Abstract
The first dedicated step in de novo cholesterol biosynthesis begins with formation of squalene and ends with the reduction of 7-dehydrocholesterol by 7-dehydrocholesterol reductase (Dhcr7) into cholesterol, which is an essential structural and signaling molecule. Mutations in the Dhcr7 gene lead to Smith-Lemli-Opitz syndrome (SLOS), which is characterized by developmental deformities, incomplete myelination, and mental retardation. To understand better the molecular consequences of Dhcr7 deficiency in neuronal tissue, we analyzed the effect of cholesterol deficiency on the transcriptome in Neuro2a cells. Transient down-regulation of Dhcr7 by siRNA led to altered expression of multiple molecules that play critical roles in intracellular signaling or vesicular transport or are inserted into membrane rafts (e.g. Egr1, Snx, and Adam19). A similar down-regulation was also observed in stable Dhrc7-shRNA-transfected cell lines, and the findings were verified by qPCR. Furthermore, we investigated the Dhcr7-deficient and control cells for the expression of several critical genes involved in lipid biosynthesis. Among these, fatty acid synthase, sterol-regulatory element binding protein 2, SREBF chaperone, site-1 protease, and squalene synthase showed a significant down-regulation, suggesting that, in a neuronal cell line, Dhcr7 is a potent regulator of lipid biosynthesis. Importantly, the gene expression changes were present in both lipid-containing and cholesterol-deficient media, suggesting that intrinsic cholesterol biosynthesis is necessary for normal neuronal function and cannot be supplemented from extrinsic sources.
Keywords: gene expression, lipid metabolism, cholesterol, neuroblastoma, SREBP
Cholesterol is a critical building block of cellular membranes (Maxfield and Tabas, 2005). Cholesterol biosynthesis is a complex cascade of events that involves at least 20 dedicated enzymes (Lutton, 1991). The rate-limiting enzyme in cholesterol biosynthesis is 3-hydroxy-3-methylglutaryl-coenzyme A reductase (Hmgcr; EC 2.3.3.10), which catalyzes conversion of Hmg-CoA to mevalonic acid (Luskey and Stevens, 1985). The cascade ends with reduction of 7-dehydrocholesterol that is catalyzed by 7-dehydrocholesterol reductase (Dhcr7; EC 1.3.1.21) enzyme (Bae et al., 1999). Liver and small intestine are the major sites of cholesterol biosynthesis for the whole body (Strandberg and Tilvis, 1988). However, all of the brain cholesterol is synthesized locally, with the highest rate of synthesis occurring during first postnatal weeks in humans and rodents (Jurevics and Morell, 1995; Jurevics et al., 1997). This peak of early postnatal biosynthesis coincides with myelination (Anitei and Pfeiffer, 2006). The expression pattern of transcripts involved in cholesterol biosynthesis and the role and regulation of cholesterol biosynthesis in the nervous system remain understudied and are mostly unknown to date (Korade and Kenworthy, 2008).
Recent studies showed that changes in cholesterol homeostasis lead to a wide variety of CNS disorders (Maxfield and Tabas, 2005). For example, deficient cholesterol biosynthesis in oligodendrocytes delays myelination (Saher et al., 2005), and defects in cholesterol trafficking lead to Niemann-Pick type C disease (Paul et al., 2004). Importantly, partial or complete lack of Dhcr7 enzymatic activity causes Smith-Lemli-Opitz syndrome (SLOS), characterized by severe developmental malformations and mental retardation (Smith et al., 1964; Tierney et al., 2000). In SLOS, mutations in Dhcr7, the last enzyme in cholesterol biosynthesis pathway, lead to accumulation of 7-dehydrocholesterol and reduced cholesterol levels (Jira et al., 2003). 7-Dehydrocholesterol differs from cholesterol in a double bond at the seventh position in the sterol ring, and this structural difference translates into a functional difference: whereas 7-dehydrocholesterol can be incorporated into the membranes just like cholesterol, its presence perturbs the protein content of lipid rafts (Keller et al., 2004).
Lipid rafts (microdomains) are cholesterol- and sphingolipid-enriched areas within cellular membranes that concentrate and segregate proteins within the membrane bilayer (Pike, 2005). Membrane rafts are present in both neurons and glial cells, and they contain different receptors and neurotransmitter transporters, regulate intramembrane proteolysis of many transmembrane proteins, and serve as organizing sites for neurotrophin signaling (Korade and Kenworthy, 2008).
In addition to its structural role as a building block of membranes, cholesterol has important role in transcription of genes containing the SRE domain (Wang et al., 1994; Horton et al., 2002). This regulation is well described in liver, where SCAP, through the sterol-sensing domain, regulates availability of SREBPs, transcription factors critical for regulation of lipid genes (Horton et al., 2002). This mechanism has not been studied in neuronal tissue. Therefore, we hypothesized that altering cholesterol biosynthesis in cells of neuronal origin will lead to changed transcription of genes belonging to multiple cellular pathways. As a result, the present study was aimed to 1) develop an in vitro model for assessment of the role and consequences of endogenous neuronal cholesterol biosynthesis, 2) uncover and validate the molecular consequences of the reduced intrinsic cholesterol biosynthesis, and 3) evaluate the dependence of the uncovered molecular changes on extrinsic vs. intrinsic sources of cholesterol.
MATERIALS AND METHODS
Cell Culture and Reagents
Neuroblastoma cell line Neuro2a was purchased from the American Type Culture Collection (Rockville, MD). Neuro2a cells were maintained in alpha modification of minimal essential medium (Eagle) with Earle’s salt and supplemented with L-glutamine, sodium bicarbonate, nonessential amino acids, sodium pyruvate, 10% fetal bovine serum (FBS; Thermo Scientific HyClone, Logan, UT), and penicillin/streptomycin at 37°C and 5% CO2. According to the vendor, the FBS contained cholesterol at concentrations of 32 mg/100 ml. This translates into a final cholesterol concentration of 32 µg/ml in our culture medium. To evaluate the role of exogenous cholesterol on gene expression, cells were also cultured with medium containing 10% cholesterol-deficient serum (Thermo Scientific HyClone Lipid Reduced FBS). This FBS medium did not have detectable cholesterol levels. The Neuro2a cells were subcultured once per week, and the culture medium was changed every 2 days. SiRNA oligonucleotides were purchased from Qiagen (Valencia, CA): AllStars Negative Control siRNA, Mm_Dhcr7_1 HP siRNA, Mm_Dhcr7_2 HP siRNA, and Mm_Dhcr7_3 HP siRNA. For stable transfections, we obtained plasmids from Open Biosystems through the Vanderbilt Microarray Shared Resource. These include OligoID V2LMM_9686 and nonsilencing GIPZ shRNAmir control.
Transient and Stable siRNA Transfections
Neuro2a cells were cultured for 2 days before transfections. Cells were transfected with three different Dhcr7 siRNA oligonucleotides (Qiagen) using a Nucleofector instrument and Nucleofector Kit V (Amaxa GmbH, Cologne, Germany) optimized for use with Neuro2a cells. Briefly, 2 × 106 cells were resuspended in 100 µl transfection buffer, siRNA was added, and cells were electroporated using program T-24. The cells were grown for 24 hr following transfection, and the expression of Dhcr7 was monitored by quantitative RT-PCR. To establish stable down-regulation of Dhcr7, after transfection of cells with pGIPZ plasmids, cells were grown in the presence of puromycin.
RNA Preparation and Array Hybridization
Total RNA was isolated from the cells using Trizol (Life Technologies, Rockville, MD). Purification of total RNA was done using RNeasy Mini Kit (Qiagen), and oncolumn digestion of DNA was performed during the RNA purification step using RNase-Free DNase Set (Qiagen). The concentration of total RNA was measured on a Nanodrop instrument (Thermo Scientific, Wilmington, DE), and the integrity of the RNA was established by electrophoresis using the Agilent Bioanalyzer 2100 instrument (Agilent Technologies, Santa Clara, CA). Sample preparation, cRNA synthesis, IVT, hybridization, and array scanning were performed as per manufacturers’ instructions. The fragmented cRNA was hybridized to Affymetrix MG-430 mouse oligonucleotide microarrays.
Data Analysis
All microarrays had exceptional quality based on present calls and 5′:3′ GAPDH integrity ratios calculated by GCOS. Segmented images were normalized and log2 transformed using robust multiarray analysis (RMA) in Expression Console (Affymetrix). Clustering and secondary data analysis was performed in GenePattern 2.0 (Reich et al., 2006).
In the microarray study, genes were considered differentially expressed between Dhcr7-deficient and control samples if they reported an absolute ALR > 0.383 (>30% change) and a groupwise statistical significance of P < 0.05. All microarray data will be publicly available at the time of publication at http://mirnicslab.vanderbilt.edu/mirnicslab/ordering.htm. Because this was an exploratory experiment to identify putative downstream effects, we did not correct for multiple testing, and the most critical data were verified by qPCR.
