Abstract
The Mycobacterium tuberculosis (MTB) membrane is rich in antigens that are potential targets for diagnostics and the development of new vaccines. To better understand the mechanisms underlying MTB virulence and identify new targets for therapeutic intervention we investigated the differential composition of membrane proteomes between virulent M. tuberculosis H37Rv (MTB) and the Mycobacterium bovis BCG vaccine strain. To compare the membrane proteomes, we used LC-MS/MS analysis in combination with label-free quantitative (LFQ) proteomics, utilizing the area-under-curve (AUC) of the extracted ion chromatograms (XIC) of peptides obtained from m/z and retention time alignment of MS1 features. With this approach, we obtained relative abundance ratios for 2,203 identified membrane-associated proteins in high confidence. Of these proteins, 294 showed statistically significant differences of at least 2 fold, in relative abundance between MTB and BCG membrane fractions. Our comparative analysis detected several proteins associated with known genomic regions of difference between MTB and BCG as being absent, which validated the accuracy of our approach. In further support of our label-free quantitative data, we verified select protein differences by immunoblotting. To our knowledge we have generated the first comprehensive and high coverage profile of comparative membrane proteome changes between virulent MTB and its attenuated relative BCG, which helps elucidate the proteomic basis of the intrinsic virulence of the MTB pathogen.
Keywords: Mycobacterium tuberculosis (MTB), Mycobacterium bovis (BCG), Label-free quantitation (LFQ), One dimensional Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (1D SDS-PAGE), Liquid chromatography mass spectrometry-mass spectrometry (LC-MS/MS), membrane proteomes, Regions of difference (RD)
Introduction
Tuberculosis remains a major world health problem with Mycobacterium tuberculosis, the bacterium responsible for this disease, claiming 1.4 million lives annually1. Exacerbating this problem is the rise in multidrug-resistant (MDR), extensively drug-resistant (XDR) and most recently, totally drug-resistant (TDR) strains of M. tuberculosis, which threaten to undermine current tuberculosis treatments2. The currently available tuberculosis vaccine, known as BCG, is an attenuated strain of Mycobacterium bovis. All M. bovis strains, including BCG, are very similar to M. tuberculosis exhibiting 99.9% identity at the DNA level3. Unfortunately, the ability of BCG vaccination to protect adults from pulmonary tuberculosis is highly variable4. Thus, there is a major need to develop new drugs and vaccines to control tuberculosis and a better understanding of M. tuberculosis biology will help achieve this goal.
Many aspects of M. tuberculosis physiology, pathogenesis, and immunity remain to be understood. Comparisons of virulent M. tuberculosis to attenuated M. bovis BCG can inform on these unknowns. Genomic comparisons reveal several regions of difference (named RDs) that are deleted in BCG but present in M. tuberculosis5. Proteomic comparisons of M. tuberculosis and BCG are another approach for identifying differences of potential importance. By reporting on protein abundance, proteomic methods have the advantage of taking into account both transcriptional and post-transcriptional effects. Further, when combined with subcellular fractionation, proteomics can report on protein localization. M. tuberculosis and BCG proteomes were initially compared using 2-dimensional gel electrophoresis (2D-GE) followed by mass spectrometry-based identification of select proteins6–8. At best, this approach led to the identification of almost 300 proteins6, 9. Since then, quantitative “shotgun” proteomics has become the choice for large scale proteome comparisons, which enables more comprehensive assessment of complex protein samples as a result of higher throughput and sensitivity associated with this method10, 11.
Proteins localized to the membrane of M. tuberculosis play critical roles in vital cell processes including nutrient transport, cell wall synthesis, energy metabolism, and signal transduction12–14. Additionally, mycobacterial membrane proteins can elicit immune responses, making the membrane proteomes of M. tuberculosis and BCG of significant interest for vaccination and diagnostic studies15. Initial efforts to identify the M. tuberculosis and BCG membrane proteome used 2D-GE; however, the high insolubility of membrane proteins poses a significant technical challenge for 2D-GE and limits the numbers of proteins that can be identified16, 17. Significantly better protein identification coverage was subsequently obtained when membrane proteins were solubilized and pre-separated by 1D SDS-PAGE followed by LC-MS/MS analysis of trypsin digested gel slices comprising the entire sample18. Using this approach in independent studies, 349 and 739 proteins out of the possible 4,015 proteins encoded by the genome were identified in M. tuberculosis membrane fractions prepared by differential centrifugation18, 19. With BCG a similar effort involving Triton X-114 fractions, which enriches for lipophilic proteins including hydrophobic membrane proteins, identified 351 proteins and 1,766 M. tuberculosis proteins were identified in Triton X-114 fractions20. Triton X-114 can be considered an alternative to differential centrifugation for enriching membrane and membrane-associated proteins20. While the number of proteins identified in mycobacterial membrane proteins has increased considerably18, 19, there has yet to be an in-depth quantitative comparison of M. tuberculosis and BCG membrane protein composition.
In the study reported here, we combined SDS-solubilization and 1D SDS-PAGE separation of membrane proteins with LC-MS/MS and label-free quantitative proteomics, to comprehensively identify and compare the membrane fraction proteomes of the virulent M. tuberculosis H37Rv strain (MTB) and M. bovis BCG. With this approach, we identified a total of 2,203 proteins associated with the mycobacterial cytoplasmic membrane. Further, label-free quantification (LFQ) revealed 294 proteins that differed significantly in relative abundance, by at least 2-fold, between MTB and BCG. Several proteins encoded by deleted RD regions of BCG were identified as missing in BCG, which validated our approach. The quantitative differences in membrane proteins identified in this work have potential to help explain the deficiencies in the BCG vaccine and to inform on virulence and immunogenic features of M. tuberculosis. The data reported here should aid efforts to develop new tuberculosis control and preventative measures.
Experimental Procedures
M. tuberculosis and M. bovis BCG growth conditions
Mycobacterium tuberculosis H37Rv (MTB) and Mycobacterium bovis BCG Pasteur were grown at 37° C in liquid Middlebrook 7H9 medium (Difco) supplemented with 0.05% Tween 80, 0.5% glycerol, and 1X ADS [>0.5% bovine serum albumin, 0.2% glucose, 0.85% NaCl]. Two independent 100 mL cultures of each strain were grown to an OD600 of 1, when cells were then harvested by centrifugation and washed with phosphate-buffered saline (PBS). For removal from BSL-3 containment, the washed pellets were sterilized by gamma-irradiation in a JL Shephard Mark I 137Cs irradiator (Dept. of Radiobiology, University of North Carolina at Chapel Hill).
Cytoplasmic membrane fraction preparation
Cytoplasmic membrane fractions were isolated by centrifugation as previously described18. Briefly, cells suspended in PBS containing a cocktail of protease inhibitors were lysed by passage through a french press cell. Unlysed cells were removed by centrifugation at 3,000 × g and the clarified whole cells lysates were spun at 27,000 × g for 30 minutes to remove the cell wall fraction. The supernatant was then spun at 100,000 × g for 2 hours to isolate the cytoplasmic membrane fraction. Membrane fractions were washed once, suspended in PBS, and proteins concentrated using an Amicon Ultra centrifugation filter with a 10 kD pore size (Millipore). Protein concentrations were determined by BCA assay (Pierce) using a BSA standard and equalized among both strains and all replicates.
Separation and in-gel digestion of membrane proteins
For label-free quantitative proteomics experiments, two biological replicates of MTB and BCG membrane proteins were separated on a 12% reducing SDS-PAGE gel. For each sample, 8.8 mg of protein was loaded in an individual lane for separation. The protein bands were visualized by Coomassie Blue R-250 staining (Bio-Rad, Hercules, CA) and protein bands for each biological sample were cut into 10 equivalent gel slices, approximately 2 mm thick. Trypsin digestion of the excised gel slices was performed as previously described21 with modifications. Briefly, each gel slice was cut into smaller pieces and placed in a single well of a 96-well U-bottom polypropylene plate. After de-staining with acetonitrile (ACN), slices were incubated in 25 mM ammonium bicarbonate (ABC) containing 20 µg/mL of sequencing-grade trypsin overnight at 37° C. The digest supernatants were transferred to a new 96-well plate. Tryptic peptides were extracted from the gel slices by two washes with 50% ACN. The ACN washes were added to the ABC wash in the fresh plate. Digested peptides were store at −80° C until lyophilization.
