Abstract
A simple, point of care, inexpensive, disposable cassette for the detection of nucleic acids extracted from pathogens was designed, constructed, and tested. The cassette utilizes a single reaction chamber for isothermal amplification of nucleic acids. The chamber is equipped with an integrated, flow-through, Flinders Technology Associates (Whatman FTA®) membrane for the isolation, concentration, and purification of DNA and/or RNA. The nucleic acids captured by the membrane are used directly as templates for amplification without elution, thus simplifying the cassette’s flow control. The FTA membrane also serves another critical role—enabling the removal of inhibitors that dramatically reduce detection sensitivity. Thermal control is provided with a thin film heater external to the cassette. The amplification process was monitored in real time with a portable, compact fluorescent reader. The utility of the integrated, single-chamber cassette was demonstrated by detecting the presence of HIV-1 in oral fluids. The HIV RNA was reverse transcribed and subjected to loop-mediated, isothermal amplification (LAMP). A detection limit of less than 10 HIV particles was demonstrated. The cassette is particularly suitable for resource poor regions, where funds and trained personnel are in short supply. The cassette can be readily modified to detect nucleic acids associated with other pathogens borne in saliva, urine, and other body fluids as well as in water and food.
Introduction
Despite global efforts to control the acquired immune deficiency syndrome (AIDS) pandemic, the human immunodeficiency virus (HIV) infection continues to spread relatively unabated in many parts of the world. The diagnosis of HIV infection at the point-of-care and in resource-poor settings poses considerable challenges due to the time delay between sample collection and diagnosis. The lack of a rapid, confirmed diagnosis leaves many individuals unaware of their condition and impedes tracking of patients by health providers.1–4 Hence, a disposable, low-cost, portable, integrated diagnostic device that can perform rapid nucleic acid testing (NAT) at the point-of-care for early detection of HIV infection is highly desirable. Although immunoassays provide rapid detection, they are unable to detect the disease during the seroconversion window when infected individuals are most contagious but lack detectable anti-HIV antibodies.5–7 In contrast, nucleic acid-based tests, which amplify a specific DNA and/or RNA target, provide high sensitivity and facilitate detection during the period of infection prior to seroconversion.8 Identification of early infection may facilitate early intervention, which, in turn, may reduce disease transmission.
Early detection of the HIV virus is frequently carried out using polymerase chain reaction (PCR).3,9,10 However, conventional PCR amplification requires a thermal cycling process, which is expensive, complex, and time consuming. The relatively newly developed Loop-mediated, isothermalAmplification (LAMP)11–14 offers an attractive alternative since the reaction takes place at a constant temperature (60–65 °C), is relatively rapid, and highly sensitive. Recently, Curtis et al.15,16 designed a set of six RT-LAMP primers and successfully detected HIV in plasma and untreated blood samples with a detection limit of better than 100 HIV-1 virions per sample. Additionally, Curtis et al.16 also developed specific fluorescent reporters for real time detection of reaction products.
In recent years, there has been a major push to integrate entire analytical/clinical chemistry processes into monolithic platforms for point-of-care tests.17,18 Compared to conventional laboratory methods, integrated microfluidic platforms offer potential advantages such as lower cost, higher speed, smaller sample and reagent volumes, and, most importantly, integrated automation of all processes from sample preparation to detection in an inexpensive portable device. The ‘‘sample-to-answer’’ concept is particularly suitable for resource-poor regions, where funds and trained personnel are in short supply.19–23 For example, Lee et al. report on a portable, point-of-care, polymer lab-on-a-chip, which integrates RT-PCR of HIV RNA with chemiluminescence-based detection.22 Wang et al. describe a polycarbonate-based, microfluidic cassette that combines on-chip PCR amplification with lateral flow (LF) detection for detecting B. cereus DNA in oral fluids.23 However, these systems do not include on-chip, nucleic acid purification and extraction steps, which limit their utility to laboratory settings.8
There are just a few reports on fully integrated microfluidic NAT chips that perform all the necessary steps from sample introduction to target detection.24–26 Easley et al. demonstrate a sample-to-answer, genetic analysis system, which integrates nucleic acid purification, PCR amplification, and electrophoresis-based detection.24 Chen et al. describe an integrated, silica membrane-based, microfluidic cassette for isolation, PCR amplification, and lateral flow detection of nucleic acids.25 The latter was further improved by incorporating on-chip reagent storage and pouches for liquid dispensing to obtain a fully self-contained, portable microfluidic cassette for HIV detection in oral fluids.26 However, all the above-mentioned, integrated, microfluidic NAT chips consist of separate, interconnected modules for nucleic acid isolation, PCR amplification, and nucleic acid detection, which necessitate the transfer of fluids from one reaction chamber to another, increase chip complexity, and complicate flow control.
In an effort to simplify system operation, a few researchers have integrated nucleic acid isolation, amplification, and detection into a single reaction chamber. For example, Lee et al. used a Laser-Irradiated Magnetic Bead System (LIMBS) for DNA extraction and real-time PCR detection in a single chamber.27 More recently, Kim et al. integrated an alumina membrane in the PCR reactor and demonstrated the feasibility of isolating and amplifying nucleic acids in a single chamber.28 The alumina membrane is, however, fragile and requires special handling.
Heretofore, most researchers have focused on detecting pathogens and antibodies in blood or plasma. Oral fluids offer, however, an attractive alternative. In many cases, oral fluids contain the same pathogens and proteins as blood, although sometimes at lower concentrations.29–33 Nevertheless, oral fluids offer a few advantages for disease diagnosis over blood.29,30 (i) Oral fluid can be collected non-invasively by individuals with little training and without a need for special equipment. (ii) It is easier to collect oral fluid samples from children and the elderly than blood samples. (iii) The collection of oral fluids is subject to fewer regulations compared with the collection of blood. (iv) Oral fluid collection reduces the risk of infection to the health care worker who collects the sample. However, since the oral fluid is a complex mixture of saliva secreted by parotid and other salivary glands, gingival cervicular fluid from the gingival crevice, and secretions from the mucous membranes, the amplification of nucleic acids in oral fluid is challenging.34
Here, we report on a simple, disposable, single-chamber, microfluidic cassette, which integrates the functional steps of viral nucleic acid capture, concentration, and purification; isothermal amplification; and real-time fluorescence detection into one chamber. We incorporated a Flinders Technology Associates (Whatman FTA®) membrane into the amplification chamber. We use the membrane in filtration mode, which is somewhat unusual and which allow us to accommodate large sample volumes and detect low-concentration targets. In addition to facilitating isolation and concentration of nucleic acids, the membrane plays a critical role in removing amplification inhibitors from oral fluids, which would have otherwise greatly compromised amplification efficiency. To further simplify cassette operation, the extracted HIV viral RNA is directly amplified in the isolation chamber without a separate step for the elution of the immobilized nucleic acids. The amplification process is monitored in real time with a compact, portable fluorescence reader. The utility of this integrated LAMP cassette was demonstrated by detecting HIV in oral fluids with a sensitivity of 10 HIV virus particles per sample. This is the first report describing the integration of a solid state membrane in a LAMP reactor and the successful isothermal amplification of pathogens in saliva without any elution process.