Northern Blotting
Northern hybridization was done using the Northern-Max system (Ambion, Austin, TX) as previously described (Korade et al., 2007). Briefly, 20 µg total RNA was loaded on a formamide gel, electrophoresed at 5 V/cm, transferred to a Bright Star nylon membrane, and cross-linked by UV light exposure. Hmgcr and Dhcr7 probes were amplified using SP6 and T7 primers from plasmid as previously described (Korade et al., 2007). Gapdh probe were generated using gene-specific primers and Neuro2a cDNA template in a standard PCR. The purified PCR product was radioactively labeled using α32P-dCTP and a Random Primed DNA labeling kit (Roche, Indianapolis, IN). The labeled probe was purified with Probe-Quant G-50 MicroColumns (Amersham Biosciences, Piscataway, NJ). The denatured probe, 106 cpm/ml, was added to the hybridization buffer (10 ml/100 cm2 membrane). The hybridization was done overnight at 42°C. The Northern membrane was washed for 3 hr at 45°C and exposed to X-ray film.
Quantitative PCR
Total RNA (100 ng) from each sample was reverse transcribed to cDNA using a High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). Real-time PCR was performed with an ABI Prism 7300 System (Applied Biosystems) using 1 ng cDNA per 50 µl reaction volume, 2×SYBR green master mix, and gene-specific primers. All samples were run in triplicate. Data from the PCRs was analyzed using the comparative cycle number determined as threshold (Ct) method (Kurrasch et al., 2004). Differential expression was calculated as ΔΔCt against expression of Pgk1 as a normalizer. We designed primers (~20 bp) to yield 85–110-bp PCR amplicons in Primer3 software (http://frodo.wi.mit.edu/) for different genes. For each gene, we designed four sets of primers. Each set was tested using no template and three different concentrations of a specific template. This translates into 16 wells for each primer set (four wells with no-template controls, four wells for each of three different template concentrations). From this PCR, we calculated the efficiency of PCR primers and R2 value (coefficient of correlation). All mouse gene-specific primers used showed a slope between −3.10 and −3.58, with R2 > 0.99 (Supp. Info. Fig. 2A). Lipid-specific qPCR primers have been described by Wang et al. (2002). All qPCR amplicons were checked by gel electrophoresis, and all of the qPCR reactions gave rise to a single product of predicted size. Furthermore, the qPCR dissociation curve of the amplicons, performed after each qPCR run (using Dissociation Curve 1.0 software; ABI), also confirmed specific amplification. The gel electrophoresis data are presented in Supporting Information Figure 2B.
Analysis of Total Cholesterol Level by Amplex Red
Total cholesterol levels were measured using an Amplex-Red Cholesterol Assay kit (Molecular Probes, Eugene, OR). Dhcr7-deficient and control cells were grown for 4 days in either regular or cholesterol-deficient culture medium and then lysed in 0.1 M phosphate lysis buffer. Cholesterol standard and samples were prepared for measurement according to the manufacturer’s instructions. Fluorescence was measured with a fluorescence microplate reader using excitation at 545 nm and fluorescence detection at 590 nm. The background fluorescence, determined for the no-cholesterol control reaction, was subtracted from each value. The total cholesterol level was determined by comparing experimental data with the data obtained for cholesterol standards. Total cholesterol values were normalized to protein values.
UV Spectrometric Analysis of 7DHC Levels
7DHC shows characteristic ultraviolet absorption maxima (λmax) at 271, 282, and 294 nm (Nes, 1985). Therefore, we used spectrophotometric measurement to detect 7DHC in stably transfected Neuro2a cells (Honda et al., 1997). Stably transfected Neuro2a cells were cultured for 3–5 days in cholesterol-deficient medium. After removal of cell culture medium, cells were washed twice in 1× PBS and scraped into PBS. Cell were spun down, PBS was removed, and 200 µl of 100% EtOH was added to the cell pellet, with incubation in an ultrasonic bath for 15 min. After addition of 200 µl water and vortexing for 10 sec, 1 ml n-hexane was added. The content was vortexed for 20 sec and spun down at 400g for 1 min. After centrifugation, the clear n-hexane layer was collected in a 1-ml quartz cuvet and used for UV measurement on a spectrophotometer. The samples were scanned using n-hexane blank in the reference beam, and the absorption maxima at 271, 282, and 294 nm were used for detection of 7DHC in lipid extracts from cells.
RESULTS
Dhcr7 siRNA Reduces Dhcr7 Transcript Expression
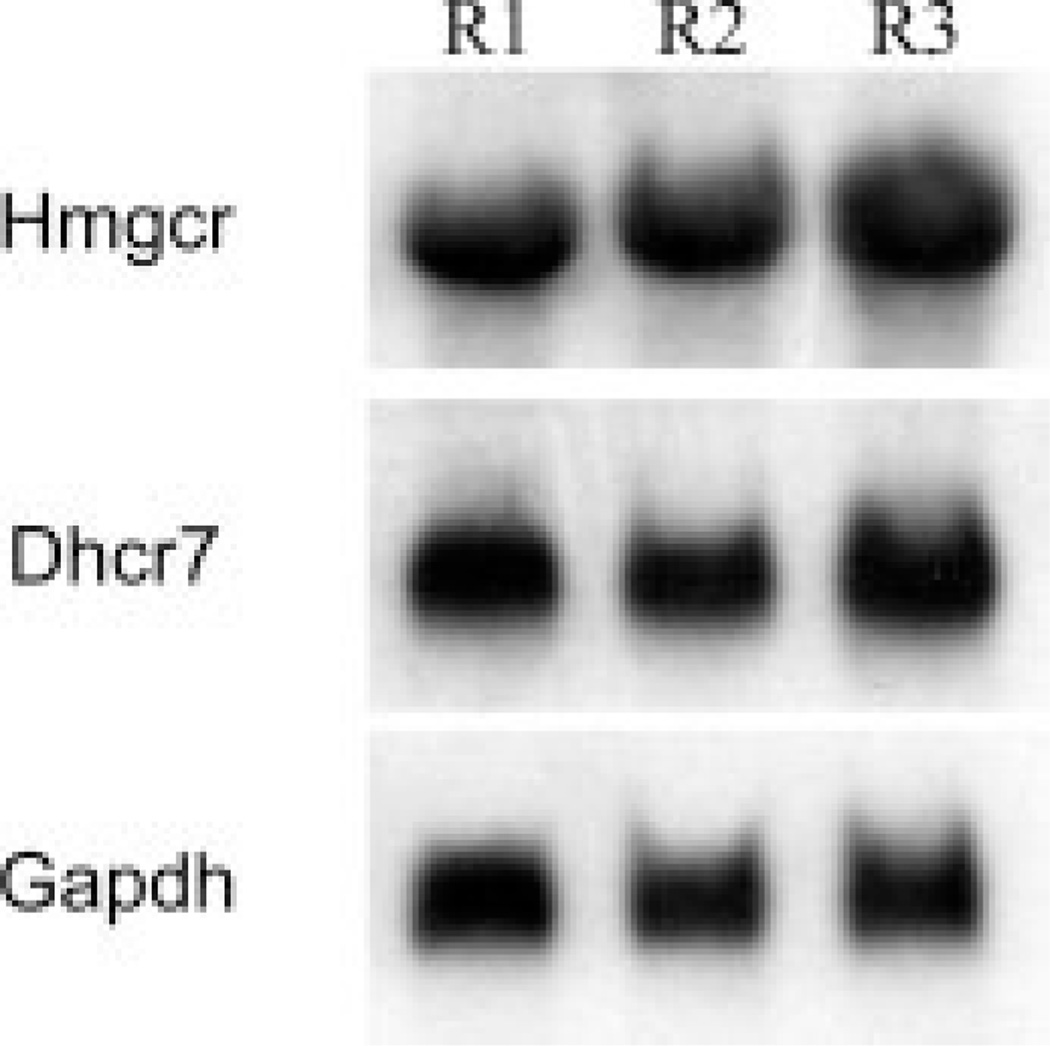
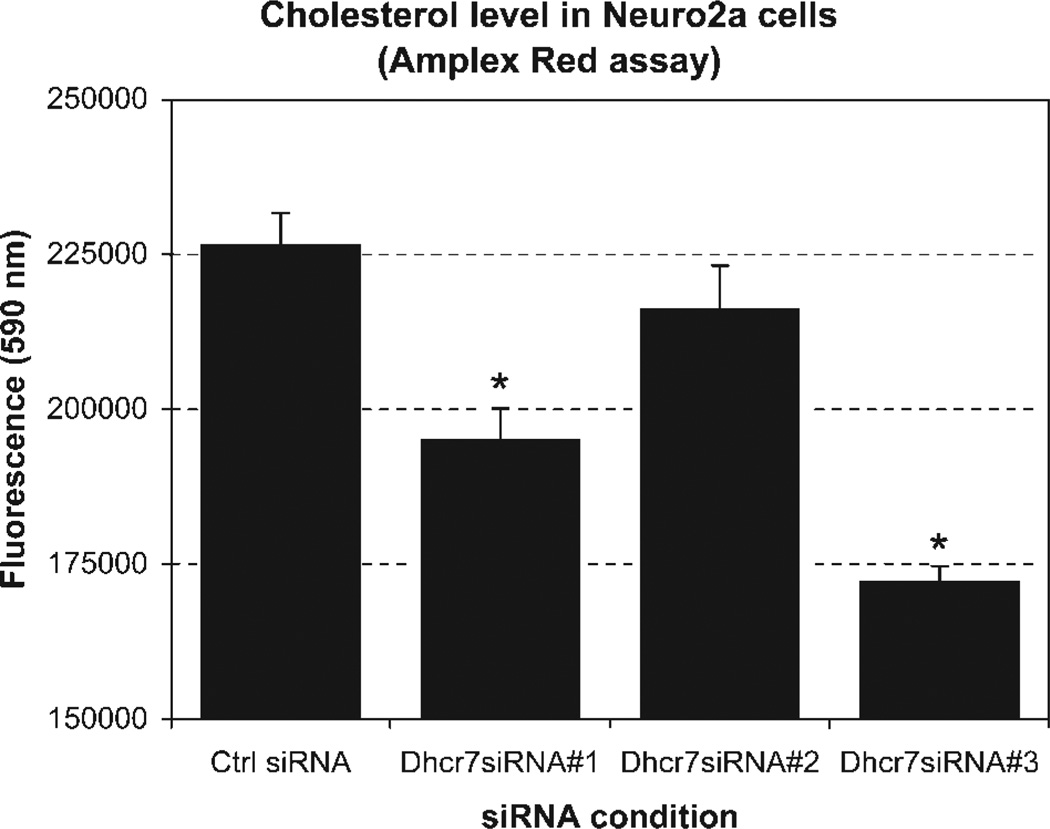
Both neurons and neuronal cell lines have the ability to synthesize cholesterol, yet the level of cholesterogenic enzyme expression varies across distinct cellular phenotypes (Korade et al., 2007). We therefore first examined the expression of the cholesterogenic enzymes in our model system, the Neuro2a neuroblastoma cell line. Northern hybridization revealed that Neuro2a cells strongly expressed the first and the last enzymes (Dhcr7 and Hmgcr, respectively) of the cholesterol biosynthesis pathway (Fig. 1). The expression of these transcripts suggested that Neuro2a cells are an appealing model to study the regulation and role of neuronal cholesterol biosynthesis. Thus, following the assessment of the expression of these two critical enzymes, we established an in vitro experimental system based on down-regulation of Dhcr7 in these cells. To achieve this, we tested the efficacy of three different siRNAs designed to down-regulate Dhcr7. Two of the three tested siRNAs successfully down-regulated Dhcr7 transcript and cholesterol levels (achieving >70% transcript reduction), whereas one failed to achieve a specific Dhcr7 downregulation (Fig. 2). Given these results, we decided to perform our DNA microarray experiments using Dhcr7-siRNA No. 3.
