Abstract
T cell responses to allogeneic major histocompatibility (MHC) antigens present a formidable barrier to organ transplantation, necessitating long-term immunosuppression to minimize rejection. Chronic rejection and drug-induced morbidities are major limitations that could be overcome by allograft tolerance induction. Tolerance was first intentionally induced in humans via combined kidney and bone marrow transplantation (CKBMT), but the mechanisms of tolerance in these patients are incompletely understood. We now establish an assay to identify donor-reactive T cells and test the role of deletion in tolerance after CKBMT. Using high-throughput sequencing of the TCRB chain CDR3 region, we define a fingerprint of the donor-reactive T cell repertoire prior to transplantation and track those clones post-transplant. We observed post-transplant reductions in donor-reactive T cell clones in three tolerant CKBMT patients; such reductions were not observed in a fourth, non-tolerant, CKBMT patient or in two conventional kidney transplant recipients on standard immunosuppressive regimens. T cell repertoire turnover due to lymphocyte-depleting conditioning only partially accounted for the observed reductions in tolerant patients; in fact, conventional transplant recipients showed expansion of circulating donor-reactive clones, despite extensive repertoire turnover. Moreover, loss of donor-reactive T cell clones more closely associated with tolerance induction than in vitro functional assays. Our analysis supports clonal deletion as a mechanism of allograft tolerance in CKBMT patients. The results validate the significance of donor-reactive T cell clones identified pre-transplant by our method, supporting further exploration as a potential biomarker of transplant outcomes.
Introduction
Chronic immunosuppression in kidney transplantation is associated with morbidities including nephrotoxicity, metabolic abnormalities, and increased risk of infection and malignancy (1). Moreover, despite recent improvements in one-year kidney allograft survival, late rejection rates remain high (2). Immune tolerance in organ transplantation, defined as the absence of rejection without immunosuppression, would avoid these morbidities. Spontaneous tolerance is rare in conventional renal transplant recipients, with frequencies estimated at less than five percent (3, 4).
Tolerance was first intentionally induced in humans via combined kidney and non-myeloablative bone marrow transplantation (CKBMT), a protocol designed to induce a mixed chimeric state in which hematopoietic elements are comprised of a mixture of host and donor cells (5, 6). Among ten patients who received CKBMT (five subjects in Immune Tolerance Network [ITN] study NKDO3; five subjects in the study ITN 036ST), seven have tolerated their allograft off immunosuppression for 4–12 years (6–8).
In the rodent regimens that led to the development of this protocol, mixed chimerism was durable and tolerance involved long-term intrathymic deletion of donor-reactive T cells (ie “central tolerance”) (9–11). In human CKBMT patients, however, mixed chimerism was transient (6, 12), suggesting that additional, likely peripheral, mechanisms are involved in maintaining long-term tolerance. Functional mechanistic studies in these CKBMT patients suggested a role for early suppression and long-term deletion of donor-reactive T cells in maintaining tolerance (6, 13). In vitro assays, however, cannot reliably distinguish anergy from deletion as mechanisms of unresponsiveness. We now establish an assay to specifically track donor-reactive T cells and test the role of deletion in maintaining long-term tolerance after CKBMT.
Tracking of donor-reactive clones in transplant patients is hampered by the large proportion (up to 10%) of T cells directly recognizing a set of MHC alloantigens (14, 15), presumably involving many specificities. We devised a deep sequencing approach to identify and track the donor-reactive T cell repertoire. Using ImmunoSEQ (Adaptive™), TCR β (TRB) CDR3 regions are amplified with primers for all 54 known expressed Vβ and all 13 Jβ regions adapted for solid phase PCR and high throughput sequencing (16–18). Each individual T cell clone has a distinct TRB CDR3 sequence. We hypothesized that CDR3 sequencing of a transplant recipient’s donor-reactive T cells, as identified by their proliferation in an anti-donor mixed lymphocyte reaction (MLR) prior to transplant, would identify donor-specific TCR sequences that could then be physically tracked in the recipient’s post-transplant peripheral blood samples to differentiate between anergy and deletion of donor-specific T cells. Using this analysis in four CKBMT and two conventional renal allograft recipients, we obtained evidence for clonal deletion as a mechanism of allograft tolerance in humans.
Results
Defining a “fingerprint” of the anti-donor T cell repertoire
Figure 1 illustrates our strategy for defining the “fingerprint” of the alloreactive repertoire for any responder-stimulator (recipient-donor) pair. An allostimulated population was generated via CFSE MLR: MLR responders and irradiated stimulator PBMCs were labeled with CFSE and violet dye respectively, co-cultured for 6 days, then FACS sorted for violet negative, CD3-positive, CFSE-low CD4+ and, in separate tubes, CD8+ cells (Fig. 1A). Deep sequencing was then performed on the genomic DNA extracted from these sorted T cell populations that had divided in response to donor antigens. To permit identification of clones expanding in the MLR, deep sequencing was also performed on unstimulated CD3+CD4+ and CD3+CD8+ FACS-sorted T cells from the same pre-transplant peripheral blood sample. To be considered donor-reactive, a clone defined by the unique nucleotide sequence of its TRB CDR3 region must have been detected above a minimum frequency threshold of 10−4 in the CFSE-low population in the stimulated (MLR) sample and have expanded at least 5-fold relative to its frequency in an unstimulated sample from the same time point, thereby excluding highly-abundant but not specifically donor-reactive clones (Fig.1B). We could thus define a “fingerprint” of the pre-transplant donor-reactive T cell repertoire for each donor-recipient pair and these clones could be tracked in unstimulated post-transplant samples.
Fig. 1. Mixed lymphocyte reaction experimental design and schematic of TCR sequencing analysis strategy to identify and track donor-reactive T cells.
A. CD3+Violet- cells, representing the responder T cells, were selected and further separated into CD4+ and CD8+ subgroups. Within each subgroup, the CFSE-low cells were isolated for DNA extraction and TCRβ CDR3 deep sequencing. Sorting strategy is indicated with boxes on the dot plots and bars on the histograms; CFSE staining in unstimulated control sample shown in Fig. S1A. B. Pre-transplant “fingerprint” of anti-donor T cell repertoire defined as all clones detected at a frequency greater than 10^−4 in the stimulated condition (CFSE-low cells in MLR) that have expanded at least 5-fold relative to their frequency in the unstimulated T cell population (unstimulated repertoire defined via TCR sequencing of CD3+CD4+ or CD3+CD8+ T cells isolated via FACS sorting of PBMCs from the same sample used for the CFSE-MLR). Each donor-reactive clone identified by its unique CDR3 nucleotide sequence could then be tracked in post-transplant unstimulated peripheral blood samples.
Reproducible detection of alloreactive TCR in blood samples obtained at different times
To validate the approach of tracking a set of alloreactive TRBs over time, we tested whether individual alloreactive T cell clones could be reproducibly detected in blood samples drawn at multiple times. Using PBMCs obtained from three healthy controls at different time points separated by two-week or one-year intervals, we set up parallel MLRs with the same responder-stimulator pairs for each time point. We then performed deep sequencing on the dividing T cells in the MLR as well as on unstimulated T cells (Fig. S1B,C). Deep CDR3 sequencing identified fewer unique clones in allostimulated versus unstimulated populations, resulting in decreased entropy and increased clonality, a trend that was most striking in the CD4 compartment (Table S1).
