Abstract
Significant gaps exist in our understanding of the causes and clinical management of glioma. One of the biggest gaps is how best to manage low grade (World Health Organization (WHO) grade II) glioma patients. Low grade glioma is a uniformly fatal disease of young adults (mean age 41 years) with survival averaging approximately 7 years. Although low grade glioma patients have better survival than patients with high grade (WHO grade III/IV) glioma, all low grade gliomas eventually progress to high grade glioma and death. Data from the Surveillance, Epidemiology and End Results (SEER) program of the National Cancer Institute suggest that for the majority of low grade glioma patients, overall survival has not significantly improved over the past three decades, highlighting the need for intensified study of this tumor. Newly published research suggests that historically utilized clinical variables are not sufficient (and are likely inferior) prognostic and predictive indicators relative to information provided by recently discovered tumor markers (e.g..1p/19q deletion and IDH1/2 mutation status), tumor expression profiles (e.g. the Proneural Profile) and/or constitutive genotype (e.g. rs55705857 on 8q24.21). Discovery of such tumor and constitutive variation may identify variables needed to improve randomization in clinical trials as well as patients more sensitive to current treatments and targets for improved treatment in the future. This manuscript reports on survival trends for patients diagnosed with low grade glioma within the United States from 1973–2011 and reviews the emerging role of tumor and constitutive genetics in refining risk stratification, defining targeted therapy, and improving survival for this group of relatively young patients.
Keywords: glioma, low grade, survival, SEER, epidemiology, genes, GWAS, treatment
Introduction
Gliomas are classified as grades I to IV based on histology and clinical criteria.45 Grade I tumors are generally benign and frequently curable with complete surgical resection, occur primarily in children and are believed to represent an entity separate from grade II-IV (seen primarily in adults). Adult grade II tumors (Low Grade Gliomas (LGG) include: 1) astrocytomas, 2) oligo-astrocytomas or mixed gliomas, and 3) oligodendrogliomas.45 Astrocytomas and oligodendrogliomas consist of astrocytes or oligodendrocytes, respectively, while mixed gliomas contain a mixture of the two cell types. Essentially all Grade II lesions eventually progress to High Grade Glioma (grade III/IV or HGG).Grade IV tumors (aka glioblastoma (GBM)) that arise from LGG are termed “secondary GBM” to differentiate them from “primary” or “de-novo” GBM as the pathway leading to these two GBM types differs by a number of genetic abnormalities and clinical characteristics.77 Most patients initially receive surgical resection/biopsy at time of diagnosis and then radiation therapy (XRT) and/or the single chemotherapeutic agent temozolamide (TMZ) at some point. However, many of these relatively uniformly treated patients advance more quickly than others to recurrence and death. Variation in the few known prognostic factors (most of which are themselves highly correlated), e.g. age, performance status, tumor size/location, extent of surgical resection, and histological subtype does not adequately explain the progression and survival differences in these patients. To date, the detection of treatment effect is limited. A surgical gross total resection appears associated with better survival for patients able to undergo such a procedure but has never been and is unlikely to be assessed in randomized clinical trials (RCT);13,32,61 the improvement may be due to biases from differential tumor aggressiveness in non-resectable versus resectable portions of the brain and from clinician predictions of the patients likely to benefit most from resection. Randomized clinical trials suggest radiation therapy prolongs time to recurrence but not overall survival38,40,66,75 and may be associated with reduction in quality of life and cognition,1,17,40,47 while the impact of the primary single chemotherapeutic agent temozolamide (TMZ) now used to treat LGG has shown benefit primarily in RCT of HGG but is not fully assessed in LGG.4,6,39,52,56,73 For LGG no RCT have compared TMZ (which is associated with blood disorders and leukemia)44 to other agents (trials are ongoing that compare TMZ to radiation therapy (XRT) as well as the combination of TMZ/XRT to XRT). A recently updated trial (RTOG 9802) comparing radiation therapy with or without procarbazine, CCNU and vincristine (PCV) reports improved progression-free as well as overall survival with addition of PCV, but ironically is infrequently used over the past decade to treat LGG.7 No comprehensive clinical prognostic or predictive classification for LGG exists that combines information on histology, tumor markers and constitutive/tumor genotype, and surgical treatment relative to outcome leading to confusion over how to best manage these patients. The goal of this review is to examine population-based survival rates for LGG within the United States by standard patient demographics and initial treatment and to then review emerging data on patient and tumor genotype relative to survival after a diagnosis of low grade glioma.
Methods
We examine data from the Surveillance, Epidemiology, and End Results Program of the National Cancer Institute from 1973–201165 that includes 2825 patients diagnosed between the ages of 20–79 years with a histologically confirmed grade II supratentorial (Topography codes C71.0–71.4) glioma (Morphology codes: mixed glioma (ICD-0 9382), oligodendroglioma (ICD-0 9450) or astrocytoma (ICD-0 9400)). In an effort to examine a homogenous study population and to reduce the probability of including individuals with metastatic lesions, individuals with more than one primary cancer (i.e. a glioma and a cancer of another site) were excluded from these analyses, as were patients diagnosed at death (autopsy only).
In addition to topography and morphology, information on sex, race, age and year of diagnosis were available as was information regarding whether the patient had received surgical resection (yes/no), radiation therapy (yes/no), and chemotherapy (yes/no) as part of the first course of treatment. Treatment parameters after the first course are not available in these data nor are specifics of chemotherapy regimes. Race was defined according to SEER categories of white, black, and other due to small sample sizes in the non-black, non-white categories. Age was utilized as a continuous variable in the proportional hazards model. The primary outcome variable was time to death as measured in years.
Comparison of cases by descriptor variables was done using a chi-square or Fisher’s exact test for discrete variables and a t-test for continuous variables. Estimates of survival probabilities (with 95% confidence intervals) were calculated using Kaplan-Meier product limit methodology and compared using a Wilcoxon log rank test. Hazard rates were computed using a Cox proportional hazards model.19 All analyses were completed using the SAS statistical software package version 9.13.63
Results
Descriptive statistics for the sample are presented in Table 1. The majority (51.6%) of the cases are classified as astrocytoma with 33.5% and 14.9% classified as oligodendroglioma and mixed glioma, respectively. The reported distributions of these three tumor types has changed significantly over time (p < 0.001) with fewer cases being classified as astrocytoma and more being identified as either oligodendroglioma or mixed gliomas.23 The majority of patients were male (58.9%) and white (89.1%). The mean age at diagnosis is 41.4 (SD 15.6) years and does not vary by sex, race or year of diagnosis. Persons with mixed glioma were diagnosed on average two years earlier than patients with other pathology. Treatment data, which includes only the first course, show the majority of LGG patients received only surgical resection at first course while only 3.7% received chemotherapy as part of the initial treatment with use of radiation at first course declining over time. Initial treatment did not vary by sex or race but did differ by age with younger patients more likely to undergo surgical resection. Treatment differed by location of the lesion (which did not vary by sex or race). As would be expected, individuals with parietal lobe lesions were more likely to receive radiation therapy and less likely to receive surgical resection than were patients with lesions located elsewhere in the brain.
Table 1.
Number of diagnoses (and deaths) for adult supratentorial low grade glioma by histologic subgroup, Surveillance, Epidemiology, and End Results (SEER) data, 1973–2011
| Astrocytoma | Oligodendroglioma | Mixed Glioma | Total | ||
|---|---|---|---|---|---|
| Age | 20–39 | 732 (442) | 445 (153) | 229 (83) | 1406 (678) |
| 40–59 | 538 (392) | 421 (248) | 161 (66) | 1120 (616) | |
| 60+ | 2188 (162) | 80 (56) | 31 (20) | 299 (238) | |
| Race | White | 1295 (889) | 849 (317) | 374 (150) | 2518 (1366) |
| Black | 92 (70) | 32 (13) | 18 (8) | 142 (91) | |
| Other | 71 (37) | 65 (27) | 29 (11) | 165 (75) | |
| Sex | Female | 591 (391) | 384 (131) | 185 (69) | 1160 (591) |
| Male | 867 (605) | 562 (226) | 236 (100) | 1665 (931) | |
| Year | Diagnosed | ||||
| 1973–1979 | 143 (133) | 7 (7) | 3 (3) | 153 (143) | |
| 1980–1989 | 414 (370) | 28 (24) | 36 (28) | 478 (422) | |
| 1990–1999 | 346 (252) | 267 (154) | 92 (64) | 705 (470) | |
| 2000–2009 | 520 (1237) | 594 (171) | 263 (74) | 1377 (482) | |
| 2010–2011 | 36 (4) | 59 (1) | 27 (0) | 112 (5) | |
The median survival for patients with astrocytoma, mixed glioma, and oligodendroglioma was 5.2, 5.6, and 7.2 years, respectively, with younger age at onset associated with an improved prognosis and use of radiation therapy at initial treatment associated with a less favorable prognosis across all three histological subtypes. Approximately 20% of patients survived for at least two decades. Female sex was associated with improved prognosis for patients with astrocytoma but not for persons diagnosed with mixed glioma or oligodendroglioma. After controlling for race (white versus non-white), age at onset, gender and initial course of treatment (surgery yes/no, radiation yes/no) there was no improvement in overall survival over time (defined as year of diagnosis before year 2000 versus diagnosis on or after the year 2000) for patients diagnosed with oligodendroglioma (HR = 1.08, (95%CI: 0.85, 1.4)), astrocytoma (HR = 0.98, (95%CI: 0.83, 1.15)) or mixed glioma (HR = 0.76, (95%CI: 0.54, 1, 07)) (Figures 1–3). Interestingly, when the time cut point is placed at 2005 rather than at 2000, the results are similar for astrocytoma and oligodendroglioma but persons diagnosed with mixed glioma on or after 2005 show improved survival versus those diagnosed prior to 2005.
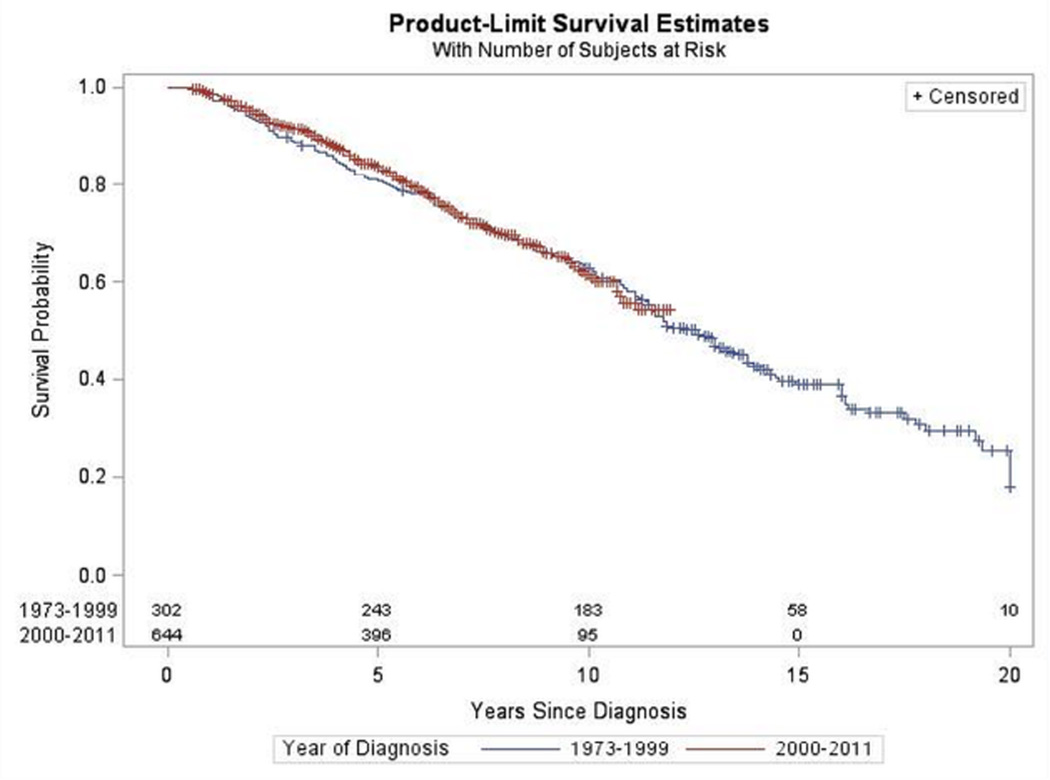
Figure 1.
Survival by Year of Diagnosis for Oligodendroglioma. SEER 1973–2011
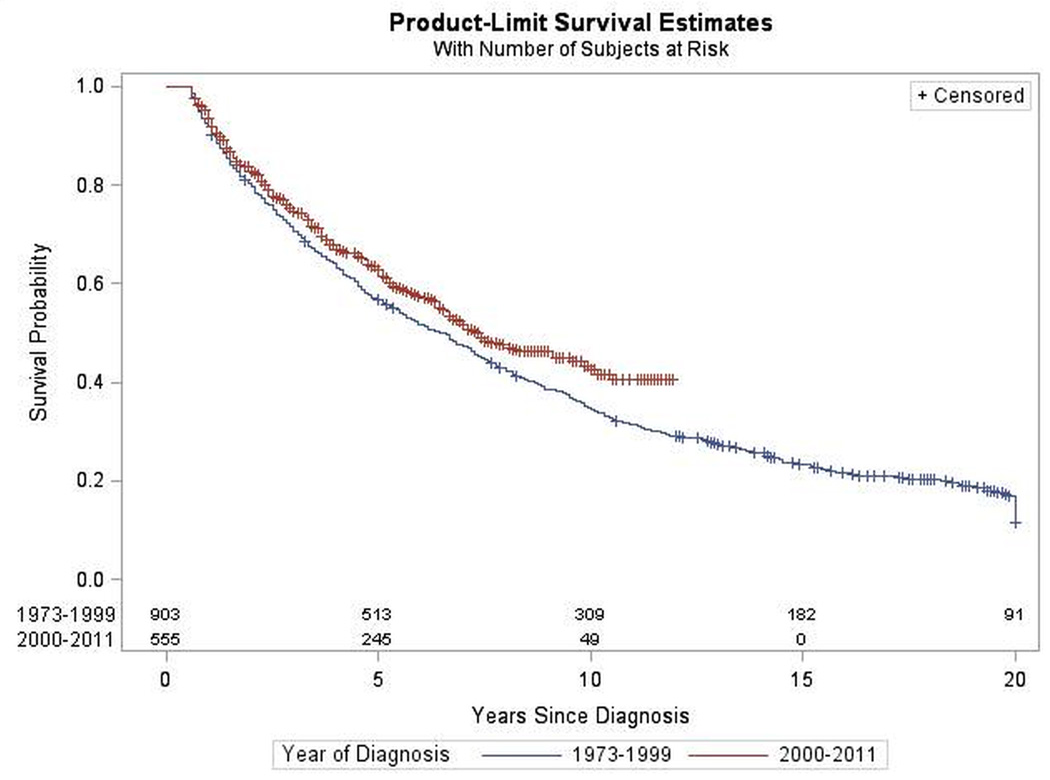
Figure 3.
Survival by Year of Diagnosis for Mixed Glioma. SEER 1973–2011
Discussion
The general lack of improvement in survival for LGG patients over the past three decades points to the need for an intensified focus on these tumors. As for HGG, several intriguing findings have emerged, many over just the past year or two, with respect to molecular tumor markers, gene expression and constitutive genotype.
Molecular Tumor Markers
A number of molecular tumor markers have been associated with LGG overall survival including 1) combined deletions of chromosomes 1p and 19q30,33 2) mutations in the Isocitrate dehydrogenase 1/2 (IDH1/2) genes30,84 and 3) methylation of the O6-methylguanine-DNA methyltransferase (MGMT) gene.30 The high rate of p53 mutation/deletion in some gliomas as well as the belief that this change represents an early step in glioma development has led investigators to examine this alteration in association with LGG survival with inconsistent results.23,28,30
Co-deletion of 1p/19q
Although present in all LGG subtypes, chromosomes 1p and 19q are deleted in 40% –90% of oligo II and are associated with increased survival as well as treatment sensitivity.33,37 The mechanism by which 1p/19q loss affects outcome and response is unknown with no gene clearly defined at either location. Recent sequencing revealed mutations in two tumor suppressor genes (homolog of Drosophiliacapicua (CIC) on 19q and far-upstream binding protein 1 (FUBP1) on 1p in 38% and 14% of 21 oligo II (and 0/15 (0%) of astro II and 1/18 (6%) of mixed II).36 Essentially all glioma with a CIC or FUBP1 mutation in that study36 also had an IDH gene mutation as well as co-deletion of 1p/19q. Jenkins et al33 found most 1p and 19q deletions in oligo II were the result of an unbalanced translocation between the whole chromosomal arms of 1p and 19q and that translocation/deletion was associated with significantly improved overall survival.
IDH1/2
A recent notable finding is that mutations in the NADP+ dependent isocitrate dehydrogenases encoded by IDH1 and IDH2 occur in the majority of grade II (all subtypes)84 and III gliomas as well as secondary GBM but in only a minority of primary GBM.53,84 IDH1 is an enzyme that catalyzes the oxidative decarboxylation of isocitrate to alpha-ketoglutarate leading to NADPH production and is thought to play a role in cellular protection from oxidative stress. IDH mutations are associated with a glioma CpG island DNA hypermethylator phenotype (G-CIMP)74 and are associated with improved LGG survival84 as well as possible LGG response to treatment.28,30 Such data suggest that IDH mutations are an early step in the development of LGG.46
MGMT
Methylation of MGMT (a DNA repair gene located on 10q) is a commonly observed change in LGG28 that predicts HGG response to treatment as well as overall survival;9,29 this change may confer chemo sensitivity in LGG28,30 by causing an altered response to TMZ (the primary agent used to treat LGG) although efforts to examine this are limited by small sample size.28,30
TP53
There is evidence to suggest that a series of ordered genetic alterations occurs when progressing from LGG to HGG with TP53 mutation an early event.84 TP53 is the most frequently mutated gene in GBM and a common event in the TCGA Proneural GBM subtype (believed to include the majority of LGG that progressed to HGG). TP53 mutation is found in all LGG subtypes28 but is highly correlated with the proportion of tumor astrocytes. Interestingly, a recent study36 examined mutations in the chromatin modifier Alpha Thalassemia/Mental Retardation Syndrome X-linked (ATRX) (as well as CIC, FUBP1, and IDH1) and noted almost complete correlation between the presence of TP53 and ATRX mutations, regardless of LGG subtype.
The extent to which any of these markers are merely indicators of the natural progression of disease or of treatment sensitivity (or both) remains ill-defined. In some instances (i.e. 1p/19q co-deletion and oligodendroglioma histology) marker and subtype are correlated leading to confusion about whether it is the marker or the subtype (or both) that is associated with outcome.28 Similarly, correlation exists between markers (i.e. IDH1 mutation and 1p/19q deletion).30 Adding to the confusion is the dynamic classification process of LGG subtype with changes in the relative reported proportions of these subtypes over time reflecting an increasing awareness of the subtleties of histopathological classification for this group of tumors.14,45 Researchers have started to elucidate the relative roles of histology and the aforementioned markers both before (thus capturing factors associated with prognosis) and after treatment (capturing factors associated with prediction). Several small studies suggest that response to TMZ41 and progression free survival (PFS)24 is associated with 1p deletion41 and low MGMT protein expression24,41 but all have a sample size <70, primarily focus on oligo II, and do not examine overall survival (OS). Recently, several groups have presented results from larger case series: Using 271 LGG drawn from the Groupe Hospitalier Pitie-Salpetriere in Paris, Houillier et al30 tested whether TP53 mutation, 1p/19q co-deletion, MGMT promoter methylation, and IDH1 mutation predicted natural course of disease or response to treatment (alkylating agent/XRT) while controlling for extent of surgical resection. In multivariate analyses only performance status and surgical resection (but neither histology nor marker) was predictive of outcome in untreated patients (PFS). IDH1 mutation and 1p/19q co-deletion was predictive of OS while IDH1 mutation, 1p/19q deletion and MGMT promoter methylation were each associated in univariate analyses with response to TMZ but small sample size precluded a multivariate analysis including the three markers simultaneously. Hartmann et al28 performed a similar analysis on139 LGG patients from the German Cancer Network. Again no marker was prognostic in patients who did not receive chemo/XRT. IDH1 mutation and 1p/19q co-deletion were predictive of OS (and of PFS in persons receiving chemo/XRT at diagnosis). As noted by the authors of both studies, insufficient sample size did not allow for examination of these markers by histological subtype.
Tumor Gene Expression
Recent Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov) analyses of high grade glioma tumors use several different technology platforms, including mutation arrays, copy number arrays, expression arrays and methylation arrays.10,54,77 Expression array data has identified molecular subtypes associated with grade and outcome and shown that expression profiles are better predictors of outcome than histological subtype.10,15,54,77 The TCGA and others10,26,54,77 have recently defined and validated four gene-expression-based classification profiles for grade IV glioma (GBM): Proneural (notable for PDGFRA alterations, IDH1 and TP53 mutations, as well as oligodendroglioma cell type), Neural (associated with a variety of neuron markers and closest to normal brain), Classical (EGFR amplification and CDKN2A alterations) and Mesenchymal (NF1 and MET alterations). Despite being constructed using only GBM tumors, intriguing findings relative to LGG are noted: 1) the Proneural profile included 3 of the 4 known secondary GBM, (believed to arise from LGG) and 2) as for LGG, the Proneural profile was notable for young age at onset as well as longer survival particularly when grade II and III gliomas from validation sets were added. The absence of LGG in the TCGA data led two groups16,27 to examine the predictive value of the TCGA profiles for LGG. Both groups16,27 utilized Affymetrix gene expression data for a small set of LGG (65 astro II, 4 mixed II, and 30 oligo II) from the Repository for Molecular Brain Neoplasia Data (Rembrandt) and reported similar findings with the TCGA profiles associated with prognostic value for LGG.16 More recently, the TCGA analyzed 293 lower (II/III) grade gliomas. Despite using a wide range of sophisticated technology platforms, the final results suggested that lower grade tumors can be simply and better characterized solely by two tumor markers, IDH1/2 and 1p/19q deletion status, than by the traditionally used histology/grade.78 The findings are considered paradigm-breaking and suggest that the decades-long classification system for glioma (focused on histology and grade) is likely inferior to a new more molecularly-based (but clinically simple and cost efficient) classification scheme. With respect to outcome, these molecular findings remain untested in a pure LGG cohort and uncorrected for an additional variable of clinical import, extent of surgical resection.13,55
Constitutive genetic polymorphisms
Glioma Risk
Genetic polymorphisms identified in association with glioma incidence are clearly of interest when considering genes associated with glioma survival.2,3,8,11,18,20–22,31,49,50,57–59,64 An emerging theme in glioma research has been that the genes/pathways identified in linkage and tumor studies are also being identified in GWAS. This demonstrates that, in addition to the rare variation associated with Mendelian disorders, common genetic variation also contributes to glioma genesis. While rare heritable loss-of-function mutations in TP53 and p16 cause glioma-associated familial cancer syndromes, inherited SNPs near both these genes also appear to contribute to glioma genesis. In total, GWAS of glioma patients have identified 9 independently significant SNP associations located in 8 genes (Table 2).34,35,62,68,71,79,81 The first two glioma GWAS.68,82 one of which included only HGG (from UCSF/Mayo)81 and the other (from the M.D. Anderson Center (MDA))68 which included HGG and some LGG, confirmed glioma risk loci in or near TERT (5p15), CDKN2A/B (9p21) (a gene region harboring p16, a tumor suppressor gene often homozygously deleted in GBM), and RTEL1 (20q13). The MDA GWAS68 which included LGG cases identified two additional loci: CCDC26 (8q24) and PHLDB1 (11q23).The top 13 SNPs in these five regions were further investigated by tumor subtype in 1446 cases and 1134 controls from UCSF/Mayo (with 224 oligo (II/III), 166 mixed (II/III) and 103 astro (II only)).34 As reported in the MDA GWAS68 CCDC26 (8q24) region loci were associated with oligo (II/III) (OR=2.05, p=8.3×10−11) but not GBM (Grade IV) risk with association with oligo II/III seen regardless of 1p/19q deletion status (although greatest risk was seen with co-deletion present). In contrast, RTEL region polymorphisms were most strongly associated with grade IV but less so with grade II/III glioma risk. The TERT region was associated with all grades and types of glioma. The CDKN2A/B region SNPs were also associated with Grade IV and grade II/III astrocytoms but not with oligo II/III. Insufficient data was available to draw conclusions about Grade II astrocytoma and Grade II oligodendroglioma independently of Grade III oligodendroglioma as well as Grade II versus Grade III mixed glioma). A similar analysis was performed in the German and French replication cohorts of the MDA GWAS again finding that CCDC26 and PHLDB1 loci were inversely and RTEL1 and TERT loci were positively correlated with grade.71 Data from a Chinese population agree as well.11 A pooled analysis of the US/UK/German/French data confirmed these findings and found evidence of an additional independent association for glioma (regardless of grade) risk with rs11979158 and rs2252586, at 7p11.2 which encompasses the EGFR gene although interestingly this gene was not associated with survival.62 The results listed above are remarkably confirmatory (in an era where GWAS results may vary widely) and strongly suggest distinct germline polymorphisms underlie different glioma subtypes (i.e. CCDC26 and PHLDB1 loci are consistently associated with LGG while other loci are either primarily associated with high grade glioma or with all glioma regardless of grade and histology. Jenkins et al35 further examine the CCDC26 (8q24) region and find strong association for a low frequency variant at 8q24.21 (rs55705857) associated with 1) oligo II/III regardless of IDH mutation status (OR=6.3, p=2.2×10−23), and 2) astro II-IV with mutated IDH1/IDH2 (OR=5.16–6.66, p=4.7×10−12 to 2.2×10−8) but not astrocytic tumors with wild type IDH1/IDH2. Their LGG specific findings are remarkable with increasing risk associated with decreasing astrocyte involvement (ORastroII=3.82 (95CI: 2.63, 5.54), p=1.7×10−12, ORmixedII=5.01 (3.48, 7.21, p=3.7×10−18 and ORoligoII=7.06 (5.10, 9.77), p=6.2×10−32). Two60,78 new reports are also of note; Using existing as well as new data from the UCSF/Mayo groups, Rice et al5,60 show that the PHLDB1 SNP is associated strictly with IDH-mutant while Walsh et al78 replicate the findings that CDKN2B SNPs are associated with low-grade astrocytomas. In summary, four of the above mentioned genes appear to contribute to development of all glioma grades and histologies (RTEL, TERT, EGFR, TP53), while the other 3 genes appear to contribute only to the development of certain glioma subtypes (Table 2). CCDC26 variants increase risk for oligodendroglial tumors regardless of IDH-mutation status and also for IDH-mutated astrocytoma. SNPs near CDKN2B/ANRIL confer increased risk for astrocytic tumors of all grades, including glioblastomas, but are not associated with oligodendroglial tumors. The histologic specificity of these SNP associations remains an area of active research.
Table 2.
Glioma-associated susceptibility variants detected by GWAS and fine-mapping
| Candidate Gene (Chromosome Location) |
Risk allele | Magnitude of Association1 |
Risk Allele Frequency2 |
Putative Functional Significance |
Associated glioma subtypes |
|---|---|---|---|---|---|
| TERC (3q26.2) | rs1920116-G | + | 0.72 | Increased telomere length | High-grade glioma |
| TERT (5p15.33) | rs2736100-C | + | 0.51 | Increased telomere length | All glioma subtypes |
| EGFR (7p11.2) | rs2252586-A | + | 0.27 | Unknown | All glioma subtypes |
| EGFR (7p11.2) | rs11979158-A | + | 0.82 | Unknown | All glioma subtypes |
| CCDC26 (8q24.21) | rs55705857-G | +++ | 0.046 | microRNA site | Oligodendroglial tumors and IDH-mutated astrocytic tumors |
| CDKN2B/ANRIL (9p21.3) | rs1412829-G | + | 0.43 | Unknown | Astrocytic tumors of all grades |
| PHLDB1 (11q23.3) | rs498872-A | + | 0.31 | Unknown | IDH-mutated gliomas |
| TP53 (17p13.1) | rs78378222-C | ++ | 0.014 | Alteration of polyadenylation signal impairs TP53 mRNA processing | All glioma subtypes |
| RTEL1 (20q13.33) | rs6010620-G | + | 0.76 | Unknown | All glioma subtypes |
Magnitude of Association: +++ represents OR>=5.0, ++ represents 2.0<=OR<5.0, and + represents 1.0<OR<2.0
Allele frequency in Caucasians, extracted from HapMap CEPH data where available.
Glioma outcome
There are few studies of genetic polymorphism and survival after diagnosis of glioma; those that exist focus on HGG (no study includes more than 50 LGG patients) with examination of SNPs in genes involved in DNA repair, cell cycle regulation, and immune function as well as in tumor markers of note.5,23,42,51,70,72,76,81,85
GWAS
No data exist specific to LGG but several informative efforts have been undertaken relative to HGG by Dr. Wrensch’s group which performed the first GBM GWAS survival analysis83,84 in uniformly treated (surgery, XRT, and TMZ) patients and found that SSBP2 (a single-stranded DNA binding protein on 5q14.1) germline variants were associated with survival (discovery (UCSF) and validation (MAYO, Glioma SE, and TCGA) sets show a combined HRrs7732320 = 1.64 (1.34, 2.00), p=1.3*10−6) and that expression of SSBP2 in GBM tumors was significantly related to reduced survival (HR=1.22, (1.09, 1.36), p=5.3 × 10−4).Interestingly, among TCGA and other GBM patients, SSBP2 expression was highest among patients with the Proneural signature, a group likely to include persons with secondary GBM (i.e. progressed from LGG) suggesting that SSBP2 germline variants and tumor expression (which were not linked to IDH mutation status) may be an important independent predictor of survival for LGG. Dr. Bondy also examined associations with GBM survival with the 100 top-ranking glioma susceptibility polymorphisms identified from the two glioma GWAS68,82 and found that polymorphisms in the LIG4, HMGA2, BTBD2, and RTEL1 genes (all involved in the double-strand break repair pathway) were associated with GBM survival in the MDA GWAS cohort (although not confirmed in the validation set).43 Preliminary analyses from the UCSF/Mayo study indicate that the variant at 8q24.21 (rs55705857) associated so strongly with glioma risk appears to also be associated with survival. The data used here to estimate survival are taken from the SEER program.65 Although an important description of “real-world” LGG practice that includes persons of all ages, race and medical status, the data are limited by 1) a lack of a uniform histological review, 2) treatment data restricted to first course (hence data on XRT and chemotherapy are limited or absent) and not adjusted for clinical factors likely to influence treatment assignment and, 3) no information on constitutive/tumor genotype, tumor markers, or patient co-morbidities.
One important reason for the lack of knowledge concerning LGG is that these patients are generally only included as a convenience sub-sample in studies of HGG with results driven by the much larger numbers of HGG in these studies.10,12,34,62,68,69,82 Furthermore, the lack of effect in randomized clinical trials is likely also due in part to the unknowing inclusion of genetically dissimilar tumors into one study arm. In the future, clarification of the tumor markers/profiles known to be associated with outcome (both natural progression as well as response to treatment) and will be required to be measured in any planned RCT to preserve randomization. The import of such markers and profiles is already recognized by organizers of the HGG trials with tumor materials retrospectively being analyzed to assess randomization. As LGG represents the first step in a multistage disease process, the need to focus efforts at the start of the disease process is clear. Large sample cohorts which will likely require the development of consortia given the relatively small numbers of these tumors. As can be seen from the literature morphological and molecular sub typing is critical to cancer genetic epidemiology35 and to date not explored specifically for LGG. Discovery of genes associated with poor outcomes will inform allow for improvement of randomization schemes in clinical trials of LGG as well as suggest novel biologic mechanisms for development of targeted therapy designed to improve survival. The time is right for researchers to take advantage of emerging genetic technology, statistical methodology and computing capability to create a new clinical paradigm for LGG.
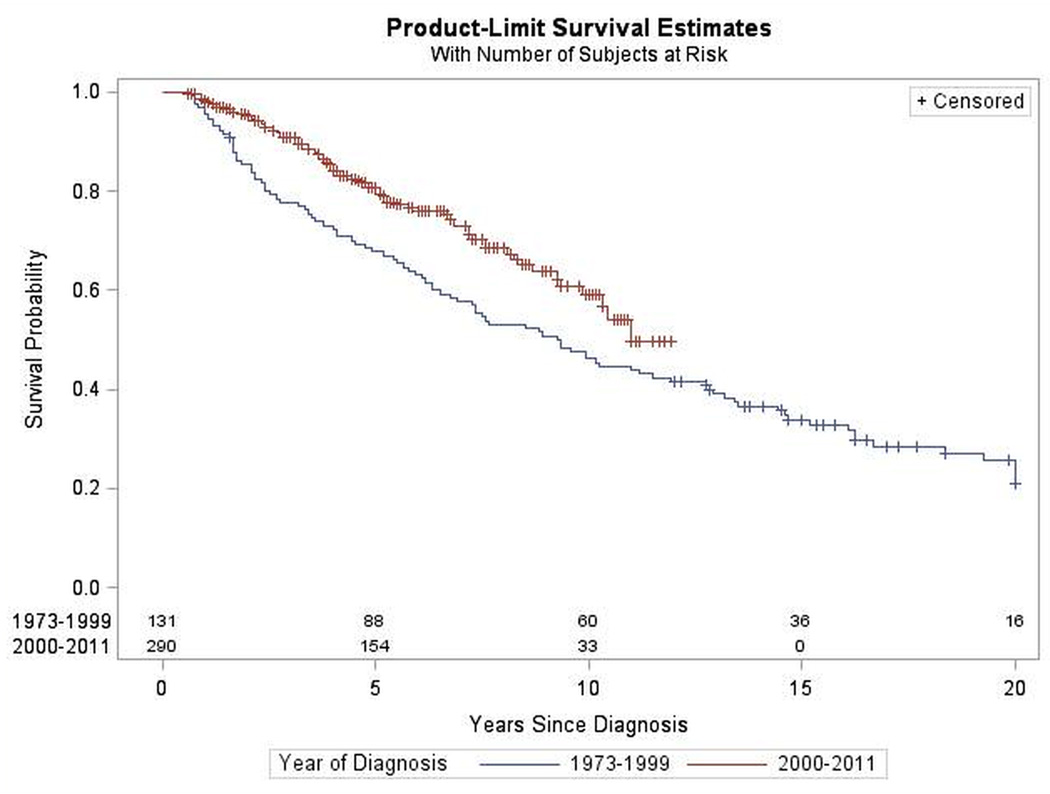
Figure 2.
Survival by Year of Diagnosis for Astrocytoma. SEER 1973–2011
Acknowledgments
This work was supported by NIH R01 grant R01 CA119215. Work at UCSF was supported by the NIH (grant numbers R25CA112355, R01CA52689, R01CA126831 and P50CA097257), as well as the National Brain Tumor Foundation, and the UCSF Lewis Chair in Brain Tumor Research.
References
- 1.Aaronsen NK, Taphoorn MJ, Heimans JJ, Postma TJ, Gundy CM, Beute GN, et al. Compromised Health-Related Quality of Life in Patients With Low-Grade Glioma. J Clin Oncol. 2011;29:4430–4435. doi: 10.1200/JCO.2011.35.5750. [DOI] [PubMed] [Google Scholar]
- 2.Bethke L, Webb E, Murray A, Schoemaker M, Johansen C, Christensen C, et al. Comprehensive analysis of the role of DNA repair gene polymorphisms on risk of glioma. Hum Molecular Genet. 2008;17:800–805. doi: 10.1093/hmg/ddm351. [DOI] [PubMed] [Google Scholar]
- 3.Bethke L, Sullivan K, Webb E, Murray A, Schoemaker M, Auvinen A, et al. The common D302H variant of CASP8 is associated with risk of glioma. Cancer Epidemiol Biomarkers Prev. 2008;17:987–989. doi: 10.1158/1055-9965.EPI-07-2807. [DOI] [PubMed] [Google Scholar]
- 4.Stege EM, Kros JM, de Brulin HG, Enting RH, van Heuvel I, Looijenga LH, et al. Successful treatment of low-grade oligodendroglial tumors with a chemotherapy regimen of procarbazine, lomustine, and vincristine. Cancer. 2005;103:802–809. doi: 10.1002/cncr.20828. [DOI] [PubMed] [Google Scholar]
- 5.Bhowmick DA, Zhuang Z, Wait SD, Weil RJ. A functional polymorphism in the EGF gene is found with increased frequency in glioblastoma multiforme patients and is associated with more aggressive disease. Cancer Res. 2004;64:1220–1223. doi: 10.1158/0008-5472.can-03-3137. [DOI] [PubMed] [Google Scholar]
- 6.Brada M, Viviers L, Abson C, Hines F, Britton J, Ashley S, et al. Phase II study of primary temozolamide chemotherapy in patients with WHO grade II gliomas. Ann Oncol. 2003;14:1715–1721. doi: 10.1093/annonc/mdg371. [DOI] [PubMed] [Google Scholar]
- 7.Bruckner J. Phase III study of radiation therapy (RT) with or without procarbazine, CCNU, and vincristine (PCV) in low-grade glioma: RTOG 9802 with Alliance, ECOG, and SWOG. J Clin Oncol. 2014;32:5s. [Google Scholar]
- 8.Caggana M, Kilgallen J, Conroy J, Wiencke J, Kelsey K, Miike R, Wrensch M. Associations between ERCC2 polymorphisms and gliomas. Cancer Epidemiol Biomarkers Prevention. 2001;10:355–360. [PubMed] [Google Scholar]
- 9.Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR, et al. Specific predictors of chemotherapeutic response and survival in patients with oligodendrogliomas. J Natl Cancer Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research Network: Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carpentier C, Laigle-Donadey F, Marie Y, Auger N, Benouaich-Amiel A, Lejeune J, et al. Polymorphism in Sp1 recognition site of the EGF receptor gene promoter and risk of glioblastoma. Neurology. 2006;67:872–874. doi: 10.1212/01.wnl.0000229927.12007.37. [DOI] [PubMed] [Google Scholar]
- 12.Chen H, Chen Y, Zhao Y, Fan W, Zhou K, Liu Y, et al. Association of sequence variants on chromosomes 20, 11, and 5 (20q13.33, 11q23.3, and 5p15.33) with glioma susceptibility in a Chinese population. Am J Epidemiol. 2011;173:915–922. doi: 10.1093/aje/kwq457. [DOI] [PubMed] [Google Scholar]
- 13.Claus EB, Horlacher A, Hsu L, Schwartz RB, Dello-Iacono D, Talos F, et al. Survival rates in patients with low-grade glioma after intraoperative magnetic resonance image guidance. Cancer. 2005;103:1227–1233. doi: 10.1002/cncr.20867. [DOI] [PubMed] [Google Scholar]
- 14.Claus EB, Black PM. Survival rates and patterns of care for patients diagnosed with supratentorial low grade gliomas: Data from the Surveillance, Epidemiology, and End Results (SEER) Program, 1973–2001. Cancer. 2006;106:1358–1363. doi: 10.1002/cncr.21733. [DOI] [PubMed] [Google Scholar]
- 15.Colman H, Zhang L, Sulman EP, McDonald JM, Shooshtari NL, Rivera A, et al. A multigene predictor of outcome in glioblastoma. Neuro Oncol. 2010;12:49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper LA, Gutman DA, Long Q, Johnson BA, Cholleti SR, Kurc T, et al. The Proneural Molecular Signature is Enriched in Oligodendrogliomas and Predicts Improved Survival Among Diffuse Gliomas. PLoS One. 2010;5:e12548. doi: 10.1371/journal.pone.0012548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Correa DD, Shi W, Thaler HT, Cheung AM, DeAngelis LM, Abrey LE. Longitudinal cognitive follow-up in low grade gliomas. J Neurooncol. 2008;86:321–327. doi: 10.1007/s11060-007-9474-4. [DOI] [PubMed] [Google Scholar]
- 18.Costa BM, Ferreira P, Costa S, Canedo P, Oliveira P, Silva A, et al. Association between functional EGF+61 polymorphism and glioma risk. Clin Cancer Res. 2007;13:2621–2626. doi: 10.1158/1078-0432.CCR-06-2606. [DOI] [PubMed] [Google Scholar]
- 19.Cox DR. Regression models and life tables. J R Stat Soc [B] 1972;34:187–202. [Google Scholar]
- 20.De Roos AJ, Rothman N, Inskip PD, Linet MS, Shapiro WR, Selker RG, et al. Genetic polymorphisms in GSTM1,-P1,-T1, and CYP2E1 and the risk of adult brain tumors. Cancer Epidemiol Biomarkers Prevention. 2003;12:14–22. [PubMed] [Google Scholar]
- 21.De Roos AJ, Rothman N, Brown M, Bell DA, Pittman GS, Shapiro WR, et al. Variation in genes relevant to aromatic hydrocarbon metabolism and the risk of adult brain tumors. Neuro Oncol. 2006;8:145–155. doi: 10.1215/15228517-2005-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egan KM, Thompson RC, Nabors LB, Olson JJ, Brat DJ, Larocca RV, et al. Cancer susceptibility variants and the risk of adult glioma in a US case-control study. J Neurooncol. 2011;104:535–542. doi: 10.1007/s11060-010-0506-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egan KM, Nabors LB, Olson JJ, Monteiro AN, Browning JE, Madden MH, et al. Rare TP53 genetic variant associated with glioma risk and outcome. J Med Genet. 2012;49:420–421. doi: 10.1136/jmedgenet-2012-100941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Everhard S, Kaloshi G, Criniere E, Benouaich-Amiel A, Lejeune J, Marie Y, et al. MGMT methylation: a marker of response to temozolamide in low-grade gliomas. Ann Neurol. 2006;60:740–743. doi: 10.1002/ana.21044. [DOI] [PubMed] [Google Scholar]
- 25.Eyre HJ, Crowley JJ, Townsend JJ, Eltringham JR, Morantz RA, Schulman SF, et al. A randomized trial of radiotherapy versus radiotherapy plus CCNU for incompletely resected low-grade gliomas. A Southwest Oncology Group study. Neurosurgery. 1993;78:909–914. doi: 10.3171/jns.1993.78.6.0909. [DOI] [PubMed] [Google Scholar]
- 26.Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 27.Guan X, Chen Y, Barnholtz-Sloan J. Association between TCGA Gene-expression-based Glioblastoma Subtypes and Lower Grade Gliomas. http://cdn0.pathable.com/abstract/28/1308951897.?1308951897. [Google Scholar]
- 28.Hartmann C, Hentschel B, Tatagiba M, Schramm J, Schnell O, Seidel C, et al. Molecular Markers in Low-Grade Gliomas: Predictive or Prognostic? Clin Cancer Res. 2011;17:4588–4599. doi: 10.1158/1078-0432.CCR-10-3194. [DOI] [PubMed] [Google Scholar]
- 29.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 30.Houillier C, Wang X, Kaloshi G, Mokhtari K, Guillevin R, Laffaire J, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. 2010;75:1560–1566. doi: 10.1212/WNL.0b013e3181f96282. [DOI] [PubMed] [Google Scholar]
- 31.Idbaih A, Boisselier B, Marie Y, El Hallani S, Sanson M, Crinière E, et al. TP53 codon 72 polymorphism, p53 expression, and 1p/19q status in oligodendroglial tumors. Cancer Genet Cytogenet. 2007;177:103–107. doi: 10.1016/j.cancergencyto.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 32.Jaloka AS, Myrmel KS, Kloster R, Torp SH, Lindal S, Unsgard G, et al. Comparison of a strategy favoring early surgical resection vs a strategy favoring watchful waiting in low-grade gliomas. J Amer Med Assoc. 2012;308:1881–1888. doi: 10.1001/jama.2012.12807. [DOI] [PubMed] [Google Scholar]
- 33.Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell