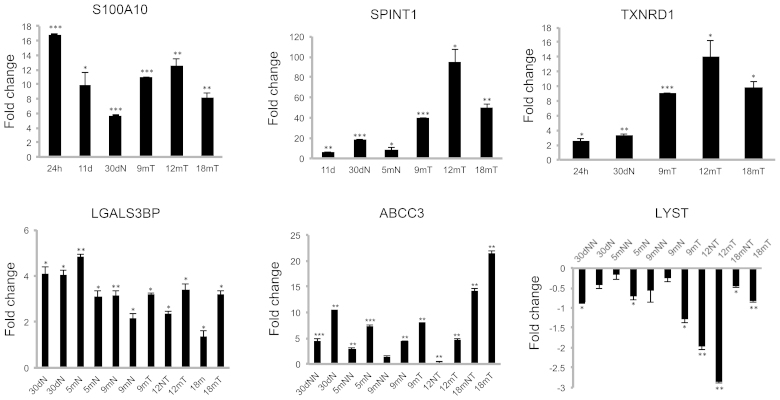
Hypothetically, the genes that show altered expression throughout cancer promotion and tumor progression are good candidates for further analyses to characterize the genetic footprint of liver cancer. In this context, LGALS3BP and ABCC3 are gene candidates for further studies. LGALS3BP was commonly expressed throughout the progression of hepatocarcinogenesis from 24 h until the end of the experiment at 18 months, and ABCC3 was highly expressed from preneoplastic progression until the 18-month time-point. The gene differential expression values were extremely statistically significant between the observed differences in LGALS3BP and ABCC3 (data not shown). Validation of the present microarrays results by RT-qPCR of S100A10, SPINT1, LGALS3BP, TXNRD1, ABCC3 and LYST presented the similar clear differential expression in the carcinogenesis process, as was observed in the microarrays results (Fig. 3). The mRNA overexpression of LGALS3BP in all the carcinogenesis processes allows the proposal of this gene as a candidate for an early biomarker. The high mRNA overexpression of ABCC3, mainly in persistent nodule, preneoplastic lesions, and tumors, and to a lesser degree in the adjacent tissue, suggests a participation in the progression of hepatocarcinogenesis that should be further studied (Fig. 3). The sustained mRNA underexpression of LYST suggests that decreased expression of this gene is required throughout the progression of the carcinogenic process.
Figure 3.
Validation by RT-qPCR of the overexpressed genes, S100A10, SPINT1, LGALS3BP, TXNRD1 and ABCC3, and the underexpressed gene, LYST. The 18S rRNA gene was used as the internal control. RT-qPCR, reverse-transcription-quantitative polymerase chain reaction.
Discussion
Animal models of hepatocarcinogenesis of liver tumorigenesis provide data for the cellular development of HCC in humans (13–15). In the present model, the progression from nodules to HCC occurs without additional carcinogen treatment. The majority of nodules undergo remodeling but a few persistent nodules show spontaneous cell proliferation and increased size (9, 16–18). These types of nodules were susceptible to dissection. In the present model, rats with persistent hepatic nodular lesions will coexist with HCC after 9 to 10 months (19). Hypothetically, persistent nodule cells may exhibit an altered genetic background that allows autonomous cell proliferation to undergo a slow evolution to cancer. The genes that were differentially expressed within the persistent nodules and commonly observed from 1 to 9 months represent preneoplastic evolution. Using a Venn diagram, the genes that were unique to each period and the genes that were shared between periods were identified. Those genes commonly expressed from the initial time analyzed through the eight time-points that followed until the last time-point at 18 months were investigated. With the suggestion that hepatocyte nodules are known precursors for HCC (18), the present analysis was directed to detect 13 genes that were commonly differentially expressed during preneoplastic evolution and during tumor progression; these are the candidates for further studies. In this context, ABCC3 and LGALS3BP are good candidates to be further studied as cancer markers.
With regards to ABCC3, multidrug resistance-associated protein 3 (MRP3) and MRP-like protein 2, upregulation of this gene has been associated with HCC progression and is negatively regulated by microRNA (20). Together with MPR2, these are components of the multidrug resistance phenotype (21) and its upregulation has been confirmed in choriocarcinoma and cervical cancer (22). The protein encoded by this gene is a tumor-associated antigen in HCC recognized by cytotoxic T cells and has been suggested as an immunogenic target for HCC immunotherapy (23).