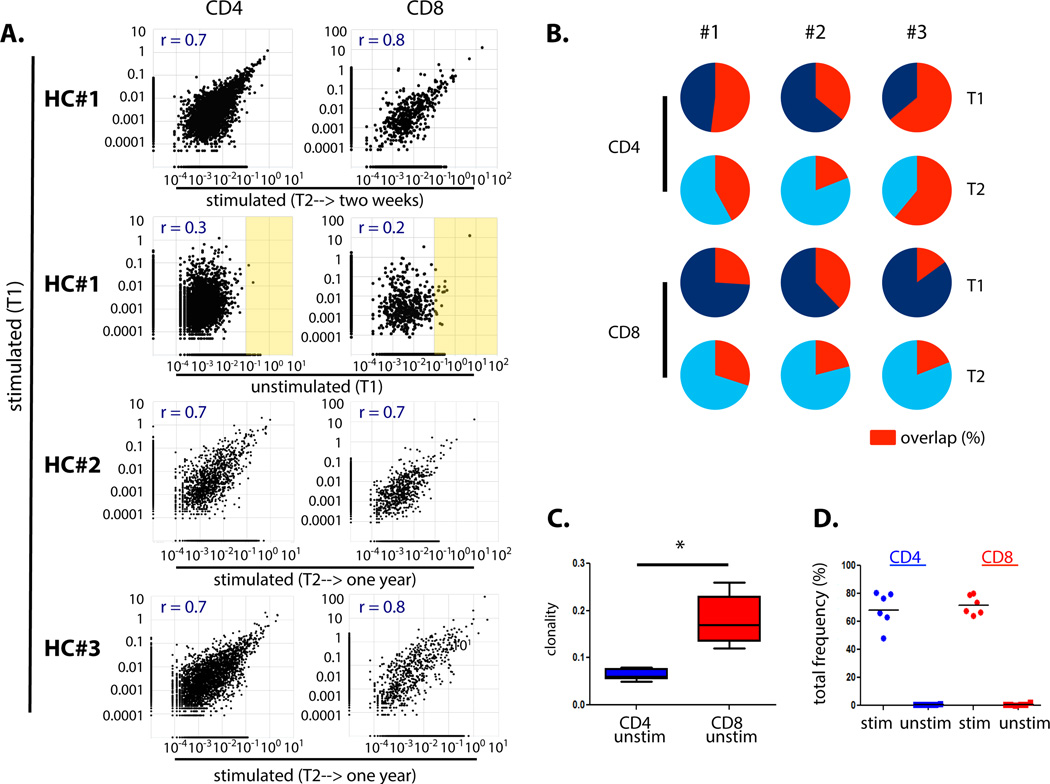
A comparison of the log clonal frequencies from the MLRs (stimulated samples) separated by two weeks showed strong linear correlations among CD4 (r = 0.7) and CD8 (r = 0.8) clones, but minimal correlation with the unstimulated repertoire (CD4: r = 0.3; CD8: r = 0.2); the linear correlation persisted in the clonal frequencies identified in MLRs from blood draws separated by a year-long interval (Fig. 2A). Figure 2B shows the relative frequency overlap of the alloreactive clones (defined in Figure 1) between the two time points for each healthy control. The degree of overlap was comparable between the three samples, including across the two week (HC#1) and year-long (HC#2, HC#3) time intervals. There was less overlap in alloreactive CD8 than CD4 T cell clones detected at different time points. This may reflect the presence of a few very high frequency clones in the unstimulated CD8 repertoires (Fig. 2A), resulting in fewer unique CD8 clones being sampled in each blood draw (Table S1). Indeed, the diversity of the unstimulated CD8 repertoires was less than that of CD4s: the clonality of a pool of identical clones is “1” while that of a pool of all unique clones is “0;” the significantly increased clonality of CD8 compared to CD4 cells in the same unstimulated samples is shown in Fig. 2C (p = 0.0062, n = 5, two-tailed paired t test). Cumulatively, all alloreactive CD4 clones comprised approximately 48–80% of clones in each MLR and less than 1.1% of the unstimulated CD4 repertoire (Table S2). For the CD8s, the alloreactive clones accounted for 64–80% of the stimulated population and less than 2.5% of the unstimulated population (Fig. 2D). Taken together, our healthy control studies showed that the alloreactive T cell populations identified via CFSE MLR that recognized a given set of alloantigens were reproducibly detectable in separate peripheral blood samples over highly disparate time intervals ranging from 2 weeks to 1 year.
Fig. 2. Overlap of the alloreactive T cell repertoire via TCR sequencing in repeat MLR assays in healthy controls.
A. Top panel: linear correlation of log frequencies of alloreactive clones detected in MLRs performed at time point 1 [stimulated (T1)] and time point 2 [stimulated (T2)] for Healthy Control (HC) #1 from blood draws separated by a two-week interval. Second panel from top: no such correlation is observed when clonal frequencies in the stimulated populations are plotted against frequencies in the unstimulated population [unstimulated (T1)]; high frequency clones in the unstimulated populations highlighted in yellow. These results are representative of similar analyses in HC#2 and HC#3 (Fig. S1D). Bottom two panels: linear correlation persists in MLRs performed from blood draws separated by a year interval (HC#2, HC#3). Same allogeneic stimulator for each HC at T1 and T2. Overlapping number of sequences detected: HC#1 T1 & T2 stim CD4 = 2944, CD8 = 465; HC#1 T1 stim & T1 unstim: CD4 = 3011, CD8 = 478; HC#2 T1 & T2 stim: CD4 =1162, CD8 = 642. HC#3 T1 & T2 stim: CD4 =2850, CD8 = 652. B. Pie charts showing relative overlap of the summed frequencies of alloreactive clones (“fingerprint” as defined in Figure 1) over time in three healthy controls. Each circle represents the cumulative frequency of all alloreactive clones identified in the sample; red segment shows the percentage of that total frequency arising from alloreactive clones identified at both T1 and T2 (tabulated values Table S2). C. Boxplot comparing the clonality of the unstimulated CD4 and CD8 T cell repertoires (* P = 0.0062, n = 5, two-tailed paired t test; tabulated values Table S1). D. Cumulative frequencies of all alloreactive clones for each pair of stimulated and unstimulated HC at all time points. Stim = stimulated; unstim = unstimulated; HC = healthy control.
Reduced circulating donor-reactive T cell clones in tolerant CKBMT patients
Our studies of alloreactive T cells in healthy controls showed that donor-reactive clones identified in MLRs could be consistently detected in peripheral blood at disparate time points. We therefore used this approach to identify donor-reactive T cells pre-transplant and track them over time post-transplant in six subjects: four CKBMT recipients (Subjects 1, 2, 4, and 5 from ITN trial ITN036ST) who were removed from immunosuppression eight months post-transplant and two kidney transplant recipients receiving conventional immunosuppression (IS#1 and IS#2). Sequencing statistics are summarized in Table S3. For each transplant recipient, we defined a “fingerprint” of the anti-donor T cell repertoire using pre-transplant PBMCs. The limit of detection of T cell clones for tracking in the unstimulated pre- and post-transplant samples was determined with a power calculation that took into account the cell number and the number of reads obtained from each sample.
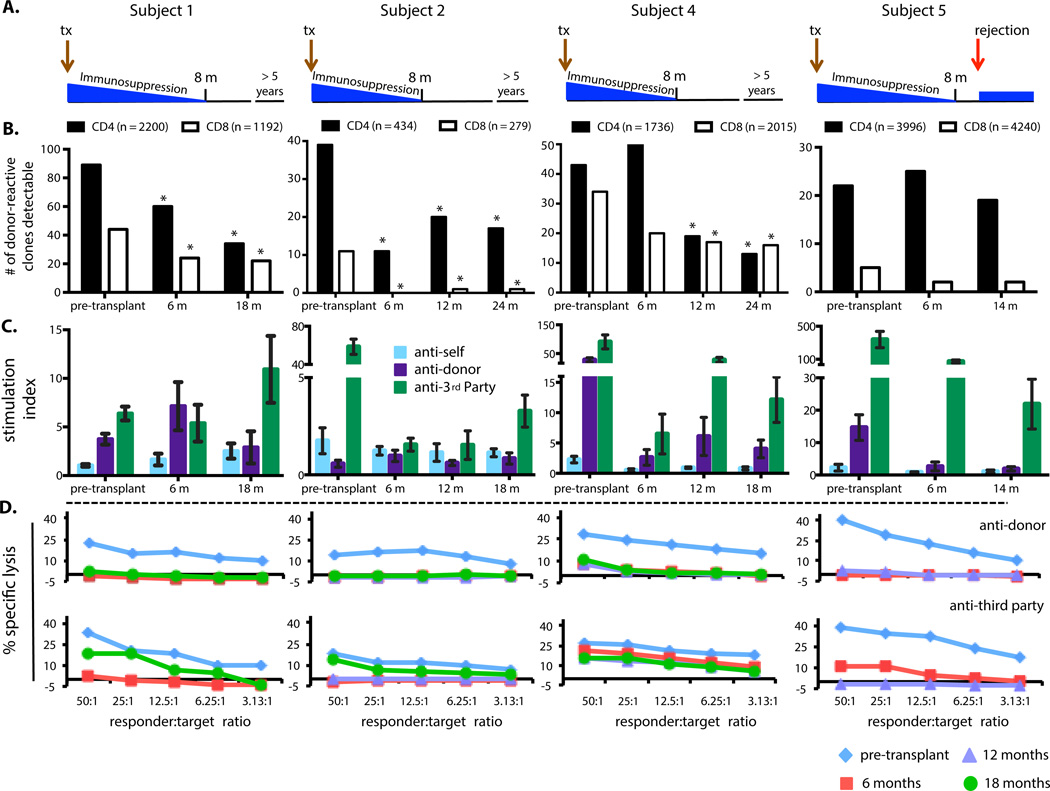
CKBMT Subject 1 has stable allograft function >5 years after stopping immunosuppression at 8 months post-transplant (Fig. 3A). All CD4 and CD8 samples permitted clonal detection at ≥10−5 frequency. We identified 2200 donor-reactive CD4+ clones and 1192 CD8+ clones as defined by our approach in Figure 1B. We then compared the number of these donor-reactive that were detectable in unstimulated pre- and post-transplant blood samples. A significant reduction in the number of circulating donor-reactive CD4+ and CD8+ clones was observed at both 6 and 18 months post-transplant compared to pre-transplant blood (Figs. 3B, S3, S4; Table S4). Results were robust to changes in definition of donor-reactive clones ranging from 5-fold to 10-fold for all subjects; the 5-fold expansion criterion included the largest number of clones while excluding clones with minimal expansion that might reflect “bystander” effects (Fig. S2).