Figure 1.
Hmgcr and Dhcr7 are highly expressed in Neuro2a cells. Total RNA was isolated from Neuro2a cells and analyzed by Northern blotting for the expression of cholesterogenic enzymes. The same membrane was probed sequentially with Hmgcr-, Dhcr7-, and Gapdh-32P-dCTP-labelled cDNA probes. The experiment was performed in triplicate. Note that Hmgcr and Dhcr7, the first and the last enzymes in the cholesterol biosynthesis pathway, are highly expressed in Neuro2a neuroblastoma cells.
Figure 2.
Cholesterol levels in the Dhcr7 siRNA-treated cultures measured by amplex red assay. Compared with the control siRNA (scrambled oligonucleotide), down-regulation of Dhcr7 by siRNAs resulted in a significant reduction of cholesterol production for two of the three specific oligonucleotides (asterisk). The most robust down-regulation was achieved by Dhcr7siRNA No. 3, so this siRNA was used in the subsequent DNA microarray experiments. *P < 0.05.
Down-regulation of Dhcr7 Transcript Leads to Transient Changes in Cellular Transcriptome
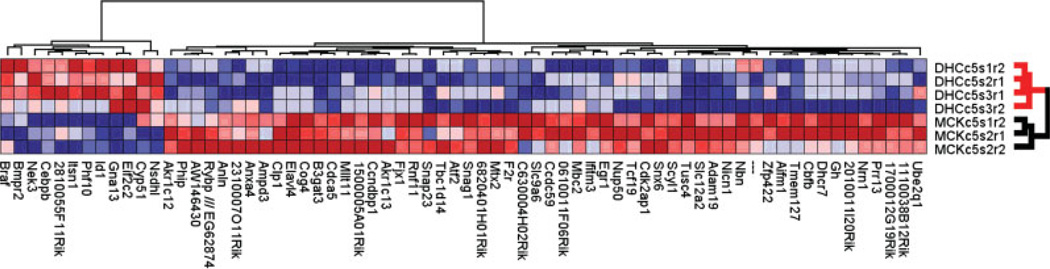
We hypothesized that reduced cholesterol biosynthesis will lead to transcriptome changes, so we used gene expression profiling by DNA microarrays to identify the Dhcr7-dependent transcript network. To down-regulate the expression of Dhcr7 in Neuro2a cells, we used the validated Dhcr7siRNA oligonucleotide or nonsilencing siRNA (control) for transient transfections. Twenty-four hours after transfections, four replicates of Dhcr7 siRNA treated and three replicates of controls were analyzed using Affymetrix Mouse 430v2 oligonucleotide arrays, establishing expression levels of >40,000 unique transcript sequences. Statistical analysis of the RMA-normalized data set revealed differential expression of 68 transcripts (Supp. Info. Fig. 1), with down-regulations greatly outnumbering transcript inductions (56 vs. 12 probe sets). The expression levels of these transcripts, when subjected to a two-way hierarchical clustering, perfectly separated the experimental and control samples (Fig. 3). Importantly, as predicted, the Dhcr7 transcript itself was the most down-regulated mRNA. Finally, among the 68 altered genes, 50 showed a high correlation with Dhcr7 expression levels (r > 0.60, P < 0.01; Supp. Info. Fig. 1), confirming a direct causal relationship between Dhcr7 down-regulation and the observed transcript changes.
Figure 3.
Dhcr7 siRNA transfection leads to strong transcriptome changes. Two-way clustering of the normalized expression levels for 68 genes showing transcript level changes in the Dhcr7 siRNA-transfected cultures. In the vertical dendrogram, each arm represents a single sample (red-Dhcr7 siRNA; black, scrambled siRNA), and rows denote gene probe sets with NCBI accession numbers and gene symbols. Each pixel corresponds to a log2-normalized expression level in a single sample. The intensity of red is proportional to transcript increase, whereas the blue intensity is proportional to transcript decrease. Based on the expression levels of these 68 probe sets, the vertical dendrogram perfectly separated out the experimental (red) and control samples (black). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Among the genes with differential expression in the microarray experiment, based on their function in neural tissue, a subset of expression changes, including Egr1 (early growth response 1) and Adam19 (a disintegrin and metallopeptidase domain 19), was of particular interest to us. Egr-1 (Zif268, NGFI-A, TIS8, or Krox24) is a transcription factor with three zinc fingers induced by various stimuli, such as growth factors, hormones, and neurotransmitters (Milbrandt, 1987; Silverman and Collins, 1999). Egr-1 is associated with both cell growth and apoptosis, and some of its downstream targets are known (Silverman and Collins, 1999). One of the Egr-1 targets is SREBP transcription factor (Fernandez-Alvarez et al., 2008). On the other hand, Adam19 (Meltrin β, FKSG34) belongs to a gene family of proteins with metalloproteinase domain that are involved in cell adhesion, cell fusion, migration, membrane protein shedding, and proteolysis (Mochizuki and Okada, 2007). Furthermore, ADAMs play a role in cell adhesion through interaction with other proteins (syndecans and fibronectin) with their cysteine-rich domains (Iba et al., 2000). Although not much is known about Adam19, the functions of some other family members are better described (Rocks et al., 2008). For example, Adam17 cleaves several transmembrane proteins, and its activity can be regulated by membrane cholesterol levels (Tellier et al., 2006, 2008). Finally, in addition to Egr1 and Adam19, the three other genes that were also chosen for follow-up included Prr13 (proline-rich 13), Snag1 (sorting nexin associated golgi protein 1), and Snx6 (sorting nexin 6) along with an Est with unknown function (2010011I20Rik).