Experimental
Materials
The HIV virus with a known concentration (which was confirmed by an independent laboratory) was purchased from ABI Advanced Biotechnologies (MD, USA). Oral fluid samples were formulated by spiking HIV-1 MN strains of various viral loads into whole mouth saliva (WMSS) obtained from healthy, consenting volunteers. For safety reasons, the HIV saliva samples were inactivated with a binding/lysis buffer (Roche Diagnostic, Indiana, USA) in a biosafety facility. The High Purity™ viral RNA Kit, which includes binding/lysis buffer, inhibitor removal buffer, and wash buffer, was provided by Roche Diagnostic (Indiana, USA). The Loopamp RNA amplification kit (RT-LAMP) was obtained from Eiken Chemical Co. Ltd. (Tochigi, Japan). SYTO-9 Green DNA binding dye was obtained from Invitrogen. Acetonitrile, ethanol, and Tris–acetate–EDTA (TAE) buffer (10×) were purchased from Sigma Aldrich and used without further purification. The FTA card was obtained from Whatman (Florham Park, NJ). A mobicol spin mini-column was purchased from MoBiTec (Göttingen, Germany). A 0.118 inch thick poly(methyl methacrylate) (PMMA) sheet and a 0.01 inch thick, PMMA film were, respectively, supplied by McMaster-Carr and Cyro Industries.
Design and fabrication of the integrated LAMP cassette
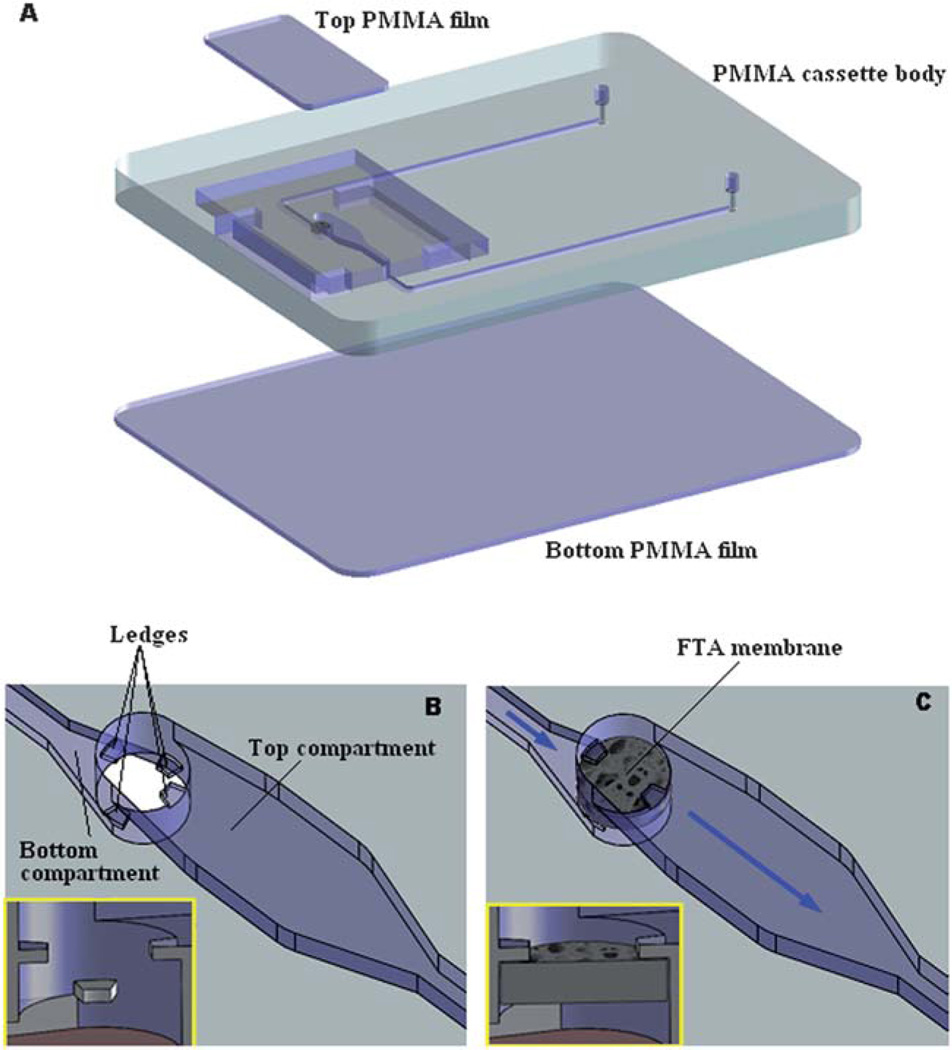
The nucleic acid amplification cassette is shown in Fig. 1. Fig. 1A is an exploded view of the cassette. The 46 mm × 36 mm × 3.50 mm cassette consists of three layers: a top made of 250 µm (0.01 inch) thick, poly(methyl methacrylate) (PMMA) film; a 3 mm (0.118 inch) thick, PMMA cassette body; and a 250 µm (0.01 inch) thick, PMMA film bottom. Both the top and bottom cover films were cut with a CO2 laser (Universal Laser Systems). The cassette body was milled with a precision, computer-controlled (CNC) milling machine (HAAS Automation Inc.) to form the reaction chamber, membrane support, and access conduits.35 The various layers were solvent-bonded with acetonitrile at room temperature.36 Residual solvent was removed by overnight heating at 50 °C.
Fig. 1.
The integrated LAMP cassette. (A) Exploded view. The cassette consists of three solvent-bonded layers of PMMA. The cassette’s features were milled in the main body of the cassette. (B) An amplified view of the reaction chamber without the FTA membrane. Two sets of protruding ledges were machined on the top and bottom of the LAMP chamber. (C) An amplified view of the reaction chamber with the installed FTA membrane, which separates the reaction chamber into a top main compartment and a bottom compartment.