Fig. 3. Clinical course, tracking of donor-reactive T cell clones, MLR, and CML results in CKBMT subjects.
A. Overview of clinical course. B. Number of donor-reactive TRB CDR3 clones (y-axis) detected in the unstimulated CD4 (black) and CD8 (white) repertoire at the indicated time points (x-axis). The “fingerprint” of the anti-donor T cell repertoire was defined for each subject as clones in the pre-transplant MLR with ≥10−4 frequency that were expanded at least 5-fold relative to their frequency in the pre-transplant unstimulated sample (total number indicated above the relevant panel). Sufficient power was obtained to consider a frequency of 10−5 as detectable in all unstimulated populations for Subjects 1, 2, 4, and 5×10−5 for Subject 5. * P < 0.05 compared to pre-transplant (P-values Table S4; two-sided Fisher’s exact test). C. MLR: proliferative responses to recipient (anti-self), donor (anti-donor) and third party (anti-3rd party) relative to proliferation of unstimulated PBMCs are shown at the indicated time points. Each bar represents the mean ±SD stimulation index of triplicate cultures. D. CML: responses at the indicated time points are shown. MLR and CML data have been previously summarized in Kawai et al. (8).
Functional MLR assays in Subject 1 (Fig. 3C) showed persistent anti-donor reactivity (greater than anti-self) at 6 months that was markedly reduced at 1.5 years (comparable to anti-self), while cell-mediated lympholysis (CML) assays (Fig. 3D) revealed donor unresponsiveness at both times; responses to extensively HLA-mismatched (from donor and recipient) third party donors recovered between 6 and 18 months post-transplant, demonstrating that immune unresponsiveness was specific to the donor. Limiting dilution analyses (LDA) quantify functional cytotoxic T lymphocyte precursors (CTLp) and IL-2-producing helper T lymphocytes (HTL) (Fig. S5). Donor-reactive CTLp were undetectable by 6 months, suggesting the remaining donor-reactive cells were anergic, as only partial deletion of donor-reactive CD8 cells was apparent (Fig. 3B). Anti-donor HTL were measureable at 6 but not 18 months.
We performed similar analyses in two additional tolerant CKBMT recipients. Subject 2 is >5 years post-CKBMT with no rejection. Her pre-transplant MLR was weak, perhaps due to extensive HLA sharing with the donor (Table S3 legend). Significantly fewer CD4+ and CD8+ donor-reactive clones were detected at 6, 12, and 24 months after CKBMT than before (Figs. 3B, S3, S4; Table S4). While TCR analysis revealed residual donor-reactive CD4 clones, functional assays (Figs. 3C,D, S5) showed minimal to no post-transplant responses to donor, with recovery of third party responses at 18 months in MLR and CML (8).
Subject 4 also shows allograft tolerance >4.5 years after CKBMT. Figure 3B shows a progressive reduction in CD8+ donor-reactive clones reaching significance at 12 months post-transplant; donor-reactive CD4+ clones showed an initial non-significant increase at 6 months post-transplant followed by significant reductions at 12 and 24 months (Figs. 3B, S3, S4; Table S4). Several functional assays, including the MLR, revealed persistent anti-donor responsiveness at 12 and 18 months (Figs. 3C,D, S5), unlike Subjects 1 and 2 and NKDO3 subjects (13). To test whether the same clones responded to the donor in post- and pre-transplant MLRs, we performed clonal analysis on one-year CD4+ and CD8+ T cells dividing in anti-donor CFSE MLR. The overlap of alloreactive clones over one year was markedly less than that observed in healthy controls over the same interval (Fig. S6A and Fig. 2B, respectively). The number of post-transplant donor-reactive CD4 and CD8 T cells defined by the 12-month MLR (clones with frequencies ≥10−4 in the 12 month post-transplant MLR expanded at least 5-fold compared to the unstimulated 12 month post-transplant sample) remained relatively constant in unstimulated pre- and post-transplant blood samples (Fig. S6B). Collectively, these data suggest that the persistent MLR in Subject 4 was largely mediated by a subset of clones that did not lead to rejection, some of which may have developed post-transplant, others of which did not expand sufficiently in the pre-transplant MLR to be defined as alloreactive, escaped deletion by conditioning treatment, and were neither expanded nor deleted in the presence of the donor graft.
No reduction in donor-reactive clones in a CKBMT recipient who failed tolerance induction
One month after discontinuing immunosuppression, CKBMT Subject 5 developed acute rejection that culminated in graft loss despite anti-rejection therapy (Fig. 3A). Of the four CKBMT recipients, only Subject 5 showed no significant reduction in numbers of circulating donor-reactive CD4 and CD8 clones after transplantation (Figs. 3B, S3,S4; Table S4). Remarkably, MLR, CML and CTLp assays showed donor-specific unresponsiveness at 6 and 12 months (Figs. 3C,D, S5). Thus, the functional assays did not distinguish the lack of clinical tolerance, whereas the clonal analysis showed a distinct pattern compared to the three tolerant subjects.
Lack of deletion of donor-reactive clones in conventional kidney transplant recipients
To provide a further comparison to tolerant CKBMT recipients, we studied two kidney transplant recipients receiving conventional immunosuppression. Subject IS#1 received a living unrelated donor kidney. Subject IS#2 received a living unrelated donor kidney after a failed prior living-related transplant, and had Banff grade 1B acute cellular rejection at 3 months post-transplant. In contrast to tolerant patients, donor-reactive CD4+ clones were significantly enriched in the post-transplant compared to pre-transplant peripheral blood in IS#1 and IS#2. Donor-reactive CD8+ clones were not significantly changed (Figs. 4A, S7; Table S4).
Fig. 4. Tracking of donor-reactive T cell clones in “conventional” kidney transplant recipients and summary of clonal analysis results for all six subjects.
A. Clinical course and number of donor-reactive TRB CDR3 clones (y-axis) detected in the unstimulated CD4 (black) and CD8 (white) repertoire at the indicated time points (x-axis). Sufficient power was obtained to consider a frequency of 10−5 as detectable in all unstimulated populations for IS#1 and 5×10−5 for IS#2. * P < 0.05 compared to pre-transplant (P-values Table S4; two-sided Fisher’s exact test). B. Change over time in detection of unstimulated T cell populations of donor-reactive CD4 and CD8 clones (defined in Fig. 3B legend). Fold change is the odds ratio of the number of donor-reactive clones detected in unstimulated post-transplant CD4 and CD8 populations relative to the number detected in unstimulated pre-transplant populations (pre-transplant = 1). Open symbol = statistically significant reduction or increase (P-value < 0.05). Tabulated results in Tables S4 and S5.
Figure 4B summarizes the fold change in the number of donor-reactive clones detected in post-transplant as compared to pre-transplant blood in all six transplant patients. In contrast to the three tolerant CKBMT subjects, there was no significant reduction in either of the two “conventional” kidney transplant subjects or the non-tolerant CKBMT subject in circulating donor-reactive CD4 and CD8 T cell clones. Of note, the limited number of donor-reactive CD8 clones tracked in Subject 5 and IS#2 limited the ability to evaluate changes over time. For non-tolerant subjects, observed expansion of donor-reactive CD4 cells was greater when the definition of donor reactivity required greater expansion in the pre-transplant MLR (Fig. S2), suggesting that the donor-reactive pre-transplant clones responding most strongly in MLR were most likely to expand post-transplant.
As additional controls, an identical analysis was performed in healthy controls in whom clones defined as alloreactive in a particular MLR were tracked in an unstimulated sample one year later. For both CD4 and CD8 T cells, there was no significant change in the number of alloreactive T cells, in contrast to the reduction seen in the tolerant CKBMT subjects and the expansion in donor-reactive CD4 cells in the “conventional” transplant patients (Fig. 4B, details Table S5).