Liquid Chromatography-Tandem Mass Spectrometry and Protein Identification
In gel digested peptides were desalted using PepClean C18 spin columns (Pierce, Rockford, IL), used according to the manufacturer’s instructions, and re-suspended in an aqueous solution of 0.1% formic acid. Identification of proteins was done using reversed-phase LC-MS/MS on a 2D-nanoLC Ultra system (Eksigent Inc, Dublin, CA) coupled to an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific, San Jose, CA). The Eksigent system was configured to trap and elute peptides via a sandwiched injection of ~ 250 fmol of sample. The trapping was performed on a 3 cm-long 100 µm i.d. C18 column while elution was performed on a 15 cm-long 75 µm i.d., 5 µm, 300Å particle ProteoPep II integraFrit C18 column (New Objective Inc, Woburn, MA). Analytical separation of the tryptic peptides was achieved with a 70-min linear gradient of 2–30% buffer B at a 200 nL/min, where buffer A is an aqueous solution of 0.1% formic acid and buffer B is a solution of 0.1% formic acid in acetonitrile.
Mass spectrometric data acquisition was performed in a data-dependent manner on a hybrid LTQ-Orbitrap velos mass spectrometer. A full scan mass analysis on an Orbitrap (externally calibrated to a mass accuracy of < 1 p.p.m and a resolution of 60,000 at m/z 400) was followed by intensity-dependent MS/MS of the 10 most abundant peptide ions. High energy collision dissociation (HCD) was used to dissociate peptides with normalized collision energy of 35 eV. The MS/MS acquisition of each precursor m/z was repeated for 30 s and subsequently excluded for 60 s. Monoisotopic precursor ion selection (MIPS) and charge state screening were enabled for triggering data-dependent MS/MS scans. All 10 gel slices of each of the two biological replicates of MTB and BCG strains were subjected to three independent LC-MS runs (three technical replicates for each of the two biological replicates), resulting in the production of 120 LC-MS runs. Mass spectra were processed, and peptide identification was performed using Andromeda search engine found in MaxQuant software ver. 2.2.1. (Max Planck Institute, Germany) All protein database searches were performed against the M. tuberculosis H37Rv protein sequence database (RefSeq NC_000962 uid57777) downloaded from National Center for Biotechnology Information (NCBI)22. This database contained 4,018 annotated proteins and the sequences were derived from the H37Rv ASM19595v2 assembly. Peptides were identified with a target-decoy approach using a combined database consisting of reverse protein sequences of the H37Rv and common repository of adventitious proteins (cRAP). The cRAP database was obtained from the Global Proteome Machine (GPM) ftp site (ftp://ftp.thegpm.org/fasta/cRAP). Peptide identification was made with a false discovery rate (FDR) of 1% (10) while peptides were assigned to proteins with a protein FDR of 5%. A precursor ion mass tolerance of 20 ppm was used for the first search that allowed for m/z retention time recalibration of precursor ions that were then subjected to a main search using a precursor ion mass tolerance of 5 ppm and a product ion mass tolerance 0.5 Da. Search parameters included up to two missed cleavages at KR on the sequence (11), and oxidation of methionine as a dynamic modification. All MTB and BCG protein identifications are reported with a posterior error probability (PEP) ≤ 0.1 and filtering of reverse and contaminant proteins.
Protein Quantitation
Label-free quantitation was based on peak area23, 24. The measured area under the curve (AUC) of m/z and retention time aligned extracted ion chromatogram (XIC) of a peptide was performed via the label-free quantitation module found in MaxQuant [ver. 1.2.2.5]. All six replicates (biological and technical replicates) of each strain, with 10 LC-MS runs each, were included in the LFQ experimental design with protein-level quantitation and normalization performed using unique and razor peptide features corresponding to identifications filtered with a peptide FDR of 0.01, and protein FDR of 0.05. The MaxQuant protein groups and evidence files were processed, stored in an MS-Access database and statistical analysis and visualization was performed using Perseus [ver. 1.2.0.17] (Max Planck Institute, Germany) and JMP genomics (SAS, Cary NC). Relative quantification changes between MTB and BCG in replicate data was performed using t-test statistics with a p-value < 0.05. The quantitative data was reported with t-test differences between MTB and BCG and corresponding p-values and estimation of false discovery rates (FDR) using q-values25 for both raw and normalized LFQ intensities and LFQ intensities that were imputed in replicated runs using a Gaussian distribution with a width of 0.3 and downshift of 1.8 as the imputation parameters26.
Immunoblotting of membrane proteins
For both replicates of MTB or BCG, 30 µg of clarified whole cell lysate (WCL), was separated on a 12% SDS-PAGE gel. Membrane fractions (MEM) obtained from 30 µg of WCL were also loaded in independent lanes to visualize subcellular localization of the protein. For the Icl1 immunoblot, 60 µg of membrane fraction was loaded, and approximately 0.5 µg of recombinant Icl1 protein27 (rIcl) was included as a control to identify the appropriate protein species. Proteins were transferred to a nitrocellulose membrane (Whatman) then detected using either anti-Icl1 (provided by David Russell) at 1:5,00027, anti-GlcB (provided by Suman Laal) at 1:2,50028, anti-SigA (provided by Murty Madiraju) at 1:20,00029, or anti-19kDa (provided by Douglas Young) at 1:20,000. Anti-mouse and anti-rabbit secondary antibodies conjugated to alkaline phosphatase (BioRad) were used as appropriate at a concentration of 1:20,000. Signal was detected using ECF reagent (GE Healthcare).
Results
Our understanding of why the BCG vaccine is attenuated or fails to elicit optimal protection against M. tuberculosis infection remains incomplete. Using quantitative “shotgun” proteomics, we set out to identify the composition of the membrane proteomes of attenuated BCG and virulent MTB and compare the relative abundance of the individual proteins in the two strains.
Identification of the MTB and BCG membrane proteome
To generate comprehensive and accurate lists of the M. tuberculosis and BCG membrane proteomes, we isolated membrane fractions from two biological replicates of M. tuberculosis H37Rv (MTB) and M. bovis BCG cultures by differential ultra-centrifugation of cell lysates. Membrane proteins were subjected to SDS-solubilization, separated by 1D SDS-PAGE, and then analyzed by LC-MS/MS.
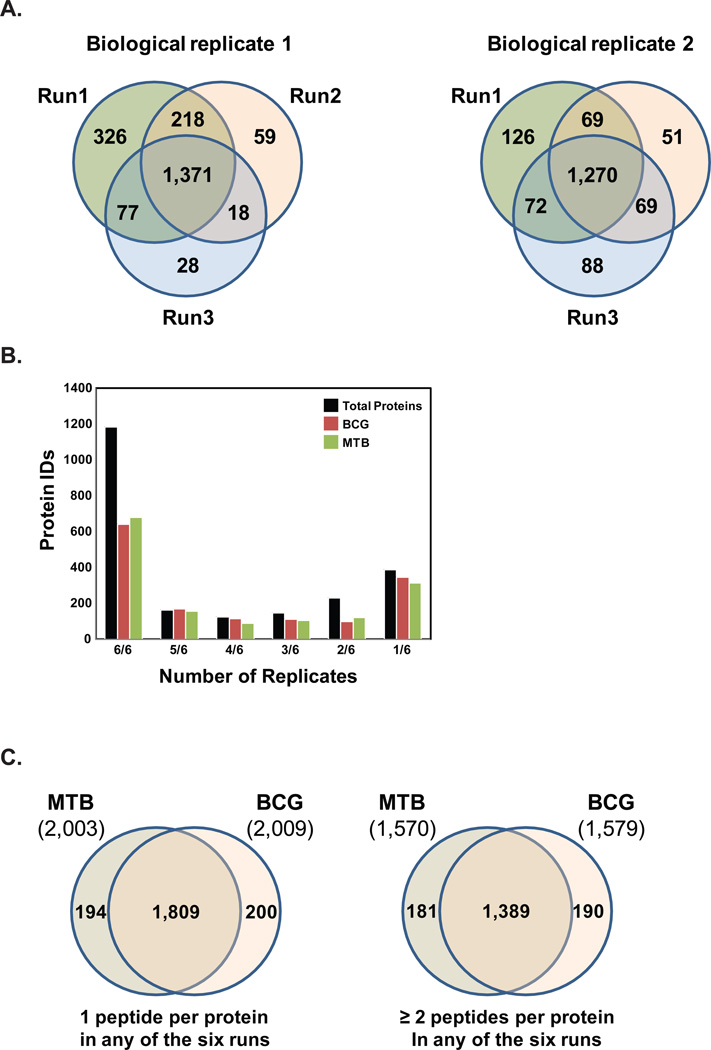
Proteins were identified in MTB and BCG membrane fractions from six replicates (two biological replicates of each species, each with three technical replicates) using MaxQuant and label-free quantification design, where each biological group was subjected to a protein sequence database search against a concatenated database consisting of H37Rv protein entries. A total of 2,203 proteins were identified in high confidence using the target-decoy approach with a peptide FDR cut-off of 0.01 (Supplemental Table 1). In addition, a protein FDR cut-off of 0.05 was employed. Figure 1A shows the reproducibility of the proteins identified between replicates. Analysis of the first set of MTB and BCG biological samples revealed 1,371 proteins that were identified in all three technical replicates, while comparison of the second set of biological samples revealed 1,270 common identifications across all technical replicates. Of the 2,203 total proteins, 1,760 were identified with a minimum of 2 peptides per protein in any one of the six runs (See Supplemental Table 1). In MTB there were 2,003 proteins identified and in BCG 2,009 proteins, with the majority of proteins identified in a strain being identified in all six replicates (Figure 1B). Figure 1C shows that a majority of the proteins (1,809 out of 2,203) were identified in both MTB and BCG while 194 proteins were only identified in MTB and 200 proteins were unique to BCG. When the same comparison is run with the proteins identified with a minimum of 2 peptides in any one of the six runs, 1,389 out of 1,760 proteins were identified in both MTB and BCG, while 181 proteins were only identified in MTB and 190 proteins only in BCG.
