Abstract
Aims: Several neurodegenerative disorders show alterations in glutamatergic synapses and increased susceptibility to excitotoxicity. Mounting evidence suggests a central role for the cellular prion protein (PrPC) in neuroprotection. Therefore, the loss of PrPC function occurring in prion disorders may contribute to the disease progression and neurodegeneration. Indeed, PrPC modulates N-methyl-d-aspartate receptors (NMDAR), thus preventing cell death. In this study, we show that PrPC and copper cooperatively inhibit NMDAR through S-nitrosylation, a post-translational modification resulting from the chemical reaction of nitric oxide (NO) with cysteines. Results: Comparing wild-type Prnp (Prnp+/+) and PrPC knockout (Prnp0/0) mouse hippocampi, we found that GluN1 and GluN2A S-nitrosylation decrease in Prnp0/0. Using organotypic hippocampal cultures, we found that copper chelation decreases NMDAR S-nitrosylation in Prnp+/+ but not in Prnp0/0. This suggests that PrPC requires copper to support the chemical reaction between NO and thiols. We explored PrPC-Cu neuroprotective role by evaluating neuron susceptibility to excitotoxicity in Prnp+/+ and Prnp0/0 cultures. We found that (i) PrPC-Cu modulates GluN2A-containing NMDAR, those inhibited by S-nitrosylation; (ii) PrPC and copper are interdependent to protect neurons from insults; (iii) neuronal NO synthase inhibition affects susceptibility in wild-type but not in Prnp0/0, while (iv) the addition of a NO donor enhances Prnp0/0 neurons survival. Innovation and Conclusions: Our results show that PrPC and copper support NMDAR S-nitrosylation and cooperatively exert neuroprotection. In addition to NMDAR, PrPC may also favor the S-nitrosylation of other proteins. Therefore, this mechanism may be investigated in the context of the different cellular processes in which PrPC is involved. Antioxid. Redox Signal. 22, 772–784.
Introduction
The cellular prion protein (PrPC) has been extensively investigated since its isoform, the prion, was identified as the causative agent of prion disorders (33). PrPC is widely expressed, reaching the highest levels in the nervous system (3, 12, 31). Comparing wild-type with PrPC-null mice (Prnp0/0) has revealed that PrPC expression at synapses contributes to hippocampus synaptic function (6, 27, 28) and exerts neuroprotection by modulating neuronal excitability (21, 34, 38). In particular, PrPC inhibits N-methyl-d-aspartate receptors (NMDAR) and requires copper for this effect (21, 40). Interestingly, Aβ oligomers induce neurotoxicity by blocking PrPC-Cu modulation of NMDAR (40). Impairments in NMDAR regulation induce excitotoxicity, the neuronal cell death pathway triggered by massive calcium influx (10). Calcium ions enter the cell through the NMDAR channel and bind several interactors, for example, calmodulin (CaM). Calcium-bound CaM activates neuronal nitric oxide synthase (nNOS) and induces copper-transporting ATPase 1 (Atp7a) translocation, thus leading to nitric oxide (NO) production and copper release in the synaptic cleft (22, 36). Copper and NO modulate the NMDAR activity through a not clearly defined mechanism: NMDAR S-nitrosylation provides a link between copper and NO inhibitory roles (7, 23). S-nitrosylation is a chemical post-translational modification targeting protein cysteines. In NMDAR, S-nitrosylation is inhibitory and addresses two residues on GluN1 and three residues on GluN2A, including Cys399, which mediates the predominant inhibitory effect. Copper released in the synaptic cleft is bound by metal-binding proteins, such as PrPC, which binds copper with high affinity and sustains Cu(II) redox cycling (5, 8, 24). In vitro results have shown that PrPC-bound Cu(II) promotes NO oxidation and is consequently reduced to Cu(I) (4). As PrPC-Cu is involved in glypican-1 heparan sulfate S-nitrosylation (29), the complex may also have a role in NMDAR S-nitrosylation.
Innovation.
A major cause of neurodegeneration is the disruption of excitatory synapse regulation. Mounting evidence points to a central neuroprotective role for the cellular prion protein (PrPC), a key player in prion disorders and Alzheimer disease (AD). Here, we describe a novel neuroprotective mechanism mediated by PrPC, copper, and nitric oxide: copper-bound PrPC modulates N-methyl-d-aspartate (NMDA)-type glutamate receptors by promoting S-nitrosylation, inhibiting the ionic channel. This mechanism is likely blocked in prion disorders and AD, thereby leading to neuron loss. PrPC function in protein S-nitrosylation may also guide the study of the numerous cellular pathways in which PrPC is involved.
Here, we show that PrPC exerts copper-dependent neuroprotection by mediating NMDAR S-nitrosylation. We define PrPC-Cu direct involvement in NMDAR S-nitrosylation comparing GluN2A and GluN1 S-nitrosylation levels in Prnp+/+ and Prnp0/0 hippocampi and cultures treated with a copper chelator. PrPC neuroprotective function is characterized comparing excitotoxicity susceptibility in different Prnp+/+ and Prnp0/0 hippocampal areas. The copper/NO-dependence of PrPC neuroprotective activity is assessed observing the effects of copper chelation, NOS inhibition, and NO donation on neuronal death in wild-type and Prnp0/0 cultures. We propose a model for this molecular mechanism based on the results presented here.
Results
PrPC is involved in NMDAR S-nitrosylation
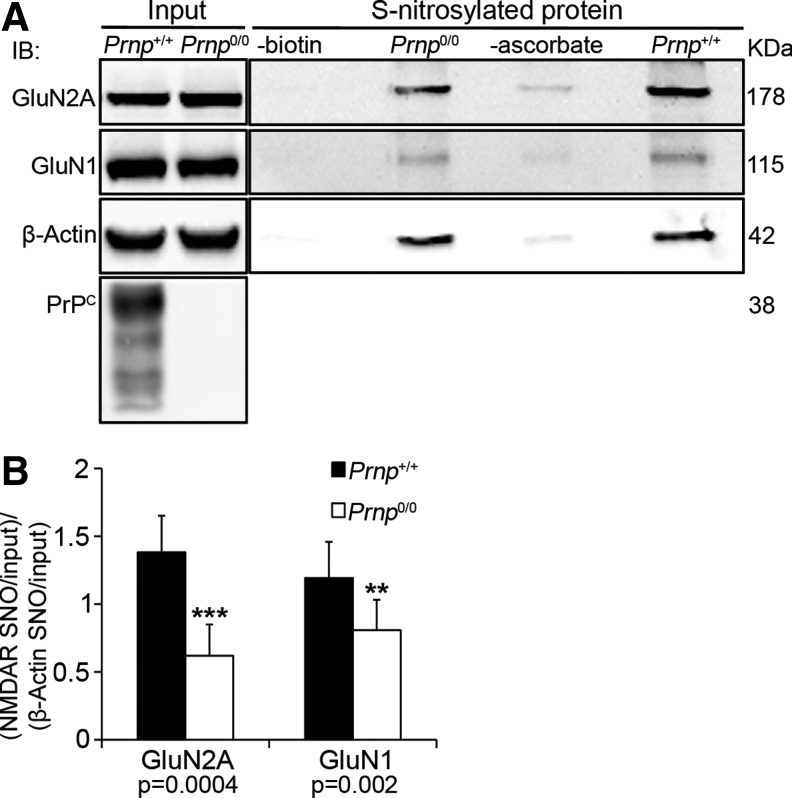
The S-nitrosylation of NMDAR extracellular cysteines may be a mechanism by which the PrPC-Cu complex inhibits ion gating. To test this hypothesis, we measured GluN2A and GluN1 S-nitrosylation levels in adult Prnp+/+ and Prnp0/0 hippocampi. To compare wild-type with PrPC-null samples, the Western blotting signals corresponding to each S-nitrosylated protein (i.e., GluN2A, GluN1 and β-Actin) were normalized against the respective input signals. Then, the obtained GluN2A and GluN1 ratio values were normalized against the corresponding β-actin ratio value, as an internal control. The resulting values for Prnp+/+ and Prnp0/0 hippocampi were compared by the Mann–Whitney test. In PrPC-null hippocampus, we detected lower GluN2A (n=9, p=0.0004) and GluN1 (n=9, p=0.00203) S-nitrosylation levels compared with those in the wild type (Fig. 1A, B). These findings show that in PrPC-null hippocampus NMDAR S-nitrosylation is reduced. We confirmed these results by using an additional S-nitrosylation-detection method (Supplementary Materials and Methods and Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/ars). This observation suggests that PrPC is involved in GluN2A and GluN1 S-nitrosylation and, possibly through this mechanism, it can modulate the NMDAR activity.
FIG. 1.
In adult Prnp0/0 hippocampus, the S-nitrosylation of GluN2A and GluN1 NMDAR subunits is lower compared to wild type. (A) Signals of S-nitrosylated fraction, controls without either biotin or ascorbate and corresponding input of GluN2A, GluN1, and β-actin resulting from the biotin switch assay of Prnp+/+ and Prnp0/0 hippocampus; controls without either biotin or ascorbate were done using Prnp+/+ samples. (B) Statistical analysis of S-nitrosylated GluN2A and GluN1 signals normalized on the corresponding input and on β-actin; all error bars indicate SD; **p<0.01; ***p<0.001. NMDAR, N-methyl-d-aspartate receptors.