T cell repertoire turnover
The loss of donor-specific clones in tolerant subjects might reflect global T cell depletion due to conditioning, leading to repertoire turnover as T cells developed de novo from thymic recovery. To quantify changes in T cell repertoire over time, we calculated the Jensen-Shannon Divergence (JSD) of the top 1000 nucleotide sequences pre- and post-transplant (Fig. 5A). A JSD of “1” reflects complete divergence of two repertoires, while the JSD of two identical repertoires is “0”. For reference, we determined JSD of pairs of samples of T cells isolated from peripheral blood one year apart from two healthy controls. While all transplant recipients showed greater repertoire divergence than healthy controls, JSD values were higher for CD4+ populations of CKBMT recipients as compared to CD8+ T cells. Among the “conventional” transplant recipients, IS#2 showed repertoire turnover close to that of the CKBMT patients while that of IS#1 was lower. The non-tolerant CKBMT recipient Subject 5 showed the highest JSD, with almost complete turnover of CD4 and CD8 repertoires (Fig. 5A).
Fig. 5. TCR repertoire turnover in kidney allograft recipients.
A. Jensen-Shannon Divergence (JSD) comparing pre- and post-transplantation (last post-transplantation time point) TCR repertoires. JSD on top 1000 nucleotide clones ranked by frequency (0 denotes identical repertoires; 1 denotes complete repertoire divergence). Healthy controls: average JSD on top 1000 nucleotide clones of two healthy controls in whom TCR sequencing was performed at two time points 1 year apart. B. Anti-donor CD4 and CD8 clonal analysis relative to overall repertoire turnover. Relative numbers of donor-reactive (as defined in Fig. 3B legend) vs non-donor-reactive clones (all other clones detected in unstimulated pre-transplant sample) detected at any level (threshold detection of assay 10−6) in post-transplant samples. Relative change is the odds ratio of the relative (post/pre) number of donor-reactive clones divided by the relative (post/pre) number of non-donor-reactive clones in unstimulated samples at the same time. A value of “1” indicates that the proportion of donor-reactive clones (defined pre-transplant as above) detected at a given timepoint was equal to that of all clones (detected pre-transplant) detected at the same timepoint. A value <1 indicates lower rate of detection of donor-reactive vs all clones and a value >1 indicates greater rate of detection of donor-reactive vs all clones. Open symbols = statistically significant reduction or increase (P-value < 0.05; two-sided Fisher’s exact test); tabulated data and P-values in Table S7. C. Anti-donor CD4 and CD8 clonal analysis relative to overall repertoire turnover in Subject 1: effect of varying definition of donor reactivity by different fold-expansion criteria (frequency in pre-transplant anti-donor MLR/frequency in unstimulated pre-transplant sample >5, 7, or 10). Red points are statistically significant (p < 0.05); tabulated data and P-values in Table S9.
We next compared the likelihood, post-transplantation, of detecting any clones detected pre-transplant compared to those defined as donor-reactive. Among tolerant subjects, no significant decrease in donor-reactive compared to all pre-transplant CD4 clones was observed post-transplant. Subject 2 showed a significant relative increase in donor-reactive CD4 cells by one year post-transplant and Subject 4 showed a significant relative increase only at 6 months post-transplant (Fig. 5B). A significant and sustained reduction in the detection of donor-reactive compared to all CD8+ clones was observed in Subjects 2 and 4, suggesting antigen-driven loss of donor-reactive CD8 clones (Fig. 5B). The notion that some, but not all, deletion of donor-reactive clones in tolerant subjects may have reflected repertoire turnover was supported by analysis of third party-reactive T cells identified pre-transplant (Fig. S8), some of which showed post-transplant reductions.
The determination of relative loss of donor-reactive compared to all pre-transplant clones was affected by the definition of “donor-reactive” in some subjects. For example, for Subject 1, when this definition required increasing levels of MLR expansion, greater relative loss of donor-reactive CD4 and CD8 clones was seen (Fig. 5C). This result suggests that more strongly donor-reactive clones were more likely to show deletion following CKBMT. Overall, post-transplant reductions in donor-reactive clones in tolerant subjects may reflect a mixture of repertoire turnover and specific deletion of donor-reactive T cells, possibly following initial antigen-driven expansion.
Reduced TCR diversity in non-tolerant compared to tolerant subjects
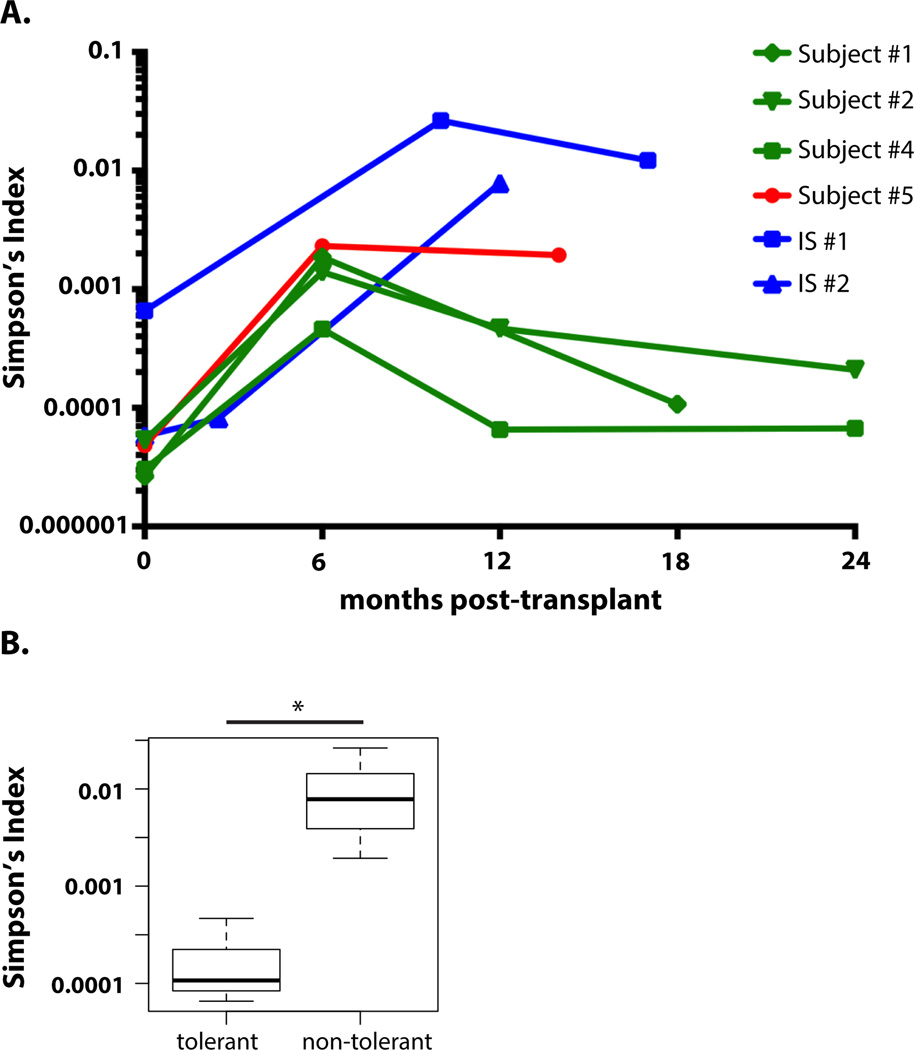
Since lymphopenia-driven proliferation (LIP) in a T cell-deficient environment (19) may reduce the repertoire diversity associated with T cell reconstitution after lymphoablative conditioning (20), we compared overall clonal diversity (by Simpson’s Index D) of post-transplant T cell populations (Fig. 6A). In contrast to tolerant subjects, in whom the CD4 T cell diversity returned to pre-transplant values, non-tolerant subjects showed persistently decreased clonal diversity (increased D) post-transplantation (Student’s t-test p-value 0.017 comparing two groups at time point nearest one year post-transplant)(Fig. 6B). No difference was seen between the groups in the diversity of CD8 repertoires.
Fig. 6. Recovery of CD4 repertoire diversity in tolerant recipients.
(A) CD4 T cell repertoire diversity (measured by Simpson’s Index D) over time after transplantation (D of “1” indicates that all clones are identical; smaller D indicates clones are more unique and therefore the repertoire is more diverse). (B) Comparison of CD4 Simpson’s Index (D) in tolerant (Subjects 1, 2, and 4) and nontolerant (Subjects 5, IS#1, and IS2) subjects near 1 year after transplant (10 months: IS#2; 12 months: Subjects, 2, 4, and IS#1; 14 months: subject 5; 18 months: subject 1). * P = 0.017; two-sided Student’s t test on logarithm of D (n = 6).