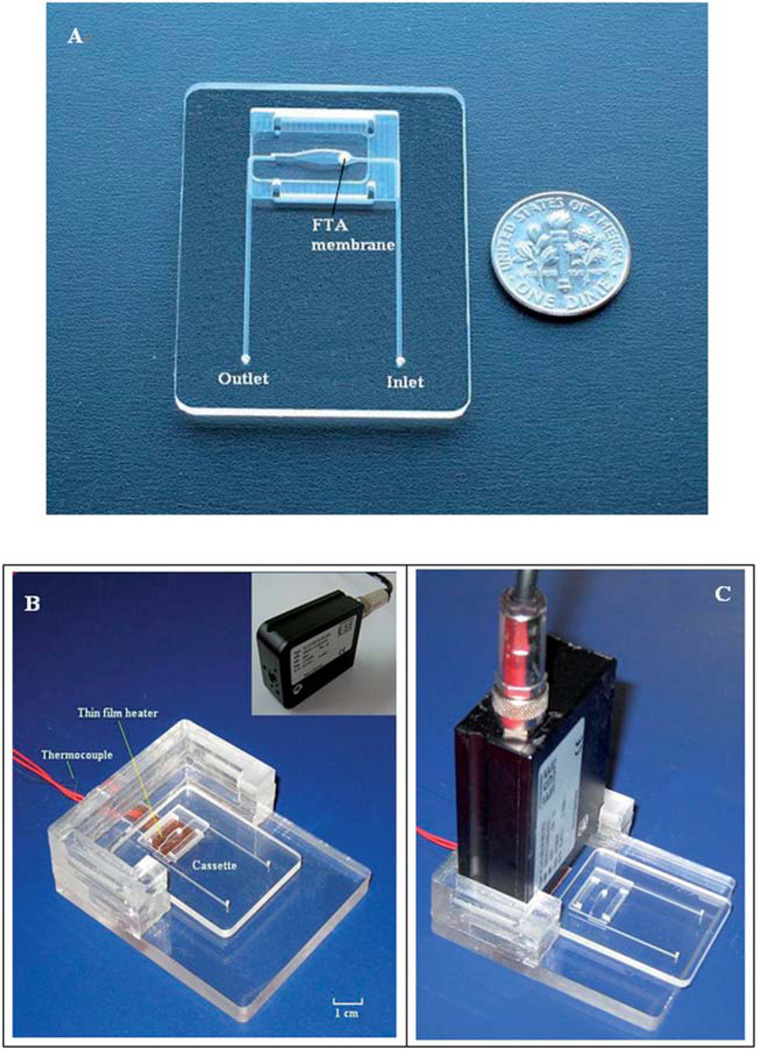
The reaction chamber was connected to the inlet and exit ports with 500 µm wide × 200 µm deep conduits. The FTA membrane disc was installed at the interface between the inlet port and the reaction chamber. To support the 500 µm thick, FTA membrane, two sets of protrusions (ledges) were machined at the bottom and top of the reaction chamber (Fig. 1B and inset). The ledges are shown in detail in the inset of Fig. 1B. The ledges protrude 500 µm from the bore surface and are 300 µm wide and 200 µm tall. The vertical distance between the lower and upper ledges is 500 µm. Fig. 1C depicts schematically an enlarged view of the reaction chamber with the installed FTA membrane. A detailed view of the membrane fixed in its supporting ledges is shown in the inset of Fig. 1C. The membrane separates the reaction chamber into a top, main compartment and a bottom compartment. The top compartment is 10.0 mm in length, 2.0 mm in width, and 1.0 mm in height. The bottom compartment is 2.5 mm in length, 1.5 mm in width, and 0.02 mm in height. The total volume of the reaction chamber is ~20 µl. The FTA membrane, which is installed between the cassette’s inlet and the reaction chamber, acts as a filter for nucleic acid purification, and blocks the introduction of air bubbles into the reaction chamber. Fig. 2A is a photograph of the completed cassette with the integrated FTA membrane.
Fig. 2.
Photographs of the microfluidic cassette and the experimental setup for real-time detection of the LAMP products. (A) A photograph of the single-chamber, microfluidic cassette with an integrated FTA membrane. (B) The cassette holder is equipped with a thin film heater, a thermocouple, and a seat for the detector. The fluorescent signal is excited and detected with the portable, compact, ESE optical detector shown in the inset. The cassette is located in its working position. (C) The optical reader is inserted in the cassette holder. The cassette is in the process of being slipped into its working position under the detector.
Operation of the integrated LAMP cassette
100 µl of the saliva sample (deactivated with lysis buffer) was pipetted into the cassette through the inlet port. The lysed saliva sample filtered through the FTA membrane, which wholly intercepted the flow path to bind nucleic acids contained in the saliva sample. In other words, the membrane isolated and concentrated the RNA molecules from the saliva sample. Next, 100 µl of Roche inhibitor removal buffer was pipetted into the cassette to remove any amplification inhibitors that may have been present in the saliva. Then, the FTA membrane was washed twice with 200 µl of wash buffer, followed by air-drying for 30 seconds. Next, 22 µl of LAMP master mixture, which contained all the reagents necessary for the RT-LAMP and fluorescent dye (SYTO® 9 Green), was injected into the reaction chamber through the inlet port. Subsequently, the inlet and outlet ports were sealed using transparent tape (Scotch tape, 3 M, St Paul, MN) to minimize evaporation during the amplification process.
Real-time RT-LAMP for HIV detection
The LAMP primers were designed by Curtis et al.15,16 at the Center for Disease Control and Prevention (CDC) and provided to New York University through a Material Transfer Agreement. The six RT-LAMP primers15,16 have been designed against the highly conserved sequences located within the p24 gene region. The primers and their respective concentrations were: outer primer F3 5′-ATTATCAGAAGGAGCCACC-3′ (0.2 µM), outer primer B3 5′-CATCCTATTTGTTCCTGAAGG-3′ (0.2 µM), loop primer F loop 5′-TTTAACATTTGCATGGCTGCTTGAT-3′ (0.8 µM), loop primer B loop 5′-GAGATCCAAGGGGAAGTGA-3′ (0.8 µM), inner primer BIP 5′-TGTTGCACCAGGCCAGATAATTTTGTACTGGTAGTTCCTGCTATG-3′ (1.6 µM) and inner primer FIP 5′-CAGCTTCCTCATTGATGGTTTCTTTTTAACACCATGCTAAACACAGT-3′ (1.6 µM).
The LAMP master reaction mix also contained 20 mM Tris–HCl (pH 8.8), 10 mM KCl, 10 mM (NH2)SO4, 8 mM MgSO4, 0.1% Tween 20, 0.8 M betaine, 8U Bst DNA polymerase (New England Biolabs, Inc., MA), 0.625U AMV reverse transcriptase (Invitrogen, Carlsbad, CA), 1.4 mM dNTPs, and 4.0 µM SYTO® 9 Green (Molecular Probes, Inc., Eugene, OR).
In addition to real-time fluorescence detection, the LAMP reaction products were subjected to gel electrophoresis. 5 µl of each LAMP-amplified product was loaded onto a lane of a 2.0% agarose gel. Electrophoresis of the amplified DNA fragment was carried out in 1× TAE buffer (Tris–acetate–EDTA) at a constant voltage of 115 V. DNA molecular mass markers (Roche Diagnostic, Indianapolis, Indiana, USA) were used to estimate the sizes of the various amplified products. The gel was stained with ethidium bromide and visualized with UV illumination.
Portable, real-time LAMP apparatus
The experimental setup for the integrated LAMP cassette is shown in Fig. 2B and C. The system consists of a cassette holder equipped with a flexible, polyimide-based, thin film heater (Model HK5572R7.5L23A, Minco Products, Inc., Minneapolis, MN), a thermocouple located at the interface between the heater and the cassette, and a portable, compact optical detector. The fluorescence excitation and detection were carried out with a minute, portable ESE optical detection system (Fluo Sens SD 003, ESE GmbH, Stockach, Germany). The ESE optical detector consists of a 470 nm, light-emitting diode as the excitation light source and a low-noise, Si-photodiode for fluorescence detection. The detector was interfaced with a computer through a USB interface. A software program was written to display a graph of the fluorescence intensity as a function of time. Fig. 2B shows the cassette holder with the cassette and Fig. 2C shows the cassette holder with the reader in place and the cassette prior to its insertion into the cassette holder.