To exclude that the results observed in Prnp0/0 hippocampus are due to different levels of either GluN2A-containing NMDAR or nNOS at synapses, we performed a co-immunoprecipitation (coIP) with the postsynaptic density protein 95 (PSD95). The results revealed comparable GluN2A (direct PSD95 interactor), GluN1 (indirect PSD95 interactor), and nNOS levels in Prnp+/+ and Prnp0/0 hippocampal synapses (Supplementary Materials and Methods and Supplementary Fig. S2A–C). Furthermore, to exclude that the results observed in Prnp0/0 hippocampus are due to altered NOS activity, first, we assessed the similarity of β-actin S-nitrosylation levels (Fig. 1A) and then measured both the consumption of NADPH, an essential cofactor for NO production, and the conversion of radiolabeled arginine to radiolabeled citrulline (Supplementary Materials and Methods). Adult Prnp+/+ and Prnp0/0 samples exhibited comparable kinetics for NADPH consumption and the same capability to convert arginine to citrulline (Supplementary Fig. S3A, C). Taken together, these results suggest that the decreased NMDAR S-nitrosylation in PrPC-null hippocampus is not due to a reduction in the levels of NMDAR and nNOS at synapses or alterations in NO production.
In the absence of PrPC, hippocampal neurons are more susceptible to NMDAR-mediated excitotoxicity
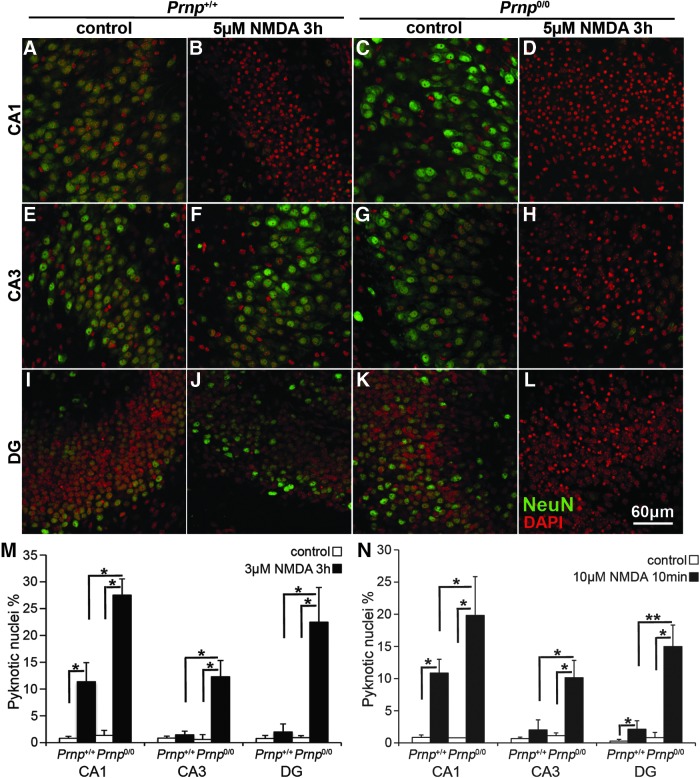
Since Prnp0/0 hippocampus shows lower NMDAR S-nitrosylation (Fig. 1 and Supplementary Fig. S1) but the same protein level at synapses (Supplementary Fig. S2A, C), PrPC-null neurons should be more susceptible to excitotoxic treatment. To investigate the neuroprotective role of PrPC through glutamate receptor inhibition, we carried out excitotoxicity studies on Prnp+/+ and Prnp0/0 organotypic hippocampal cultures (OHC) by analyzing specifically neuronal death in Cornus Ammonis 1 (CA1), Cornus Ammonis 3 (CA3), and dentate gyrus (DG). We exposed Prnp+/+ and Prnp0/0 OHC to two toxic treatments with NMDA as selective NMDAR agonist: 5 μM NMDA for 3 h (prolonged insult) and 10 μM NMDA for 10 min (acute insult). We chose these two protocols after observing that they induce low neuronal cell death levels in wild-type samples, hence they can clearly reveal a different susceptibility of Prnp0/0 samples. Neuronal cell death was evaluated as pyknotic nuclei percentage over the total number of nuclei in CA1, CA3, and DG, respectively. The concomitant staining with the anti-neuronal nuclei marker antibody (NeuN) allowed us to localize the pyknotic event in neurons and clearly distinguished glia cells. Figure 2A–M shows the different responses of Prnp+/+ and Prnp0/0 OHC to the prolonged treatment (n=4 OHC, 5 slices per treatment in each culture). In CA1, the NMDA exposure significantly induced neuronal cell death in both Prnp+/+ (p=0.03038) and Prnp0/0 (p=0.03038) OHC (Fig. 2A–D, M), but at the same time, it revealed a higher susceptibility of Prnp0/0 OHC (p=0.03038). The NMDA treatment did not induce significant neuronal cell death in wild-type CA3 and DG, whereas it yielded high toxicity for both regions in PrPC-null cultures (CA3, p=0.03038; DG, p=0.03038), highlighting a significant difference between Prnp+/+ and Prnp0/0 (CA3, p=0.03038; DG, p=0.01996) (Fig. 2E–M). The same quantitative differences between Prnp+/+ and Prnp0/0 OHC were observed following the acute treatment (Fig. 2N; n=5 OHC, 5 slices per treatment in each culture). In CA1, 10 μM NMDA for 10 min induced neuronal cell death in both Prnp+/+ (p=0.01996) and Prnp0/0 (p=0.01996) OHC, but it revealed a higher susceptibility of Prnp0/0 compared with Prnp+/+ (p=0.02157). In CA3, the acute treatment was toxic only for Prnp0/0 OHC (p=0.03038), confirming their higher susceptibility compared with Prnp+/+ (p=0.01421). The wild-type DG showed low levels of neuronal cell death in response to the acute insult (p=0.02518), whereas the same treatment caused a greater cell death percentage (p=0.01421) in Prnp0/0 DG, leading to a higher damage compared to Prnp+/+ (p=0.00507). The enhanced susceptibility of Prnp0/0 neurons to the excitotoxic stimulus confirms findings previously reported in the literature, obtained by stimulating neurons with kainate (34) and NMDA (21).
FIG. 2.
Wild-type and PrPC-null OHC show different regional susceptibility to NMDA treatment, under both prolonged and acute insult. Images from three hippocampal regions are reported in rows: (A–D) CA1, (E–H) CA3, and (I–L) DG. The four different combinations of treatment and samples are reported in columns: (A, E, I) Prnp+/+ control, (B, F, J) Prnp+/+ treated with 5 μM NMDA for 3 h, (C, G, K) Prnp0/0 control, and (D, H, L) Prnp0/0 treated with 5 μM NMDA for 3 h. NeuN staining is displayed in green and DAPI in red. Confocal microscope fluorescence images were acquired using a 40×/1.30 NA oil objective. Comparison of the neuronal pyknotic nuclei percentage induced by 5 μM NMDA for 3 h (M; n=4 OHC, 5 slices per treatment in each culture) and by 10 μM NMDA for 10 min (N; n=5 OHC, 5 slices per treatment in each culture), calculated over the total nuclei number, between Prnp+/+ (white bars) and Prnp0/0 (black bars) OHC in CA1, CA3, and DG; all error bars indicate SD; *p<0.05, **p<0.01. CA1, Cornus Ammonis 1; CA3, Cornus Ammonis 3; DAPI, 4′,6-diamidino-2-phenylindole; DG, dentate gyrus; OHC, organotypic hippocampal cultures. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
To evaluate the early changes in neuronal cell viability, the MTT (thiazolyl blue tetrazolium bromide) mitochondrial toxicity assay was performed on Prnp+/+ and Prnp0/0 OHC after exposure to 5 μM NMDA for 3 h. Results revealed that Prnp0/0 OHC are more susceptible to excitotoxicity also in the early phases after insult (Supplementary Materials and Methods and Supplementary Fig. S4).
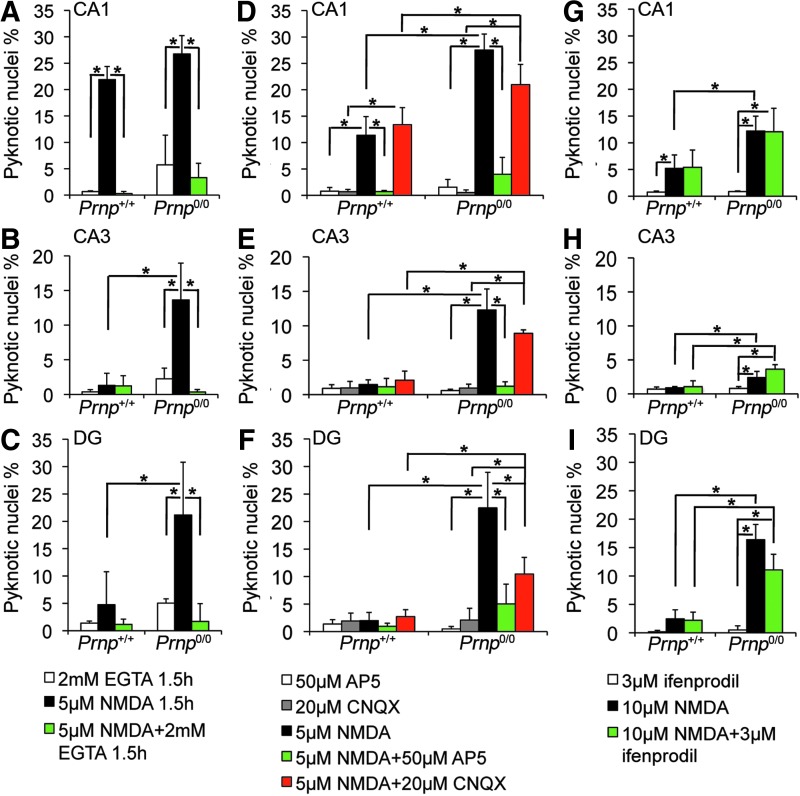
The excitotoxic mechanism induced by NMDAR overactivation is triggered by calcium ion influx into the neuronal cell (10). To verify that the excitotoxicity observed was a calcium-mediated effect, the same treatments were performed adding a calcium chelator, ethylene glycol tetraacetic acid (EGTA; n=3 OHC, 4 slices per treatment in each culture). It should be noted that the treatment with 5 μM NMDA was shortened to 1.5 h since calcium deprivation for 3 h resulted to be toxic per se. As expected, adding EGTA to NMDA abolished neuronal cell death in all the analyzed regions and under both prolonged and acute insults (Fig. 3A–C and Supplementary Fig. S5A–I; Prnp+/+ CA1: p=0.03038 and p=0.03038; Prnp0/0 CA1: p=0.01996 and p=0.03038; Prnp0/0 CA3: p=0.01894 and p=0.0294; Prnp0/0 DG: p=0.01945 and p=0.01996). Shortening the exposure time to 5 μM NMDA did not affect the differences between Prnp+/+ and Prnp0/0 OHC.
FIG. 3.
Neuronal cell death induced by NMDA treatment is prevented by calcium chelation and NMDAR antagonist, unaltered by AMPA/kainate receptor and GluN2B-containing NMDAR antagonists. Comparison of NMDA-induced neuronal pyknotic nuclei percentages between EGTA-treated Prnp+/+ and Prnp0/0 OHC in the (A) CA1, (B) CA3, and (C) DG; n=3 OHC, 5 slices per treatment in each culture. Comparison of NMDA-induced neuronal pyknotic nuclei percentages between NMDAR inhibitors (AP5 and/or CNQX)-treated Prnp+/+ and Prnp0/0 OHC in the (D) CA1, (E) CA3, and (F) DG; n=4 OHC, 5 slices per treatment in each culture. Comparison of NMDA-induced neuronal pyknotic nuclei percentages between GluN2B inhibitor (ifenprodil)-treated Prnp+/+ and Prnp0/0 OHC in the (G) CA1, (H) CA3, and (I) DG; n=4 OHC, 5 slices per treatment in each culture. All error bars indicate SD; *p<0.05. AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid; AP5, (2R)-amino-5-phosphonovaleric acid; CNQX, 6-cyano-7-nitroquinoxaline-2,3-dione; EGTA, ethylene glycol tetraacetic acid. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Next, we assessed the primary involvement of NMDAR in the neurotoxic mechanism (n=4 OHC, 5 slices per treatment in each culture). Adding the NMDAR blocker AP5 to 5 μM NMDA for 3 h abolished cell death (Fig. 3D–F and Supplementary Fig. S6A–F; Prnp+/+ CA1: p=0.03038; Prnp0/0 CA1: p=0.03038; Prnp0/0 CA3: p=0.03038; Prnp0/0 DG: p=0.03038). In contrast, adding the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)/kainate receptor blocker 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX) to 5 μM NMDA for 3 h did not diminish cell death in Prnp+/+ CA1, Prnp0/0 CA1, and Prnp0/0 CA3 (Fig. 3D, E and Supplementary Fig. S6A–D). However, adding CNQX to NMDA did reduce the pyknosis percentage in Prnp0/0 DG (p=0.03038), suggesting a partial AMPA/kainate receptor involvement (Fig. 3F and Supplementary Fig. S6E, F). Overall, despite the AMPA/kainate receptor inhibition, the higher cell death levels in PrPC-null were maintained, confirming a primary involvement of NMDAR. To assess whether the neurotoxic effect was mainly due to either GluN2A- or GluN2B-containig NMDAR, we used ifenprodil, a selective inhibitor of GluN2B. The excitotoxic condition applied was 10 μM NMDA for 10 min since ifenprodil has an aspecific toxic effect if added to lower NMDA concentrations (20). In both Prnp+/+ and Prnp0/0 OHC, ifenprodil did not reduce the cell death levels observed with NMDA alone, suggesting that the overactivation of GluN2B-containing receptor is not the leading cause for neuronal death (Fig. 3G–I and Supplementary Fig. S7). A major involvement of GluN2A-containing receptor is therefore a more likely hypothesis.
Copper ions are required for PrPC to support NMDAR S-nitrosylation
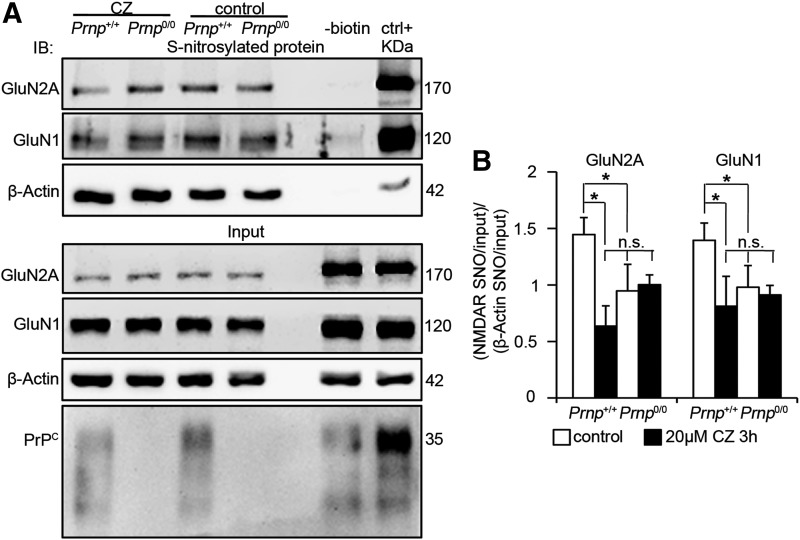
Copper ions are fundamental in the S-nitrosylation reaction because their reduction from Cu(II) to Cu(I) allows for the oxidation of NO and its ensuing reaction with cysteine thiols (11, 17). Because PrPC-bound Cu(II) promotes NO oxidation (4), PrPC is likely to promote NMDAR S-nitrosylation through the copper ions it binds. To verify this mechanism, we measured GluN2A and GluN1 S-nitrosylation levels in Prnp+/+ and Prnp0/0 OHC upon treatment with cuprizone (CZ), a selective copper chelator that binds Cu2+. Since Cu2+ promotes S-nitrosylation while Cu+ triggers denitrosylation, CZ treatment specifically blocks S-nitrosylation induction. CZ does not affect neuronal viability and cannot cross plasma membranes (2); hence, it should not affect cytoplasmic protein S-nitrosylation. Indeed, β-actin S-nitrosylation levels were comparable among control and CZ-treated wild-type and PrPC-null OHC, providing us with a reliable internal control for the normalization of NMDAR S-nitrosylation signal. Moreover, we used serum-free medium to avoid copper content variability and thereby ensure uniform copper chelation. In serum-free medium, CZ binds copper ions released in the synaptic cleft (36), thus affecting the copper-mediated S-nitrosylation in the extracellular environment. Prnp+/+ and Prnp0/0 OHC were exposed to 20 μM CZ for 3 h, immediately frozen, and then processed for the detection of S-nitrosylated proteins. As already observed in the hippocampus, GluN2A and GluN1 S-nitrosylation was decreased in Prnp0/0 samples (Fig. 4A, B; n=4 slice pools, p=0.03038 and p=0.03038, respectively). Consistent with the hypothesized mechanism, CZ addition reduced GluN2A and GluN1 S-nitrosylation in Prnp+/+ OHC (p=0.03038 and p=0.03038, respectively) but not in Prnp0/0 ones (Fig. 4A, B). Furthermore, NMDAR S-nitrosylation levels in CZ-treated wild-type OHC were not different from either control or CZ-treated Prnp0/0 samples. This result shows that CZ addition affects NMDAR S-nitrosylation only in wild type but not in Prnp0/0 OHC; hence, it strongly suggests that the lower S-nitrosylation of NMDAR subunits in Prnp0/0 samples is due to the lack of the PrPC-Cu complex and that PrPC-bound copper ions favor the S-nitrosylation of NMDAR. Residual GluN1 and GluN2A S-nitrosylation is due to PrPC-independent mechanisms.
FIG. 4.
Copper chelation decreases S-nitrosylation of GluN2A and GluN1 in wild type but not in Prnp0/0 OHC. (A) Signals of S-nitrosylated fraction and corresponding input of GluN2A, GluN1, and β-actin resulting from the biotin switch assay of Prnp+/+ and Prnp0/0 OHC, treated and untreated with CZ; adult wild-type hippocampus was used as negative (−biotin) and positive control. (B) Statistical analysis of S-nitrosylated GluN2A and GluN1 signals normalized on the corresponding input and on β-actin; all error bars indicate SD; sample size n=4; *p<0.05, n.s., not statistically significant difference. CZ, cuprizone.
To rule out that the results observed in Prnp0/0 OHC are due to different levels of GluN2A-containing NMDAR and nNOS at synapses, we performed coIP assays with PSD95 using hippocampi from postnatal day 20 (P20) mice. This age was selected because it is close to the developmental stage of OHC at the time of the treatment. The results revealed comparable GluN2A, GluN1, and nNOS levels in P20 Prnp+/+ and Prnp0/0 hippocampal synapses (Supplementary Fig. S8A–C). Also, at this developmental stage, we reasoned to exclude that the results observed in Prnp0/0 OHC were due to different levels of NOS activity. Like in the adult hippocampus, we observed that β-actin S-nitrosylation levels were similar in control and CZ-treated Prnp+/+ and Prnp0/0 OHC (Fig. 4A). Next, we measured the consumption of NADPH and the conversion of radiolabeled arginine to radiolabeled citrulline in P20 samples and found comparable results in Prnp+/+ and Prnp0/0 hippocampi (Supplementary Fig. S3B, C).
The neuroprotective function of PrPC is copper-dependent
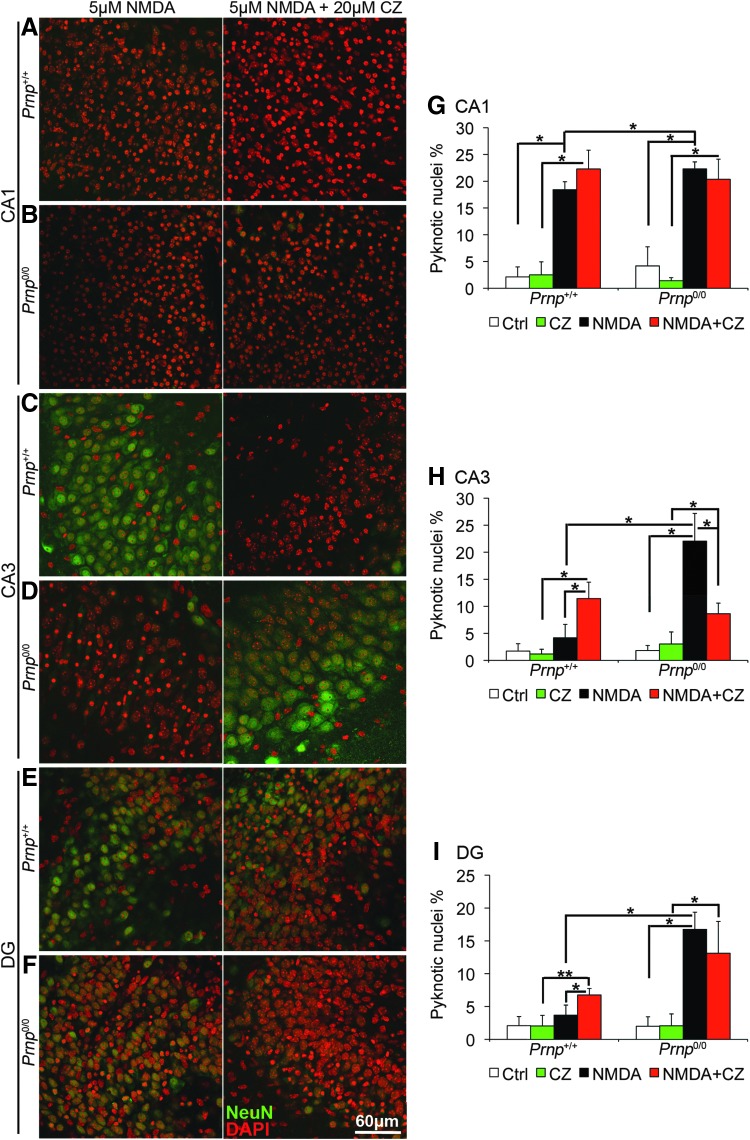
So far, we have shown that in Prnp0/0 mouse OHC, NMDAR S-nitrosylation is not mediated by copper since it is not lowered by CZ addition. However, CZ treatment decreases NMDAR S-nitrosylation in Prnp+/+ mouse OHC to the level detected in Prnp0/0 mouse OHC (Fig. 4). Since extracellular cysteine S-nitrosylation inhibits NMDAR by lowering the frequency of channel opening (23), we measured intracellular calcium [Ca2+]i waves in wild-type mouse hippocampal neurons upon treatment with 20 μM CZ, 5 μM NMDA, and 5 μM NMDA+20 μM CZ (Supplementary Materials and Methods). We found that both CZ and NMDA triggered [Ca2+]i waves, but the addition of CZ to NMDA induced a much higher [Ca2+]i increase (Supplementary Fig. S9). These results indicate that copper chelation enhances NMDAR channel opening. Consistent with this finding, it has been reported that, upon copper chelation, wild-type hippocampal neurons exhibit nondesensitizing NMDAR currents comparable to those registered in Prnp0/0 (40). Therefore, we studied the neurotoxic effects of NMDA exposure in Prnp+/+ and Prnp0/0 OHC in the presence of CZ (n=4 OHC, 5 slices per treatment in each culture). CZ treatment without NMDA did not induce neuronal cell death in neither Prnp+/+ nor Prnp0/0 OHC (Supplementary Fig. S10A–F). Our results confirm the neuroprotective function of PrPC through a copper-dependent inhibition of NMDAR (Fig. 5). In wild-type OHC CA1, copper chelation did not enhance significantly the pyknosis, but it abolished the difference with PrPC-null OHC CA1 (Fig. 5A, G). In wild-type OHC CA3 and DG, CZ addition to NMDA significantly increased neuronal cell death (Fig. 5C, E, H, I; p=0.01219, p=0.02157). In contrast, in Prnp0/0 CA1 and DG, the copper chelator did not alter the susceptibility of neurons to NMDA (Fig. 5 B, F, G, I), whereas in Prnp0/0 CA3, it did decrease neuronal cell death (Fig. 5D, H; p=0.01219). This partial neuroprotection exerted by CZ in Prnp0/0 CA3 may be related to a toxic effect triggered by copper ions released after NMDAR activation in the absence of PrPC. These results indicate that copper is neuroprotective in wild-type mouse OHC; on the contrary, this effect is greatly reduced in PrPC-null OHC, thus suggesting that PrPC and copper can contribute to neuroprotection only if simultaneously present.
FIG. 5.
Copper chelation increases neuronal cell death in Prnp+/+ OHC but not in Prnp0/0. Images from Prnp+/+ and Prnp0/0 OHC areas are reported in rows: (A) Prnp+/+ CA1; (B) Prnp0/0 CA1; (C) Prnp+/+ CA3; (D) Prnp0/0 CA3; (E) Prnp+/+ DG; (F) Prnp0/0 DG. The different treatments are reported in columns: 5 μM NMDA for 3 h, left column; 5 μM NMDA+20 μM CZ for 3 h, right column. NeuN staining is displayed in green and DAPI in red. Confocal microscope fluorescence images were acquired using a 40×/1.30 NA oil objective. Graphs show the comparison of the neuronal pyknotic nuclei percentage, calculated over the total nuclei number, between Prnp+/+ and Prnp0/0 OHC in CA1 (G), CA3 (H), and DG (I); all error bars indicate SD; sample size n=4 OHC, 5 slices per treatment in each culture; *p<0.05, **p<0.01. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
The neuroprotective function of PrPC is NO-dependent
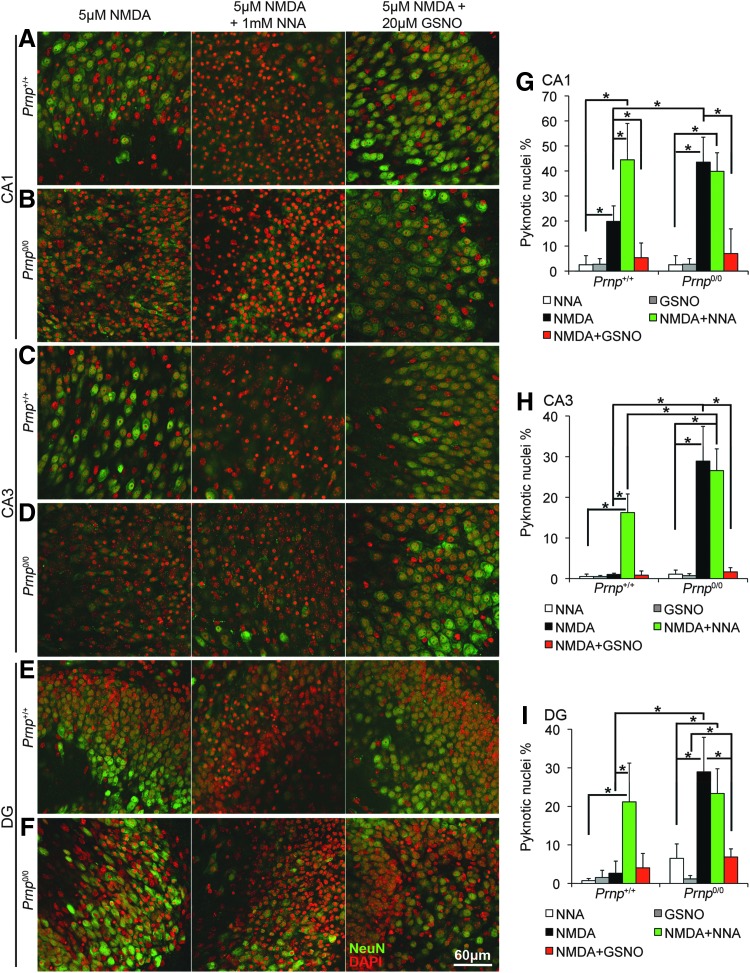
Next, we verified the involvement of NO in PrPC neuroprotective function. To this aim, we studied the neurotoxic effect of NMDA exposure in Prnp+/+ and Prnp0/0 OHC in the presence of either Nω-nitro-l-arginine (NNA), which blocks NO production by all NO synthase isoforms (i.e., nNOS, endothelial NOS, and inducible NOS) or S-nitrosoglutathione (GSNO), a NO donor (n=4 OHC, 5 slices per treatment in each culture). Both NNA and GSNO had no toxic effect (Supplementary Fig. S10A–F). NOS inhibition by NNA significantly enhanced cell death in all the analyzed regions in wild-type hippocampal cultures (Fig. 6A, C, E, G–I; p=0.03038, p=0.03038, p=0.03038), while it had no effect in PrPC-null OHC (Fig. 6B, D, F, G–I). GSNO addition to NMDA significantly increased neuronal cell survival in all the analyzed regions in PrPC-null OHC (Fig. 6B, D, F, G–I; p=0.01996, p=0.03038, p=0.03038) and also in wild-type CA1 region (Fig. 6A, G; p=0.03038).
FIG. 6.
NOS inhibition increases neuronal cell death in Prnp+/+ OHC but not in Prnp0/0, while NO addition enhances neuron survival upon NMDA exposure. Images from Prnp+/+ and Prnp0/0 OHC areas are reported in rows: (A) Prnp+/+ CA1; (B) Prnp0/0 CA1; (C) Prnp+/+ CA3; (D) Prnp0/0 CA3; (E) Prnp+/+ DG; (F) Prnp0/0 DG. The different treatments are reported in columns: 5 μM NMDA for 3 h, left column; 5 μM NMDA+1 mM NNA for 3 h, central column; 5 μM NMDA+20 μM GSNO for 3 h, right column. NeuN staining is displayed in green and DAPI in red. Confocal microscope fluorescence images were acquired using a 40×/1.30 NA oil objective. Graphs show the comparison of the neuronal pyknotic nuclei percentage, calculated over the total nuclei number, between Prnp+/+ and Prnp0/0 OHC in CA1 (G), CA3 (H), and DG (I); all error bars indicate SD; sample size n=4 OHC, 5 slices per treatment in each culture; *p<0.05. GSNO, S-nitrosoglutathione; NNA, Nω-nitro-L-arginine; NO, nitric oxide. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
These results indicate that NOS production of NO is necessary to mediate PrPC neuroprotective function.
Discussion
Determining the function of PrPC is of utmost importance to understand different neurodegeneration mechanisms, in particular those underlying prion disorders and Alzheimer disease (AD). In prion disorders, the loss of PrPC function upon protein aggregation may contribute to the pathology progression (1). PrPC has been found to play a role also in AD (19), where it has been involved in pathological mechanisms together with copper and NMDAR. Recent results indicate that the interaction with Aβ oligomers prevents the PrPC-Cu complex-mediated inhibition of NMDAR, leading to overactivation of the ion channel (40). Zamponi and colleagues propose that PrPC bound to copper limits excessive NMDAR activity by reducing the receptor affinity for glycine. In the present work, we put forward an additional mechanism in which PrPC-Cu modulates NMDAR together with NO. The novelty of this study is represented by the description of the functional interaction of PrPC, copper, and NO. The relevance of copper, NO, and NMDAR overactivation in processes that lead to synapse loss in AD is widely accepted (15). In light of these premises, the mechanism we present here may help our understanding of the pathological processes involved in the progression of both AD and prion disorders.
Here, we argue that PrPC protects glutamatergic synapses from excitotoxic insults by promoting NMDAR S-nitrosylation. Figure 7 shows the proposed model for NMDAR inhibition by PrPC. Glutamate released from the presynaptic terminal activates NMDAR with ensuing calcium influx. In the intracellular compartment, calcium binds to CaM, and the Ca2+/CaM complex activates, among others, nNOS and Atp7a (22, 36); NO and copper are thus released in the synaptic cleft. Because of PrPC high affinity to copper ions (8) and high expression levels in hippocampal synapses (31, 35), it is likely to bind copper released in the synaptic cleft. In this milieu, PrPC-bound Cu(II) can oxidize NO to NO+ and be reduced to Cu(I) (4). This reaction enables the electrophilic attack of NO+ on GluN2A and GluN1 extracellular cysteine thiol groups, resulting in the S-nitrosylation of NMDAR. coIP experiments of PrPC with NMDAR subunits have yielded positive results (40), confirming the proximity of PrPC and NMDAR subunits in lipid rafts (9, 32). The experimental evidence we present here supports this molecular mechanism.
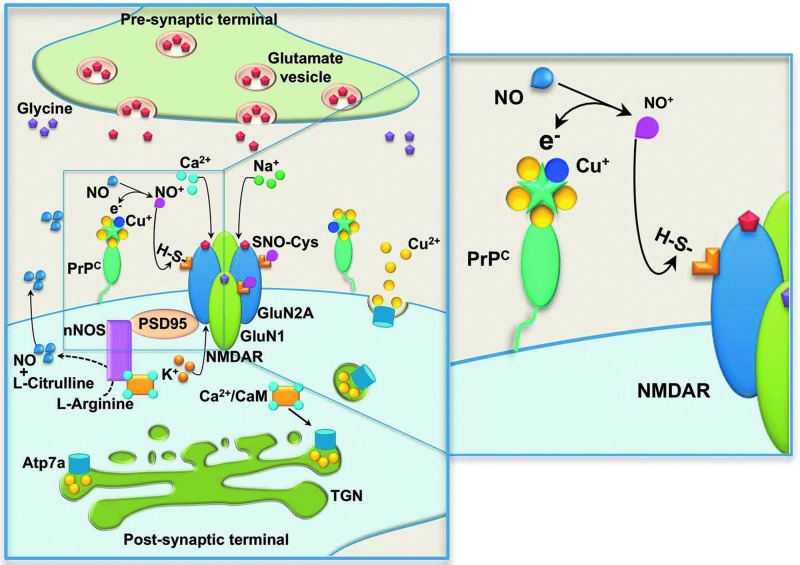
FIG. 7.
Mechanism of PrPC-mediated S-nitrosylation of NMDAR. Glutamate released from the presynaptic terminal activates NMDAR on the postsynaptic terminal. NMDAR activation triggers Na+ and Ca2+ influx, together with K+ efflux. In the intracellular compartment, Ca2+ ions bind different proteins, including calmodulin (CaM). The Ca2+/CaM complex activates, among others, neuronal nitric oxide synthase (nNOS) and copper-transporting ATPase 1 (Atp7a): nNOS activation results in NO release in the synaptic cleft; Atp7a activation in the trans-Golgi network (TGN) results in Cu2+ release in the synaptic cleft. Released Cu2+ ions are immediately bound by copper-binding proteins: PrPC is highly expressed in both presynaptic and postsynaptic terminals, and it can be included in lipid raft domains that also contain NMDARs, and it has high affintiy for both Cu2+ and Cu+. Released NO can react with extracellular cysteines thiols of NMDAR subunits GluN1 and GluN2A, leading to cysteines S-nitrosylation (SNO-Cys). The S-nitrosylation inhibits NMDAR activation by closing the channel. The chemical reaction between NO and the cystein thiol requires the presence of an electron acceptor, often represented by Cu2+. According to this model, PrPC brings the Cu2+ ions that support the reaction of NO with thiols, leading to the S-nitrosylation of GluN1 and GluN2A, thus inhibiting NMDAR. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
NMDAR regulation operated by NO reduces significantly the neurotoxic effect induced by the overactivation of the NMDAR-coupled ionic channel and the ensuing excessive calcium entry (30). S-nitrosylation diminishes the amplitude of NMDAR-evoked currents by reducing the number of channel openings (23). By comparing GluN2A and GluN1 S-nitrosylation levels in wild-type and PrPC-null mouse hippocampi, we found that, in absence of PrPC, NMDAR S-nitrosylation levels are decreased. S-nitrosylation can proceed through different routes not involving copper (14), and PrPC-Cu is not the only complex able to support the copper-mediated reaction; otherwise, no S-nitrosylated NMDAR signal would be detectable in PrPC-null samples. However, the extent of the reduction observed is remarkable, indicating that PrPC-Cu is a key player in this post-translational modification on NMDAR.
Since S-nitrosylation limits NMDAR overactivation, thus staving off apoptosis, PrPC-null neurons should be more susceptible to excitotoxicity. Consistent with this observation and with results reported in the literature (21), we found that PrPC ablation strongly impairs neuronal survival upon toxic exposure to NMDA. By applying a pool of glutamate receptors antagonists, we found that GluN2A-containing NMDAR were mainly responsible for increasing the cell death level in PrPC-null neurons. These results contrast with the notion that GluN2B-enriched extrasynaptic receptors may be key mediators of neuron death (16). However, growing evidence supports an involvement of both GluN2A and GluN2B in synaptic and extrasynaptic NMDAR-mediated excitotoxicity (39, 41, 42). Therefore, it is plausible that a dysfunction in GluN2A inhibition causes higher neuron death in PrPC-null mouse OHC. Interestingly, GluN2A is the subunit containing the cysteine residue that mediates the predominant inhibitory effect of NO (7, 23). This regulation provides a clear link between the increased neuron death level and the lower NMDAR S-nitrosylation detected in PrPC-null mouse hippocampus. In situ hybridization experiments have shown that PrPC knockout mouse hippocampus has higher GluN2A mRNA levels (27). However, we excluded the possibility that the higher susceptibility of PrPC-null mouse OHC was due to an overexpression of NMDAR at synapses, as we revealed comparable levels of interaction between PSD95 and GluN2A, and indirectly with GluN1, in wild-type and PrPC-null mouse hippocampi.
By measuring GluN2A and GluN1 S-nitrosylation levels in CZ-treated wild-type and PrPC-null mouse OHC by CZ addition, we found that copper chelation lowers GluN2A and GluN1 S-nitrosylation in wild-type cultures but not in PrPC-null ones. The residual signal detected in wild-type upon copper deprivation was not different from that in treated and untreated PrPC knockout cultures.
In line with previously published electrophysiological recordings of NMDAR currents (40), our results on S-nitrosylation highlight that PrPC regulation of NMDAR activity is copper dependent. Consistent with this fact, we found that also the neuroprotective effect of PrPC requires copper. These results convincingly indicate that in glutamatergic synapses, the neuroprotective action of PrPC depends on copper ions and that, vice versa, the neuroprotective action of copper relies on PrPC expression. This finding has relevant implications in defining copper-mediated modulation of NMDAR and the involvement of copper in neuroprotection, as these functions are impaired in both AD and prion diseases. Moreover, it is known that copper metabolism dysfunction is a primary cause of other pathologies, such as Menkes' disease (MD)—a disorder caused by a loss-of-function mutation in the Atp7a-encoding gene that results in altered copper trafficking (26). Indeed, hippocampal neurons derived from a MD murine model have shown altered NMDAR current and increased damage upon excitotoxic stimulus (37). Strikingly, this phenotype is very similar to that observed in PrPC-null models (21, 34, 38).
Our findings indicate that PrPC, copper, and NO regulate NMDAR and impart neuroprotection through a common mechanism that is likely to be lost in AD, MD, and prion diseases, thus leading to synapse degeneration. In this mechanism, PrPC-bound copper ions act as electron acceptors in the reaction between NO and NMDAR cysteine thiols. The ensuing S-nitrosylation inhibits the receptor and reduces the neurotoxic effect caused by its overactivation. This mechanism is neuroprotective and crucial for normal synaptic functionality.
As in the case of NMDAR, PrPC-Cu may also promote the S-nitrosylation of other membrane proteins. Indeed, inducing protein S-nitrosylation may be an overarching function of PrPC. The novel mechanism presented here may provide a platform to investigate additional and previously unforeseen cellular processes in which PrPC may be involved.
Materials and Methods
Animals
All experiments were performed in accordance with European regulations (European Community Council Directive, November 24, 1986 [86/609/EEC]). Experimental procedures were notified to and approved by the Italian Ministry of Health, Directorate General for Animal Health. All experiments were approved by the local authority veterinary service of Trieste, Italy, and by the Ethics Committee of the Scuola Internazionale Superiore di Studi Avanzati (SISSA), Trieste. All efforts were made to minimize animal suffering and to reduce the number of animals used. Inbred FVB/N (Friend virus B-type susceptibility-NIH) wild-type and FVB Prnp0/0 mice were used in these experiments. The FVB Prnp0/0 mice were obtained from George A. Carlson, McLaughlin Research Institute (Great Falls, MT) and were bred by backcrossing with the original Prnp0/0 mice at least 20 times (25).
S-nitrosylated protein detection: biotin switch assay
The biotin switch assay was performed, as described by Jaffrey and Snyder (18) with a few modifications. Mouse hippocampal samples (age-matched samples from 6-, 10-, and 12-month-old male animals) were dissected, immediately frozen in liquid nitrogen, and stored at −80°C. Each hippocampus was homogenized in 400 μl HEN buffer: 250 mM HEPES pH 7.5 (H3375; Sigma-Aldrich, St Louis, MO), 1 mM EDTA (E6758; Sigma-Aldrich), 0.1 mM neocuproine (N1501; Sigma-Aldrich), protease inhibitors cocktail (Roche Diagnostics Corp., Mannheim, Germany), and centrifuged at 2000 g, 10 min at 4°C. To evaluate the role of copper in NMDAR S-nitrosylation, Prnp+/+ and Prnp0/0 OHC were treated for 3 h with 20 μM CZ (14690; Sigma-Aldrich) from the 4 mM stock in 50% ethanol, then immediately frozen. For each experiment, at least 60 slices per group were collected and homogenized in 100 μl HEN buffer by sonication. Protein concentration in the supernatant was determined by bicinchoninic acid assay and the same amount of proteins per sample was used for the further steps of the protocol (1 mg for hippocampal samples and 0.7–1 mg for OHC depending on the lowest sample concentration in each experiment). All the reagents were used proportionally. Samples were diluted to 0.8 μg/μl in HEN buffer+0.04% CHAPS (C9426; Sigma-Aldrich) and incubated at 50°C for 1 h in 4 volumes of blocking solution: 9 volumes of HEN, 1 volume of 25% w/v SDS (L3771; Sigma-Aldrich) in ddH2O, and 20 mM S-methyl thiomethanesulfonate (64306; Sigma-Aldrich) from the 2 M stock solution in dimethylformamide. S-methyl thiomethanesulfonate was removed by acetone precipitation and the protein pellet was resuspended in 100 μl of HENS buffer (HEN+1%SDS) per mg of starting proteins. To selectively and efficiently reduce S-nitrosylated thiols, ascorbate (A4034; Sigma-Aldrich), prepared as 180 mM in ddH2O, was added to 30 mM final concentration in the samples, and the reaction was carried out for 3 h at 25°C (14). A negative control without ascorbate was performed on Prnp+/+ samples to check the efficiency of thiols blocking with S-methyl thiomethanesulfonate. Ascorbate was removed by acetone precipitation and the protein pellet was resuspended in 100 μl of HENS buffer per mg of starting proteins. EZ-Link HPDP-biotin (21341; Thermoscientific, Waltham, MA) was used to bind free thiols, corresponding to the previously S-nitrosylated residues. It was added 1:3 to the samples from the 4 mM stock in dimethylformamide. A negative control without biotin was performed on Prnp+/+ samples to check the selective binding to the resin. After 1 h incubation at 25°C, HPDP-biotin was removed by dialysis in HENS buffer. To purify biotinylated proteins, 2 volumes of neutralization buffer (20 mM HEPES pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.5% Triton X-100 [X100; Sigma-Aldrich]) and 50 μl of wet Immobilized NeutrAvidin Agarose (29200; Thermoscientific) were added to the samples and incubated for 2 h at room temperature (RT). Resin was washed 5 times with neutralization buffer adjusted to 600 mM NaCl; biotinylated proteins were eluted directly in SDS-PAGE sample buffer, boiled, and processed for Western blot detection. For each sample, 30 μg of protein extract were loaded as input. The following primary antibodies were used: anti-GluN1 1:500–1:2000 (G8913; Sigma-Aldrich); anti-GluN2A 1:500–1:2000 (G9038; Sigma-Aldrich); monoclonal anti-PrP SHA31 1 μg/ml (A03213; BertinPharma, Montigny le Bretonneux, France); anti-β-actin Peroxidase (AC-15) 1:10,000 (A3854; Sigma-Aldrich). After incubation with the secondary antibody, membranes were developed with the ECL detection reagent (GE Healthcare, Waukesha, WI) and recorded by the digital imaging system Alliance 4.7 (UVITEC, Cambridge, United Kingdom). Band quantification was performed with Uviband 15.0 software (UVITEC) obtaining an optical density (OD) value. To normalize OD values, the following formula was applied: (NMDAR subunit S-nitrosylation/input)/(β-actin S-nitrosylation/input). Basically, S-nitrosylation signals were normalized on the corresponding input. Then, NMDAR subunit values were normalized on β-actin value. Prnp+/+ and Prnp0/0 samples were compared by the Mann–Whitney test setting F(x)<>G(x) as alternate hypothesis.
Organotypic hippocampal cultures preparation and treatments
OHC preparation protocol was set up based on Gahwiler's protocol (13). Briefly, in aseptic condition, P5 mouse hippocampus was dissected in the dissection medium (Gey's balanced salt solution, 5.6 mM d-glucose [G8270; Sigma-Aldrich], 1 mM kynurenic acid [K3375; Sigma-Aldrich]) and sliced by means of a tissue chopper into 300-μm-thick sections. After washing in the dissection medium for 40 min at 4°C, slices showing an intact hippocampal cytoarchitecture were selected, singularly attached to a coverslip by embedding in a chicken plasma (P3266; Sigma-Aldrich) and thrombin clot (112374; Merck KGaA, Darmstadt, Germany) and maintained at 10 rotations per hour at 37°C in Nunc cell culture tubes (156758; Thermoscientific) with a medium composed of 50% basal medium Eagle (41010026; Gibco, Carlsbad, CA), 25% horse serum (26050-088; Gibco), 25% Hank's balanced salt solution (24020141; Gibco), 5.6 mM d-glucose, 2 mM l-glutamine (25030-032; Gibco). After 13 days in vitro, OHC were treated in the serum-free medium, as described in Supplementary Table S1. The drug stock solutions were prepared as follows: 1 mM NMDA (0114; Tocris Biosciences, Missouri, United Kingdom) in ddH2O stored at −20°C; 250 mM EGTA (E4378; Sigma-Aldrich) in phosphate-buffered saline (PBS), pH 7.4, stored at RT; 40 mM CNQX (C239; Sigma-Aldrich) in dimethyl sulfoxide (DMSO, D8418; Sigma-Aldrich) stored at −20°C; 10 mM (2R)-amino-5-phosphonovaleric acid (AP5, A5282; Sigma-Aldrich) in ddH2O stored at −20°C; 5 mM threo ifenprodil hemitartrate (2892; Tocris Biosciences) in ddH2O stored at −20°C; 10 mM GSNO (N4148; Sigma-Aldrich) in ddH2O stored at −20°C; freshly prepared three NNA (N5501; Sigma-Aldrich) in serum-free medium; freshly prepared 4 mM CZ in 50% ethanol. To induce excitotoxicity, OHC were exposed either to 5 μM NMDA for 3 h or to 10 μM NMDA for 10 min. Experiments with EGTA were performed using 2 mM EGTA in combination with either 5 μM NMDA for 1.5 h (EGTA treatment for 3 h was toxic for OHC) or 10 μM NMDA for 10 min. Treatments with 5 μM NMDA in combination with either 50 μM AP5 or 20 μM CNQX or 20 μM GSNO or 1 mM NNA or 20 μM CZ were incubated for 3 h. To obtain a more complete inhibition, AP5, CNQX, and NNA were preincubated for 30 min before adding NMDA. Treatments with NMDA and ifenprodil were carried out differently because of the toxic effect of ifenprodil when associated to low NMDA concentrations (20). For this reason, OHC were exposed to 10 μM NMDA and 3 μM ifenprodil for 10 min. To evaluate neuronal cell death, all the described treatments were followed by a 24 h washout before processing OHC for immunofluorescence.
Immunofluorescence
To evaluate neuronal cell death, slices were fixed in 4% paraformaldehyde (P6148; Sigma-Aldrich) in PBS, pH 7.4, O/N at 4°C. To improve the nuclear staining, slices were treated as follows: 3×5 min washes in PBS with 1% Triton X-100, 10 min in 0.1 N HCl at 4°C, 10 min in 0.2 N HCl at RT, 20 min in 0.2 N HCl at 37°C, 12 min in borate buffer pH 8.4 at RT, 3×5 min washes in PBS with 1% Triton X-100. Slices were incubated for 2 h at RT in the blocking solution (1 M glycin [G8898; Sigma-Aldrich], 5% normal goat serum [005-000-121; Jackson ImmunoResearch] in PBS with 1% Triton X-100) and O/N at 4°C with anti-neuronal nuclei (NeuN) clone A60 antibody 1:500 (mab377; Millipore, Billerica, MA) in the blocking solution. After washing with PBS and 1% Triton X-100, slices were incubated with Alexa fluorophore-conjugated secondary antibodies (1:500, R37120; Invitrogen, Carlsbad, CA) and 4′,6-diamidino-2-phenylindole (DAPI, 1:500) in the blocking solution for 2 h at RT. Slices were washed in PBS, rinsed in water, and mounted with VectaShield (Vector Laboratories, Burlingame, CA).
Confocal microscopy, image analysis, and statistics of OHC experiments
Immunofluorescence images were acquired on a Leica DMIRE2 confocal microscope (Leica Microsystem GmbH, Wetzlar, Germany) equipped with DIC and fluorescent optics, diode laser 405 nm, and Ar/ArKr 488 nm lasers. The fluorescence images (1024×1024 pixels) were acquired with a 40×/1.30 NA oil objective, additionally zoomed 1.5-fold, with 200 Hz acquisition speed. Stacks of z-sections with an interval of 3 μm were sequentially scanned. CA1, CA3, and DG were the regions selected for the analysis. A protocol for automatic count of the total amount of nuclei was set up for each hippocampal zone with the Volocity 5.4 3D imaging software (PerkinElmer, Coventry, United Kingdom). The same software was used to manually count the pyknotic nuclei. The dead cell count was performed as a blind analysis. The CA1, CA3, and DG regions were clearly distinguishable in OHC used in these experiments (Supplementary Fig. S11A) and the staining protocol allowed for the exact identification of neuronal pyknotic nuclei (Supplementary Fig. S11B). Pyknotic nuclei count results for Prnp+/+ and Prnp0/0 OHC were compared by performing the Mann–Whitney test setting F(x)<>G(x) as alternate hypothesis. Each culture preparation (resulting from three to four mice dissection) was considered as one sample (n=1). In each culture, the average pyknotic nuclei percentage among the hippocampal slices (four to five slices) of the same treatment group was calculated, grouped with values obtained from other OHC of the same genotype with the same treatment for a final sample size of four (n=4) and compared with the other treatments and genotype by means of the Mann–Whitney test.
Supplementary Material
Abbreviations Used
- AD
Alzheimer disease
- AP5
(2R)-amino-5-phosphonovaleric acid
- Atp7a
copper-transporting ATPase 1
- CA1
Cornus Ammonis 1
- CA3
Cornus Ammonis 3
- CaM
calmodulin
- CNQX
6-cyano-7-nitroquinoxaline-2,3-dione
- coIP
co-immunoprecipitation
- CZ
cuprizone
- DAPI
4′,6-diamidino-2-phenylindole
- DG
dentate gyrus
- DMSO
dimethyl sulfoxide
- EGTA
ethylene glycol tetraacetic acid
- GluN1
NMDAR subunit GluN1
- GluN2A
NMDAR subunit GluN2A
- GSNO
S-nitrosoglutathione
- MD
Menkes' disease
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NeuN
anti-neuronal nuclei marker antibody
- NMDA
N-methyl-d-aspartate
- NMDAR
N-methyl-d-aspartate receptors
- NNA
Nω-nitro-l-arginine
- nNOS
neuronal nitric oxide synthase
- NO
nitric oxide
- OD
optical density
- OHC
organotypic hippocampal cultures
- Prnp+/+, Prnp0/0
PrPC wild type, PrPC knockout
- PrPC
cellular prion protein
- PSD95
postsynaptic density protein 95
- SNO-Cys
cysteine S-nitrosylation
Acknowledgments
This work was supported by a grant from the Italian Ministry for Education, University and Research to G.L. (Grant MIUR-PRIN 2010/2011–A.AC.NSCI.780). F.B. gratefully acknowledges the International School for Advanced Studies (SISSA), Trieste, for partially supporting this work through the “Young SISSA Scientists' Research Projects” 2011–2012 scheme. The authors are grateful to Diego Favretto, Sara De Iudicibus, and Prof. Giuliana Decorti (Department of Life Sciences, University of Trieste) for their expertise in the analytical measurement of the NOS activity with radioactive substrate. The authors thank Prof. Enrico Cherubini for helpful scientific discussion and suggestions, Roberta Antonelli for her assistance in coIP experiments, and Erica Sarnataro for editing and proofreading the article.
Author Disclosure Statement
The authors declare no competing financial interests exist.
References
- 1.Aguzzi A, Baumann F, and Bremer J. The prion's elusive reason for being. Annu Rev Neurosci 31: 439–477, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Benetti F, Ventura M, Salmini B, Ceola S, Carbonera D, Mammi S, Zitolo A, D'Angelo P, Urso E, Maffia M, Salvato B, and Spisni E. Cuprizone neurotoxicity, copper deficiency and neurodegeneration. Neurotoxicology 31: 509–517, 2010 [DOI] [PubMed] [Google Scholar]
- 3.Benvegnu S, Poggiolini I, and Legname G. Neurodevelopmental expression and localization of the cellular prion protein in the central nervous system of the mouse. J Comp Neurol 518: 1879–1891, 2010 [DOI] [PubMed] [Google Scholar]
- 4.Bonomo RP, Pappalardo G, Rizzarelli E, Tabbi G, and Vagliasindi LI. Studies of nitric oxide interaction with mono- and dinuclear copper(II) complexes of prion protein bis-octarepeat fragments. Dalton Trans 3805–3816, 2008 [DOI] [PubMed] [Google Scholar]
- 5.Brown DR, Qin K, Herms JW, Madlung A, Manson J, Strome R, Fraser PE, Kruck T, von Bohlen A, Schulz-Schaeffer W, Giese A, Westaway D, and Kretzschmar H. The cellular prion protein binds copper in vivo. Nature 390: 684–687, 1997 [DOI] [PubMed] [Google Scholar]
- 6.Caiati MD, Safiulina VF, Fattorini G, Sivakumaran S, Legname G, and Cherubini E. PrPC controls via protein kinase A the direction of synaptic plasticity in the immature hippocampus. J Neurosci 33: 2973–2983, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi YB, Tenneti L, Le DA, Ortiz J, Bai G, Chen HS, and Lipton SA. Molecular basis of NMDA receptor-coupled ion channel modulation by S-nitrosylation. Nat Neurosci 3: 15–21, 2000 [DOI] [PubMed] [Google Scholar]
- 8.D'Angelo P, Della Longa S, Arcovito A, Mancini G, Zitolo A, Chillemi G, Giachin G, Legname G, and Benetti F. Effects of the pathological Q212P mutation on human prion protein non-octarepeat copper-binding site. Biochemistry 51: 6068–6079, 2012 [DOI] [PubMed] [Google Scholar]
- 9.Delint-Ramirez I, Fernandez E, Bayes A, Kicsi E, Komiyama NH, and Grant SG. In vivo composition of NMDA receptor signaling complexes differs between membrane subdomains and is modulated by PSD-95 and PSD-93. J Neurosci 30: 8162–8170, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong Z, Saikumar P, Weinberg JM, and Venkatachalam MA. Calcium in cell injury and death. Annu Rev Pathol 1: 405–434, 2006 [DOI] [PubMed] [Google Scholar]
- 11.Foster MW, McMahon TJ, and Stamler JS. S-nitrosylation in health and disease. Trends Mol Med 9: 160–168, 2003 [DOI] [PubMed] [Google Scholar]
- 12.Fournier JG, Escaig-Haye F, Billette de Villemeur T, Robain O, Lasmezas CI, Deslys JP, Dormont D, and Brown P. Distribution and submicroscopic immunogold localization of cellular prion protein (PrPc) in extracerebral tissues. Cell Tissue Res 292: 77–84, 1998 [DOI] [PubMed] [Google Scholar]
- 13.Gahwiler BH, Capogna M, Debanne D, McKinney RA, and Thompson SM. Organotypic slice cultures: a technique has come of age. Trends Neurosci 20: 471–477, 1997 [DOI] [PubMed] [Google Scholar]
- 14.Gow AJ. and Ischiropoulos H. Nitric oxide chemistry and cellular signaling. J Cell Physiol 187: 277–282, 2001 [DOI] [PubMed] [Google Scholar]
- 15.Greenough MA, Camakaris J, and Bush AI. Metal dyshomeostasis and oxidative stress in Alzheimer's disease. Neurochem Int 62: 540–555, 2013 [DOI] [PubMed] [Google Scholar]
- 16.Hardingham GE. and Bading H. Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat Rev Neurosci 11: 682–696, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hess DT, Matsumoto A, Kim SO, Marshall HE, and Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol 6: 150–166, 2005 [DOI] [PubMed] [Google Scholar]
- 18.Jaffrey SR. and Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001: pl1, 2001 [DOI] [PubMed] [Google Scholar]
- 19.Kellett KA. and Hooper NM. Prion protein and Alzheimer disease. Prion 3: 190–194, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kew JN, Trube G, and Kemp JA. A novel mechanism of activity-dependent NMDA receptor antagonism describes the effect of ifenprodil in rat cultured cortical neurones. J Physiol 497 (Pt 3): 761–772, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khosravani H, Zhang Y, Tsutsui S, Hameed S, Altier C, Hamid J, Chen L, Villemaire M, Ali Z, Jirik FR, and Zamponi GW. Prion protein attenuates excitotoxicity by inhibiting NMDA receptors. J Cell Biol 181: 551–565, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kone BC, Kuncewicz T, Zhang W, and Yu ZY. Protein interactions with nitric oxide synthases: controlling the right time, the right place, and the right amount of nitric oxide. Am J Physiol Renal Physiol 285: F178–F190, 2003 [DOI] [PubMed] [Google Scholar]
- 23.Lipton SA, Choi YB, Takahashi H, Zhang D, Li W, Godzik A, and Bankston LA. Cysteine regulation of protein function—as exemplified by NMDA-receptor modulation. Trends Neurosci 25: 474–480, 2002 [DOI] [PubMed] [Google Scholar]
- 24.Liu L, Jiang D, McDonald A, Hao Y, Millhauser GL, and Zhou F. Copper redox cycling in the prion protein depends critically on binding mode. J Am Chem Soc 133: 12229–12237, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lledo PM, Tremblay P, DeArmond SJ, Prusiner SB, and Nicoll RA. Mice deficient for prion protein exhibit normal neuronal excitability and synaptic transmission in the hippocampus. Proc Natl Acad Sci U S A 93: 2403–2407, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lutsenko S. and Petris MJ. Function and regulation of the mammalian copper-transporting ATPases: insights from biochemical and cell biological approaches. J Membr Biol 191: 1–12, 2003 [DOI] [PubMed] [Google Scholar]
- 27.Maglio LE, Perez MF, Martins VR, Brentani RR, and Ramirez OA. Hippocampal synaptic plasticity in mice devoid of cellular prion protein. Brain Res Mol Brain Res 131: 58–64, 2004 [DOI] [PubMed] [Google Scholar]
- 28.Mallucci GR, Ratte S, Asante EA, Linehan J, Gowland I, Jefferys JG, and Collinge J. Post-natal knockout of prion protein alters hippocampal CA1 properties, but does not result in neurodegeneration. EMBO J 21: 202–210, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mani L, Cheng F, Havsmark B, Jonsson M, Belting M, and Fransson L. Prion, amyloid-derived Cu(II) ions, or free Zn(II) ions support S-nitroso-dependent autocleavage of glypican-1 heparan sulfate. J Biol Chem 278: 38956–38965, 2003 [DOI] [PubMed] [Google Scholar]
- 30.McBain CJ. and Mayer ML. N-methyl-D-aspartic acid receptor structure and function. Physiol Rev 74: 723–760, 1994 [DOI] [PubMed] [Google Scholar]
- 31.Moya KL, Sales N, Hassig R, Creminon C, Grassi J, and Di Giamberardino L. Immunolocalization of the cellular prion protein in normal brain. Microsc Res Tech 50: 58–65, 2000 [DOI] [PubMed] [Google Scholar]
- 32.Naslavsky N, Stein R, Yanai A, Friedlander G, and Taraboulos A. Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. J Biol Chem 272: 6324–6331, 1997 [DOI] [PubMed] [Google Scholar]
- 33.Prusiner SB. Shattuck lecture—neurodegenerative diseases and prions. N Engl J Med 344: 1516–1526, 2001 [DOI] [PubMed] [Google Scholar]
- 34.Rangel A, Burgaya F, Gavin R, Soriano E, Aguzzi A, and Del Rio JA. Enhanced susceptibility of Prnp-deficient mice to kainate-induced seizures, neuronal apoptosis, and death: Role of AMPA/kainate receptors. J Neurosci Res 85: 2741–2755, 2007 [DOI] [PubMed] [Google Scholar]
- 35.Sales N, Rodolfo K, Hassig R, Faucheux B, Di Giamberardino L, and Moya KL. Cellular prion protein localization in rodent and primate brain. Eur J Neurosci 10: 2464–2471, 1998 [DOI] [PubMed] [Google Scholar]
- 36.Schlief ML, Craig AM, and Gitlin JD. NMDA receptor activation mediates copper homeostasis in hippocampal neurons. J Neurosci 25: 239–246, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlief ML, West T, Craig AM, Holtzman DM, and Gitlin JD. Role of the Menkes copper-transporting ATPase in NMDA receptor-mediated neuronal toxicity. Proc Natl Acad Sci U S A 103: 14919–14924, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spudich A, Frigg R, Kilic E, Kilic U, Oesch B, Raeber A, Bassetti CL, and Hermann DM. Aggravation of ischemic brain injury by prion protein deficiency: role of ERK-1/-2 and STAT-1. Neurobiol Dis 20: 442–449, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Wroge CM, Hogins J, Eisenman L, and Mennerick S. Synaptic NMDA receptors mediate hypoxic excitotoxic death. J Neurosci 32: 6732–6742, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.You H, Tsutsui S, Hameed S, Kannanayakal TJ, Chen L, Xia P, Engbers JD, Lipton SA, Stys PK, and Zamponi GW. Abeta neurotoxicity depends on interactions between copper ions, prion protein, and N-methyl-D-aspartate receptors. Proc Natl Acad Sci U S A 109: 1737–1742, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X, Ding Q, Chen Z, Yun H, and Wang H. Involvement of the GluN2A and GluN2B subunits in synaptic and extrasynaptic N-methyl-D-aspartate receptor function and neuronal excitotoxicity. J Biol Chem 288: 24151–24159, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou X, Hollern D, Liao J, Andrechek E, and Wang H. NMDA receptor-mediated excitotoxicity depends on the coactivation of synaptic and extrasynaptic receptors. Cell Death Dis 4: e560, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.