Discussion
T cell responses to allogeneic MHC molecules are orders of magnitude stronger than other responses (14, 15, 21, 22), presumably involving myriads of T cell receptors. We have developed an approach using deep TCR sequencing to identify, prior to transplant, and track, following transplantation, human transplant recipients’ donor-reactive T cell repertoires. We demonstrate the feasibility of this approach and use it to address mechanisms of tolerance in CKBMT recipients.
Previous studies in ITN CKBMT trial NKDO3 (6, 13) could not distinguish between anergy and deletion of donor-reactive T cells in maintaining long-term tolerance. Our new method allowed specific assessment of clonal deletion. Circulating donor-reactive CD4+ and CD8+ clones, identified prior to transplantation by CFSE-MLR, decreased after transplantation in all tolerant subjects. Deletion was either partial or not apparent at 6 months and frequently evolved over time. In the only CKBMT subject who failed to achieve operational tolerance, significant reductions of donor-specific clones were not observed. These studies provide evidence for a role for deletion in the maintenance of allograft tolerance in humans. While we cannot exclude the possibility that donor-reactive clones moved from the circulation into the allograft, this would be inconsistent with protocol biopsies showing no rejection and minimal cellular infiltrates, which are enriched for Foxp3+ cells, in tolerant CKBMT recipients (6, 7).
Conventional transplant recipients showed persistent expansion of donor-reactive T cell clones following transplant, despite considerable repertoire turnover, suggesting that expansion was allograft-driven and showing that pre-transplant MLRs identify biologically relevant donor-reactive clones. A role in graft-versus-host-disease was identified for a CD4 clone recognizing a recipient minor histocompatibility antigen in MLR following HLA-identical hematopoietic cell transplantation (23), but our studies are the first to examine the entire alloreactive repertoire against HLA antigens. Further evidence from our studies that biologically significant clones are identified in pre-transplant MLR includes the following: 1) The donor-reactive clones expanding most strongly in pre-transplant MLR were most likely to be expanded in post-transplant blood in non-tolerant subjects (Fig. S2); 2) The donor-reactive clones expanding most strongly in pre-transplant MLR were more likely than other clones to be deleted in post-transplant blood of tolerant subjects (Fig. 5C); 3) Donor-reactive clones that were insufficiently dominant to be identified as donor-reactive in pre-transplant MLR persisted in constant numbers and contributed to a post-transplant MLR in Subject 4, but did not cause rejection. In contrast, Subject 5 failed to show deletion of donor-reactive clones identified in pre-transplant MLR and rejected the graft. These data suggest that non-dominant donor-reactive clones that can produce a post-transplant MLR when dominant clones are deleted may be of minimal biological significance, whereas those that dominate in the pre-transplant MLR are of major importance.
Deletion of donor-reactive clones in CKBMT recipients was partially explained by global T cell depletion induced by conditioning (6, 7, 12, 13). The initial recovery of T cells in CKBMT recipients is most likely driven by LIP, as most T cells express an effector/memory phenotype in the first 3–6 months post-transplant (12), as observed for rapid LIP (19), which is largely antigen-driven (24, 25). Therefore, residual donor-reactive clones may expand initially in response to bone marrow and/or kidney alloantigens. However, donor-reactive T cell deletion is unlikely to reflect only repertoire turnover, given the similar, high level of repertoire turnover in all CKBMT recipients and lack of donor-reactive clonal deletion only in the subject who failed tolerance. The persistence of some pre-existing donor-reactive clones along with the sometimes progressive deletion observed over time in tolerant patients is consistent with initial LIP/antigen-driven expansion of surviving donor-reactive clones. Indeed, two tolerant subjects, while showing an absolute loss of donor-reactive CD4 clones, showed an increase in these clones relative to all preexisting clones, one at 6 months and another persisting longer post-transplant (Fig. 5B). Thus, the relative numbers of persisting donor-reactive and non-donor-reactive clones may be the net effect of T cell-depleting conditioning (affecting any clone), antigen-driven expansion (donor-reactive clones) and antigen-driven deletion (donor-reactive clones). During rejection, entry of donor-reactive T cells, especially CD8 clones, into the graft may also reduce circulating clonal frequencies, perhaps mitigating the detection of expanded anti-donor clones in the rejecting CKBMT Subject 5. Ultimately, the relative loss of donor-reactive T cell clones was greater than that of all clones in several instances in the tolerant subjects. Thus, our data suggest that expanded donor-reactive clones are gradually deleted in tolerant subjects, resulting in specific clonal reduction, especially among clones with strongest anti-donor reactivity.
The reduction of donor-reactive clones in tolerant subjects is consistent with our hypothesis that donor-reactive T cells are slowly deleted in response to repeated encounter with donor antigens on quiescent, accepted allografts (13). Late deletion seems less likely to occur intrathymically, as hematopoietic chimerism was short-lived (12), making long-term intrathymic antigen presenting cells (APC) chimerism unlikely. However, peripheral APCs presenting intact or processed donor antigen might migrate to the thymus (26) and mediate ongoing deletion.
Clonal analysis detected persistent donor-reactive clones with greater sensitivity than in vitro assays that revealed donor-specific unresponsiveness in CKBMT Subjects 1, 2, and 5. Donor-specific unresponsiveness was particularly surprising in the rejector, Subject 5. Unlike the functional assays, TCR clonal analysis is not affected by anergy or suppression. The absence of a post-transplant MLR in Subject 5 despite the lack of deletion of dominant donor-reactive clones suggests that these clones may have been anergic and that anergy could have been broken by the renal infection that preceded rejection (8). The poor predictive value of MLR and CML with respect to graft outcomes is consistent with prior studies in animals (27, 28), patients receiving conventional transplants (29–33) and patients receiving a different CKBMT protocol for HLA-mismatched kidney allograft tolerance induction (34). Overall, clonal analysis distinguished between tolerance and non-tolerance among the six subjects, suggesting a new and specific method of assessing transplantation tolerance.
The similar clonal expansion in two conventional kidney transplant recipients despite disparate clinical outcomes (IS#2 but not IS#1 had a rejection) may reflect the limited cell numbers available for IS#2, resulting in a higher threshold frequency to declare a clone “present”. Moreover, important differences in donor-reactive T cell clone numbers might be present in the kidney graft and not the circulation and future studies of allograft biopsies and urine will be important. Future studies will include analyses of indirect anti-donor alloreactivity, which also contributes to rejection (35–37).
Repertoire turnover was greater for CKBMT patients compared to the non-rejecting conventional transplant recipient, consistent with more potent T cell-depleting treatments in conditioning for CKBMT. However, both conventional transplant recipients also showed markedly greater TCR turnover than healthy controls over one year, demonstrating the potential of our approach to provide insight into the effects of immunosuppressive regimens. In a recent report (38) on TCR repertoire in multiple sclerosis (MS) patients receiving conditioning and autologous hematopoietic cell transplantation, T cell diversity recovered more quickly in MS patients who responded to treatment compared to non-responders. It is interesting, therefore, that CD4 T cell diversity returned to baseline levels more rapidly in tolerant than in non-tolerant subjects (Fig. 6A). This return of diversity did not correlate with recovery of naïve-type CD4 cells, which was more rapid in CKBMT Subjects 4 and 5 than in Subjects 1 and 2 (12).
Our study is limited by the small number of these tolerant patients available and will require further validation in larger cohorts as transient chimerism protocols are evaluated in additional subjects. While differences in immunosuppression between the CKBMT and conventional patients may have affected the observed clonal behavior, re-establishment of a new T cell repertoire due to T cell depleting conditioning is insufficient to account for deletion of donor-reactive clones in tolerant patients. Further exploration of how various induction and maintenance immunosuppression regimens differentially affect donor-reactive clones after transplant will be of interest.
In conclusion, we have described a novel method whereby donor-reactive recipient T cell clones are identified pre-transplant, then tracked after transplantation. We obtained evidence for a role for deletion of donor-reactive CD4 and CD8 T cells in maintaining tolerance in CKBMT recipients with transient chimerism. A recent study (39) used high throughput CDR3 sequencing to detect donor-reactive T cells in the graft and urine of a patient with allograft dysfunction. However, that study relied on a measurable post-transplant MLR to identify donor-reactive clones, and did not assess the fate of pre-existing donor-reactive T cells. Our approach of identifying donor-reactive clones prior to transplantation and tracking them prospectively avoids dependence on functional assays post-transplant, which correlate poorly with outcomes. This strategy has revealed the biological importance of donor-reactive T cell clones detected in a pre-transplant MLR, demonstrating post-transplant expansion of these clones even in the face of global T cell repertoire turnover and has provided new mechanistic insights into tolerance achieved via transient chimerism, implicating eventual peripheral deletion of donor-reactive T cells in response to an accepted renal allograft.
