Abstract
Streptococcus mutans and Aggregatibacter actinomycetemcomitans are oral pathogens associated with dental caries and periodontitis, respectively. The aim of this study was to determine the colonization of these two microorganisms in the dental plaque of a group of Haitian adolescents using two different polymerase chain reaction (PCR) methods, standard PCR, and quantitative real-time PCR (qPCR) assays. Fifty-four pooled supra-gingival plaque samples and 98 pooled sub-gingival plaque samples were obtained from 104 12- to 19-year-old rural-dwelling Haitians. The total genomic DNA of bacteria was isolated from these samples, and all participants also received caries and periodontal examinations. Caries prevalence was 42.2%, and the mean decayed, missing, and filled surface (DMFS) was 2.67±5.3. More than half of the adolescents (53.3%) experienced periodontal pockets (Community Periodontal Index score ≥:3). S. mutans was detected in 67.3% by qPCR and 38.8% by PCR of the supra-gingival plaque samples (p<0.01), and 36.6% by qPCR and 8.1% by PCR of the sub-gingival samples (p<0.01). A. actinomycetemcomitans was detected in 85.1% by qPCR and 44.0% by PCR of the sub-gingival samples (p< 0.01), but the prevalence was similar, 67.3% by qPCR and 59.2% by PCR, in the supra-gingival plaque samples. Neither age nor gender was significantly correlated to the bacterial colonization. The results demonstrated a moderate-to-high prevalence of S. mutans and A. actinomycetemcomitans in the Haitian adolescent population, and qPCR is more sensitive than standard PCR in field conditions. These findings suggest that qPCR should be considered for field oral epidemiologic studies and may be necessary in investigations having major logistic challenges.
Keywords: Haitian adolescents, PCR, qPCR, Dental caries, Periodontitis, Streptococcus mutans, Aggregatibacter actinomycetemcomitans
Introduction
The microbial population in dental plaque is known to be highly complex. A number of microorganisms are significantly associated with the development of human dental caries and periodontitis. Specifically, Streptococcus mutans and Aggregatibacter actinomycetemcomitans are two oral pathogenic bacteria which have been extensively studied for their positive associations with dental caries [1 ,2] and periodontal diseases [3–5], respectively. Traditionally, determination of these two microorganisms in the oral cavity has been based on bacterial cultivation methods that are impractical for field epidemiological studies. In recent years, molecular approaches, such as standard polymerase chain reaction (PCR) and quantitative real-time PCR (qPCR) with species-specific primers, have been developed, commonly used, and demonstrated a high degree of efficiency and accuracy in the detection of target micro- organisms and the evaluation of bacterial colonization in saliva and dental plaque samples [6–9).
Although numerous studies have qualitatively and quantitatively described the microbiology of caries and periodontitis in various populations [10, 11], studies on the colonization of S. mutans and A. actinomycetemcomitans in Haitian adolescents have not been conducted, mainly due to the lack of comprehensive microbiology laboratories for oral bacterial cultivation and quantification in Haiti, a situation applicable to other developing countries. In the present study, we aimed to qualitatively and quantitatively determine, using both standard PCR and qPCR assays, the colonization of S. mutans and A. actinomycetemcomitans in the dental plaque of a group of Haitian adolescents. Meanwhile, we sought to determine the feasibility of utilizing PCR techniques for an epidemiological field study, especially in remote rural areas. The information generated from the study will provide baseline data and validated microbiological approaches for future studies in Haiti and elsewhere.
Methods
Study population
The present study was pa11 of a larger investigation to determine whether early childhood (<60 months of age) protein-energy malnutrition (ECPEM) is related to oral diseases and conditions in adolescents [12, 13]. The original study included a total of I ,058 Haitian adolescents in the rural Jeremie area of Haiti [12]. All subjects were selected from a list of children maintained by the Haitian Health Foundation on whom anthropometric data were collected during 1988–1996. For this study, 104 adolescents aged 12–19 years old who were in the original ECPEM study and from two road-accessible villages, Fr. Torbeck and Carretour-Prince, were re-examined. The subjects had a range of ECPEM levels from malnourished to normaL Informed consent was obtained from each parent and assent from adolescents. The study proposal was approved by the New York University School of Medicine Institutional Review Board. Field data collection was conducted between May and August 2005.
Clinical examination
Each examination included a dental examination and dental plaque sample collection. Dental caries examinations were completed by four study examiners and were scored for DMFS, decayed (D), missing due to caries (M), and filled (F) surface (S), using World Health Organization (WHO) diagnostic criteria. Two trained and calibrated examiners performed all periodontal examinations and plaque sampling. Periodontal pockets were measured using a pressure-sensitive periodontal probe (Vivadent, Lichtenstein), and disease was classified using the WHO Community Periodontal Index (CPI).
Bacterial sample collection and transportation
From the 104 subjects, a total of 152 viable pooled dental plaque samples were randomly collected: 54 were supragingival plaque samples defmed as supra-gingival group, and 98 were sub-gingival plaque samples defmed as sub-gingival group. For the supra-gingival plaque sample collection, all teeth present in the mouth were sampled using a modified plaque sample collection method for field studies [14,15]. Briefly, a sterile toothpick was rubbed across all upper and lower teeth buccaVlabial gingival margin swfaces. Tn order to maximize plaque sample collection, a second toothpick was placed into each interproximal tooth swface. After carefully removing supra-gingival plaque, sterile periodontal curettes were used to collect sub-gingival plaque from all available gingival sulci and periodontal pockets [16]. The toothpicks and the curettes were immediately immersed in separate DNase- and RNase-free 2-ml pre-labeled Eppendorf tubes containing 500 μl lysis buffer (Epicentre, Madison, WI, USA).