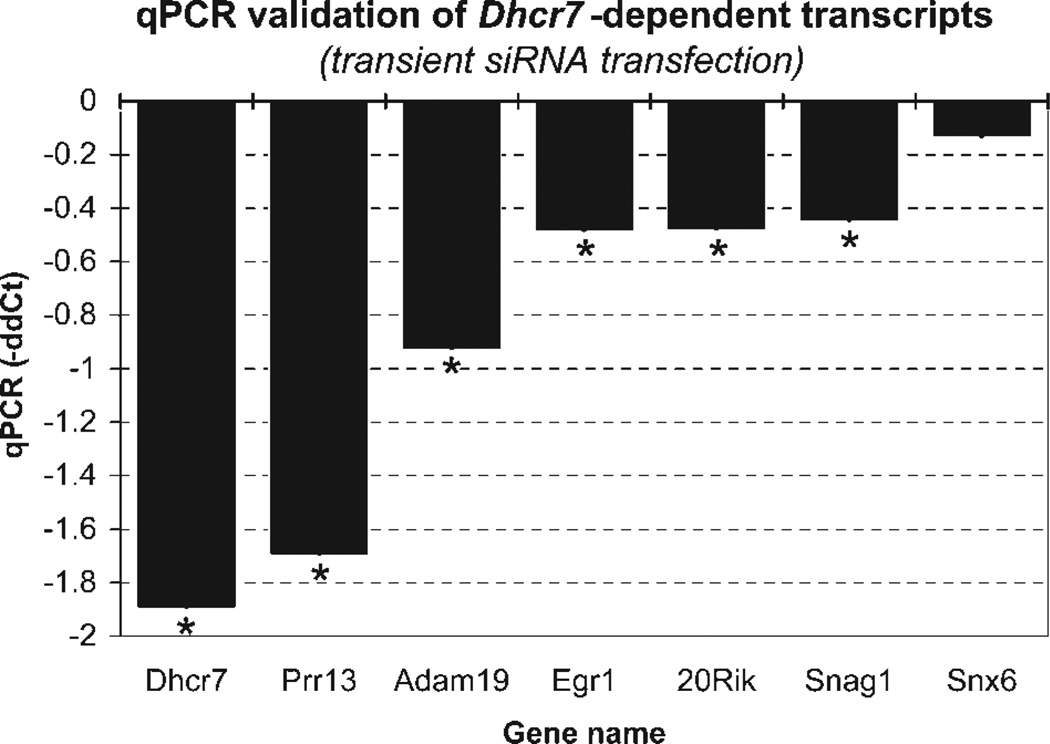
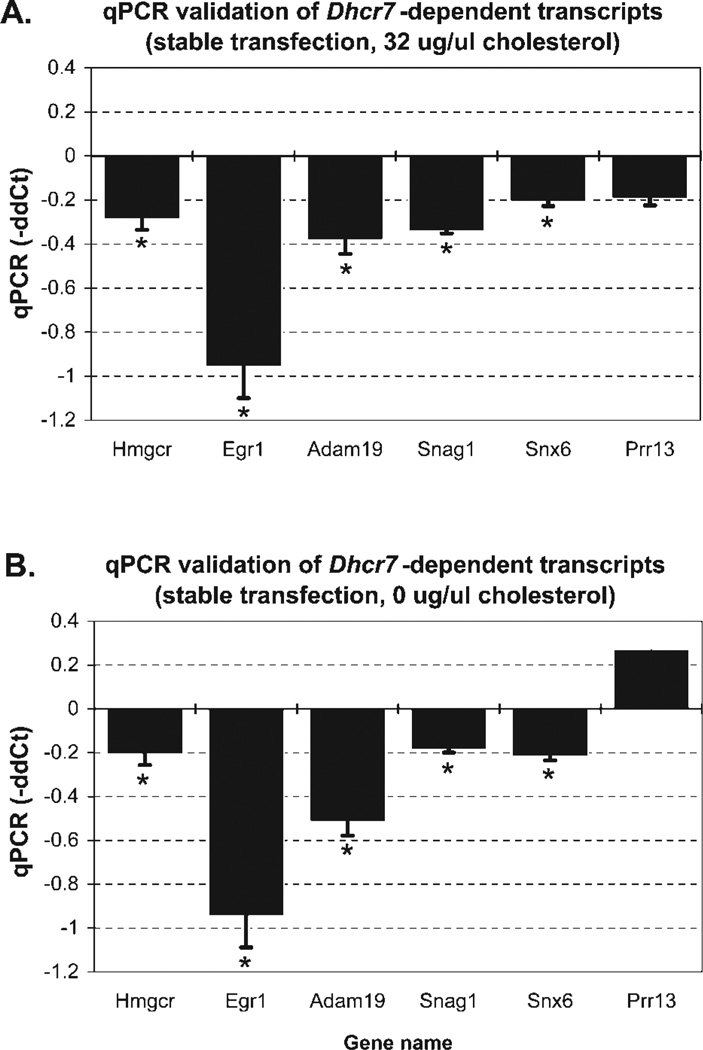
To verify microarray findings, we developed qPCR primers and verified the differential expression of these six putatively changed genes (Supp. Info. Fig. 1, Fig. 4). For these genes, the Sybr green, ddCt-based qPCR findings were highly correlated with the microarray data (r = 0.89, P < 0.01).
Figure 4.
Transient transfection with Dhcr7 siRNA leads to reproducible gene expression changes. Genes are plotted on the x-axis, and the y-axis denotes the average −ΔΔCt from three independent experiments, two independent reverse transcriptions for each experiment, and four replicates for each reverse transcription. qPCR assays confirmed the DNA microarray findings for all tested genes: Prr13 (proline-rich 13), Adam19 (a disintegrin and metallopeptidase domain 19), Egr1 (early growth response1), 20Rik (EST 2010011I20Rik), Snag1 (sorting nexin-associated Golgi protein 1), and Snx6 (sorting nexin 6). *P < 0.05.
Transcriptome Changes Persist in Neuro2a Cells With Stable Dhcr7shRNA Transfection
Although the transient transfection experiments identified and validated Dhcr7-dependent transcript network, two critical questions remained. First, we wanted to verify that a different Dhcr7siRNA can produce an effect similar to that observed in our initial experiment. Second, whereas the initial experiment suggested an acute response to the Dhcr7siRNA exposure, it did not provide insight into whether these changes persist over time. To address both of these two concerns simultaneously, we developed a stably transfected Neuro2a cell line that utilized a different Dhcr7shRNA inserted in pGIPZ plasmid. Again, we used as our control a Neuro2a cell line with a stable transfection of a nonsilencing pGIPZ plasmid. Both constructs contained a puromycin selection cassette, and the presence of green fluorescent protein (GFP) allowed monitoring the expression of the specific shRNA. Both Dhcr7-deficient and control cell lines had comparable growth rates and morphologies (Figs. 5, 6). However, using UV spectrophotometry (Honda et al., 1997), we detected a greater than tenfold rise in 7-dehydrocholesterol (7DHC) levels in Dhcr7-deficient cells compared with the control cells (Supp. Info. Fig. 3), suggesting that silencing of the Dhcr7 enzyme strongly prevented 7DHC conversion into mature cholesterol, thus leading to 7DHC accumulation.
Figure 5.
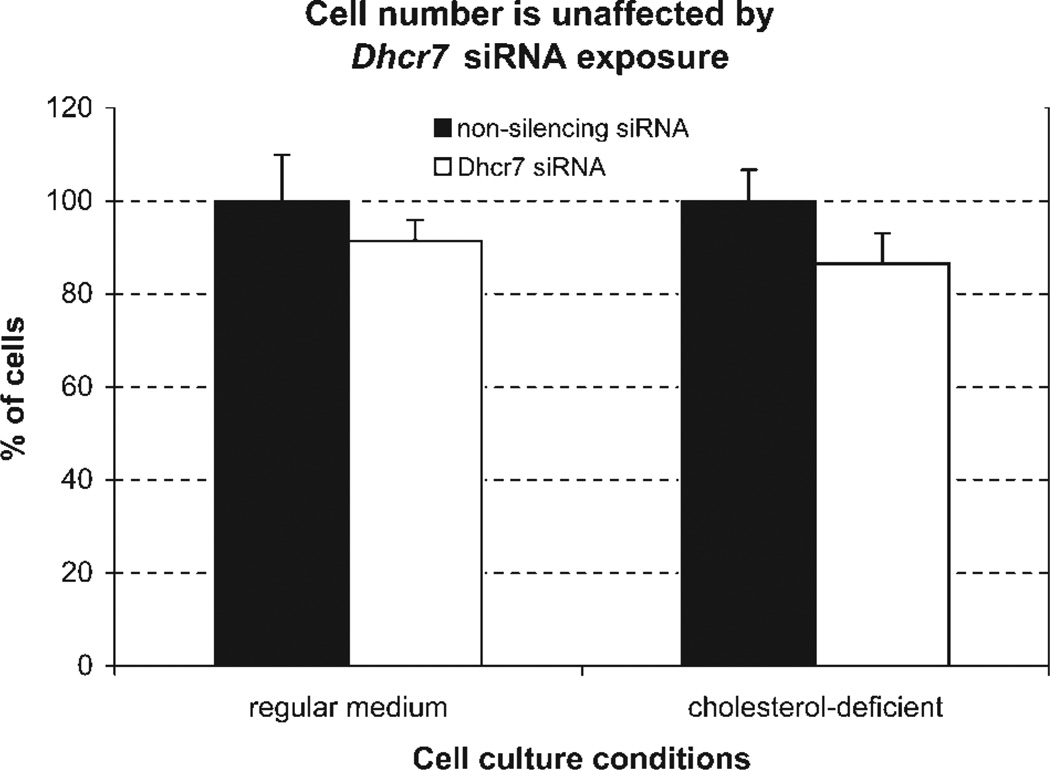
Cell number is unaffected by Dhcr7 siRNA transfection. The total number of cells was counted after 4 days in culture. Cells grown in regular medium proliferated more than cells grown in cholesterol-deficient serum. However, there was no significant difference in the growth between Dhcr7-deficient and control cells regardless of culture media (P = 0.235 in lipid-containing and P = 0.072 in cholesterol-deficient media).