Materials and Methods
Study Design
The object of this study was to study the fate of pre-transplant donor-reactive T cells in transplant patients and to provide insight into the mechanisms of long-term tolerance in CKBMT. Laboratory investigations were performed on four of five CKBMT patients (Subjects 1, 2, 4, and 5) in study ITN036ST, which included for-protocol PBMC collections pre- and post- transplant, and two conventional transplant patients from Columbia’s Center for Translational Immunology Biobank Core of transplant recipient clinical specimens. CKBMT Subject 3 was removed from ITN036T after early allograft loss due to thrombotic microangiopathy and post-transplant PBMCs were therefore not available for analysis. The conventional transplant patients studied were those for whom sufficient post-transplant PBMC were available to perform the required in vitro assays and for whom one or more post-transplant kidney transplant biopsies, clearly indicating rejection or lack thereof, were available. There was no randomization or blinding.
Subjects
CKBMT subjects: clinical outcomes in five CKBMT patients in study ITN036ST have been described (7, 8). Conventional transplant recipients: Subject IS#1 had end-stage renal disease (ESRD) secondary to focal segmental glomerulosclerosis (FSGS) and received a renal transplant from a living related donor. Subject IS#1 received thymoglobulin and methylprednisolone for induction therapy and was subsequently maintained on tacrolimus and mycophenolate. Allograft biopsies performed at 10 and 17 months after transplantation to evaluate acute rises in serum creatinine showed no evidence of cellular or antibody-mediated rejection, and were consistent with calcineurin inhibitor toxicity. Subject IS#2 also had ESRD due to FSGS, and received a renal transplant from a living unrelated donor several years after a prior living-related transplant had failed. Because the patient was highly sensitized, he received plasmapheresis and IVIg preoperatively, and received rituximab, basiliximab, and methylprednisolone as induction therapy. A three-month protocol biopsy revealed Banff grade 1B acute cellular rejection (ACR), which was treated with thymoglobulin and corticosteroids; subsequent protocol biopsies at 6 months and 1 year were suspicious for ongoing rejection. Informed consent was obtained from all subjects. The study protocols were approved by the Massachusetts General Hospital and Columbia University Medical Center Institutional Review Boards.
Mixed Lymphocyte Reactions
Preparation of carboxyfluorescein succinimidyl ester (CFSE)-labeled responders: For HC#1 (T1 and T2 stim samples) and CKBMT Subjects 1 and 2, MLRs were set up using purified T-cells as responders. Previously frozen pre-transplant PBMCs were thawed, washed, and resuspended in MACS buffer. MACS beads (Pan T cell Isolation Kit II, Miltenyi Biotec catalog # 130-091-156) were used to generate “untouched” T cells. These T cells were resuspended in PBS at 1×106 cells/mL, labeled with CFSE at a concentration of 0.2uM–0.5uM (CellTrace CFSE Proliferation Kit, Molecular Probes™ (catalog # C34554), washed 3 times, and resuspended in MLR medium (AIM-V supplemented with 5% AB heat-inactivated human serum, 0.01M HEPES, and 50 uM 2-ME at a concentration of 2×106 cells/mL. For HC #2 and #3, CKBMT Subjects 4 and 5, the two conventional transplant recipients, and the anti-third party responses, whole PBMC were used as responders instead of purified T cells. PBMCs were labeled with CFSE as above and resuspended at 2×106 cells/mL. Preparation of violet dye-labeled stimulators: Cryopreserved donor (or healthy control) PBMCs were thawed, washed, resuspended in PBS, and labeled with BD Horizon™ Violet Proliferation Dye 450 (catalog # 562158). After labeling, cells were washed twice, resuspended in MLR medium at 2×106 cells/mL, and irradiated at 30–35 Gray. Plating of cells: One million CFSE-labeled pre-transplant responder cells and one million violet dye-labeled irradiated stimulators were plated in each well of a 24-well plate (total well volume 1mL). For HC#2, HC#3, IS#1, IS#2, and the anti-third party MLRs, we used 96-well plates with each well containing 200,000 responder PBMC and 200,000 stimulators (total well volume 200uL). MLR cultures were incubated at 37C for 5–6 days.
Flow Cytometry
MLR wells were harvested after 6 days of culture. Cells were resuspended in FACS buffer, stained for 30 minutes with fluorochome-conjugated antibodies against CD3 (BD Pharmingen™ clone SP34-2, catalog # 552852), CD4 (BiolegendR clone OKT4, catalog # 317426), and CD8 (BD Pharmingen™ clone DK1, catalog # 557834), washed and filtered before FACS sorting on a BD Influx™ cell sorter to isolate two discrete cell populations (violet−CD3+CD4+CFSElo and violet−CD3+CD8+CFSElo) representing the CD4+ and CD8+ recipient-derived donor-reactive T cells. For unstimulated cell populations, PBMC were thawed and stained with anti-CD3, CD4, and CD8, then FACS sorted into CD3+CD4+ and CD3+CD8+ populations.
DNA isolation and sequencing: Genomic DNA was isolated from sorted cell populations using the Qiagen DNeasy Blood and Tissue Kit. DNA was frozen at −20C and shipped on dry ice to Adaptive Biotechnologies for high-throughput TCRB CDR3 sequencing. The TCR sequencing data were retrieved from Adaptive’s ImmunoSeq software.
In Vitro Immunologic Assays
Standard MLR, CML, and LDA assays were performed using the methods detailed previously (5, 40).
Computational and Statistical Analysis
Mapping of the reads, identification of CDR3 regions and V/J genes, and bias adjustment were performed by Adaptive (41) through their proprietary software. We receive tabulated TRB sequencing data from Adaptive, including CDR3 nucleotide and amino acid sequences, raw copy number (read counts), adjusted copy number and frequency, V/J genes and gene families, inferred insertions and deletions in V-D-J junctions etc.
Repertoire diversity
We measured the diversity of each repertoire by two approaches: (a) entropy (42) (H ≡ ∑pi log2 pi, where pi is the frequency of clone i) and clonality (S ≡ 1 − Hobs/Hmax), where Hmax is the entropy of a repertoire with the same number of clones, each having exactly the same frequency; (b) Simpson’s Index (, where pi is the frequency of clone i). Compared to entropy (and clonality), Simpson's index is more sensitive to changes in frequency of dominant clones.
Comparison of repertoires
We measured the difference between two repertoires using Jensen-Shannon divergence (43) and Pearson correlation, both of which ranges from 0 to 1. We defined expanded clones in MLR by a minimum frequency in stimulated samples (f; f is set at 0.01%) and fold change (C = frequency in stimulated pre-transplant samples/frequency in unstimulated pre-transplant samples; C is conventionally set at 5). We define a clone as detectable if the frequency is larger than a threshold (m; m is usually 0.001% for samples with ≥106 T cells sequenced with 2 million reads). This threshold was set based on power estimation. We model the TCR sequencing procedure by two random processes: the first (P1) is a sample of T cells randomly taken from the entire repertoire; the second (P2) is multiplexed PCR cloning of CDR3 regions from the cells in a sample. Assuming the total number of cells from P1 is N, the total number of sequence reads is R (usually R > 2N), and the “quantum efficiency” in P2 (defined as the average chance of a cell being cloned in PCR) is q, then the expected total number of cells cloned in P2 is Nq, and the number of reads per cell follows a Poisson distribution with . If q is in the order of 40%, then the λ is usually larger than 5, which means about 95% of sampled cells will be represented by at least two reads, a detection threshold in Adaptive’s analytical pipeline. For a clone with a frequency f in the entire repertoire, the number of such cells in P2 follows a Poisson distribution with λ’ = Nfq. Any clone with λ’ > 4 will have a 90% of chance of detection. If N is 1 million and q is 40%, then to achieve 85% detection power requires f greater than 0.001%.
Testing expansion and deletion
To test expansion or deletion of clones, we first identified donor-reactive unique clones (defined as above) in pre-transplant MLR, counted their number (N), and tested if these clones are equally likely to be detectable in unstimulated pre- and post-transplant samples. Specifically, we generated a 2×2 contingency table: [Dpre, N − Dpre; Dpost, N − Dpost], where Dpre is the number of detectable pre-identified donor- (or third party-) reactive clones in unstimulated pre-transplant and Dpost is the number of detectable clones in unstimulated post-transplant samples. Two-sided Fisher’s exact test was performed and p-values and odds ratios reported.