When the integrated LAMP cassette, filled with LAMP master mixture, was inserted into the cassette holder, the reaction chamber formed a thermal contact with the thin film heater positioned in the cassette holder. In our experiments, the heater was powered with a DC power supply (Model 1611, B&K Precision Corporation, CA) and the reaction chamber’s temperature was controlled in an open-loop mode. To calibrate the device, we constructed a mock cassette, filled the reaction chamber with water, and inserted a type-K thermocouple (Omega Engr., each wire 75 µm in diameter, and a junction diameter of ~170 µm) into the chamber. The thermocouple reading was monitored with a HH506RA multilogger thermometer (Omega Engr., Stamford, CT, USA) and correlated with the power input.
To further determine the reaction’s specificity, we determined the amplicon’s melting curve. A custom software program directed the power supply to gradually increase the amplification chamber’s temperature from 55 °C to 90 °C. Both the chamber’s temperature and the fluorescent signal intensity were continuously recorded and the fluorescent intensity was displayed as a function of the reaction chamber’s temperature.
Benchtop LAMP experiments
Prior to testing the isothermal amplification in our cassettes, we carried out a sequence of benchtop studies. In the first set of experiments (without filtration and purification), the saliva samples were directly mixed with the RT-LAMP master mixture in a PCR vial, and then incubated at 60 °C for 60 min followed by 5 min at 80 °C to inactivate the polymerase activity in the thermal cycler (PTC-220 DNA Engine Dyad™ Peltier Thermal Cycler).
In the second set of experiments, we tested the utility of a FTA membrane for isolating RNA from saliva and removing substances that inhibit enzymatic amplification. To this end, we fitted a FTA membrane into a Mobicol spin mini-column. The 100 µl of spiked saliva samples were spun through the membrane at 14 000 rpm for 30 s, followed by 100 µl of inhibitor removal buffer, and additional centrifugation (10 000 rpm, 30 s). The membrane was then washed twice with 200 µl wash-buffer. Each wash was followed by centrifugation (10 000 rpm, 30 s). The column was then spun for one minute to dry the membrane at 14 000 rpm. Next, the FTA membrane was removed from the spin mini-column and inserted in a PCR vial together with the RT-LAMP reagents, and incubated at 60 °C for 60 min followed by 5 min at 80 °C. Subsequent to the amplification process, each amplification product was subjected to gel electrophoresis in a 2% agarose gel.
Results and discussion
Inhibition effect of saliva on RT-LAMP
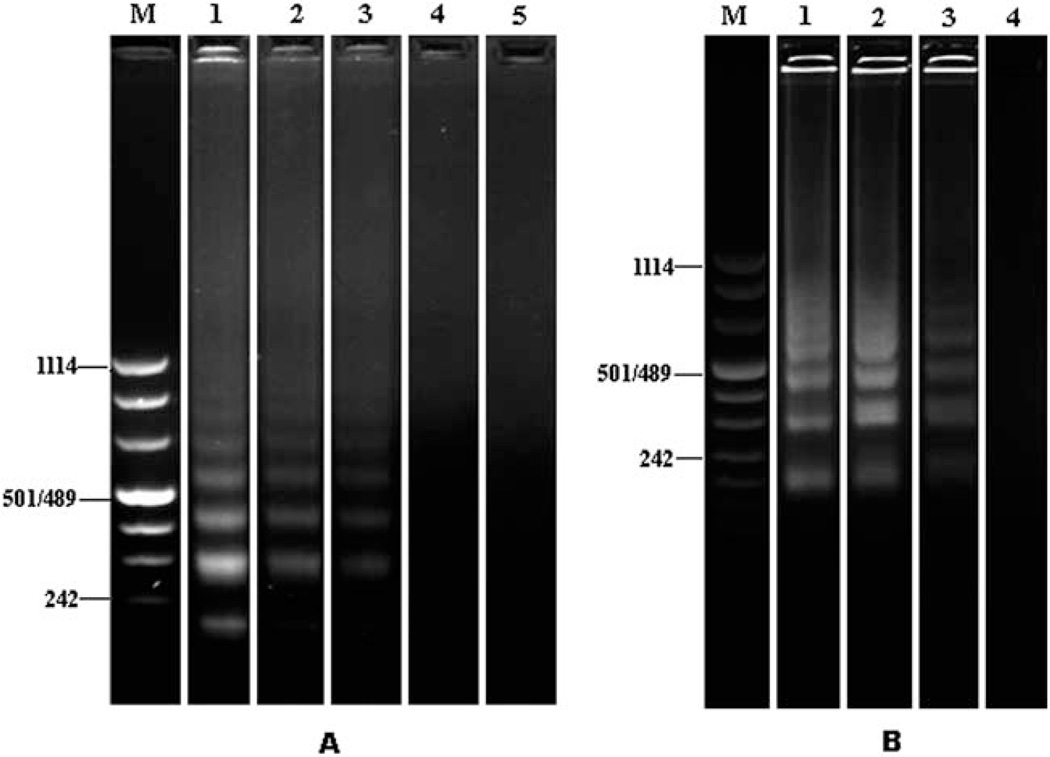
Prior studies34 revealed the presence of PCR inhibitors in saliva. To test the potential impact of salivary inhibitors on the LAMP process, a benchtop dilution study (107 to 104 virions per ml) was performed. The resulting electropherograms are shown in Fig. 3A. Lane M is the DNA ladder (Roche DNA Molecular Weight Marker VIII). Lanes 1, 2,3,4, and 5 correspond, respectively, to 107, 106, 105, 104, and 0 (negative control) HIV virus particles per ml. Due to the sample size, the actual number of virus particles in the vial was ten-fold smaller. The experiment was repeated in triplicate with similar results. LAMP amplification products consist of stem-loop DNA structures with inverted repeats of the target and cauliflower-like structures with multiple loops. Consequently, the LAMP amplicons have different lengths and the corresponding electropherograms feature a characteristic, ladder-like pattern, which consists of many bands of different lengths all the way up to the loading well.37 Lanes 1, 2, and 3 exhibit the characteristic pattern of LAMP electropherograms. The bands of the shorter segments are located approximately at positions corresponding to n × 115 bp, where n = 2, 3,….
Fig. 3.
Electropherograms of HIV RT-LAMP amplicons obtained with saliva samples spiked with HIV without purification (A) and with FTA membrane-based purification (B). (A) Lane M: DNA marker; lane 1: 107 HIV particles per ml; lane 2: 106 HIV particles per ml; lane 3: 105 HIV virus particles per ml; lane 4: 104 HIV particles per ml; and lane 5: negative control. (B) Lane M: DNA markers; lane 1: 104 HIV particles per ml; lane 2: 103 HIV particles per ml; lane 3: 102 HIV particles per ml; and lane 4: negative control. The electrophoresis separation processes (A) and (B) were carried out for different lengths of time.