All specimen vials were packed in coolers with cold packs supplemented with ice. The plaque samples were transfered from the field sites to a field office with a 4°C refrigerator within a maximum of 8 h. Flights were scheduled so that the plaque samples were transferred from the field office in Haiti to the microbiology laboratory at the NYU College of Dentistry within a maximum of 24 h packed in coolers with ice refreshed after 8 h if necessary, and then stored immediately in a −80°C freezer.
PCR evaluation methods of bacterial colonization
Total bacterial genomic DNA was extracted from each plaque sample using the Epicentre method (Epicentre, Madison, WI, USA) as described previously [17, 18]. Briefly, a MasterPure DNA purification kit was used for bacterial cells lysis and total DNA precipitation. The final quantity and quality of the DNA was evaluated using DU-7400 UV- VIS spectrophotometer at OD260/0D280. A standard concentration of 10 ng/μl was prepared for each individual sample for all PCR assays.
Initially, a standard PCR was performed using a standardized protocol and GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA, USA). The S. mutans species-specific primer set generated a 479-bp amplicon: forward, 5’-TCG CGA AAA AGA TAA ACA AAC A-3’, and reverse, 5’-GCC CCT TCA CAG TTG GTT AG-3’ [19]. The PCR reaction mixture for S. mutans contained a total volume of 25 μl, which consisted of 1 Ox buffer, 2.5 mM/ea dNTPs, 10 pmol/μl primers, 5 U/μl Taq, 50 mm MgCI2, and 10 ng/μl DNA template. The positive control for S. mutans was UA159 (ATCC700610). The standard PCR conditions were performed at 95°C for 5 min for initial denaturing, followed by 35 cycles at 95°C for 15 s, 56°C for 30 s, 72°C for 1 min, and a 72°C extension for 7 min. The minimum detectable level was found to be 16 fg or 7 copies/μl for the S. mutans genomic DNA.
The species-specific primers for A. actinomycetemcomitans were selected based on published literature. The primer set generated a 194-bp amplicon: forward, 5’-ATT GGG GTT TAG CCC TGG T-3’, and reverse, 5’-GGC ACA AAC CCA TCT CTG A-3’ [20]. The PCR reaction mixture for A. actinomycetemcomitans contained a total vo lume of 25 μl, which consisted of 10× buffer, 2.5 mM/ea dNTPs, lO pmol/μl primers, 5 U /μl Taq, 50 mm MgCI2, and 10 ng/μl DNA template. The positive control for A. actinomycetemcomitans was ATCC29522. The standard PCR conditions were performed at 95°C for 4 min for initial denaturing, followed by 40 cycles at 95°C for 15 s, 58°C for 20 s, 72°C for 40 s, and a 72°C extension for 4 min. The minimum detectable level was found to be 10 fg or 4 copies/μl for the A. actinomycetemcomitans genomic DNA.
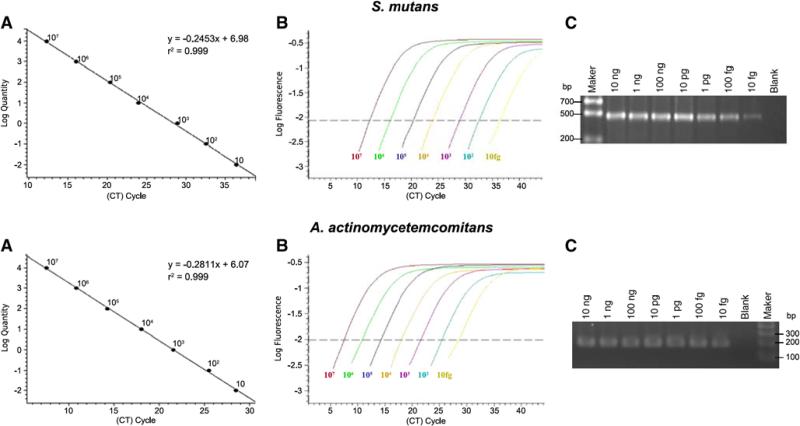
As the standard PCR cannot provide quantitative measurement of the two bacterial colonizations, we further performed quantitative real-time PCR to determine the amounts of S. mutans and A. actinomycetemcom itans DNA in the plaque samples, using DNA Engine Opticon™ 2 System (MJ Research, San Francisco, CA, USA) and the SYBR Green T fluorescent dye detection kit (Qiagen, Valencia, CA, USA). The same sets of primers used for standard PCR were used for qPCR. Tn order to validate the qPCR assay, standard curves were first obtained by externally quantified standards of UA159 for S. mutans and ATCC29522 for A. actinomycetemcomitans. The amount of DNA template for the standard curve was tenfold serial dilutions from 107 to 10 fg/μl. The minimum detectable level by using qPCR was 10 fg or 4 copies/μl of genomic DNA for both S. mutans and A. actinomycetemcomitans ATCC type strains, respectively (Fig. 1).
Fig. 1.
Quantitative real-time polymerase chain reaction (qPCR) validation. a Standard curves for Streptococcus mutans (UAI59) and Aggregatibacter actinomycetemcomitans (ATCC29522) generated from a plot of threshold cycle (C,) values against log value for the tenfold serial dilutions of known concentrations (107–10 fg/μl). b The amplification plot obtained using SYBR Green I assay for S. mutans and A. actinomycetemcomitans. c The confirmations of the qPCR amplification detected on 1.5% agarose gels. As expected, the sizes of S. mutans and A. actinomycetemcomiwns amplicons are 479 and 194 bp, respectively
For detecting S. mutans in the plaque samples, the qPCR reaction mixture contained a total volume of 25 μl, which consisted of SYBR master Mix 2X (12.5 μl; QuantiTect SYBR Green PCR Kits, Qiagen), 10 pmool/μl of each primer (2 μl), and 10 ng/μl DNA template (10.5 μl). The standard curve mixture contained a total volume of 25 μl, which consisted of SYBR master Mix 2X (12.5 μl), 10 pmol/μl of each primer (2 μl), 3d H20 (9.5 μl), and UA159 genomic DNA from 10 ng/μl to 10 fg/μl (1 μl). The reagents and reaction conditions used in the plaque samples mixtures were the same as those used for the standard curve to ensure reaction consistency. The qPCR conditions were performed at 95°C for 15 min, followed by 44 cycles at 95°C for 15 min, 56°C for 30 s, and 72°C for 30 s. For detecting A. actinomycetemcomitans, the qPCR reaction mixture of the plaque sample DNA and the standard strain DNA used were the same as for the qPCR for S. mutans. The qPCR conditions were performed at 95°C for 15 min, followed by 44 cycles at 94°C for 15 s, 58°C for 20 s, and 72°C for 40 s. Each plate was read after each cycle and then incubated at 72°C for 4 min. The melting curves were performed from 60°C to 90°C and read every 0.5°C for l s, and fmally, the plate was incubated at 4°C.
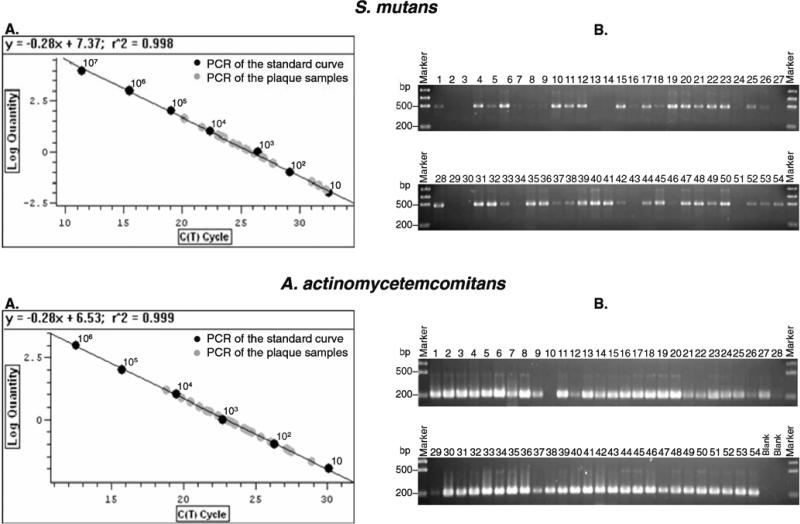
All output data were analyzed using Opticon Monitor™ software (BioRad, Hercules, CA, USA). The qPCR results demonstrated that both standard curves, representative of typical experiments, were linear generated from a plot of Ct against log concentration for the known standard DNA for S. mutans (r2 = 0.998) and A. actinomycetemcomitans (r2= 0.999; Fig. 2), both melting curves for S. mutans at 78°C and A. actinomycetemcomitans at 83°C demonstrated homogeneous products without primer-dimers (data not shown), and all the amplifications of clinical samples were linear generated within the standard range (Fig. 2).
Fig. 2.
Quantitative real-time polymerase chain reaction (qPCR) for plaque samples. a The linear standard curve was generated based on the log concentration of known DNA samples of Streptococcus mutans (UA 159) and Aggregatibacter actinomycetemcomitans (ATCC 29522) (black circle) and the plaque samples (gray circle). b The qPCR final results were confirmed on a 1.5% agarose gel
All qPCR for the standards and the plaque samples were performed in duplicate to eliminate variation between tubes with the same templates. The final analysis was based on the mean of the two reactions, and the specificity of all PCR and q PCR products of clinical samples was confirmed for correct molecular size by electrophoresis in a 1.5% agarose gel. The gel was then stained with ethidium bromide (l ug/ml) for 15 min and washed with water for 5 min. The gels were photographed and recorded with Alphalmager 3300 imaging system (Alpha lnnotech Corp., San Leandro, CA, USA).
Statistical analysis
The PCR and qPCR results were organized and analyzed using the SPSS 17.0 statistical software package (SPSS Inc., Chicago, IL, USA). Descriptive statistics including prevalence, mean, range, standard deviation, and variance were examined. Nonparametric Mann- Whitney U test was used to examine the significance of the distribution among continuous variables. The Pearson Chi-square test was used to examine the significance of the distribution among categorical variables. McNemar's test was used to examine the significance of the association between matched paired categorical variables. The p value of the SEgnificance probability less than or equal to 0.05 was ,considered statistically significant.
Results
The mean age of the pat1tctpants was 14.9 years (SD = 1.7 years, median=l4.7; range, 12.2 to 18.7 years), and 54.6% of the adolescents were male. The caries prevalence was 42.2%, 48.1% for males and 33.3% for females (p= 0.37), and the DMFS averaged 2.67 (SD =5.3; Table 1). The distribution of DMFS scores was skewed, the Kurtosis value was 7.5, and the median values were 0 for both males and females. In terms of the gingival-periodontal complex, we found that more than half of the adolescents (53.3%), 62.8% for males and 43.6% for females (p= 0.09), exhibited gingival or periodontal pockets (CPI score 3 or 4; Table 1). The distribution of periodontal pocket depth was also skewed, the Kurtosis value was 5.8, and the median values were 1.5 and 0 for males and females, respectively. Therefore, all statistical analyses were conducted by using nonparametric statistical tests. The mean values were presented in Table 1 sole for clinical relevance. Though male Haitian adolescents had more dental caries and higher CPI scores, the differences were not statistically significant. Additionally, age was not significantly related to either caries status or CPT score (Table 1).
Table 1.
Demographic characteristics and clinical examination results of the study population
| Study group | Demography |
Clinical examinations |
||||||
|---|---|---|---|---|---|---|---|---|
| Gendera |
Age (years)a |
Prevalence (%)b |
Mean±SDc |
|||||
| % | Median | Mean±SD | Subject with caries | Subject with CPI≥3 | DMFS | Pocket depth (mm) | ||
| Supra-gingival | ||||||||
| N=54 | All | 100 | 14.7 | 14.9±1.7 | 42.2 | 2.67±5.3 | ||
| Male | 57.1 | 15.0 | 15.0±1.8 | 48.1 | 2.59±4.6 | |||
| Female | 42.9 | 14.5 | 14.8±1.6 | 33.3 | 2.78±6.3 | |||
| Sub-gingival | ||||||||
| N=98 | All | 100 | 14.7 | 150± 1.6 | 53.3 | 2.33±3.4 | ||
| Male | 56.0 | 15.0 | 15.0±1.7 | 62.8 | 3.00±4.0 | |||
| Female | 44.0 | 14.6 | 14.9±1.5 | 43.6 | 1.49±2.1 | |||
Pearson Chi-square test and nonparametric Mann-Whitney U test. No statistically significant differences in the gender and the age distributions were found between the two study groups
Pearson Chi-square test. No statistically significant differences in the prevalence of caries and periodontal pockets were found between the males and the females
Nonparametrie Mann-Whitney test for statistical testing the distributions of DMFS score and the depth of periodontal pockets. No statistically significant differences were found between the males and the Females. Because the distributions of DMFS score and the depth of periodontal pockets were skewed, the mean values presented in this table are for clinical relevance, not for statistical analysis
Our results showed that the prevalence of S. mutans was statistically significantly higher in supra-gingival samples compared to that in sub-gingival samples using either the standard PCR (p<0.001) or qPCR (p<0.001) method. Using the qPCR method, S. mutans was detected in 67.3% of the total supra-gingival samples and 38.8% of the total supra-gingival samples by using standard PCR (p< 0.00 1; Table 2). For the sub-gingival plaque samples, S. mutans was found in 36.6% and 8.1% of the samples using the qPCR and the standard PCR, respectively (p<0.001; Table 2). Hence, the prevalence of bacteria detected by qPCR was significantly higher than the prevalence determined by standard PCR (McNemar's test (p<0.001)).
Table 2.
Comparison of standard polymerase chain reaction (PCR) and quantitative real-time PCR (qPCR) in detection of Streptococcus nutans and Aggregatibacter actinomycetemcomitans in dental plaque of a Haitian adolescent population
| Study group | Gender | PCRa |
qPCRb |
|||
|---|---|---|---|---|---|---|
| % positive for S. mutans or A. actinomycetemcomitans |
% positive for S. mutans or A. actinomycetemcomitans |
|||||
| % | S. mutants c | A. actinomycetemcomitans d | S. mutans c | A. actinomycetemcomitans d | ||
| Supra-gingival | ||||||
| N=54 | All | 100 | 38.8 | 59.2 | 67.3 | 67.3 |
| Male | 57.1 | 42.9 | 67.9 | 67.9 | 75.0 | |
| Female | 42.9 | 33.3 | 47.6 | 66.7 | 57.1 | |
| Sub-gingival | ||||||
| N=98 | All | 100 | 8.l | 44.0 | 36.6 | 85.1 |
| Male | 56.0 | 7.0 | 43.1 | 39.0 | 83.7 | |
| Female | 44.0 | 9.7 | 45.0 | 33.3 | 87.1 | |
Pearson Chi-square test showed no differences in the prevalence of S. mutuns and A. actinomycetemcomitans detected by PCR between the males and the females. However, the prevalence were statistically significant higher in supra-gingival samples compared to that in sub-gingival samples for cither S. mutans (p< 0.001) or A. actinomycetemcomitans (p=0.031)
Pearson Chi-squarc test showed no differences in the prevalence of S. mutans and A. actinomycetemcomitans detected by qPCR between the males and the females. However, the prevalence was statistically significant higher in supra-gingival samples compared to that in sub-gingival samples for S. mutuns (p<0.001), but marginally higher for A. actinomycetemcomitans (p 0.055)
The prevalence of S. mutans was significantly higher when detected by qPCR compared to the results detected by standard PCR for cither supra-gingival samples (McNcmar's test, p<0.00l) or sub-gingival samples (McNcmar's test, p<0.001)
The prevalence of A. actinomycetemcomitans was significantly higher when detected by qPCR compared to the results detected by standard PCR for sub-gingival samples (McNcmar's test, p<0.001), but not for supra-gingival samples (McNcmar's test, p=0.219)
The results also showed that A. actinomycetemcomitans was detected in 44.0% of the total sub-gingival samples using standard PCR, 85.1% by using the qPCR (McNemar's test, p<0.001 Table 2). When examining only the supra-gingival plaque samples, the percentage of A. actinomycetemcomitans-positjve was 59.2% and 67.3% using the standard PCR and qPCR, respectively (McNemar's test, p = 0.219). When comparing a single PCR assay for the detection of A. actinomycetemcomitans in different samples types (supra- vs. sub-gingival), we observed using standard PCR resulted in a prevalence that was significantly higher in the supra-gingival samples (59.2%) compared to that in the sub-gingival samples (44.0%; Pearson Chi-square test, p =0.031). However, in the samples analyzed using qPCR group, the prevalence in the sub-gingival samples was higher (85.1 %) compared to that in the suprd-gingival samples (67.3%; Pearson Chi-square test, p=0.055; Table 2).
There were no statistically significant relationships found between the prevalence of colonization and gender, age, ECPEM levels, caries status, and CPI score (data not shown).
Discussion
Information on the oral health status of Haitian adolescents is lacking. Tn the 1999 Oral Health National Survey of Haiti [21], Psoter et al. reported a mean DMFS of 2.53 and a caries prevalence of 46% in 12- to 15-year-old urban and rural Haitian adolescents. The present study sampled two rural villages and found a mean DMFS of 2.67 and a caries prevalence of 42.2%, which was similar to the national data, suggesting that the caries status found in this study population may be comparable to that generally found in rural Haiti.
To date, there are no published reports on oral bacterial colonization in this population. As part of a major epidemiological study on ECPEM and its relationship to oral diseases and conditions [12, 13,22], the present study further assessed the colonization of S. mutans, known as a key cariogenic bacteria associated with caries development, and A. actinomycetemcomitans, known as a key periodontal pathogen associated with periodontal disease, in a subsample of the Haitian subjects. We found a moderate level of dental caries and periodontal disease and also detected a moderate-to-high prevalence of S. mutans and A. actinomycetemcomitans among these adolescents. Neither age nor gender was significantly related to the colonization of these two microorganisms. Although a trend indicated a positive relationship between bacterial colonization and caries and periodontitis, this present study was not statistically powered to test for those correlations. As the adolescent population under consideration was recruited from only two rural vi llages, our microbiologic findings may limit the generalizability of these findings, although life-style conditions are generally similar throughout rural Haiti. Importantly, however, as the present study employed validated molecu lar techniques, these findings can be used as baseline data for future microbiology and epidemiologic studies in the Haitian as well as other populations.
One of the major objectives of this study was to conduct a field ep idemiological study with a microbiological component in rural areas of Haiti. Traditionally, laboratory-based bacterial cultivation is used as the “gold standard” to determine the presence, absence, and levels of bacterial colonization. Qualitative and quantitative findings from enriched or species-specific selective culture media are then analyzed for associations with oral disease. Although bacteriological methods have been the method of choice for the evaluation of bacterial colonization for decades, PCR with S. mutans-specific and A. actinomycetemcomitans-specific primers has become a more commonly used and important technique for more rapid and reliable detection of these two oral pathogens in the saliva and dental plaque. PCR-based assay is particularly useful and cost-effective when bacterial pure culture is not required or a lab-based operation is not available. In this study, we first obtained the prevalence of S. mutans or A. actinomycetemcomitans rapidly by using standard PCR assays with limited expenses. However, it has been recognized that the presence of S. mutans or A. actinomycetemcomitans in the saliva and dental plaque may not be necessarily correlated with the development and severity of dental caries or periodontal disease, respectively [1 ,23). Consequently, we took a second approach to conduct qPCR to determine the colonization level of S. mutans and A. actinomycetemcomitans in dental plaque. Although significant differences in the DNA levels were not found, all results consistently demonstrated higher detection rate by qPCR compared to that by standard PCR.
Using the standard PCR method, we found that the prevalence of S. mutans was 38.8% in the supra-gingival samples and 8.1% in the sub-gingival samples. When qPCR assay was performed, the prevalence of S. mutans increased from 38.8% to 67.3% in the supra-gingival plaque sample of Haitian adolescents. Overall, this prevalence is lower than the 90% prevalence reported by Linossier (culture method) in 1989 for 9- to 13-year-old Chilean children, but higher than the prevalence reported in Colombian and Venezuelan children (culture method) [24–26]. The observed difference in prevalence could be possibly attributable to the different methods used. Meanwhile, the low prevalence of S. mutans in the sub-gingival samples was not unexpected, since the presence of S. mutans in subgingival samples is associated with root caries and the prevalence of root caries was likely low in the study population.
Based on previously published cross-sectional studies, the prevalence of A. actinomycetemcomitans varies from 20% to 75%, depending on the population and stage of periodontitis [11 ,23). Tinoco et al., for instance, reported that A. actinomycetemcomitans was detected in 80% of young patients with localized juvenile periodontitis, but also found in 43.9% of all siblings who did not have the disease and 35.3% of their parents [27]. Cortelli et at. reported an A. actinomycetemcomitans prevalence of 41.6% among subjects with chronic periodontitis and 72% among subjects with aggressive periodontitis [28]. In the present study, the prevalence of A. actinomycetemcomitans detected by qPCR in Haitian adolescents is similar to the prevalence reported among American adolescents and young adults [29], but higher than that reported from some other countries using different methods [30,31]. As the association between A. actinomycetemcomitans and aggressive fonns of periodontitis is well established [5,32,33 ], the high prevalence of A. actinomycetemcomitans may serve as a risk indicator for future initiation of aggressive periodontitis in rural Haitian adolescents.
Interestingly, we fotmd that PCR-positive detection rate for A. actinomycetemcomitans was higher in the supragingival plaque (59.2%) than in the sub-gingival plaque samples (44.0%), but 85. 1% in sub-gingival plaque samples and 67.3% in supra-gingival samples by using qPCR. We speculate that the difference found between the supra- and sub-gingival samples may be associated with diminished salivary gland antimicrobial function resulting of early childhood malnutrition (ECPEM) [13). The differences observed between the PCR-based methods could also attribute to the fact that the colonization level of A. actinomycetemcomitans is higher and concentrated at subgingival level than in the saliva, but collecting sufficient amount of sub-gingival plaque samples for bacterial DNA isolation and consequently PCR experiment is often challenging when there is little periodontal pocketing. As qPCR is more sensitive and less bacterial DNA samples are required for pe1forming the experiment, the qPCR results, therefore, are likely to be accurate for reporting Uevels of A. actinomycetemcomitans colonization in both supra- and sub-gingival samples.
Furthermore, the different findings of S. mutans and A. actinomycetemcomitans colonization between the standard PCR and qPCR were statistically significant, which may be due to the difference in the detection technique designed for each method. Standard PCR method detects PCR amplification at the end-point of PCR reaction. The use of agarose gels tor the detection presents low sensitivity and low resolution. Ethidium bromide for staining is not quantitative and cannot delineate differences less than tenfold among samples. In addition, these results are based on size discrimination, wh ich is not precise. Quantitative real-time PCR, on the other hand, invented in the mid- 1990s [6], combines standard PCR with the use of fluorescent markers to measure the amount of PCR-amplified bacterial DNA during the exponential amplification phase. The technique can be used to determine the amount of sta1ting material, as little as a twofold change can be detected. Thus, qPCR provides much precise, accurate, and reliable results compared to standard PCR [6,34]. Since its introduction, qPCR has been used increasingly in the evaluation of oral microbial infection and colonization (35–41]. The results of our study further support the contention that qPCR is more sensitive than standard PCR and should be considered as a preferred method to detect and quantify dental pathogens in clinical and field epidemiologic studies, at least under the difficult conditions of storage and transpottation as reported here.
In summary, the findings (1) demonstrate a moderate-to-high prevalence of S. mutans and A. actinomycetemcomitans in a Haitian adolescent population and (2) suggest that qPCR is more sensitive than standard PCR in difficult field conditions. Thus, qPCR should be considered for field oral epidemiologic studies and may be necessary in investigations having major logistic challenges.
Acknowledgment
Supported by NIH/NIDCR grants R03 DE15706,, RO1 DEO14708, T32 DE007255, and Dean's Faculty Research Fund of New York University College of Dentistry.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest to the present materials.
Contributor Information
Walter J. Psoter, Department of Epidemiology and Health Promotion, New York University College of Dentistry, 345 E. 24th Street, New York, NY 100104086, USA
Yao Ge, Department of Basic Science and Craniofacial Biology, New York University College of Dentistry, New York, NY, USA.
Stefanic L. Russell, Department of Epidemiology and Health Promotion, New York University College of Dentistry, 345 E. 24th Street, New York, NY 100104086, USA
Zhou Chen, Department of Basic Science and Craniofacial Biology, New York University College of Dentistry, New York, NY, USA.
Ralph V. Katz, Department of Epidemiology and Health Promotion, New York University College of Dentistry, 345 E. 24th Street, New York, NY 100104086, USA
Germain Jean-Charles, Department of Epidemiology and Health Promotion, New York University College of Dentistry, 345 E. 24th Street, New York, NY 100104086, USA.
Yihong Li, Department of Basic Science and Craniofacial Biology, New York University College of Dentistry, New York, NY, USA.
References
- 1.Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Houte J. Role of micro-organisms in caries etiology. J Dent Res. 1994;73:672–681. doi: 10.1177/00220345940730031301. [DOI] [PubMed] [Google Scholar]
- 3.Slots J, Ting M. Actinobacillus actinomycetemcomitans and Porphyromonas gingiva/is in human periodontal disease: occurrence and treatment. Periodontal. 2000;20:82–121. doi: 10.1111/j.1600-0757.1999.tb00159.x. 1999. [DOI] [PubMed] [Google Scholar]
- 4.Henderson B, Nair SP, Ward JM, Wilson M. Molecular pathogenicity of the oral opportunistic pathogen Actinobacillus actinomycetemcomitans. Annu Rev Microbial. 2003;57:29–55. doi: 10.1146/annurev.micro.57.030502.090908. [DOI] [PubMed] [Google Scholar]
- 5.Fine DH, Markowitz K, Furgang D, Fairlie K, Ferrandiz J, Nasri C, McKiernan M, Gunsolley J. Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: longitudinal coholt study of initially healthy adolescents. J Clin Microbial. 2007;45:3859–3869. doi: 10.1128/JCM.00653-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heid CA, Stevens J, Livak KJ, Williams PM. Real-time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 7.Rupf S, Merte K, Eschrich K. Quantification of bacteria in oral samples by competitive polymerase chain reaction. J Dent Res. 1999;78:850–856. doi: 10.1177/00220345990780040501. [DOI] [PubMed] [Google Scholar]
- 8.Rupf S, Me11e K, Kneist S, Al-Robaiy S, Eschrich K. Comparison of different techniques of quantitative PCR for determination of Streptococcus mutans counts in saliva samples. Oral Microbial Immunol. 2003;18:50–53. doi: 10.1034/j.1399-302x.2003.180108.x. [DOI] [PubMed] [Google Scholar]
- 9.Nonnenmacher C, Dalpke A, Mutters R, Heeg K. Quantitative detection of periodontopathogens by real-time PCR. J Microbial Methods. 2004;59(1):17–125. doi: 10.1016/j.mimet.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Thenisch NL, Bachmann LM, Imfeld T, Leisebach Minder T, Steurer J. Are mutans streptococci detected in preschool children a reliable predictive factor for dental canies risk? A systematic review. Caries Res. 2006;40:366–374. doi: 10.1159/000094280. [DOI] [PubMed] [Google Scholar]
- 11.Rylev M, Kilian M. Prevalence and distribution of principal periodontal pathogens worldwide. J Clin Periodontal. 2008;35:346–361. doi: 10.1111/j.1600-051X.2008.01280.x. [DOI] [PubMed] [Google Scholar]
- 12.Psoter W, Gebrian B, Prophete S, Reid B, Katz R. Effect of early childhood malnutrition on tooth eruption in Haitian adolescents. Community Dent Ora l Epidemiol. 2008;36:179–189. doi: 10.1111/j.1600-0528.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- 13.Psoter WJ, Spielman AL, Gebrian B, St Jean R, Katz RV. Effect of childhood malnutrition on salivary flow and pH. Arch Oral Bioi. 2008;53:231–237. doi: 10.1016/j.archoralbio.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wennerholm K, Lindquist B, Emilson CG. The toothpick method in relation to other plaque sampling techniques for evaluating mutans streptococci. Eur J Oral Sci. 1995;103:36–41. doi: 10.1111/j.1600-0722.1995.tb00008.x. [DOI] [PubMed] [Google Scholar]
- 15.Kazor C, Taylor GW, Loesche WJ. The prevalence of SANA-hydrolyzing periodontopathic bacteria in smokers. J Clin Periodontal. 1999;26:814–82. doi: 10.1111/j.1600-051x.1999.tb02526.x. [DOI] [PubMed] [Google Scholar]
- 16.Tanner AC, Goodson JM. Sampling of microorganisms associated with periodontal disease. Oral Microbial Immunol. 1986;1:15–22. doi: 10.1111/j.1399-302x.1986.tb00310.x. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Ge Y, Saxena D, Caufield PW. Genetic profiling of the oral microbiota associated with severe early-childhood caries. J Clin Microbial. 2007;45:81–87. doi: 10.1128/JCM.01622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Ku CY, Xu J, Saxena D, Caufield PW. Survey of oral microbial diversity using PCR-based denaturing gradient gel electrophoresis. J Dent Res. 2005;84:559–564. doi: 10.1177/154405910508400614. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z, Saxena D, Caufield PW, Ge Y, Wang M, Li Y. Development of species-specific primers for detection of Streptococcus mutans in mixed bacterial samples. FEMS Microbial Lett. 2007;272:154–162. doi: 10.1111/j.1574-6968.2007.00756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudney JO, Chen R, Pan Y. Endpoint quantitative PCR assays for Bacteroides jorsythus, POrphyromonas gingivalis, and Actinobacillus actinomycetemcomitans. J Periodontal Res. 2003;38:465–470. doi: 10.1034/j.1600-0765.2003.00670.x. [DOI] [PubMed] [Google Scholar]
- 21.Psoter WJ, Saint Jean HL, Morse DE, Prophte SE, Joseph JR, Katz RV. Dental caries in twelve-and fifteen-year-olds: results from the basic oral health survey in Haiti. J Public Health Dent. 2005;65:209–214. doi: 10.1111/j.1752-7325.2005.tb03020.x. [DOI] [PubMed] [Google Scholar]
- 22.Psoter WJ, Gebrian B, Katz RV. Reliability of a sugar consumption questionnaire forural Haiti. P R Health Sci J. 2008;27:69–74. [PubMed] [Google Scholar]