Repertoire turnover analysis
To test whether post-transplant reductions in donor-reactive clones were distinguishable from general repertoire turnover, we set the null hypothesis to be that a donor-reactive clone is equally likely to be present as any other pre-transplant (pre-tx) clones in the post-transplant (post-tx) samples. We defined donor reactive clones as described above, and set the threshold of detectability at 10−6 (there is no need to adjust for detection power, because the comparison is internally controlled within each post-tx sample). For each post-tx sample, we generated a 2×2 contigency table: [Nl, N0; D1, D0], where Nl is the number of detected pre-tx clones, N0 is the number of undetected pre-tx clones, D1 is the number of detected donor-reactive clones and D0 is the number of undetected donor-reactive clones. We performed Fisher’s exact test to assess significance, and report odds ratio as relative change.
Supplementary Material
Acknowledgments
We thank Drs. Donna Farber and Jenny Sims for helpful review of the manuscript and Ms. Susan Allen for assistance with submission. We thank the Immune Tolerance Network for providing patient PBMC samples. Funding: The work was supported in part by NIH grants ROI #AI084074, PO1 grant #P01 AI106697, and Immune Tolerance Network contract N01 AI15416. S.D. was supported by a Kidney Research Student Scholar Grant from the American Society of Nephrology. FACS instrumentation was purchased with NIH grant #S10RR027050.
Footnotes
Supplementary Materials
Fig. S1. Validation study design and analysis details of the tracking of alloreactive clones in healthy controls.
Fig. S2. Anti-donor clonal analysis at increasing fold-expansion criteria.
Fig. S3. Frequencies of donor-reactive clones pre- and post-transplant in CKBMT recipients (CD4).
Fig. S4. Frequencies of donor-reactive clones pre- and post-transplant in CKBMT recipients (CD8).
Fig. S5. Limiting dilution analysis (CTLp and HLT) for Subjects 1, 2, 4, and 5.
Fig. S6. Comparison of pre- and post- transplant anti-donor MLR in Subject 4.
Fig. S7. Frequencies of donor-reactive clones pre- and post-transplant in the “conventional” kidney transplant recipients.
Fig. S8. Detection of third party-reactive TCR pre- and post-transplant.
Table S1. Cell counts, entropy, and clonality for healthy control experiments.
Table S2. Sum frequency of alloreactive clones in healthy controls (%).
Table S3: Cell numbers, number of unique clones, and total number of reads for each patient sample.
Table S4: Tabulated data from clonal analysis in Subject 1, 2, 4, 5, IS#1, and IS#2 (Figs. 3B, 4).
Table S5: Tabulated data from clonal analysis in healthy controls (Fig. 4B).
Table S6: Sum frequency of alloreactive clones in Subject 4 (%).
Table S7: Relative turnover analysis in tolerant subjects (Fig. 5B).
Table S8: Cell numbers, number of unique clones, and total number of reads for anti-third-party responses.
Table S9: Relative turnover analysis at increasing fold-expansion criteria: Subject 1 (Fig. 5C).
Author contributions: H.M. and S.D. designed and performed experiments, analyzed and interpreted data, and wrote the paper; B.S., S.A.L., B.A.S. processed samples, performed experiments, and analyzed and interpreted data; H.R. and J.Z. supervised analysis and edited the manuscript. T.K. designed, supervised, and carried out the clinical protocol and care of the CKBMT patients and participated in sample procurement. W.W. identified donors and participated in sample procurement from “conventional” transplant patients. S.Y. participated in sample procurement for healthy control experiments. Y.S. analyzed and interpreted data and wrote the paper. M.S. participated in the design of the clinical protocol of the CKBMT patients, designed and oversaw the laboratory studies, interpreted the data, and wrote the paper.
Competing interests: H.R. owns stock and receives consulting fees from Adaptive Biotechnologies. All other authors declare no competing financial interests. M.S. and H.R. have filed a patent, “Tracking donor-reactive TCR as a biomarker in transplantation,” based on findings presented herein.
References and Notes
- 1.Fehr T, Sykes M. Tolerance induction in clinical transplantation. Transplant immunology. 2004;13:117–130. doi: 10.1016/j.trim.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Lamb KE, Lodhi S, Meier-Kriesche HU. Long-term renal allograft survival in the United States: a critical reappraisal. American journal of transplantation. 2011;11:450–462. doi: 10.1111/j.1600-6143.2010.03283.x. [DOI] [PubMed] [Google Scholar]
- 3.Brouard S, Le Bars A, Dufay A, Gosselin M, Foucher Y, Guillet M, Cesbron-Gautier A, Thervet E, Legendre C, Dugast E, Pallier A, Guillot-Gueguen C, Lagoutte L, Evanno G, Giral M, Soulillou JP. Identification of a gene expression profile associated with operational tolerance among a selected group of stable kidney transplant patients. Transpl Int. 2011;24:536–547. doi: 10.1111/j.1432-2277.2011.01251.x. [DOI] [PubMed] [Google Scholar]
- 4.Cippa PE, Fehr T. Spontaneous tolerance in kidney transplantation--an instructive, but very rare paradigm. Transpl Int. 2011;24:534–535. doi: 10.1111/j.1432-2277.2011.01260.x. [DOI] [PubMed] [Google Scholar]
- 5.Fudaba Y, Spitzer TR, Shaffer J, Kawai T, Fehr T, Delmonico F, Preffer F, Tolkoff-Rubin N, Dey BR, Saidman SL, Kraus A, Bonnefoix T, McAfee S, Power K, Kattleman K, Colvin RB, Sachs DH, Cosimi AB, Sykes M. Myeloma responses and tolerance following combined kidney and nonmyeloablative marrow transplantation: in vivo and in vitro analyses. American journal of transplantation. 2006;6:2121–2133. doi: 10.1111/j.1600-6143.2006.01434.x. [DOI] [PubMed] [Google Scholar]
- 6.Kawai T, Cosimi AB, Spitzer TR, Tolkoff-Rubin N, Suthanthiran M, Saidman SL, Shaffer J, Preffer FI, Ding R, Sharma V, Fishman JA, Dey B, Ko DS, Hertl M, Goes NB, Wong W, Williams WW, Jr, Colvin RB, Sykes M, Sachs DH. HLA-mismatched renal transplantation without maintenance immunosuppression. N Engl J Med. 2008;358:353–361. doi: 10.1056/NEJMoa071074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawai T, Sachs DH, Sykes M, Cosimi AB N. Immune Tolerance. HLA-mismatched renal transplantation without maintenance immunosuppression. N Engl J Med. 2013;368:1850–1852. doi: 10.1056/NEJMc1213779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawai T, Sachs DH, Sprangers B, Spitzer TR, Saidman SL, Zorn E, Tolkoff-Rubin N, Preffer F, Crisalli K, Gao B, Wong W, Morris H, LoCascio SA, Sayre P, Shonts B, Williams WW, Jr, Smith RN, Colvin RB, Sykes M, Cosimi AB. Long-term results in recipients of combined HLA-mismatched kidney and bone marrow transplantation without maintenance immunosuppression. American journal of transplantation. 2014;14:1599–1611. doi: 10.1111/ajt.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manilay JO, Pearson DA, Sergio JJ, Swenson KG, Sykes M. Intrathymic deletion of alloreactive T cells in mixed bone marrow chimeras prepared with a nonmyeloablative conditioning regimen. Transplantation. 1998;66:96–102. doi: 10.1097/00007890-199807150-00015. [DOI] [PubMed] [Google Scholar]
- 10.Sharabi Y, Sachs DH. Mixed chimerism and permanent specific transplantation tolerance induced by a nonlethal preparative regimen. The Journal of experimental medicine. 1989;169:493–502. doi: 10.1084/jem.169.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomita Y, Khan A, Sykes M. Role of intrathymic clonal deletion and peripheral anergy in transplantation tolerance induced by bone marrow transplantation in mice conditioned with a nonmyeloablative regimen. Journal of immunology (Baltimore, Md.: 1950) 1994;153:1087–1098. [PubMed] [Google Scholar]