When the HIV concentration was smaller than 105 particles per ml, no amplification products were visible. This relatively poor detection limit is particularly surprising since some of us have successfully obtained detectable amplicons with less than 100 virions in blood using identical LAMP reagents.15 We speculate that the relatively low efficiency of our saliva assay is due to the presence of yet uncharacterized inhibitors in saliva.
A solid state membrane (FTA) fitted into a spin column format was used to test the ability to remove inhibitors from the saliva sample prior to amplification. The electropherogram of the resulting LAMP amplicons is shown in Fig. 3B. Lane M is the DNA ladder. Lanes 1, 2, 3, and 4 correspond, respectively, to 104, 103, 102, and 0 (negative control) HIV particles per ml. The experiment was repeated in triplicate with similar results. When we used a FTA membrane as a solid phase extraction (SPE) matrix to purify viral RNA from oral fluid, we were able to consistently detect down to 102 virions per ml, which corresponds to 10 virus particles in the sample. In other words, with the FTA membrane-based purification, we improved the detection limit a thousand fold. The experiment demonstrates that the FTA membrane effectively purified nucleic acids, allowed the removal of potential inhibitors from the saliva sample, and dramatically improved the detection sensitivity of the RT-LAMP process. Moreover, the experiment demonstrates the compatibility of FTA with the LAMP amplification process. In other words, the presence of the FTA membrane in the reaction vial does not adversely impact the efficiency of the LAMP process. We conclude that to obtain high detection sensitivity of HIV in saliva, it is necessary to purify the nucleic acids from the raw oral fluid.
Integrated LAMP cassette
The LAMP cassette body was fabricated using CNC machining technology, which can mill a variety of complex three-dimensional (3-D) microstructures. In mass production, it is anticipated that the cassette would be fabricated by injection molding. To enhance the nucleic acid extraction efficiency, a flow-through mode was adopted in our design. The saliva sample, mixed with binding/lysis buffer, flows through the membrane, and nucleic acids from the sample bind to the membrane (Fig. 1C). Once wet, the FTA membrane expands and seals against the chamber walls, preventing the sample from bypassing the membrane.
Because amplification occurs at a constant temperature, the integrated LAMP cassette does not require precise thermal control as in conventional PCR.38–41 A single, thin film heater and open loop control were sufficient for performing the LAMP reaction. Although the heater could have been patterned on the cassette directly, this alternative design was not selected out of cost concerns.
Real-time, RT-LAMP for HIV detection
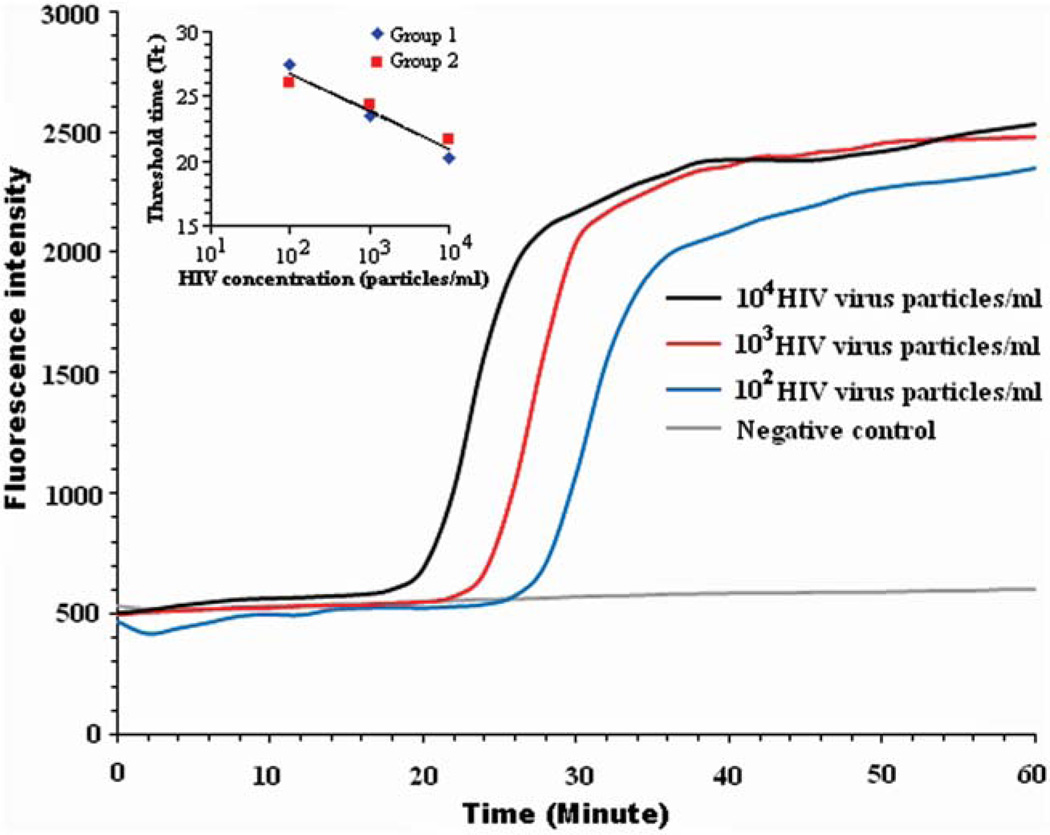
Fig. 4 depicts the real-time fluorescence intensity as a function of time detected from our cassette when the saliva test sample contained 104, 103, 102, and 0 (negative control) HIV particles per ml corresponding to 103, 102, 10, and 0 (negative control) HIV particles in the sample. The fluorescent intensity of the negative control (no target) remains nearly level throughout the entire detection time, indicating negligible amplicon formation, if any, of primer–dimers and the absence of any significant contamination. When HIV was present, the signal intensity increased from a baseline of about 500 relative fluorescent units (RFU) to the saturation level of the detection system. The higher the target concentration was, the earlier the intensity curve increased above the baseline. The LAMP amplicons from the lowest sample concentration (10 HIV particles in the reactor) registered a visible increase in emitted fluorescence intensity at approximately 27 min. Beyond this time, the signal intensity increased rapidly, reaching a saturation level at about 36 min. Thus, in a point of care setting, the test could have terminated in less than 40 min. The results clearly indicate that as few as 10 HIV particles can be detected with relatively unsophisticated equipment. The tests were repeated twice with nearly identical results.
Fig. 4.
Real-time monitoring of RT-LAMP amplification of saliva samples containing 104, 103, 102, and 0 (negative control) HIV particles per ml. Inset: the threshold time Tt (min) as a function of the HIV concentration (particles per ml).