- 23.Mombelli A, Casagni F, Madianos PN. Can presence or absence of periodontal pathogens distinguish between subjects with chronic and aggressive periodontitis? A systematic review. J Clin Periodontal. 2002;29(Suppl 3):10–21. doi: 10.1034/j.1600-051x.29.s3.1.x. discussion 37-18. [DOI] [PubMed] [Google Scholar]
- 24.Acevedo AM, Ray MY, Socorro M, Rajas-Sanchez F. Frequency and distribution of mutans streptococci in dental plaque from caries-free and caries-affected Venezuelan children. Acta Odontol Latinoam. 2009;22:15–20. [PubMed] [Google Scholar]
- 25.Linossier A, Gajardo M, Silva N, Larroque C, Aspillaga E, Charmin B, Pizarro F. Prevalence of Streptococcus mutans in pehuenche children, Chilean ethnic group. Rev Med Chilean ethnic group. 1989;117:872–878. [PubMed] [Google Scholar]
- 26.Thomson LA, Little WA, Bowen WH, Sierra LI, Aguirrer M, Gillespie G. Prevalence of Streptococcus mutans serotypes, Actinomyces, and other bacteria in the plaque of children. J Dent Res. 1980;59:1581–1589. doi: 10.1177/00220345800590100501. [DOI] [PubMed] [Google Scholar]
- 27.Tinoco EM, Beldi MI, Loureiro CA, Lana M, Carnpedelli F, Tinoco NM, Gjermo P, Preus HR. Localized juvenile periodontitis and Actinobacillus actinomycetemcomitans in a Brazilian population. Eur J Oral Sci. 1997;105:9–14. doi: 10.1111/j.1600-0722.1997.tb00174.x. [DOI] [PubMed] [Google Scholar]
- 28.Cortelli JR, Cortelli SC, Jordan S, Haraszthy VT, Zambon JJ. Prevalence of periodontal pathogens in Brazilians with aggressive or chronic periodontitis. J Clin Periodontal. 2005;32:860–866. doi: 10.1111/j.1600-051X.2005.00777.x. [DOI] [PubMed] [Google Scholar]
- 29.Lamell CW, Griffen AL, McClellan DL, Leys EJ. Acquisition and colonization stability of Actinobacillus actinomycetemcomitans and Potphyromonas gingivalis in children. J Clin Microbial. 2000;38:1196–1199. doi: 10.1128/jcm.38.3.1196-1199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zambon JJ, Christersson LA, Slots J. Actinobacillus actinomycetemcomitans in human periodontal disease. Prevalence in patient groups and distribution of biotypes and serotypes within families. J Periodontal. 1983;54:707–711. doi: 10.1902/jop.1983.54.12.707. [DOI] [PubMed] [Google Scholar]
- 31.Paolantonio M, di Bonaventura G, di Placido G, Tumini V, Catamo G, di Donato A, Piceolomini R. Prevalence of Actinobacillus actinomycetemcomitans and clinical conditions in children and adolescents from rural and urban areas of central Italy. J Clin Periodontal. 2000;27:549–557. doi: 10.1034/j.1600-051x.2000.027008549.x. [DOI] [PubMed] [Google Scholar]
- 32.Wilson M, Henderson B. Virulence factors of Actino-bacillus actinomycetem comitans relevant to the pathogenesis of inflammatory periodontal diseases. FEMS Microbiol Rev. 1995;17:365–379. doi: 10.1111/j.1574-6976.1995.tb00220.x. [DOI] [PubMed] [Google Scholar]
- 33.Slots J, Reynolds HS, Genco RJ. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect Immun. 1980;29:1013–1020. doi: 10.1128/iai.29.3.1013-1020.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, Yao JD, Wengenack NL, Rosenblatt JE, Cockerill FR, 3rd, Smith TF. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyons SR, Griffen AL, Leys EJ. Quantitative real-time PCR for Potphyromonas gingivalis and total bacteria. J Clin Microbiol. 2000;38:2362–2365. doi: 10.1128/jcm.38.6.2362-2365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shelburne CE, Prabhu A, Gleason RM, Mullally BH, Coulter WA. Quantitation of Bacteroides forsythus in subgingival plaque comparison of immunoassay and quantitative polymerase chain reaction. J Microbiol Methods. 2000;39:97–107. doi: 10.1016/s0167-7012(99)00106-2. [DOI] [PubMed] [Google Scholar]
- 37.Asai Y, Jinno T, Igarashi H, Ohyama Y, Ogawa T. Detection and quantification of oral treponemes in subgingival plaque by real-time PCR. J Clin Microbiol. 2002;40:3334–3340. doi: 10.1128/JCM.40.9.3334-3340.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakamoto M, Umeda M, Benno Y. Molecular analysis of human oral microbiota. J Periodontal Res. 2005;40:277–285. doi: 10.1111/j.1600-0765.2005.00793.x. [DOI] [PubMed] [Google Scholar]
- 39.Orru G, Marini MF, Ciusa ML, Isola D, Cotti M, Baldoni M, Piras V, Pisano E, Montaldo C. Usefulness of real time PCR for the differentiation and! quantification of 652 and JP2 Actino-bacillus actinomycetemcomitans genotypes in dental plaque and saliva. BMC Infect Dis. 2006;6:98. doi: 10.1186/1471-2334-6-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jervoe-Storrn PM, AlAhdab H, Semaan E, Fimmers R, Jepsen S. Microbiological outcomes of quadrant versus full-mouth root planing as monitored by real-time PCR. J Clin Periodontol. 2007;34:156–163. doi: 10.1111/j.1600-051X.2006.01035.x. [DOI] [PubMed] [Google Scholar]
- 41.Kishi M, Abe A, Kishi K, Ohara-Nemoto Y, Kimura S, Yonemitsu M. Relationship of quantitative salivary levels of Streptococcus mutans and S. sobrinus in mothers to caries status and colonization of mutans streptococci in plaque in their 2.5-year-old children. Community Dent Oral Epidemiol. 2009;37:241–249. doi: 10.1111/j.1600-0528.2009.00472.x. [DOI] [PubMed] [Google Scholar]