- 12.LoCascio SA, Morokata T, Chittenden M, Preffer FI, Dombkowski DM, Andreola G, Crisalli K, Kawai T, Saidman SL, Spitzer TR, Tolkoff-Rubin N, Cosimi AB, Sachs DH, Sykes M. Mixed chimerism, lymphocyte recovery, and evidence for early donor-specific unresponsiveness in patients receiving combined kidney and bone marrow transplantation to induce tolerance. Transplantation. 2010;90:1607–1615. doi: 10.1097/TP.0b013e3181ffbaff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreola G, Chittenden M, Shaffer J, Cosimi AB, Kawai T, Cotter P, Locascio SA, Morokata T, Dey BR, Tolkoff-Rubin NT, Preffer F, Bonnefoix T, Kattleman K, Spitzer TR, Sachs DH, Sykes M. Mechanisms of donor-specific tolerance in recipients of haploidentical combined bone marrow/kidney transplantation. American journal of transplantation. 2011;11:1236–1247. doi: 10.1111/j.1600-6143.2011.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherman LA, Chattopadhyay S. The molecular basis of allorecognition. Annual review of immunology. 1993;11:385–402. doi: 10.1146/annurev.iy.11.040193.002125. [DOI] [PubMed] [Google Scholar]
- 15.Suchin EJ, Langmuir PB, Palmer E, Sayegh MH, Wells AD, Turka LA. Quantifying the frequency of alloreactive T cells in vivo: new answers to an old question. Journal of immunology (Baltimore, Md.: 1950) 2001;166:973–981. doi: 10.4049/jimmunol.166.2.973. [DOI] [PubMed] [Google Scholar]
- 16.Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH, Carlson CS. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robins H, Desmarais C, Matthis J, Livingston R, Andriesen J, Reijonen H, Carlson C, Nepom G, Yee C, Cerosaletti K. Ultra-sensitive detection of rare T cell clones. Journal of immunological methods. 2012;375:14–19. doi: 10.1016/j.jim.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM, Steen MS, LaMadrid-Herrmannsfeldt MA, Williamson DW, Livingston RJ, Wu D, Wood BL, Rieder MJ, Robins H. Using synthetic templates to design an unbiased multiplex PCR assay. Nature communications. 2013;4:2680. doi: 10.1038/ncomms3680. [DOI] [PubMed] [Google Scholar]
- 19.Onoe T, Kalscheuer H, Chittenden M, Zhao G, Yang Y-G, Sykes M. Homeostatic expansion and phenotypic conversion of human T cells depend on peripheral interactions with APCs. Journal of immunology (Baltimore, Md.: 1950) 2010;184:6756–6765. doi: 10.4049/jimmunol.0901711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenblum JM, Kirk AD. Recollective homeostasis and the immune consequences of peritransplant depletional induction therapy. Immunological reviews. 2014;258:167–182. doi: 10.1111/imr.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guillet M, Sebille F, Soulillou J. TCR usage in naive and committed alloreactive cells: implications for the understanding of TCR biases in transplantation. Current opinion in immunology. 2001;13:566–571. doi: 10.1016/s0952-7915(00)00260-0. [DOI] [PubMed] [Google Scholar]
- 22.Smith C, Miles JJ, Khanna R. Advances in direct T-cell alloreactivity: function, avidity, biophysics and structure. American journal of transplantation. 2012;12:15–26. doi: 10.1111/j.1600-6143.2011.03863.x. [DOI] [PubMed] [Google Scholar]
- 23.Michalek J, Collins RH, Hill BJ, Brenchley JM, Douek DC. Identification and monitoring of graft-versus-host specific T-cell clone in stem cell transplantation. Lancet. 2003;361:1183–1185. doi: 10.1016/S0140-6736(03)12917-0. [DOI] [PubMed] [Google Scholar]
- 24.Min B, Yamane H, Hu-Li J, Paul WE. Spontaneous and homeostatic proliferation of CD4 T cells are regulated by different mechanisms. Journal of immunology (Baltimore, Md.: 1950) 2005;174:6039–6044. doi: 10.4049/jimmunol.174.10.6039. [DOI] [PubMed] [Google Scholar]
- 25.Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Bonasio R, Scimone ML, Schaerli P, Grabie N, Lichtman AH, von Andrian UH. Clonal deletion of thymocytes by circulating dendritic cells homing to the thymus. Nat Immunol. 2006;7:1092–1100. doi: 10.1038/ni1385. [DOI] [PubMed] [Google Scholar]
- 27.Streilein JW, Strome P, Wood PJ. Failure of in vitro assays to predict accurately the existence of neonatally induced H-2 tolerance. Transplantation. 1989;48:630–634. [PubMed] [Google Scholar]
- 28.Murase N, Kim DG, Todo S, Cramer DV, Fung J, Starzl TE. FK506 suppression of heart and liver allograft rejection. II: The induction of graft acceptance in rats. Transplantation. 1990;50:739–744. doi: 10.1097/00007890-199011000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cullen PR, Lester S, Rouch J, Morris PJ. Mixed lymphocyte reaction and graft survival in forty cadaveric renal transplants. Clin Exp Immunol. 1977;28:218–222. [PMC free article] [PubMed] [Google Scholar]
- 30.Goulmy E, Persijn G, Blokland E, D'Amaro J, van Rood JJ. Cell-mediated lympholysis studies in renal allograft recipients. Transplantation. 1981;31:210–217. doi: 10.1097/00007890-198103000-00014. [DOI] [PubMed] [Google Scholar]
- 31.Goulmy E, Stijnen T, Groenewoud AF, Persijn GG, Blokland E, Pool J, Paul LC, van Rood JJ. Renal transplant patients monitored by the cell-mediated lympholysis assay. Evaluation of its clinical value. Transplantation. 1989;48:559–563. doi: 10.1097/00007890-198910000-00004. [DOI] [PubMed] [Google Scholar]
- 32.Kahn D, du Toit E, Jacobson JE, Creemers P. Induction of donor-specific tolerance or sensitization as measured by sequential MLC reactivity up to 24 months after renal transplantation. Transpl Int. 1994;7(Suppl 1):S287–S289. doi: 10.1111/j.1432-2277.1994.tb01369.x. [DOI] [PubMed] [Google Scholar]
- 33.Najafian N, Albin MJ, Newell KA. How can we measure immunologic tolerance in humans? J Am Soc Nephrol. 2006;17:2652–2663. doi: 10.1681/ASN.2005070707. [DOI] [PubMed] [Google Scholar]
- 34.Leventhal J, Abecassis M, Miller J, Gallon L, Tollerud D, Elliott MJ, Bozulic LD, Houston C, Sustento-Reodica N, Ildstad ST. Tolerance induction in HLA disparate living donor kidney transplantation by donor stem cell infusion: durable chimerism predicts outcome. Transplantation. 2013;95:169–176. doi: 10.1097/TP.0b013e3182782fc1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ali JM, Bolton EM, Bradley JA, Pettigrew GJ. Allorecognition pathways in transplant rejection and tolerance. Transplantation. 2013;96:681–688. doi: 10.1097/TP.0b013e31829853ce. [DOI] [PubMed] [Google Scholar]
- 36.Baker RJ, Hernandez-Fuentes MP, Brookes PA, Chaudhry AN, Cook HT, Lechler RI. Loss of direct and maintenance of indirect alloresponses in renal allograft recipients: implications for the pathogenesis of chronic allograft nephropathy. Journal of immunology (Baltimore, Md. : 1950) 2001;167:7199–7206. doi: 10.4049/jimmunol.167.12.7199. [DOI] [PubMed] [Google Scholar]
- 37.Gokmen MR, Lombardi G, Lechler RI. The importance of the indirect pathway of allorecognition in clinical transplantation. Current opinion in immunology. 2008;20:568–574. doi: 10.1016/j.coi.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Muraro PA, Robins H, Malhotra S, Howell M, Phippard D, Desmarais C, de Paula Alves Sousa A, Griffith LM, Lim N, Nash RA, Turka LA. T cell repertoire following autologous stem cell transplantation for multiple sclerosis. The Journal of clinical investigation. 2014;124:1168–1172. doi: 10.1172/JCI71691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dziubianau M, Hecht J, Kuchenbecker L, Sattler A, Stervbo U, Rodelsperger C, Nickel P, Neumann AU, Robinson PN, Mundlos S, Volk HD, Thiel A, Reinke P, Babel N. TCR repertoire analysis by next generation sequencing allows complex differential diagnosis of T cell-related pathology. American journal of transplantation. 2013;13:2842–2854. doi: 10.1111/ajt.12431. [DOI] [PubMed] [Google Scholar]
- 40.Kraus AB, Shaffer J, Toh HC, Preffer F, Dombkowski D, Saidman S, Colby C, George R, MCafee S, Sackstein R, Dey B, Spitzer TR, Sykes M. Early host CD8 T-cell recovery and sensitized anti-donor IL-2-producing and cytolytic T-cell responses associated with marrow graft rejection following nonmyeloablative bone marrow transplantation. Exp.Hematol. 2003;31:609–621. doi: 10.1016/s0301-472x(03)00082-1. [DOI] [PubMed] [Google Scholar]
- 41.Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, Riddell SR, Warren EH, Carlson CS. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hill MO. Diversity and Evenness: A Unifying Notation and Its Consequences. Ecology. 1973;54:427–432. [Google Scholar]
- 43.Manning CD, Schütze H. Foundations of statistical natural language processing. Cambridge, Mass: MIT Press; 1999. p. xxxvii.p. 680. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.