The threshold time (Tt) is defined as the reaction time that elapses until the florescent signal increases ~20% above the baseline level. Fig. 4 indicates that Tt increases as the target molecule’s concentration C (HIV virons per ml) decreases. Fig. 4 (inset) depicts the threshold time Tt (min) as a function of C on a semi-log plot. In the range 10 < C < 1000 viral particles per reaction chamber, the threshold time Tt decreases linearly as a function of log (C). The data can be correlated with the formula Tt ≈ 32.6 − 2.9 log (C), where Tt is expressed in min.
The amplification results were monitored in real time with a compact, portable fluorescent detector, which simplifies cassette design and eliminates the need to transfer the reaction products from the amplification chamber to a detection chamber. Real-time detection also reduces the analysis time since the test can be terminated as soon as the threshold time (Tt) is determined.
Specificity of the RT-LAMP
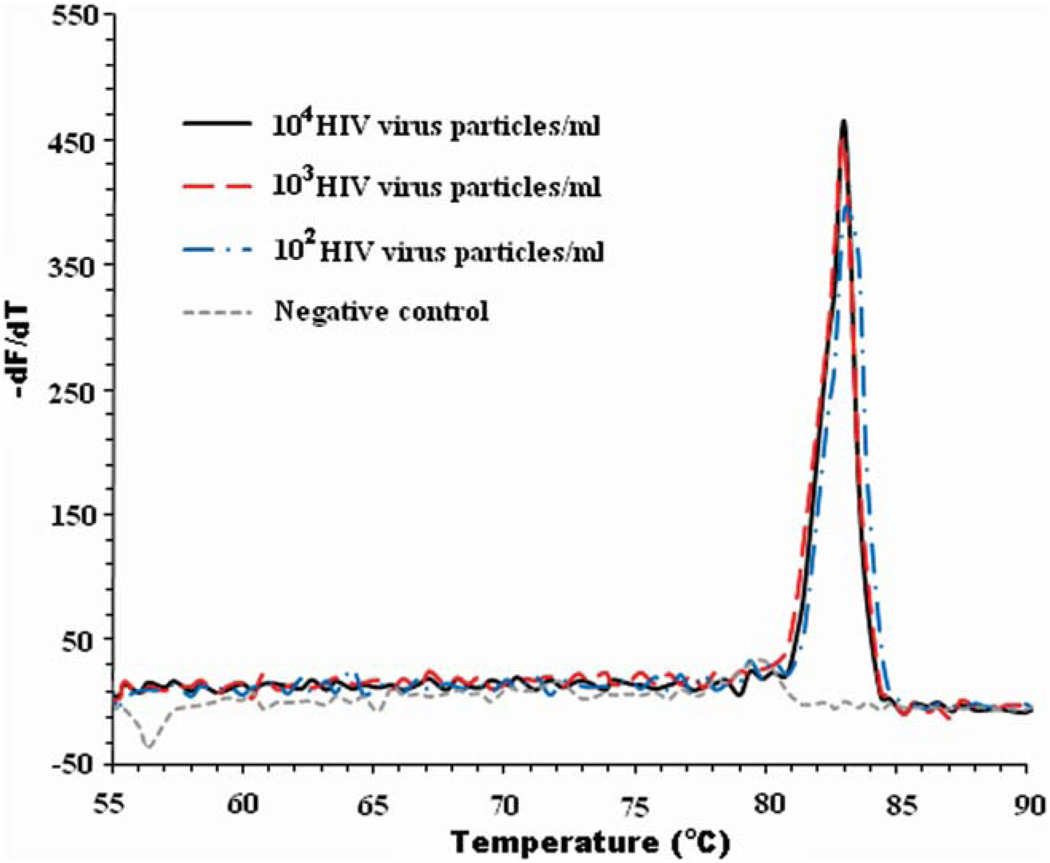
To determine the reaction specificity, we obtained the melting curve shown in Fig. 5. At the conclusion of the RT-LAMP amplification process, the reaction chamber’s temperature was increased gradually from 55 °C to 90 °C while monitoring the fluorescence emission. When the temperature reached the melting temperature of the amplicons, the fluorescent signal intensity diminished greatly. Although the LAMP amplicons have different lengths, as is evident from the electropherograms (Fig. 3), in the absence of non-specific binding, the double stranded segments of each amplicon have the same length and thus a single melting temperature (as we observed). Using finite differences, we calculated the derivative of the emission intensity with respect to the temperature dF/dT, where F is the emission intensity expressed in RFU and T is the reactor temperature expressed in °C. Fig. 5 depicts dF/dT as a function of T when there are 104, 103, 102 and 0 (negative control) HIV particles per ml. In the absence of target analytes (negative control), −dF/dT remains level for the entire temperature range. When target analyte and amplification products are present, all the curves feature sharp peaks centered at a melting temperature (Tm) ~83 °C.
Fig. 5.
The derivative of the fluorescence intensity with respect to the temperature, −dF/dT, is depicted as a function of the temperature when the analyte consisted of 104, 103, 102 and 0 (negative control) HIV particles per ml.
Using the Nearest-Neighbor module of Oligo Calc: Oligonucleotide Properties Calculator (http://www.basic.northwestern.edu/biotools/oligocalc.html), we estimated the melting temperature of the amplicon to be 83.5 °C, which is in good agreement with the measured value of 83 °C (Fig. 5).
To assess whether the melting temperature measurement could reveal the presence of primer–dimers that could potentially lead to a false positive result, we estimated the melting temperatures of various hypothetical primer–dimers that could form in our LAMP process. We assumed that the hypothetical primer–dimers are formed by end-to-end joining of forward and reverse primers. As there are a total of three forward and three reverse primers used in LAMP, there are nine distinct potential primer–dimer combinations. Listed below are the possible primer–dimers and their estimated melting temperatures (Tm) as calculated using the Nearest-Neighbor module of the Oligo Calc. These are FIP–BIP (92 bp, estimated Tm = 77.5 °C), FIP–LoopB (66 bp, Tm = 74.0 °C), FIP–B3 (68 bp, est. Tm = 72 °C), LoopF–BIP (70 bp, est. Tm = 74 °C), LoopF–LoopB (44 bp, est. Tm = 68 °C), LoopF–B3 (46 bp, est. Tm = 68 °C), F3–BIP (64 bp, est. Tm = 75 °C), F3–LoopB (38 bp, est. Tm = 68 °C), and F3–B3 (40 bp, est. Tm = 67.5 °C). The melting temperatures of the various hypothetical primer–dimers are more than 5 °C below the melting point of the target amplicon. Thus, the melting temperature curve is capable of distinguishing between target amplicon and primer–dimers. No primer–dimer formation was detected in our experiments.