Figure 6.
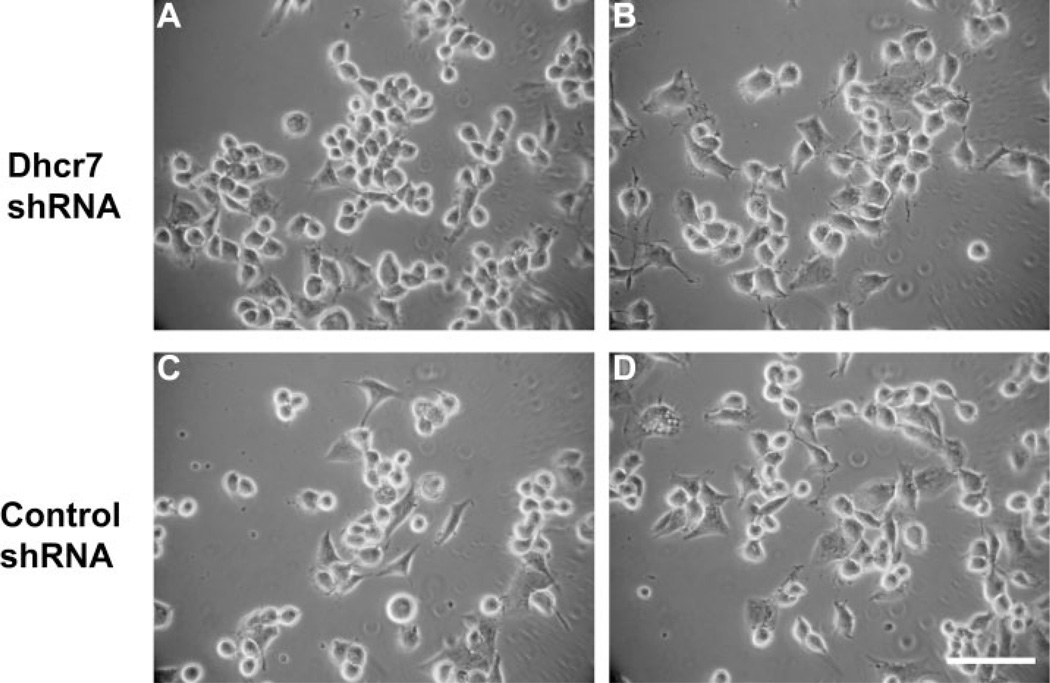
Stable transfection with Dhcr7 shRNA does not affect morphology of Neuro2a cells. A and B show representative phase-contrast micrographs of Neuro2a cells with stable expression of Dhcr7shRNA, whereas C and D depict two different cultures of Neuro2a cells with stable expression of nonsilencing shRNA. Note that the morphologies of the experimental and control cells are comparable. Scale bar = 50 µm.
We tested the expression of the same genes (Dhcr7, Egr1, Adam19, Prr13, Snag1, Snx6, and clone 2010011I20Rik) in the stably transfected Dhcr7-deficient and control Neuro2a cells by qPCR. We found that these transcripts, initially identified in transiently transfected cells, had a similarly altered expression in stably transfected cells (Fig. 7A). Importantly, this result was achieved using two different Dhcr7siRNAs and cell culture models, suggesting that Dhcr7shRNA effects on specific gene transcriptions persist over time.
Figure 7.
Stable transfection with Dhcr7 shRNA leads to expression changes similar to those observed in transiently transfected cells. Genes are plotted on the x-axis, whereas the y-axis denotes the average −ΔΔCt from three independent experiments, two independent reverse transcriptions for each experiment, and four replicates for each reverse transcription. qPCR assays confirmed that Dhcr7 deficiency leads to long- lasting transcriptome changes for cells grown in both regular medium (A) and cholesterol-deficient serum (B). For abbreviations see Figure 3 legend. *P < 0.05.
Exogenous Cholesterol Has No Effect on Dhcr7shRNA-Mediated Expression Changes
Cultured cells can obtain cholesterol from extrinsic sources in the absence of intrinsic cholesterol production. To test whether cholesterol can reverse the observed expression changes, we cultured cells in either regular medium (containing cholesterol in 10% FBS) or medium with cholesterol-deficient serum (cholesterol-deficient 10% FBS). Stably transfected control and Dhcr7-deficient cells did not differ in their growth in the presence of either regular or cholesterol-deficient serum (Figs. 5, 6). Furthermore, the gene expression changes of interest between the Dhcr7-deficient and the control cells were present and were highly correlated in both cholesterol-containing and cholesterol-deficient media (r = 0.98, P < 0.01; Fig. 7B), suggesting that addition of exogenous cholesterol cannot replace the homeostatic need for intrinsic cholesterol biosynthesis.
Dhcr7 Down-Regulation Leads to Transcript Reduction of Genes Involved in Sterol and Fatty Acid Metabolism
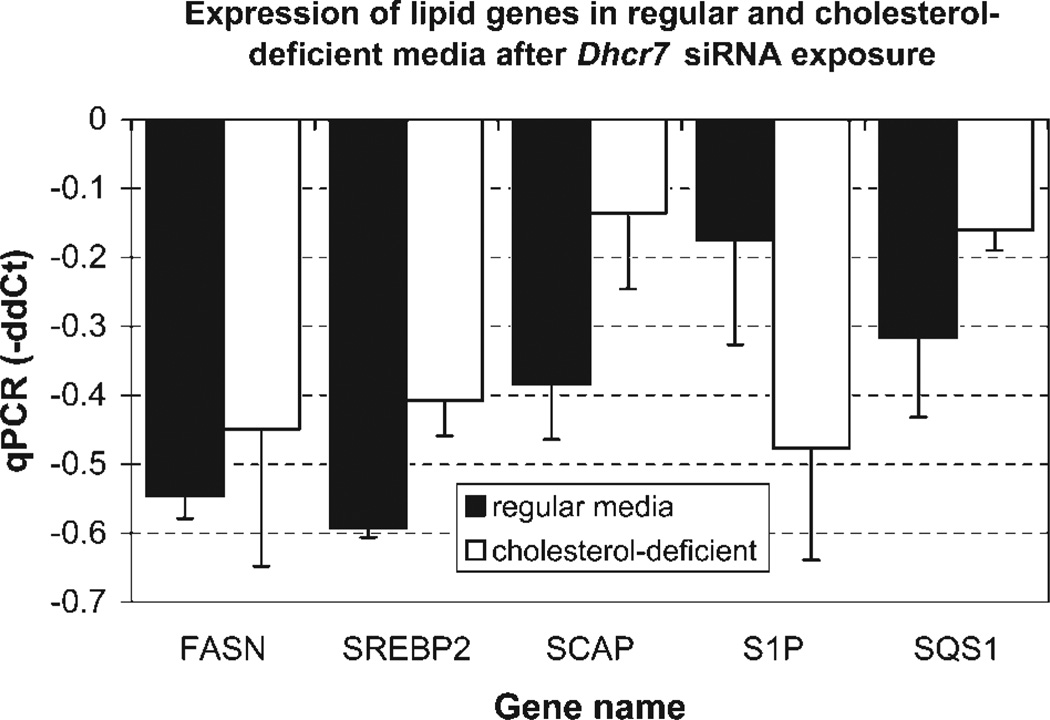
DNA microarrays, although very good at providing leads, often suffer from type II errors (false-negative observations; Mirnics et al., 2006; Mirnics and Pevsner, 2004). Thus, once the initial data are obtained, a hypothesis-driven follow-up is usually beneficial. Sterol-regulatory element binding proteins (SREBPs) are transcription factors that bind to DNA sterol-regulatory element (SRE) sequences that are found in the promoter regions of a number of genes involved in cholesterol and fatty acid biosynthesis (Yokoyama et al., 1993) and regulate their expression and cholesterol homeostasis (Eberle et al., 2004). We hypothesized that down-regulation of Dhcr7 will also lead to significant changes in the expression of multiple SREBP-related genes. Thus, by using qPCR, we compared the expression of fatty acid synthase (FASN), sterol-regulatory element binding protein 2 (SREBP2), SREBF chaperone (SCAP; Korn et al., 1998), site-1 protease (S1P, MBTPS1), and squalene synthase (SQS1) between the cells transfected with Dhcr7shRNA and nonsilencing control shRNA (Supp. Info. Fig. 2). All of these genes showed a significant (P < 0.05) down-regulation (Fig. 8). Again, this was true both in the cholesterol-containing and in the cholesterol-deficient media. This suggests that Dhcr7 expression is a strong regulator of the SREBP system in cells of neuronal origin, and that the endogenous cholesterol production is an important part of neuronal homeostasis.