Figure 1. Total proteins identified in the M. tuberculosis (MTB) and M. bovis BCG membrane fractions.
(A) Mass spectrometry analysis of membrane fractions derived from either M. tuberculosis (MTB) or M. bovis BCG (BCG) resulted in the high-level of reproducibility in three technical runs for each of the two biological replicates with biological replicate 1 showing 1,371 identifications in all three technical replicates while biological replicate 2 shows 1,270 identifications in all three technical replicates. (B) Proteins identified in all six runs were categorized on the basis of the number of replicates they were identified, for all proteins in MTB, BCG, or both strains. (C) Protein identifications in MTB and BCG using 1 peptide per protein criteria in any of the six runs and ≥ 2 peptides per protein criteria
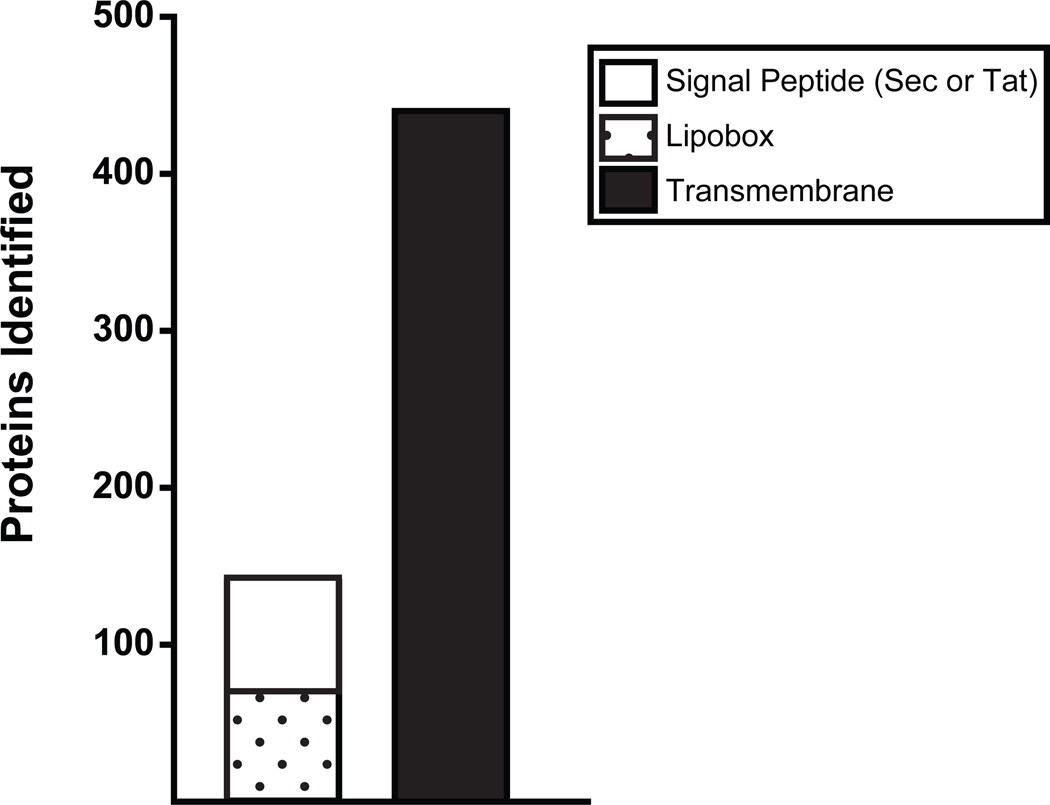
Of the 2,203 total proteins identified in the membrane fractions of MTB and/or BCG, 26% (580) are predicted to have a transmembrane domain for integration into the cytoplasmic membrane or a cleavable signal peptide for export across the cytoplasmic membrane (Figure 2). 440 proteins have putative transmembrane domains predicted by the TMHMM algorithm30. 140 proteins are predicted by SignalP31 or TatP32 to contain either Sec or Tat export signal peptides, which would target them for export across the cytoplasmic membrane by one of the corresponding conserved protein export systems33. Of the proteins with signal peptides, 54% contain a lipobox in the signal peptide that predicts the proteins become lipidated during the process of export34. We identified 75 out of the almost 100 predicted lipoproteins of M. tuberculosis H37Rv35. The relatively high number of potential lipoproteins detected in the membrane fractions is consistent with the established role of lipid modification in anchoring proteins to the membrane36.
Figure 2. Membrane-associated proteins with predicted export signal peptides or transmembrane domains.
The primary amino acid sequence of the 2,203 total proteins identified in the membrane fractions of MTB and BCG were analyzed for the presence of aminoterminal export signal peptides or transmembrane domains. A total of 440 proteins contain probable transmembrane domains predicted by the TMHMM 2.030 (omitting TMHMM transmembrane predictions likely representing N-terminal signal peptides). Additionally, 140 proteins are predicted by SignalP31 or TatP32 to contain either Sec or Tat export signal peptides, which would target them for export out of the cytoplasm by conserved bacterial secretion machinery. Of the 140 proteins with putative export signals, 54% are possible lipoproteins with lipobox motifs in their signal peptides predicted by LipoP34 or by using a lipoprotein pattern search against the M. tuberculosis H37Rv genome35.
Interestingly, however, the majority of proteins identified in the membrane fraction lacked transmembrane domains and/or signal peptides. Previous mass spectrometry studies have similarly identified a population of proteins lacking export signals in mycobacterial cytoplasmic membrane fractions prepared by the differential centrifugation method we used here, as well as with Triton X-114 fractionation19, 37. Some of these proteins lacking export signals and/or transmembrane domains may be cytoplasmic proteins that are peripherally associated with phospholipids in the membrane or integral membrane proteins. Alternatively, they may represent mycobacterial membrane proteins that lack predicted signals for localization. A final possibility is that these proteins lacking predicted export signals are cytoplasmic contaminants that are detected by the high sensitivity of mass spectrometry.
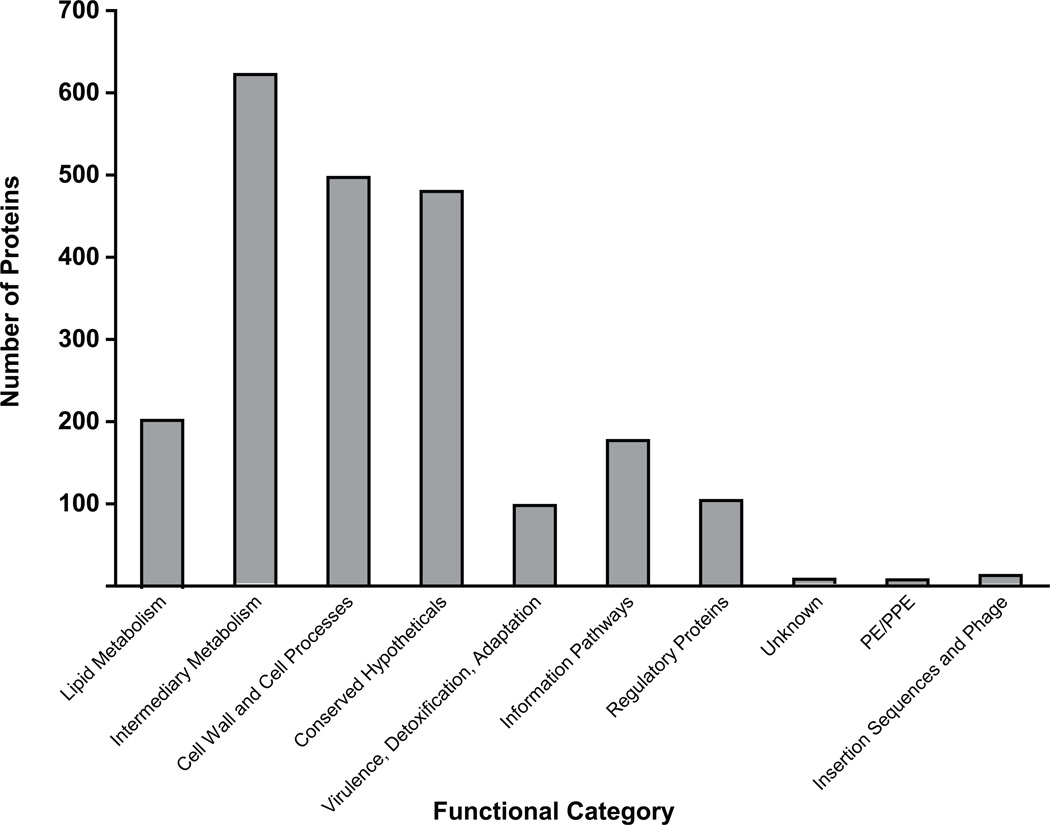
These 2,203 proteins that we identified can be broadly divided into several functional categories (Figure 3)38. In addition to a large list of conserved hypothetical proteins of unknown function, the majority of the proteins we identified in the membrane have predicted roles in cell wall processes, lipid metabolism, and intermediary metabolism. This pattern of functional category enrichment is similar to a previous proteomic analysis of the M. tuberculosis membrane19, 20, 39.
Figure 3. Functional categories of proteins identified in MTB and/or BCG.
The 2,203 proteins identified in both biological replicates of either M. tuberculosis H37Rv or M. bovis BCG represented several functional categories.
Label-free quantitation and comparison of M. tuberculosis and BCG membrane proteomes
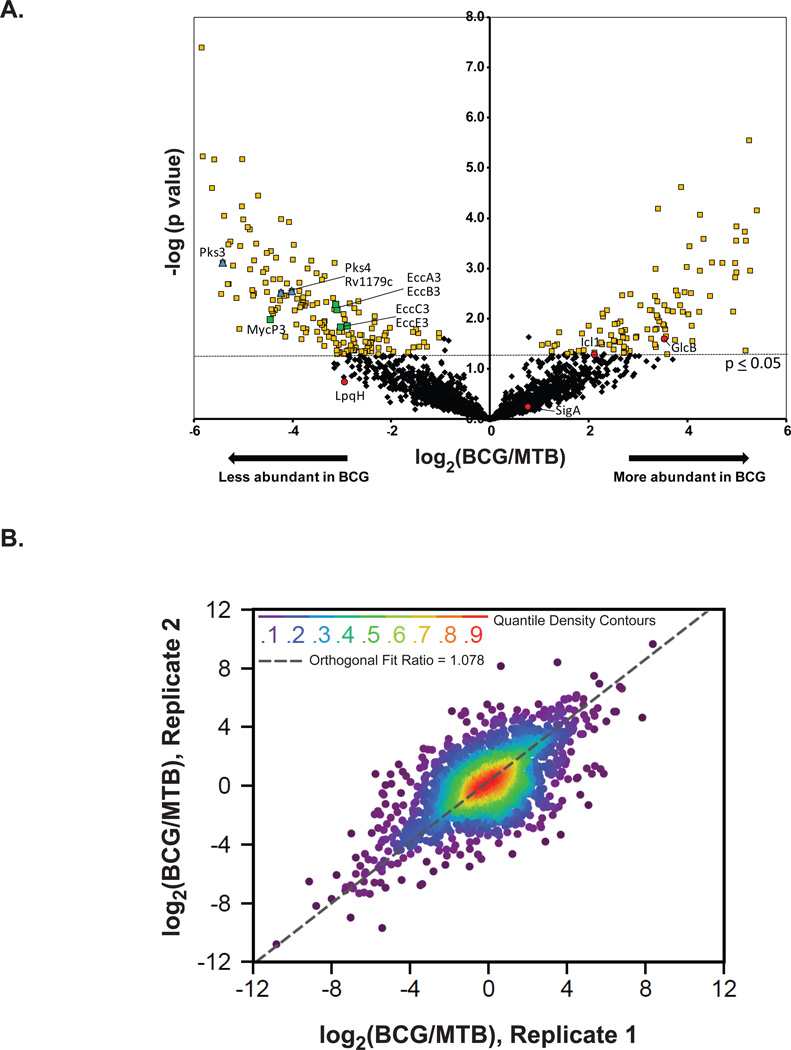
Label-free quantification (LFQ) was performed across all biological samples and replicates via alignment of retention-time and m/z features that were detected at the MS1 level. Using the MaxQuant software, the area-under-the curve (AUC) of each extracted peptide ion chromatogram was used to compute the LFQ intensities at the peptide and protein levels for each biological replicate. This analysis led to relative abundance ratios for 1,788 proteins. We also analyzed the data with imputation of missing values in order to generate abundance ratios and statistics for all 2,203 proteins identified. Imputation provides a means of statistical correction for missing values by calculating what could have been derived experimentally26. We defined significant differences (p ≤ 0.05) in protein levels in MTB versus BCG using a t-test. Using this criterion, the majority of proteins showed no significant difference in relative abundance between MTB and BCG (Figure 4A). Supplement Table 4 lists the 294 proteins with significant differences and a log2 BCG/MTB ratio of ±1 or greater (representing differences > 2-fold) before and after missing value imputation (191 of these proteins were significant without imputation). These 294 proteins include 188 proteins (139 without imputation) that were lower in BCG and 106 after missing value imputation (85 without imputation) that were present at higher levels in BCG compared to MTB. Figure 4B shows a prototypical correlation analysis between the quantitative ratios between BCG and MTB. The good orthogonal fit between the data reveals good reproducibility between replicate runs with a Pearson’s correlation coefficient of 0.8
Figure 4. Label-free quantification of proteins from M. tuberculosis (MTB) and M. bovis BCG.
(A) To identify proteins with differences in abundance between the membranes of MTB and BCG, a label-free quantification analysis based on peak area was performed. The measured area under the curve of m/z and retention time aligned extracted ion chromatogram of each peptide was performed via the label-free quantitation module found in MaxQuant [ver. 1.2.2.5]. We obtained relative abundance ratios for 2,203 proteins (black diamonds), shown here plotted by BCG/MTB ratio (log2 scale) and pvalue (−log10 scale). Of these shared proteins, 294 were statistically different (p≤0.05, t-test) in abundance between strains by 2-fold or more yellow squares). Proteins with LFQ ratios confirmed by immunoblot are shown as red circles (SigA-levels unchanged) and (LpqH-lower level in BCG, albeit not significant). Also labeled are proteins under-represented in the BCG that correspond to PAT synthesis (blue diamonds) and the ESX-3 secretion system (green squares). (B) The correlation between log2 (BGG/MTB) ratios obtained from two biological replicates of MTB and BCG. Nonparemetric quantile densities are shown between the two replicates with an orthogonal fit
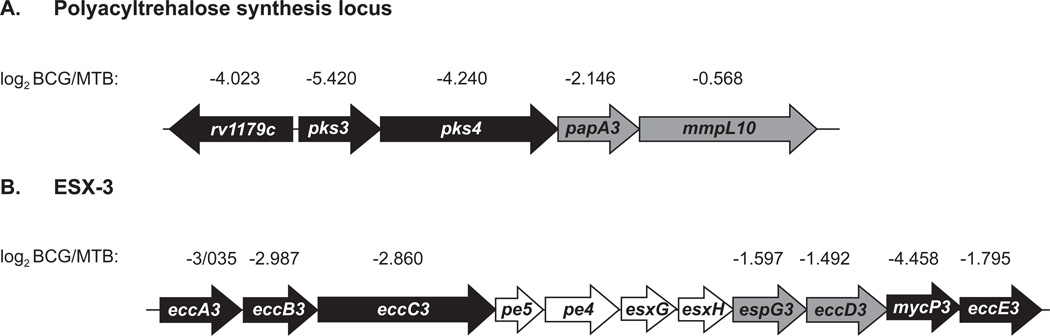
Given the genomic identity between M. tuberculosis and BCG of >99.9%, 294 proteins (after missing value imputation) with statistically significant differences of at least 2-fold may seem high. However, it is not too much of a surprise when considering the deleted genomic RD regions of BCG5, 40 (discussed below) and >2000 SNPs between BCG and M. tuberculosis3. In addition, a prior study comparing differential gene expression at the level of transcripts between M. tuberculosis and M. bovis (the virulent parent of BCG) reported 6% of the total genome showing differential expression5, 40, 41. Further, the potential for a single protein alteration/disruption to affect numerous proteins in a pathway is a real possibility. The proteins on our stringent lists of differences include interesting examples of proteins with roles in lipid and intermediary metabolism, cell wall processes, and transport systems (Tables 1). There are additionally many conserved hypothetical proteins that differ between the strains. Review of the proteins on these lists also revealed cases in which the relative abundance of multiple proteins encoded in the same genomic region was altered similarly. In these cases, where the corresponding genes are located in operons or under the control of the same promoter it is likely the proteomic differences reflect transcriptional differences between strains. Among such examples, is an interesting group of three proteins encoded by neighboring genes that are part of a polyacyltrehalose (PAT) biosynthesis locus (rv1180–rv1183) and a genomic locus encoding components of the ESX-3 specialized secretion system (rv0282–rv0292) (Figure 5).
Table 1.
Proteins identified that correspond to RD regions (regions of difference), which are genes absent in the M. bovis BCG genome but present in the M. tuberculosis H37Rv.
| Protein ID | Locus | Gene Name |
Description/Function | Regions of deletion (RD) | Filter1: ≥ 2 peptides in 4 of 6 runs for BCG |
Filter1: ≥ 2 peptides in 4 of 6 runs for Rv |
Total number of BCG peptides in all runs |
Total number of Rv peptides in all runs |
|---|---|---|---|---|---|---|---|---|
| NP_215143.1 | Rv3871 | hypothetical protein | 1RD | 0 | 6 | 0 | 66 | |
| NP_218391.1 | Rv3874 | esxB | protein EsxB | 1RD | 0 | 0 | 0 | 2 |
| NP_216214.1 | Rv3876 | hypothetical protein | 1RD | 0 | 6 | 0 | 14 | |
| NP_216388.1 | Rv3877 | transmembrane protein | 1RD | 0 | 6 | 0 | 94 | |
| NP_214806.1 | Rv1978 | hypothetical protein | 2RD/7RD | 0 | 6 | 0 | 82 | |
| NP_217821.1 | Rv1987 | chitinase | 2RD | 0 | 0 | 0 | 5 | |
| NP_216094.1 | Rv1578c | phiRv1 phage protein | 3RD | 0 | 0 | 0 | 8 | |
| NP_216099.1 | Rv1583c | phiRv1 phage protein | 3RD | 0 | 4 | 0 | 10 | |
| NP_215204.1 | Rv1586c | phiRv1 integrase | 3RD | 0 | 3 | 0 | 1 | |
| NP_216806.1 | Rv1508c | hypothetical protein | 4RD | 0 | 6 | 0 | 20 | |
| NP_216329.1 | Rv1509 | hypothetical protein | 4RD | 0 | 6 | 0 | 26 | |
| YP_177770.1 | Rv1513 | hypothetical protein | 4RD | 0 | 6 | 0 | 23 | |
| NP_215777.1 | Rv1516c | sugar transferase | 4RD | 0 | 6 | 0 | 18 | |
| NP_217894.1 | Rv1514c | hypothetical protein | 4RD | 0 | 4 | 0 | 26 | |
| NP_216031.1 | Rv1515c | hypothetical protein | 4RD | 0 | 2 | 0 | 1 | |
| NP_216028.1 | Rv1512 | epiA | nucleotide-sugar epimerase epiA | 4RD | 0 | 0 | 0 | 4 |
| NP_216863.1 | Rv2347c | esxP | ESAT-6 like protein | 5RD | 0 | 0 | 0 | 1 |
| NP_216865.1 | Rv2349c | plcC | phospholipase C 3 | 5RD | 0 | 0 | 0 | 2 |
| NP_216866.1 | Rv2350c | plcB | membrane-associated phospholipase C | 5RD | 0 | 1 | 0 | 1 |
| NP_216867.1 | Rv2351c | plcA | membrane-associated phospholipase C | 5RD | 0 | 0 | 0 | 1 |
| NP_216588.1 | Rv2072c | cobL | precorrin-6y methyltransferase CobL | 9RD | 0 | 0 | 0 | 3 |
| NP_218371.1 | Rv2073c | short-chain dehydrogenase | 9RD | 0 | 5 | 0 | 23 | |
| NP_216350.1 | Rv2074 | hypothetical protein | 9RD | 0 | 6 | 0 | 31 | |
| NP_216388.1 | Rv1977 | hypothetical protein | 7RD | 0 | 6 | 0 | 37 | |
| NP_218134.1 | Rv3617 | ephA | epoxide hydrolase | 8RD | 0 | 0 | 0 | 1 |
| NP_214798.1 | Rv3623 | lpqG | lipoprotein LpqG | 8RD | 0 | 5 | 0 | 28 |
| NP_214735.1 | Rv0221 | hypothetical protein | 10RD | 0 | 6 | 0 | 64 | |
| NP_214736.1 | Rv0222 | echA1 | enoyl-CoA hydratase | 10RD | 0 | 0 | 0 | 1 |
| NP_217174.1 | Rv2658c | prophage protein | 11RD | 0 | 2 | 0 | 6 | |
| NP_217175.1 | Rv2659c | phiRv2 prophage integrase | 11RD | 0 | 1 | 0 | 4 | |
| NP_216645.1 | Rv3120 | hypothetical protein | 12RD | 0 | 5 | 0 | 12 | |
| NP_215771.1 | Rv1255c | transcriptional regulatory protein | 13RD | 0 | 0 | 0 | 1 | |
| NP_215772.1 | Rv1256c | cyp130 | cytochrome P450 130 CYP130 | 13RD | 0 | 0 | 0 | 2 |
| YP_177976.1 | Rv1769 | hypothetical protein | 14RD | 0 | 6 | 0 | 31 | |
| NP_215612.1 | Rv1770 | hypothetical protein | 14RD | 0 | 6 | 0 | 129 | |
| NP_218405.1 | Rv1771 | oxidoreductase | 14RD | 0 | 6 | 0 | 68 |
Figure 5. Genomic loci encoding proteins less abundant in the M. bovis BCG membrane compared to M. tuberculosis H37Rv (MTB) membrane.
Among the differences observed between the MTB and BCG membrane proteomes were proteins involved in polyacyltrehalose (PAT) biosynthesis (A) and specialized protein secretion via the ESX-3 system (B). Genes encoding proteins that were determined by LFQ analysis to be under-represented in BCG are shaded in black or grey and the corresponding log2(BCG/MTB) ratio is provided (imputed values). Those genes shaded black encode proteins whose LFQ ratios had p values of 0.05 or less.
Validation of differences detected between MTB and BCG membrane proteomes
As one way to validate the accuracy of our proteomic approach to identify proteins that differ between MTB and BCG, we searched our datasets for proteins encoded by the RD deletion regions of BCG. From a total of 132 genes that have been mapped to the 14 RD deletion regions of BCG Pasteur40, we identified peptides mapping to 36 RD proteins in MTB, 24 of which were identified with ≥2 peptides per protein. As expected, all RD-associated proteins identified in MTB were absent in BCG, reflecting deletion of the corresponding genes in the BCG strain (Table 1). When quantified with LFQ, the RD proteins had extremely low values with imputation as expected (Supplement Table 4).
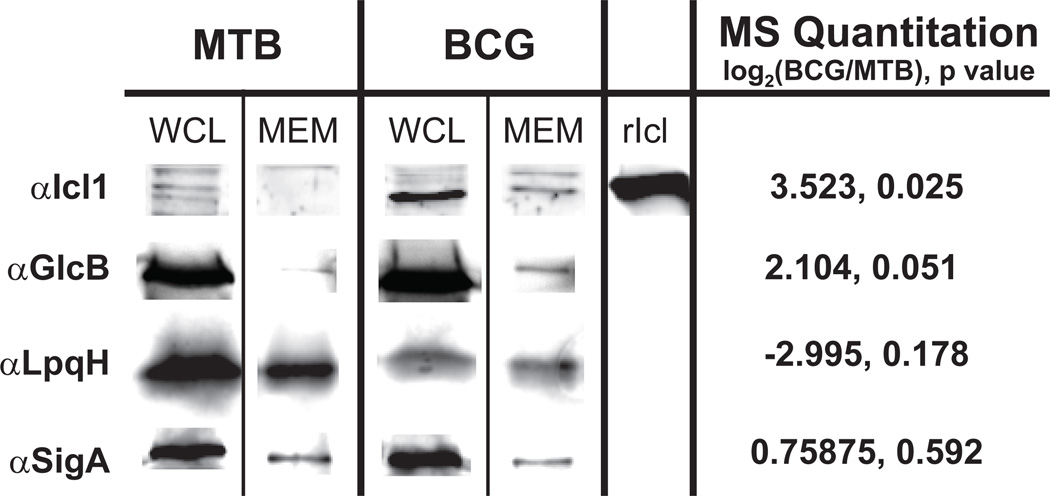
We also experimentally validated differences in protein abundance by immunoblotting. However, this analysis was limited to proteins for which we could obtain antibodies. We evaluated a set of representative proteins that were either increased in BCG, decreased in BCG, or equal between the BCG and MTB membrane proteomes. We analyzed whole cell lysates (WCL) and the corresponding membrane (MEM) fractions prepared from MTB and BCG, to look for protein level differences comparable to our mass spectrometry data (Figure 6).
Figure 6. Immunoblot confirmation of relative protein abundances between the M. tuberculosis H37Rv and M. bovis BCG membrane fractions.
The same membrane fractions (MEM) used for MS analysis were also analyzed by immunoblot using antibodies against four representative proteins. Also included are the whole cell lysates (WCL) from which the corresponding membrane fractions were derived. The isocitrate lysase Icl1 and malate synthase GlcB represent proteins that were significantly reduced in the BCG membrane as determined by our MS analysis. The sigma factor SigA produced a label-free quantitation log2(BCG/MTB) ratio of 0.759 with a p value of 0.555, indicating no difference in abundance between the strains. The LpqH lipoprotein was determined to be lower in abundance in the BCG membrane than MTB (−2.995) albeit with a p value of 0.178.
The M. tuberculosis housekeeping sigma factor SigA (rv2703) is primarily a cytoplasmic protein, but small amounts were detected in membrane fractions. This protein was relatively unchanged between the MTB and BCG membranes using our label-free analysis (log2 BCG/MTB = 0.759, p = 0.555 after imputation). Using an anti-SigA antibody we found similar amounts of SigA protein in membrane fractions from both strains, confirming the mass spectrometry finding (Figure 6). As an example of a protein under-represented in the BCG membrane in comparison to MTB, we chose the LpqH lipoprotein (19kDa lipoprotein, Rv3763), which is a potent M. tuberculosis antigen42. Our LFQ data for LpqH, although not significant (p = 0.178) had a BCG/MTB log2 ratio of −2.955. Immunoblotting was consistent and revealed that the amount of LpqH was reduced in the BCG membrane compared to the MTB membrane. The reduced amount of LpqH in BCG was also evident when comparing whole cell lysates of the two strains.
We also evaluated two proteins that were significantly increased in abundance in the BCG membrane: isocitrate lysase Icl1 [rv0467, log2(BCG/MTB) = 3.52, p = 0.025] and malate synthase GlcB [rv1837c, log2(BCG/MTB) = 2.10, p = 0.051]. Our immunoblotting analysis detected Icl1 and GlcB in the membrane of BCG but little to no protein in the membrane of MTB (Figure 6). Interestingly, the amount of Icl1 was also higher in the whole cell lysate of BCG but the levels of GlcB in whole cell lysates appeared equivalent. These data highlight an advantage of proteomic comparisons of subcellular fractions, which is the ability to identify both localization differences of proteins that appear equally expressed between strains, such as GlcB, as well as changes in total protein abundance that consequently results in less protein at the membrane, such as LpqH and Icl1. Taken together, our validation efforts support our results using a label free quantitative proteomic approach to compare mycobacterial proteomes.
Discussion
Tuberculosis remains a major world health problem. Unfortunately, BCG is the only tuberculosis vaccine currently available and it demonstrates highly variable and incomplete protection against adult pulmonary tuberculosis. There is a need to develop improved vaccines and there is also a need to better understand the virulence properties of M. tuberculosis in order to develop new drugs to treat tuberculosis. A better understanding of the differences between the attenuated BCG vaccine and M. tuberculosis will aid these efforts.
Comparative genomics has revealed several differences at the chromosomal level between BCG and MTB, but these strains have yet to be compared using large scale comparative proteomics strategies. Quantitative “shotgun” proteomics allows for determining the relative abundance of large numbers of proteins between strains. Furthermore, the use of label-free quantification is a simple method for assessing relative quantities of proteins between samples that does not require additional chemistries or sample preparation. This is particularly ideal for slow-growing organisms such as M. tuberculosis, which may not be easily amendable to metabolic labeling.
So far, label-free quantitative proteomics have only been applied in a handful of studies of M. tuberculosis, but are producing encouraging results with enhanced coverage of the mycobacterial proteome37, 43, 44. However, this approach had yet to be extended to a direct comparison of the BCG vaccine and virulent MTB. In applying this method to our study here, we quantified significantly more proteins (2,203) than past studies involving 2D gel comparisons of M. tuberculosis and BCG, which yielded between 200 and 300 proteins6, 8, 45. We measured relative abundances for a large number of proteins, including 191 (296 after imputation) proteins with statistically significant (p≤0.05) and protein abundance differences of 2-fold or more. It is also interesting to compare the total number of proteins we identified in this study to the total number of proteins identified in a Triton X-114 fraction of M. tuberculosis by Malen et al. (2011)37, which represents an alternate membrane protein enrichment strategy to the fractionation scheme we used. Of the 1,575 M. tuberculosis proteins identified in the Triton X-114 fraction, 71% of them were also identified in our study. However, 44% of the 2,003 H37Rv proteins we identified were not identified in the prior Malen et al. study (data not shown). Thus, our report provides important confirmation for many proteins as being membrane associated while also identifying additional examples. The differences can likely be attributed to the different membrane enrichment protocols used. Importantly, the differential centrifugation method is widely used to prepare membrane fractions in M. tuberculosis research46.
Importantly, all our efforts to validate our proteomic analysis gave consistent results, which provide confidence in the previously unknown differences identified in our study. We were able to detect changes in LpqH, Icl1, and GlcB proteins, which were predicted by the mass spectrometry data to differ between BCG and MTB membranes. We also identified RD region-associated proteins in the MTB membrane that were undetected in the BCG membrane as expected. Along with the proteins encoded in RD regions, SigK (rv0445) is another protein we expected to find significantly reduced in BCG. SigK is a sigma factor known to have a point mutation in its start codon in BCG47. In MTB the SigK start codon is ATG, but in BCG it is ATA which reduces translation efficiency in BCG. Our LFQ analysis showed that SigK was reduced in the BCG membrane [log2 (BCG/MTB) = −4.872, p = 1.6×10−4], which is consistent with the protein being reduced in abundance in BCG. It is important to recognize that label-free quantification is a discovery tool for initial hypothesis generation. For proteins that do not have antibodies or other orthogonal testing methods that are highly quantitative, accurate quantification will require stable isotope dilution mass spectrometry methods such as SRM (selected reaction monitoring), which could be used as a verification assay.
When quantitated by LFQ, the RD region-associated proteins had extremely low values with imputation. Other proteins with very large changes in LFQ abundance (either low or high) could represent other examples of strain-specific proteins. In fact, there were 178 proteins for which ≥ 2 peptides was identified in at least 4 out of 6 replicates of the BCG membranes samples, but no corresponding peptides were identified in any MTB replicate (Supplemental Table 2). Alternatively, 186 proteins were only identified in MTB using the same criteria (Supplemental Table 3).
It will also be particularly interesting to follow up on proteins that were significantly more abundant or only identified in the pathogenic MTB membrane (i.e. less abundant or undetected in BCG), as candidates for virulence factors, antigens, or explaining physiologic differences between the strains. We identified 294 proteins that were at least 2-fold difference in abundance in the MTB membrane at p ≤ 0.05 (Supplemental Table 4). Furthermore, there were 186 proteins that were only identified in MTB by at least 2 peptides in 4 of 6 runs with no corresponding peptides identified in any BCG replicate (Supplemental Table 3). These proteins would be leading candidates for roles in these processes.
Among the interesting examples of proteins with reduced levels in BCG versus MTB were a group of three proteins encoded by neighboring genes that are part of a polyacyltrehalose (PAT) biosynthesis locus (rv1180–rv1183). For Rv1179c, Rv1180/Pks3 and Rv1181 Pks4, these proteins were present at significantly lower levels in BCG versus MTB (Figure 6A). Rv1180/Pks3 and Rv1181/Pks4 (also known as Msl3) comprise a polyketide synthase needed to synthesize the mycolipenic side chains of PAT.48, 49Interestingly, PAT is a surface lipid only found in pathogenic mycobacteria49, 50 but the explanation for the PAT deficiency in BCG Pasteur has remained unknown3. Our data suggests it is due to deficiency in the PAT biosynthetic machinery. Rv1179c is a protein of unknown function and not currently thought to function in PAT biosynthesis but its similar behavior in BCG suggests it might also have a role in PAT biosynthesis.
Another set of proteins that are reduced or absent in the BCG membrane proteome versus that of MTB are components of the ESX-3 specialized secretion system, which is encoded by a genomic locus, including Rv0282/EccA3, Rv2083/EccB3, Rv0284/EccC3, Rv0289/EspG3, Rv0290/EccD3, Rv0291, and Rv0292/EccE3 (Figure 6B). The ESX-3 system is essential in M. tuberculosis with an apparent function in iron and zinc homeostasis51, 52. In addition, a role for ESX-3 in eliciting protective immunity against M. tuberculosis was recently reported. In mouse studies, vaccination with a non-pathogenic M. smegmatis strain engineered to express the M. tuberculosis ESX-3 locus elicited better protection against M. tuberculosis challenge than BCG53. While the role of ESX-3 in this protection is not clear, one possibility is that some ESX-3 components are protective antigens. To our knowledge, our study is the first to show that levels of ESX-3 in BCG are reduced compared to M. tuberculosis, which raises the interesting possibility that the vaccination efficacy of BCG could be improved by increasing the levels of the ESX-3 system.
Also of interest are proteins that were more abundant in the BCG membrane compared to that of MTB, as they may provide insight into the adaptive changes that BCG has undergone that influence virulence and immunogenicity. We identified several proteins involved in the TCA and glyoxylate shunt pathways that were statistically more abundant in the BCG membrane. The significance of this result is unclear although it suggests a difference in metabolic activity between BCG and M. tuberculosis, at least in our growth conditions. Among the group of proteins that are more abundant in BCG were both of the mycobacterial isocitrate lyases: Icl1 [Rv0467, log2 (BCG/MTB) = 3.523, p = 0.025] and AceA [Rv1915/Rv1916, log2 (BCG/MTB) = 2.104, p = 0.051]. In MTB, both of these isocitrate lyases are required for survival in a mouse model of infection54. In line with our observations of higher Icl levels in BCG, we also detected increased GlcB malate synthase, an enzyme that participates along with isocitrate lyase in the glyoxylate shunt. Because the glyoxylate shunt is required for mycobacteria to utilize fatty acids as carbon sources, which are believed to be the primary carbon source for MTB during infection, it would be interesting to determine if these observations reflect any differences in fatty acid metabolism between BCG and MTB, which could influence growth in the host.
In conclusion, we used a label-free quantitative approach to compare protein abundance in different species of mycobacteria. Not only did this work greatly expand the number of proteins that have been compared between BCG and M. tuberculosis, it identified many differences at the protein level that were not predicted from genomic comparisons. Comparison to the RD regions of the genome and immunoblot analysis supports the accuracy of our data for proteins showing significant differences in abundance between MTB and BCG. Among the proteins identified are new candidates for understanding the well-established but poorlyunderstood differences between M. tuberculosis and BCG.
Supplementary Material
Acknowledgements
We thank Murty Madiraju, Suman Laal, and Douglas Young for providing antibodies. We also thank David Russell for providing both the Icl1 antibody and purified Icl1 protein. Support to XC was provided by NIH/NIAID 1R01A1064806 and support to MB was provided by NIAID AI054540.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.WHO. Global Tuberculosis Report 2012. Geneva, Switzerland: World Health Organization Press; 2012. [Google Scholar]
- 2.Nguyen L. Targeting antibiotic resistance mechanisms in Mycobacterium tuberculosis: recharging the old magic bullets. Expert Rev Anti Infect Ther. 2012;10(9):963–965. doi: 10.1586/eri.12.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brosch R, Gordon SV, Garnier T, Eiglmeier K, Frigui W, Valenti P, Dos Santos S, Duthoy S, Lacroix C, Garcia-Pelayo C, Inwald JK, Golby P, Garcia JN, Hewinson RG, Behr MA, Quail MA, Churcher C, Barrell BG, Parkhill J, Cole ST. Genome plasticity of BCG and impact on vaccine efficacy. Proc Natl Acad Sci U S A. 2007;104(13):5596–5601. doi: 10.1073/pnas.0700869104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colditz GA, Brewer TF, Berkey CS, Wilson ME, Burdick E, Fineberg HV, Mosteller F. Efficacy of BCG vaccine in the prevention of tuberculosis. Metaanalysis of the published literature. JAMA. 1994;271(9):698–702. [PubMed] [Google Scholar]
- 5.Brosch R, Pym AS, Gordon SV, Cole ST. The evolution of mycobacterial pathogenicity: clues from comparative genomics. Trends Microbiol. 2001;9(9):452–458. doi: 10.1016/s0966-842x(01)02131-x. [DOI] [PubMed] [Google Scholar]
- 6.Jungblut PR, Schaible UE, Mollenkopf HJ, Zimny-Arndt U, Raupach B, Mattow J, Halada P, Lamer S, Hagens K, Kaufmann SH. Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG strains: towards functional genomics of microbial pathogens. Mol Microbiol. 1999;33(6):1103–1117. doi: 10.1046/j.1365-2958.1999.01549.x. [DOI] [PubMed] [Google Scholar]
- 7.Mattow J, Jungblut PR, Schaible UE, Mollenkopf HJ, Lamer S, Zimny-Arndt U, Hagens K, Muller EC, Kaufmann SH. Identification of proteins from Mycobacterium tuberculosis missing in attenuated Mycobacterium bovis BCG strains. Electrophoresis. 2001;22(14):2936–2946. doi: 10.1002/1522-2683(200108)22:14<2936::AID-ELPS2936>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 8.Mattow J, Schaible UE, Schmidt F, Hagens K, Siejak F, Brestrich G, Haeselbarth G, Muller EC, Jungblut PR, Kaufmann SH. Comparative proteome analysis of culture supernatant proteins from virulent Mycobacterium tuberculosis H37Rv and attenuated M. bovis BCG Copenhagen. Electrophoresis. 2003;24(19–20):3405–3420. doi: 10.1002/elps.200305601. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt F, Donahoe S, Hagens K, Mattow J, Schaible UE, Kaufmann SH, Aebersold R, Jungblut PR. Complementary analysis of the Mycobacterium tuberculosis proteome by two-dimensional electrophoresis and isotope-coded affinity tag technology. Mol Cell Proteomics. 2004;3(1):24–42. doi: 10.1074/mcp.M300074-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Bantscheff M, Lemeer S, Savitski MM, Kuster B. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal Bioanal Chem. 2012;404(4):939–965. doi: 10.1007/s00216-012-6203-4. [DOI] [PubMed] [Google Scholar]
- 11.Cox J, Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem. 2011;80:273–299. doi: 10.1146/annurev-biochem-061308-093216. [DOI] [PubMed] [Google Scholar]
- 12.Niederweis M. Nutrient acquisition by mycobacteria. Microbiology. 2008;154(Pt 3):679–692. doi: 10.1099/mic.0.2007/012872-0. [DOI] [PubMed] [Google Scholar]
- 13.Crick DE, Quadri L, Brennan PJ. Biochemistry of the cell envelope of Mycobacterium tuberculosis. In: S.H.E K, Rubin EJ, editors. Handbook of Tuberculosis: Molecular Biology and Biochemistry. WILEY-VCH Verlag GmbH & Co; 2008. pp. 1–20. [Google Scholar]
- 14.Calva E, Oropeza R. Two-component signal transduction systems, environmental signals, and virulence. Microb Ecol. 2006;51(2):166–176. doi: 10.1007/s00248-005-0087-1. [DOI] [PubMed] [Google Scholar]
- 15.Kunnath-Velayudhan S, Salamon H, Wang HY, Davidow AL, Molina DM, Huynh VT, Cirillo DM, Michel G, Talbot EA, Perkins MD, Felgner PL, Liang X, Gennaro ML. Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proc Natl Acad Sci U S A. 2010;107(33):14703–14708. doi: 10.1073/pnas.1009080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinha S, Arora S, Kosalai K, Namane A, Pym AS, Cole ST. Proteome analysis of the plasma membrane of Mycobacterium tuberculosis. Comp Funct Genomics. 2002;3(6):470–483. doi: 10.1002/cfg.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sinha S, Kosalai K, Arora S, Namane A, Sharma P, Gaikwad AN, Brodin P, Cole ST. Immunogenic membrane-associated proteins of Mycobacterium tuberculosis revealed by proteomics. Microbiology. 2005;151(Pt 7):2411–2419. doi: 10.1099/mic.0.27799-0. [DOI] [PubMed] [Google Scholar]
- 18.Gu S, Chen J, Dobos KM, Bradbury EM, Belisle JT, Chen X. Comprehensive proteomic profiling of the membrane constituents of a Mycobacterium tuberculosis strain. Mol Cell Proteomics. 2003;2(12):1284–1296. doi: 10.1074/mcp.M300060-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Mawuenyega KG, Forst CV, Dobos KM, Belisle JT, Chen J, Bradbury EM, Bradbury AR, Chen X. Mycobacterium tuberculosis functional network analysis by global subcellular protein profiling. Mol Biol Cell. 2005;16(1):396–404. doi: 10.1091/mbc.E04-04-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malen H, Berven FS, Softeland T, Arntzen MO, D'Santos CS, De Souza GA, Wiker HG. Membrane and membrane-associated proteins in Triton X-114 extracts of Mycobacterium bovis BCG identified using a combination of gel-based and gelfree fractionation strategies. Proteomics. 2008;8(9):1859–1870. doi: 10.1002/pmic.200700528. [DOI] [PubMed] [Google Scholar]
- 21.Jyoti SC, Lu Y, Mark OC. Analysis protein complexes by 1D-SDS-PAGE and tandem mass spectrometry. 2008 [Google Scholar]
- 22.Wolfe LM, Mahaffey SB, Kruh NA, Dobos KM. Proteomic definition of the cell wall of Mycobacterium tuberculosis. J Proteome Res. 2010;9(11):5816–5826. doi: 10.1021/pr1005873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chelius D, Bondarenko PV. Quantitative profiling of proteins in complex mixtures using liquid chromatography and mass spectrometry. J Proteome Res. 2002;1(4):317–323. doi: 10.1021/pr025517j. [DOI] [PubMed] [Google Scholar]
- 24.Bondarenko PV, Chelius D, Shaler TA. Identification and relative quantitation of protein mixtures by enzymatic digestion followed by capillary reversed-phase liquid chromatography-tandem mass spectrometry. Anal Chem. 2002;74(18):4741–4749. doi: 10.1021/ac0256991. [DOI] [PubMed] [Google Scholar]
- 25.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deeb SJ, D'Souza RC, Cox J, Schmidt-Supprian M, Mann M. Super-SILAC allows classification of diffuse large B-cell lymphoma subtypes by their protein expression profiles. Mol Cell Proteomics. 2012;11(5):77–89. doi: 10.1074/mcp.M111.015362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honer Zu Bentrup K, Miczak A, Swenson DL, Russell DG. Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis. J Bacteriol. 1999;181(23):7161–7167. doi: 10.1128/jb.181.23.7161-7167.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Florczyk MA, McCue LA, Stack RF, Hauer CR, McDonough KA. Identification and characterization of mycobacterial proteins differentially expressed under standing and shaking culture conditions, including Rv2623 from a novel class of putative ATP-binding proteins. Infect Immun. 2001;69(9):5777–5785. doi: 10.1128/IAI.69.9.5777-5785.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu S, Howard ST, Lakey DL, Kipnis A, Samten B, Safi H, Gruppo V, Wizel B, Shams H, Basaraba RJ, Orme IM, Barnes PF. The principal sigma factor sigA mediates enhanced growth of Mycobacterium tuberculosis in vivo. Mol Microbiol. 2004;51(6):1551–1562. doi: 10.1111/j.1365-2958.2003.03922.x. [DOI] [PubMed] [Google Scholar]
- 30.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 31.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 32.Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. Prediction of twin-arginine signal peptides. BMC Bioinformatics. 2005;6:167. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Natale P, Bruser T, Driessen AJ. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane--distinct translocases and mechanisms. Biochim Biophys Acta. 2008;1778(9):1735–1756. doi: 10.1016/j.bbamem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 34.Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12(8):1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutcliffe IC, Harrington DJ. Lipoproteins of Mycobacterium tuberculosis: an abundant and functionally diverse class of cell envelope components. FEMS Microbiol Rev. 2004;28(5):645–659. doi: 10.1016/j.femsre.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Hutchings MI, Palmer T, Harrington DJ, Sutcliffe IC. Lipoprotein biogenesis in Gram-positive bacteria: knowing when to hold 'em, knowing when to fold 'em. Trends Microbiol. 2009;17(1):13–21. doi: 10.1016/j.tim.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Malen H, De Souza GA, Pathak S, Softeland T, Wiker HG. Comparison of membrane proteins of Mycobacterium tuberculosis H37Rv and H37Ra strains. BMC Microbiol. 2011;11:18. doi: 10.1186/1471-2180-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Camus JC, Pryor MJ, Medigue C, Cole ST. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology. 2002;148(Pt 10):2967–2973. doi: 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
- 39.Xiong Y, Chalmers MJ, Gao FP, Cross TA, Marshall AG. Identification of Mycobacterium tuberculosis H37Rv integral membrane proteins by one-dimensional gel electrophoresis and liquid chromatography electrospray ionization tandem mass spectrometry. J Proteome Res. 2005;4(3):855–861. doi: 10.1021/pr0500049. [DOI] [PubMed] [Google Scholar]
- 40.Brosch R, Gordon SV, Pym A, Eiglmeier K, Garnier T, Cole ST. Comparative genomics of the mycobacteria. Int J Med Microbiol. 2000;290(2):143–152. doi: 10.1016/S1438-4221(00)80083-1. [DOI] [PubMed] [Google Scholar]
- 41.Rehren G, Walters S, Fontan P, Smith I, Zarraga AM. Differential gene expression between Mycobacterium bovis and Mycobacterium tuberculosis. Tuberculosis (Edinb) 2007;87(4):347–359. doi: 10.1016/j.tube.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brightbill HD, Libraty DH, Krutzik SR, Yang RB, Belisle JT, Bleharski JR, Maitland M, Norgard MV, Plevy SE, Smale ST, Brennan PJ, Bloom BR, Godowski PJ, Modlin RL. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science. 1999;285(5428):732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 43.de Souza GA, Fortuin S, Aguilar D, Pando RH, McEvoy CR, van Helden PD, Koehler CJ, Thiede B, Warren RM, Wiker HG. Using a label-free proteomics method to identify differentially abundant proteins in closely related hypo- and hypervirulent clinical Mycobacterium tuberculosis Beijing isolates. Mol Cell Proteomics. 2010;9(11):2414–2423. doi: 10.1074/mcp.M900422-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelkar DS, Kumar D, Kumar P, Balakrishnan L, Muthusamy B, Yadav AK, Shrivastava P, Marimuthu A, Anand S, Sundaram H, Kingsbury R, Harsha HC, Nair B, Prasad TS, Chauhan DS, Katoch K, Katoch VM, Chaerkady R, Ramachandran S, Dash D, Pandey A. Proteogenomic analysis of Mycobacterium tuberculosis by high resolution mass spectrometry. Mol Cell Proteomics. 2011;10(12) doi: 10.1074/mcp.M111.011627. M111 011627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mattow J, Jungblut PR, Muller EC, Kaufmann SH. Identification of acidic, low molecular mass proteins of Mycobacterium tuberculosis strain H37Rv by matrix-assisted laser desorption/ionization and electrospray ionization mass spectrometry. Proteomics. 2001;1(4):494–507. doi: 10.1002/1615-9861(200104)1:4<494::AID-PROT494>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 46.Rezwan M, Laneelle MA, Sander P, Daffe M. Breaking down the wall: fractionation of mycobacteria. J Microbiol Methods. 2007;68(1):32–39. doi: 10.1016/j.mimet.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 47.Charlet D, Mostowy S, Alexander D, Sit L, Wiker HG, Behr MA. Reduced expression of antigenic proteins MPB70 and MPB83 in Mycobacterium bovis BCG strains due to a start codon mutation in sigK. Mol Microbiol. 2005;56(5):1302–1313. doi: 10.1111/j.1365-2958.2005.04618.x. [DOI] [PubMed] [Google Scholar]
- 48.Dubey VS, Sirakova TD, Kolattukudy PE. Disruption of msl3 abolishes the synthesis of mycolipanoic and mycolipenic acids required for polyacyltrehalose synthesis in Mycobacterium tuberculosis H37Rv and causes cell aggregation. Mol Microbiol. 2002;45(5):1451–1459. doi: 10.1046/j.1365-2958.2002.03119.x. [DOI] [PubMed] [Google Scholar]
- 49.Hatzios SK, Schelle MW, Holsclaw CM, Behrens CR, Botyanszki Z, Lin FL, Carlson BL, Kumar P, Leary JA, Bertozzi CR. PapA3 is an acyltransferase required for polyacyltrehalose biosynthesis in Mycobacterium tuberculosis. J Biol Chem. 2009;284(19):12745–12751. doi: 10.1074/jbc.M809088200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guilhot C, Daffe M. Handbook of Tuberculosis: Molecular Biology and Biochemistry. WILEY-VCH Verlag GmbH & Co; 2008. Polyketides and Polyketide-containing Glycolipids of Mycobacterium tuberculosis: Structure, Biosynthesis, and Biological Activities; pp. 21–52. [Google Scholar]
- 51.Serafini A, Boldrin F, Palu G, Manganelli R. Characterization of a Mycobacterium tuberculosis ESX-3 conditional mutant: essentiality and rescue by iron and zinc. J Bacteriol. 2009;191(20):6340–6344. doi: 10.1128/JB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siegrist MS, Unnikrishnan M, McConnell MJ, Borowsky M, Cheng TY, Siddiqi N, Fortune SM, Moody DB, Rubin EJ. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci U S A. 2009;106(44):18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sweeney KA, Dao DN, Goldberg MF, Hsu T, Venkataswamy MM, Henao-Tamayo M, Ordway D, Sellers RS, Jain P, Chen B, Chen M, Kim J, Lukose R, Chan J, Orme IM, Porcelli SA, Jacobs WR., Jr A recombinant Mycobacterium smegmatis induces potent bactericidal immunity against Mycobacterium tuberculosis. Nat Med. 2011;17(10):1261–1268. doi: 10.1038/nm.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Munoz-Elias EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11(6):638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.