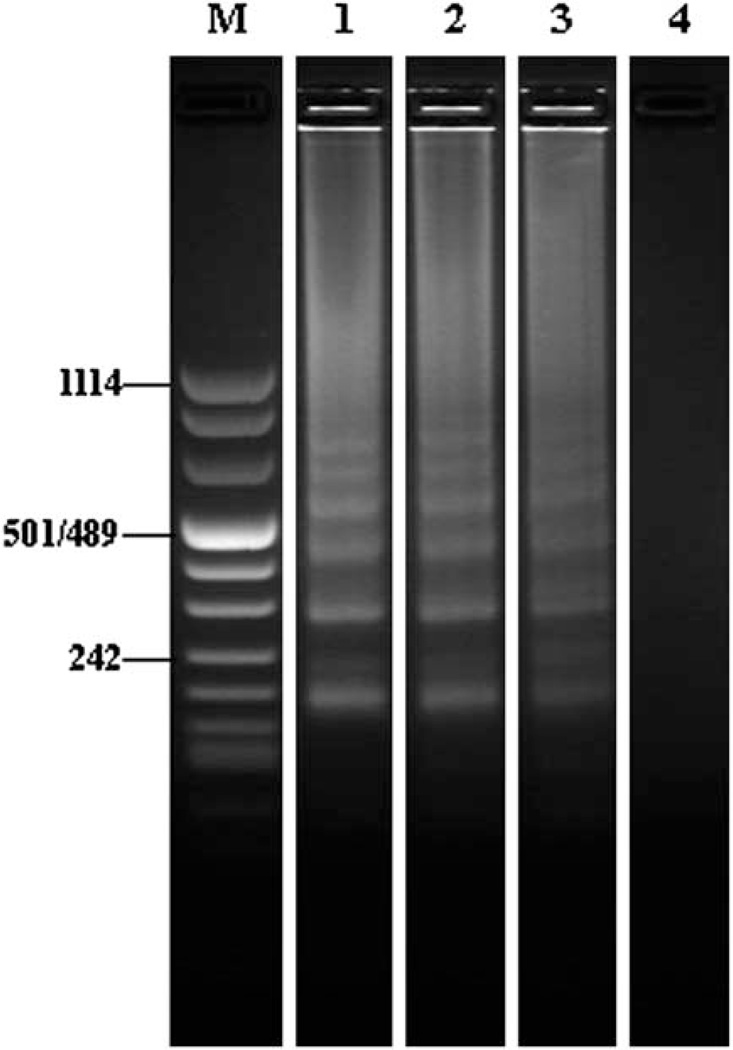
The results of the real-time LAMP measurements were further confirmed by agarose gel electrophoresis (Fig. 6). Lane M is DNA ladder markers. Lanes 1, 2, 3, and 4 correspond, respectively to 103, 102, 10 and 0 (negative control) viral particles in the reaction chamber. The negative control exhibited no signal. The electropherograms of the three positive samples showed band patterns characteristic of LAMP amplicons and consistent with the benchtop results (Fig. 3B). Thus, Fig. 6 indicates that our integrated LAMP cassette produced amplification results comparable with the benchtop experiments. The lack of bands corresponding to amplicons shorter than 220 kb indicates that no significant amounts of primer–dimer were formed, which is consistent with the observed melting curve (Fig. 5).
Fig. 6.
Electropherograms of RT-LAMP products obtained with integrated LAMP cassette. Lane M is the DNA ladder markers and lanes 1, 2, 3, and 4 correspond, respectively, to 103, 102, 10, and 0 (negative control) HIV particles per reaction chamber.
Conclusions and outlook
A single-use, low-cost, disposable, integrated, single-chamber LAMP cassette that utilizes a FTA membrane for nucleic acid isolation, purification, and concentration was designed, constructed, and tested. The nucleic acids, which were captured on the FTA membrane, were directly used as templates for nucleic acid amplification without any need for a special elution and transfer of nucleic acids, which, in turn, greatly simplified chip design and flow control. To demonstrate the system’s utility for point of care detection of virus, we carried out a series of experiments in which we detected HIV particles suspended in raw saliva. The experiments indicated that our single-chamber LAMP cassette system could detect HIV-1 in oral fluid with the sensitivity of 10 HIV particles per reaction chamber within less than an hour, while PCR assays required well over an hour to complete the amplification process.42–44 To the best of our knowledge, this is the first demonstration of a LAMP reactor with an integrated flow-through membrane.
Future modifications and improvements of the single-chamber cassette may include dry storage of the LAMP/RT-LAMP reagents in the reaction chamber. This can be achieved by encapsulating the dry reagents with low melting point paraffin,45 which melts on heating the reaction chamber to the desired incubation temperature of 60 °C and releases the LAMP reagents for amplification. Another improvement may include storing the buffers for the isolation and purification of nucleic acid in the cassette, possibly in a pouch format as previously reported.26,35 With appropriate modifications of the reagents, the system can be used to detect other infectious diseases, monitor the health of individuals, provide a trigger for the administration of expensive or dangerous medications, handle samples of body fluids other than saliva, and also facilitate the monitoring of water and food quality.
Acknowledgements
The work was supported by NIH/NIDCR Grant U01DE017855. The primers for the LAMP process were designed and provided by the Center for Disease Control and Prevention (CDC) through a Material Transfer Agreement with New York University.
References
- 1.Malnati MS, Scarlatti G, Gatto F, Salvatori F, Cassina G, Rutigliano T, Volpi R, Lusso P. Nat. Protoc. 2008;3:1240–1248. doi: 10.1038/nprot.2008.108. [DOI] [PubMed] [Google Scholar]
- 2.Cheng X, Gupta A, Chen C, Tompkins R, Rodriguez W, Toner M. Lab Chip. 2009;9:1357–1364. doi: 10.1039/b818813k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rouet F, Ekouevi DK, Chaix ML, Burgard M, Inwoley A, Tony TD, Danel C, Anglaret X, Leroy V, Msellati P, Dabis F, Rouzioux C. J. Clin. Microbiol. 2005;43:2709–2717. doi: 10.1128/JCM.43.6.2709-2717.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jokerst JV, Floriano PN, Christodoulides N, Simmons GW, McDevitt JT. Lab Chip. 2008;8:2079–2090. doi: 10.1039/b817116e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donovan BJ, Rublein JC, Leone PA, Pilcher CD. Ann. Pharmacother. 2004;38:670–676. doi: 10.1345/aph.1D314. [DOI] [PubMed] [Google Scholar]
- 6.Branson BM. J. Lab. Clin. Med. 2003;27:288–295. [Google Scholar]
- 7.Owen SM, Yang C, Spira T, Ou CY, Pau CP, Parekh BS, Candal D, Kuehl D, Kennedy MS, Rudolph D, Luo W, Delatorre N, Masciotra S, Kalish ML, Cowart F, Barnett T, Lal R, McDougal JS. J. Clin. Microbiol. 2008;46:1588–1595. doi: 10.1128/JCM.02196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dineva MA, MahiLum-Tapay L, Lee H. Analyst. 2007;132:1193–1199. doi: 10.1039/b705672a. [DOI] [PubMed] [Google Scholar]
- 9.Mehta N, Trzmielina S, Nonyane BA, Eliot MN, Lin R, Foulkes AS, McNeal K, Ammann A, Eulalievyolo V, Sullivan JL, Luzuriaga K, Somasundaran M. PLoS One. 2009;4:e5819. doi: 10.1371/journal.pone.0005819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibellini D, Vitone F, Gori E, Placa ML, Re MC. J. Virol. Methods. 2004;115:183–189. doi: 10.1016/j.jviromet.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 11.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thai HTC, Le MQ, Vuong CD, Parida M, Minekawa H, Notomi T, Hasebe F, Morita K. J. Clin. Microbiol. 2004;42:1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill J, Beriwal S, Chandra I, Paul VK, Kapil A, Singh T, Wadowsky RM, Singh V, Goyal A, Jahnukainen T, Johnson JR, Tarr PI, Vats A. J. Clin. Microbiol. 2008;46:2800–2804. doi: 10.1128/JCM.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang X, Liu Y, Kong J, Jiang X. Anal. Chem. 2010;82:3002–3006. doi: 10.1021/ac1000652. [DOI] [PubMed] [Google Scholar]