Figure 8.
Dhcr7 shRNA transfection results in decreased expression of critical lipid biosynthesis genes. The experiment was performed on three + three parallel experimental and control cultures, with eight reactions replicates/sample. ΔΔCt was calculated against Pgk1 as a normalizer and plotted on the y-axis; genes are denoted on the x-axis. Note that in response to Dhcr7 shRNA transfection lipid transcripts showed a significantly reduced expression. The results were concordant in lipid-containing and cholesterol-deficient media. FASN, fatty acid synthase; SREBP2, sterol regulatory element-binding protein 2; SCAP, SREBF chaperone; S1P, MBTPS1, site-1 protease; and SQS1, squalene synthase.
DISCUSSION
The results of our study can be summarized as follows: 1) reduction in Dhcr7 expression leads to strong transcriptome changes in neuroblastoma cells; 2) the critical transcript changes, which included reduction in Dhcr7, Egr1, Adam19, Prr13, Snag1, and clone 2010011I20Rik, were present in Neuro2a cells with both transient and permanent reduction of Dhcr7; 3) the siRNA-induced Dhcr7 reduction in the expression of these genes was observed in both cholesterol-containing and cholesterol-free cell culture media; and 4) Dhcr7 expression is a strong regulator of the SREBP system in cells of neuronal origin, suggesting that the endogenous cholesterol production is an important part of neuronal homeostasis. These findings are also in concordance with the report by Waage-Baudet et al. (2005), whose data suggest a systemic cholesterol biosynthesis disturbance in an in vivo knockout model of Dhcr7. However, insofar as the ablation of the Dhcr7 gene leads to a lethal phenotype, our model system (in addition to mimicking the changes that occur in vivo) provided an easy to use, relevant, and rapid experimental assay allowing comprehensive assessment of the Dhcr7-dependent cellular events.
It has been previously reported that disturbances of Dhcr7 expression lead to altered ratio of 7-DHC and cholesterol (Batta et al., 1995; Tint et al., 1995). In this context, our results raise several interesting questions. Namely, how do the observed expression changes and endogenous cholesterol production influence normal cellular function? We propose, based on our results and previous literature findings, that Dhcr7 depletion in cells of neuronal origin has a simultaneous effect on three cellular mechanisms. First, reduced cholesterol and increased 7-DHC levels may change the function of sterol-sensing domain (SSD)-containing proteins, which will change the expression of SREBP gene family, leading to further downstream expression changes (Kuwabara and Labouesse, 2002). Second, inasmuch as cholesterol constitutes a substrate for synthesis of neurosteroids, oxysterols, and potentially other molecules, reduced levels of cholesterol may impair the biosynthesis of these molecules (Stoffel-Wagner, 2001; Tsutsui et al., 2004; Tsutsui, 2008). Finally, altered cholesterol levels may alter organization of cellular membranes and/or composition of lipid rafts (Keller et al., 2004).
Proteins can bind sterols through their SSD, and some SSD proteins are responsible for sterol-dependent proteolysis in the Golgi and transcriptional regulation of proteins containing an SRE domain (Kuwabara and Labouesse, 2002; Weber et al., 2004). For example, SCAP, through its SSD, regulates transcription of target genes through SREBPs, by either retaining them in the ER or escorting them to the Golgi (Brown and Goldstein, 1999; Brown et al., 2002; Horton et al., 2002; Goldstein et al., 2006). Dhcr7-siRNA transfection introduces an imbalance between the amount of “mature” cholesterol (decreased) and 7-DHC (increased). It is known that different sterols have slightly different effects on specific proteins, putatively altering their structure and function (Song et al., 2005). For example, the cholesterol intermediate lanosterol is more efficient in stimulating the ubiquitination and degradation of Hmgcr than cholesterol (Song et al., 2005), and previous studies led to the proposal that 7-DHC may perturb the function of other SSD-containing proteins in SLOS (Wassif et al., 2002). Interestingly, 7-DHC leads to increased proteolysis of Hmgcr, thus further exacerbating the deficit in SLOS (Fitzky et al., 2001). Finally, fibroblast cultures of patients with CHILD syndrome (an inborn error of the cholesterol biosynthesis pathway) report an accumulation of sterol precursors leading to formation of lipid vacuoles with a lamellar appearance (Hashimoto et al., 1998). In agreement with these findings, the expression changes observed in our experiments also strongly support the notion that 7-DHC accumulation has a critical action on the function of proteins with SSD. We observed reduced transcripts of both SSD-containing proteins (SCAP) and SRE element-containing genes (FASN, SREBP2, S1P-MBTPS1, and SQS1). Therefore, we hypothesize that altered balance between 7-DHC and cholesterol will change the structure and function of SSD-containing protein SCAP, which will change the expression of multiple SREBP gene family members. More specifically, we believe that Srebp2 (one of the most down-regulated genes in our experiment) has a critical role in this process. Srebp2 is a transcription factor, and it controls the transcription of multiple lipid biosynthesis genes (Goldstein et al., 2006). Interestingly, Srebp2 contains three Egr1 binding sites in its promoter region (McMullen et al., 2005). Thus, we propose that Egr1 (another transcription factor with changed expression in the Dhcr7-siRNA treated cells) is likely to be an important upstream regulator of neuronal cholesterol biosynthesis.
Dhcr7 siRNA treatment reduces “mature” cholesterol levels. Because cholesterol is a substrate for biosynthesis of neurosteroids and oxysterols, reduction of cholesterol levels will cause use of 7-DHC for neurosteroid synthesis. Although literature data suggest that 7-DHC can serve as a substrate for neurosteroid synthesis, it is not clear whether the efficacy of the 7-DHC-derived neurosteroid synthesis is comparable to that generated from cholesterol or whether the activities of the 7-DHC-generated neurosteroids are similar to those of “normal” neurosteroids (Marcos et al., 2004). Support for this hypothesis is provided by the observation that, in Niemann-Pick disease, reduced cholesterol levels lead to reduced alopregnanole synthesis (Chen et al., 2007), suggesting that normal cholesterol level likely are required for maintenance of normal CNS neurosteroid levels.
Lipid rafts are a critical place for receptor insertion, neurotrophin signaling, neurotransmitter release, and regulated intramembrane proteolysis of transmembrane proteins (Pike, 2005). Reduced Dhcr7 levels will lead to accumulation of 7-dehydrocholesterol, which is inserted into the rafts instead of cholesterol (Keller et al., 2004). However, because the structures of 7-dehydrocholesterol and cholesterol are different, the structure and composition of the lipid rafts will be altered. Thus, altered insertion of many receptors could change signaling from the cell surface (Suzuki et al., 2004; Freeman et al., 2007). Although our experimental data do not provide direct evidence for such a mechanism, this hypothesis is supported by several critical, literature-based lines of evidence: 1) cholesterol content of rafts affects membrane thickness, elasticity, and curvature (Allende et al., 2004; Bacia et al., 2005); 2) cholesterol content of rafts affects ion conductance and excitability of membranes, trafficking of ionotropic receptors to and from the cell membrane, size and number of some postsynaptic receptor clusters, and neurotransmitter signaling through G-protein coupled receptors (Korade and Kenworthy, 2008); 3) protein activity may be modulated by the composition of lipid rafts (Pike, 2003); and 4) presence of 7-dehydrocholesterol in hippocampal membranes impairs ligand-binding activity of the serotonin 1A receptor (Singh et al., 2007).