LGALS3BP is also known as Mac-2 binding protein (Mac-2BP or MAC2BP), Cyp-C-associated protein (CyCAP), and protein 90K. This protein has been detected as a ligand of dendritic cell (DC)-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN), suggesting that it binds to LGALS3BP-bearing tumor-associated Le glycans. Of note, Mac-2BP was detected as a predominant DC-SIGN ligand expressed in several primary colorectal cancer tissues in patients in comparison with CEAs from other areas, and may become a novel potential colorectal cancer biomarker for certain patients, rather than CEA (24). Notably, the LGALS3BP from the cell matrix of neuroblastoma cells was observed to act as a secreted protein that stimulates interleukin-6 expression in bone marrow stromal cells, which raises the question of whether LGALS3BP-binding protein could be a valuable target for therapeutic intervention in metastatic neuroblastoma (25).
In conclusion, a comparative profiling during preneoplastic and tumor progression provided an unbiased selection of a set of genes that should be analyzed as candidates for cancer markers or therapeutic targets.
Acknowledgements
The present study was supported by a scholarship, a grant contribution (no. 39525-M) from CONACYT and a postdoctoral scholarship for multidisciplinary project 3 from the SVT grant of CINVESTAV. The authors would like to acknowledge the animal technical support at UPEAL-Cinvestav, including M. Vet. Rafael Leyva-Muñoz, M. Vet. Ricardo Gaxiola-Centeno and Dr Jorge Fernandez.
Glossary
Abbreviations
- NN
no nodule
- N
nodule
- NT
no tumor
- T
tumor
- GEP
gene expression profiles
- ABCC3
ATP-binding cassette sub-family C member 3
- LGALS3BP
lectin galactoside-binding soluble 3-binding protein
- Lyst
lysosomal trafficking regulator
- MRHM
modified resistant hepatocyte model
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Blum HE. Hepatocellular carcinoma: therapy and prevention. World J Gastroenterol. 2005;11:7391–7400. doi: 10.3748/wjg.v11.i47.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunn L, Demichele A. Genomic predictors of outcome and treatment response in breast cancer. Mol Doagn Ther. 2009;13:73–90. doi: 10.1007/BF03256317. [DOI] [PubMed] [Google Scholar]
- 4.Woo HG, Park ES, Cheon JH, et al. Gene expression-based recurrence prediction of hepatitis B virus-related human hepatocellular carcinoma. Clin Cancer Res. 2008;14:2056–2064. doi: 10.1158/1078-0432.CCR-07-1473. [DOI] [PubMed] [Google Scholar]
- 5.Ge X, Yamamoto S, Tsusumi S, et al. Interpreting expression profiles of cancers by genome-wide survey of breath expression in normal tissues. Genomics. 2005;86:127–141. doi: 10.1016/j.ygeno.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Chuma M, Sakamoto M, Yamazaki K, et al. Expression profiling in multistage hepatocarcinogenesis: identification of HSP70 as a molecular marker of early hepatocellular carcinoma. Hepatology. 2003;37:198–207. doi: 10.1053/jhep.2003.50022. [DOI] [PubMed] [Google Scholar]
- 7.Nojima M, Maruyama R, Yasui H, et al. Genomic screening of genes silenced by DNA methylation revealed an association between RASD1 inactivation and dexamethasone resistance in multiple myeloma. Clin Cancer Res. 2009;15:4356–4364. doi: 10.1158/1078-0432.CCR-08-3336. [DOI] [PubMed] [Google Scholar]
- 8.Lee JS, Grisham JW, Thorgeirsson SS. Comparative functional genomics for identifying models of human cancer. Carcinogenesis. 2005;26:1013–1020. doi: 10.1093/carcin/bgi030. [DOI] [PubMed] [Google Scholar]
- 9.Farber E. Pre-cancerous steps in carcinogenesis. Their physiological adaptive nature. Biochim Biophys Acta. 1984;738:171–180. doi: 10.1016/0304-419x(83)90002-1. [DOI] [PubMed] [Google Scholar]
- 10.Carrasco-Legleu CE, Márquez-Rosado L, Fattel-Fazenda S, et al. Chemoprotective effect of caffeic acid phenethyl ester on promotion in a medium-term rat hepatocarcinogenesis assay. Int J Cancer. 2004;108:488–492. doi: 10.1002/ijc.11595. [DOI] [PubMed] [Google Scholar]
- 11.Marche-Cova A, Fattel-Fazenda S, Rojas-Ochoa A, et al. Follow-up of GST-p during hepatocarcinogenesis with DEN-2AAF in F344 rats. Arch Med Res. 1995;26:S169–S173. [PubMed] [Google Scholar]
- 12.Rutemburg AM, Kim H, Fischbein JW, et al. Histochemical and structural demonstration of gamma-glutamyl transpeptidase activity. J Histochem Cytochem. 1969;17:517–526. doi: 10.1177/17.8.517. [DOI] [PubMed] [Google Scholar]
- 13.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 14.Lee JS, Chu IS, Heo J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 15.Feitelson MA, Pan J, Lian Z. Early molecular and genetic determinants of primary liver malignancy. Surg Clin North Am. 2004;84:339–354. doi: 10.1016/S0039-6109(03)00226-3. [DOI] [PubMed] [Google Scholar]
- 16.Enomoto K, Farber E. Kinetics of phenotypic maturation of remodeling of hyperplastic nodules during liver carcinogenesis. Cancer Res. 1982;42:2330–2335. [PubMed] [Google Scholar]
- 17.Tatematsu M, Nagamine Y, Farber E. Redifferenciation as a basis for remodeling of carcinogen-induced hepatocyte nodules to normal appearing liver. Cancer Res. 1983;1:5049–5058. [PubMed] [Google Scholar]
- 18.Farber E. Experimental induction of hepatocellular carcinoma as paradigm for carcinogenesis. Clin Physiol Biochem. 1987;5:152–159. [PubMed] [Google Scholar]
- 19.Pérez-Carreón JI, López-García C, Fattel-Fazenda S, et al. Gene expression profile related to the progression of preneoplastic nodules toward hepatocellular carcinoma in rats. Neoplasia. 2006;8:373–383. doi: 10.1593/neo.05841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borel F, Han R, Visser A, et al. Adenosine triphosphate-binding cassette transporter genes up-regulation in untreated hepatocellular carcinoma is mediated by cellular micro RNAs. Hepatology. 2012;55:821–832. doi: 10.1002/hep.24682. [DOI] [PubMed] [Google Scholar]
- 21.Nies AT, König J, Pfannschmidt M, et al. Expression of the multidrug resistance proteins MRP2 and MRP3 in human hepatocellular carcinoma. Int J Cancer. 2001;94:492–499. doi: 10.1002/ijc.1498. [DOI] [PubMed] [Google Scholar]
- 22.Serrano MA, Macias RI, Briz O, et al. Expression in human trophoblast and choriocarcinoma cell lines, BeWo, Jeg-3 and JAr of genes involved in the hepatobiliary-like excretory function of the placenta. Placenta. 2007;28:107–117. doi: 10.1016/j.placenta.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 23.Mizukoshi E, Honda M, Arai K, et al. Expression of multidrug resistance-associated protein 3 and cytotoxic T cell responses in patients with hepatocellular carcinoma. J Hepatol. 2008;49:946–954. doi: 10.1016/j.jhep.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 24.Nonaka M, Ma BY, Imaeda H, et al. Dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN) recognizes a novel ligand, Mac-2-binding protein, characteristically expressed on human colorectal carcinomas. J Biol Chem. 2011;286:22403–22413. doi: 10.1074/jbc.M110.215301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukaya Y, Shimada H, Wang LC, Zandi E, DeClerck YA. Identification of galectin-3-binding protein as a factor secreted by tumor cells that atimulates interleukin-6 expression in the bone marrow stroma. J Biol Chem. 2008;283:18573–18581. doi: 10.1074/jbc.M803115200. [DOI] [PubMed] [Google Scholar]