- 15.Curtis KA, Rudolph DL, Owen SM. J. Virol. Methods. 2008;151:264–270. doi: 10.1016/j.jviromet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 16.Curtis KA, Rudolph DL, Owen SM. J. Med. Virol. 2009;81:966–972. doi: 10.1002/jmv.21490. [DOI] [PubMed] [Google Scholar]
- 17.Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 18.Herr AE, Hatch AV, Throckmorton DJ, Tran HM, Brennan JS, Giannobile WV, Singh AK. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5268–5273. doi: 10.1073/pnas.0607254104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagally ET, Emrich CA, Mathies RA. Lab Chip. 2001;1:102–107. doi: 10.1039/b109031n. [DOI] [PubMed] [Google Scholar]
- 20.Liu RH, Yang JN, Lenigk R, Bonanno J, Grodzinski P. Anal. Chem. 2004;76:1824–1831. doi: 10.1021/ac0353029. [DOI] [PubMed] [Google Scholar]
- 21.Dimov IK, Garcia-Cordero JL, O’Grady J, Poulsen CR, Viguier C, Kent L, Daly P, Lincoln B, Maher M, O’Kennedy R, Smith TJ, Ricco AJ, Lee LP. Lab Chip. 2008;8:2071–2078. doi: 10.1039/b812515e. [DOI] [PubMed] [Google Scholar]
- 22.Lee SH, Kim SW, Kang JY, Ahn CH. Lab Chip. 2008;8:2121–2127. doi: 10.1039/b811131f. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Chen Z, Corstjens PLAM, Mauk MG, Bau HH. Lab Chip. 2006;6:46–53. doi: 10.1039/b511494b. [DOI] [PubMed] [Google Scholar]
- 24.Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, Roper MG, Feldman SH, Hughes MA, Hewlett EL, Merkel TJ, Ferrance JP, Landers J. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19272–19277. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, Mauk MG, Wang J, Abrams WR, Corstjens PLAM, Niedbala RS, Malamud D, Bau HH. Ann. N. Y. Acad. Sci. 2007;1098:429–436. doi: 10.1196/annals.1384.024. [DOI] [PubMed] [Google Scholar]
- 26.Chen D, Mauk M, Qiu X, Liu C, Kim J, Ramprasad S, Ongagna S, Abrams WR, Malamud D, Corstjens PLAM, Bau HH. Biomed. Microdevices. 2010;12:705–719. doi: 10.1007/s10544-010-9423-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, Cheong K, Huh N, Kim1 S, Choi J, Ko C. Lab Chip. 2006;6:886–895. doi: 10.1039/b515876a. [DOI] [PubMed] [Google Scholar]
- 28.Kim J, Mauk M, Chen D, Qiu X, Kim J, Gale B, Bau HH. Analyst. 2010;135:2408–2414. doi: 10.1039/c0an00288g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malamud D. Br. Med. J. 1992;305:207–208. doi: 10.1136/bmj.305.6847.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malamud D, Bau HH, Niedbala S, Corstjens P. Adv. Dent. Res. 2005;18:12–16. doi: 10.1177/154407370501800104. [DOI] [PubMed] [Google Scholar]
- 31.Segal A, Wong DT. Eur. J. Dent. Educ. 2008;12:22–29. doi: 10.1111/j.1600-0579.2007.00477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malamud D. Am. J. Med. 1997;102:9–14. doi: 10.1016/s0002-9343(97)00032-6. [DOI] [PubMed] [Google Scholar]
- 33.Ziober BL, Mauk MG, Falls EM, Chen Z, Ziober AF, Bau HH. Head Neck. 2008;30:111–121. doi: 10.1002/hed.20680. [DOI] [PubMed] [Google Scholar]
- 34.Ochert AS, Boulter AW, Birnbaum W, Johnson NW, Teo CG. Genome Res. 1994;3:365–368. doi: 10.1101/gr.3.6.365. [DOI] [PubMed] [Google Scholar]
- 35.Liu C, Qiu X, Ongagna S, Chen D, Chen Z, Abrams WR, Malamud D, Corstjens PLAM, Bau HH. Lab Chip. 2009;9:768–776. doi: 10.1039/b814322f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou P, Young L, Chen Z. Biomed. Microdevices. 2010;12:821–832. doi: 10.1007/s10544-010-9436-z. [DOI] [PubMed] [Google Scholar]
- 37.Tomita N, Mori Y, Kanda H, Notomi T. Nat. Protoc. 2009;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 38.Lee CS, Moon HJ, Yang JS. J. Virol. Methods. 2007;139:39–43. doi: 10.1016/j.jviromet.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 39.Westh H, Lisby G, Breysse F, Boddinghaus B, Chomarat M, Gant V, Goglio A, Raglio A, Schuster H, Stuber F, Wissing H, Hoeft A. Clin. Microbiol. Infect. 2009;15:544–551. doi: 10.1111/j.1469-0691.2009.02736.x. [DOI] [PubMed] [Google Scholar]
- 40.Shen F, Du W, Davydova EK, Karymov MA, Pandey J, Ismagilov RF. Anal. Chem. 2010;82:4606–4612. doi: 10.1021/ac1007249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Legendre LA, Bienvenue JM, Roper MG, Ferrance JP, Landers JP. Anal. Chem. 2006;78:1444–1451. doi: 10.1021/ac0516988. [DOI] [PubMed] [Google Scholar]
- 42.Butler SL, Hansen MS, Bushman FD. Nat. Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 43.Palmer S, Wiegand AP, Maldarelli F, Bazmi H, Mican JM, Polis M, Dewar RL, Planta A, Liu S, Metcalf JA, Mellors JW, Coffin JM. J. Clin. Microbiol. 2003;41:4531–4536. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saha BK, Tian B, Bucy RP. J. Virol. Methods. 2001;93:33–42. doi: 10.1016/s0166-0934(00)00288-3. [DOI] [PubMed] [Google Scholar]
- 45.Kim J, Byun D, Mauk MG, Bau HH. Lab Chip. 2009;9:606–612. doi: 10.1039/b807915c. [DOI] [PMC free article] [PubMed] [Google Scholar]