Finally, our findings suggest that the role of endogenous cholesterol biosynthesis in neuronal cells deserves equal consideration. The human brain contains as much as 25% of total cholesterol, and the CNS cholesterol pool is regulated independently of the peripheral cholesterol pool (Brown and Goldstein, 1999; Dietschy and Turley, 2001). Although the role, origin, metabolism, and regulation of the CNS cholesterol pool remain understudied and mostly unknown to date (Korade and Kenworthy, 2008), it has been historically assumed that cholesterol of nonneuronal origin is the only important source of cholesterol for the brain cells (Pfrieger, 2003). Recent evidence that 1) cholesterogenic enzymes are coexpressed at high level in neurons across various brain structures, 2) hippocampal and cholinergic neurons coexpress the highest levels of cholesterogenic enzymes, and 3) growth factors regulate cholesterol biosynthesis in a neuroblastoma cell line suggest that, in at least some of the neurons, endogenous neuronal cholesterol synthesis is likely to be an important homeostatic factor (Korade et al., 2007; Suzuki et al., 2007). Our current findings strongly underscore this notion. That the expression changes we observed were present in both cholesterol-containing and cholesterol-free cell culture media strongly argues that endogenous cholesterol biosynthesis is critical for neuronal homeostasis, and extrinsic supplementation of cholesterol cannot reverse the effects of reduced intrinsic neuronal cholesterol biosynthesis. This finding may also contribute to our understanding of the pathophysiological mechanism underlying some of the CNS-related disturbances in SLOS. SLOS, characterized by a range of developmental brain abnormalities, is due to inactivation of the Dchr7 enzyme and a consequent reduction in cholesterol biosynthesis (Jira et al., 2003). Unfortunately, cholesterol supplementation has only a limited therapeutic effect in these patients, and we believe that this is due to the need of some neurons for an endogenous neuronal cholesterol biosynthesis that cannot be supplemented from sources outside the cell (Sikora et al., 2004). Similarly, our findings warrant further follow-up studies on the role of statins (inhibitors of cholesterol biosynthesis) in neuronal cholesterol biosynthesis and their effects on neuronal homeostasis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Charles Asher Day for help with the UV spectrometry assay.
Contract grant sponsor: NIH; Contract grant number: K02 MH070786 (to K.M.); Contract grant number: R01 MH079299 (to K.M.); Contract grant number: R01 GM073846 (to A.K.K.); Contract grant sponsor: VU Kennedy Center for Research on Human Development (to Ž.K.).
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Allende D, Vidal A, McIntosh TJ. Jumping to rafts: gatekeeper role of bilayer elasticity. Trends Biochem Sci. 2004;29:325–330. doi: 10.1016/j.tibs.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Anitei M, Pfeiffer SE. Myelin biogenesis: sorting out protein trafficking. Curr Biol. 2006;16:R418–R421. doi: 10.1016/j.cub.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Bacia K, Schwille P, Kurzchalia T. Sterol structure determines the separation of phases and the curvature of the liquid-ordered phase in model membranes. Proc Natl. Acad Sci U S A. 2005;102:3272–3277. doi: 10.1073/pnas.0408215102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae SH, Lee JN, Fitzky BU, Seong J, Paik YK. Cholesterol biosynthesis from lanosterol. Molecular cloning, tissue distribution, expression, chromosomal localization, and regulation of rat 7-dehydrocholesterol reductase, a Smith-Lemli-Opitz syndrome-related protein. J. Biol Chem. 1999;274:14624–14631. doi: 10.1074/jbc.274.21.14624. [DOI] [PubMed] [Google Scholar]
- Batta AK, Salen G, Tint GS, Shefer S. Identification of 19-nor-5,7,9(10)-cholestatrien-3 beta-ol in patients with Smith-Lemli-Opitz syndrome. J Lipid. Res. 1995;36:2413–2418. [PubMed] [Google Scholar]
- Brown AJ, Sun L, Feramisco JD, Brown MS, Goldstein JL. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol Cell. 2002;10:237–245. doi: 10.1016/s1097-2765(02)00591-9. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci U S A. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Li HM, Chen YR, Gu XS, Duan S. Decreased estradiol release from astrocytes contributes to the neurodegeneration in a mouse model of Niemann-Pick disease type C. Glia. 2007;55:1509–1518. doi: 10.1002/glia.20563. [DOI] [PubMed] [Google Scholar]
- Dietschy JM, Turley SD. Cholesterol metabolism in the brain. Curr Opin Lipidol. 2001;12:105–112. doi: 10.1097/00041433-200104000-00003. [DOI] [PubMed] [Google Scholar]
- Eberle D, Hegarty B, Bossard P, Ferre P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Fernandez-Alvarez A, Tur G, Lopez-Rodas G, Casado M. Reciprocal regulation of the human sterol regulatory element binding protein (SREBP)-1a promoter by Sp1 and EGR-1 transcription factors. FEBS Lett. 2008;582:177–184. doi: 10.1016/j.febslet.2007.11.083. [DOI] [PubMed] [Google Scholar]
- Fitzky BU, Moebius FF, Asaoka H, Waage-Baudet H, Xu L, Xu G, Maeda N, Kluckman K, Hiller S, Yu H, Batta AK, Shefer S, Chen T, Salen G, Sulik K, Simoni RD, Ness GC, Glossmann H, Patel SB, Tint GS. 7-Dehydrocholesterol-dependent proteolysis of HMG-CoA reductase suppresses sterol biosynthesis in a mouse model of Smith-Lemli-Opitz/RSH syndrome. J Clin Invest. 2001;108:905–915. doi: 10.1172/JCI12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MR, Cinar B, Kim J, Mukhopadhyay NK, Di Vizio D, Adam RM, Solomon KR. Transit of hormonal and EGF receptor-dependent signals through cholesterol-rich membranes. Steroids. 2007;72:210–217. doi: 10.1016/j.steroids.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Prada S, Lopez AP, Hoyos JG, Escobar M. CHILD syndrome with linear eruptions, hypopigmented bands, and verruciform xanthoma. Pediatr Dermatol. 1998;15:360–366. doi: 10.1046/j.1525-1470.1998.1998015360.x. [DOI] [PubMed] [Google Scholar]
- Honda A, Batta AK, Salen G, Tint GS, Chen TS, Shefer S. Screening for abnormal cholesterol biosynthesis in the Smith-Lemli-Opitz syndrome: rapid determination of plasma 7-dehydrocholesterol by ultraviolet spectrometry. Am J Med Genet. 1997;68:288–293. doi: 10.1002/(sici)1096-8628(19970131)68:3<288::aid-ajmg8>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba K, Albrechtsen R, Gilpin B, Frohlich C, Loechel F, Zolkiewska A, Ishiguro K, Kojima T, Liu W, Langford JK, Sanderson RD, Brakebusch C, Fassler R, Wewer UM. The cysteine-rich domain of human ADAM 12 supports cell adhesion through syndecans and triggers signaling events that lead to beta1 integrin-dependent cell spreading. J. Cell Biol. 2000;149:1143–1156. doi: 10.1083/jcb.149.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jira PE, Waterham HR, Wanders RJ, Smeitink JA, Sengers RC, Wevers RA. Smith-Lemli-Opitz syndrome and the DHCR7 gene. Ann Hum Genet. 2003;67:269–280. doi: 10.1046/j.1469-1809.2003.00034.x. [DOI] [PubMed] [Google Scholar]
- Jurevics H, Morell P. Cholesterol for synthesis of myelin is made locally, not imported into brain. J Neurochem. 1995;64:895–901. doi: 10.1046/j.1471-4159.1995.64020895.x. [DOI] [PubMed] [Google Scholar]
- Jurevics HA, Kidwai FZ, Morell P. Sources of cholesterol during development of the rat fetus and fetal organs. J Lipid Res. 1997;38:723–733. [PubMed] [Google Scholar]
- Keller RK, Arnold TP, Fliesler SJ. Formation of 7-dehydrocholesterol-containing membrane rafts in vitro and in vivo, with relevance to the Smith-Lemli-Opitz syndrome. J Lipid Res. 2004;45:347–355. doi: 10.1194/jlr.M300232-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korade Z, Kenworthy A. Lipid rafts, cholesterol, and the brain. Neuropharmacology. 2008 doi: 10.1016/j.neuropharm.2008.02.019. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korade Z, Mi Z, Portugal C, Schor NF. Expression and p75 neurotrophin receptor dependence of cholesterol synthetic enzymes in adult mouse brain. Neurobiol Aging. 2007;28:1522–1531. doi: 10.1016/j.neurobiolaging.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Korn BS, Shimomura I, Bashmakov Y, Hammer RE, Horton JD, Goldstein JL, Brown MS. Blunted feedback suppression of SREBP processing by dietary cholesterol in transgenic mice expressing sterol-resistant SCAP(D443N) J Clin Invest. 1998;102:2050–2060. doi: 10.1172/JCI5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurrasch DM, Huang J, Wilkie TM, Repa JJ. Quantitative real-time polymerase chain reaction measurement of regulators of G-protein signaling mRNA levels in mouse tissues. Methods Enzymol. 2004;389:3–15. doi: 10.1016/S0076-6879(04)89001-3. [DOI] [PubMed] [Google Scholar]
- Kuwabara PE, Labouesse M. The sterol-sensing domain: multiple families, a unique role? Trends Genet. 2002;18:193–201. doi: 10.1016/s0168-9525(02)02640-9. [DOI] [PubMed] [Google Scholar]
- Luskey KL, Stevens B. Human 3-hydroxy-3-methylglutaryl coenzyme A reductase. Conserved domains responsible for catalytic activity and sterol-regulated degradation. J Biol Chem. 1985;260:10271–10277. [PubMed] [Google Scholar]
- Lutton C. Dietary cholesterol, membrane cholesterol and cholesterol synthesis. Biochimie. 1991;73:1327–1334. doi: 10.1016/0300-9084(91)90097-k. [DOI] [PubMed] [Google Scholar]
- Marcos J, Guo LW, Wilson WK, Porter FD, Shackleton C. The implications of 7-dehydrosterol-7-reductase deficiency (Smith-Lemli-Opitz syndrome) to neurosteroid production. Steroids. 2004;69:51–60. doi: 10.1016/j.steroids.2003.09.013. [DOI] [PubMed] [Google Scholar]
- Maxfield FR, Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438:612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- McMullen MR, Pritchard MT, Wang Q, Millward CA, Croniger CM, Nagy LE. Early growth response-1 transcription factor is essential for ethanol-induced fatty liver injury in mice. Gastroenterology. 2005;128:2066–2076. doi: 10.1053/j.gastro.2005.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbrandt J. A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science. 1987;238:797–799. doi: 10.1126/science.3672127. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Pevsner J. Progress in the use of microarray technology to study the neurobiology of disease. Nat Neurosci. 2004;7:434–439. doi: 10.1038/nn1230. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Levitt P, Lewis DA. Critical appraisal of DNA microarrays in psychiatric genomics. Biol Psychiatry. 2006;60:163–176. doi: 10.1016/j.biopsych.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Mochizuki S, Okada Y. ADAMs in cancer cell proliferation and progression. Cancer Sci. 2007;98:621–628. doi: 10.1111/j.1349-7006.2007.00434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nes WR. A comparison of methods for the identification of sterols. Methods Enzymol. 1985;111:3–37. doi: 10.1016/s0076-6879(85)11003-7. [DOI] [PubMed] [Google Scholar]
- Paul CA, Boegle AK, Maue RA. Before the loss: neuronal dysfunction in Niemann-Pick Type C disease. Biochim Biophys Acta. 2004;1685:63–76. doi: 10.1016/j.bbalip.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Pfrieger FW. Outsourcing in the brain: do neurons depend on cholesterol delivery by astrocytes? Bioessays. 2003;25:72–78. doi: 10.1002/bies.10195. [DOI] [PubMed] [Google Scholar]
- Pike LJ. Lipid rafts: bringing order to chaos. J Lipid Res. 2003;44:655–667. doi: 10.1194/jlr.R200021-JLR200. [DOI] [PubMed] [Google Scholar]
- Pike LJ. Growth factor receptors, lipid rafts and caveolae: an evolving story. Biochim Biophys Acta. 2005;1746:260–273. doi: 10.1016/j.bbamcr.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Rocks N, Paulissen G, El Hour M, Quesada F, Crahay C, Gueders M, Foidart JM, Noel A, Cataldo D. Emerging roles of ADAM and ADAMTS metalloproteinases in cancer. Biochimie. 2008;90:369–379. doi: 10.1016/j.biochi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Saher G, Brugger B, Lappe-Siefke C, Mobius W, Tozawa R, Wehr MC, Wieland F, Ishibashi S, Nave KA. High cholesterol level is essential for myelin membrane growth. Nat Neurosci. 2005;8:468–475. doi: 10.1038/nn1426. [DOI] [PubMed] [Google Scholar]
- Sikora DM, Ruggiero M, Petit-Kekel K, Merkens LS, Connor WE, Steiner RD. Cholesterol supplementation does not improve developmental progress in Smith-Lemli-Opitz syndrome. J Pediatr. 2004;144:783–791. doi: 10.1016/j.jpeds.2004.02.036. [DOI] [PubMed] [Google Scholar]
- Silverman ES, Collins T. Pathways of Egr-1-mediated gene transcription in vascular biology. Am J Pathol. 1999;154:665–670. doi: 10.1016/S0002-9440(10)65312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Paila YD, Chattopadhyay A. Differential effects of cholesterol and 7-dehydrocholesterol on the ligand binding activity of the hippocampal serotonin(1A) receptor: implications in SLOS. Biochem Biophys Res Commun. 2007;358:495–499. doi: 10.1016/j.bbrc.2007.04.135. [DOI] [PubMed] [Google Scholar]
- Smith DW, Lemli L, Opitz JM. A newly recognized syndrome of multiple congenital anomalies. J Pediatr. 1964;64:210–217. doi: 10.1016/s0022-3476(64)80264-x. [DOI] [PubMed] [Google Scholar]
- Song BL, Javitt NB, DeBose-Boyd RA. Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 2005;1:179–189. doi: 10.1016/j.cmet.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Stoffel-Wagner B. Neurosteroid metabolism in the human brain. Eur J Endocrinol. 2001;145:669–679. doi: 10.1530/eje.0.1450669. [DOI] [PubMed] [Google Scholar]
- Strandberg TE, Tilvis RS. Physiological and pharmacological regulation of small intestinal cholesterol synthesis. Gen Pharmacol. 1988;19:321–329. doi: 10.1016/0306-3623(88)90024-9. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Numakawa T, Shimazu K, Koshimizu H, Hara T, Hatanaka H, Mei L, Lu B, Kojima M. BDNF-induced recruitment of TrkB receptor into neuronal lipid rafts: roles in synaptic modulation. J Cell Biol. 2004;167:1205–1215. doi: 10.1083/jcb.200404106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Kiyosue K, Hazama S, Ogura A, Kashihara M, Hara T, Koshimizu H, Kojima M. Brain-derived neurotrophic factor regulates cholesterol metabolism for synapse development. J Neurosci. 2007;27:6417–6427. doi: 10.1523/JNEUROSCI.0690-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellier E, Canault M, Rebsomen L, Bonardo B, Juhan-Vague I, Nalbone G, Peiretti F. The shedding activity of ADAM17 is sequestered in lipid rafts. Exp Cell Res. 2006;312:3969–3980. doi: 10.1016/j.yexcr.2006.08.027. [DOI] [PubMed] [Google Scholar]
- Tellier E, Canault M, Poggi M, Bonardo B, Nicolay A, Alessi MC, Nalbone G, Peiretti F. HDLs activate ADAM17-dependent shedding. J Cell Physiol. 2008;214:687–693. doi: 10.1002/jcp.21265. [DOI] [PubMed] [Google Scholar]
- Tierney E, Nwokoro NA, Kelley RI. Behavioral phenotype of RSH/Smith-Lemli-Opitz syndrome. Ment Retard Dev Disabil Res Rev. 2000;6:131–134. doi: 10.1002/1098-2779(2000)6:2<131::AID-MRDD7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Tint GS, Seller M, Hughes-Benzie R, Batta AK, Shefer S, Genest D, Irons M, Elias E, Salen G. Markedly increased tissue concentrations of 7-dehydrocholesterol combined with low levels of cholesterol are characteristic of the Smith-Lemli-Opitz syndrome. J Lipid Res. 1995;36:89–95. [PubMed] [Google Scholar]
- Tsutsui K. Progesterone biosynthesis and action in the developing neuron. Endocrinology. 2008 doi: 10.1210/en.2007-1592. (in press). [DOI] [PubMed] [Google Scholar]
- Tsutsui K, Sakamoto H, Shikimi H, Ukena K. Organizing actions of neurosteroids in the Purkinje neuron. Neurosci Res. 2004;49:273–279. doi: 10.1016/j.neures.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Waage-Baudet H, Dunty WC, Jr, Dehart DB, Hiller S, Sulik KK. Immunohistochemical and microarray analyses of a mouse model for the Smith-Lemli-Opitz syndrome. Dev Neurosci. 2005;27:378–396. doi: 10.1159/000088453. [DOI] [PubMed] [Google Scholar]
- Wang L, Schuster GU, Hultenby K, Zhang Q, Andersson S, Gustafsson JA. Liver X receptors in the central nervous system: from lipid homeostasis to neuronal degeneration. Proc Natl Acad Sci U S A. 2002;99:13878–13883. doi: 10.1073/pnas.172510899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- Wassif CA, Vied D, Tsokos M, Connor WE, Steiner RD, Porter FD. Cholesterol storage defect in RSH/Smith-Lemli-Opitz syndrome fibroblasts. Mol Genet Metab. 2002;75:325–334. doi: 10.1016/S1096-7192(02)00010-0. [DOI] [PubMed] [Google Scholar]
- Weber LW, Boll M, Stampfl A. Maintaining cholesterol homeostasis: sterol regulatory element-binding proteins. World J Gastroenterol. 2004;10:3081–3087. doi: 10.3748/wjg.v10.i21.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama C, Wang X, Briggs MR, Admon A, Wu J, Hua X, Goldstein JL, Brown MS. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.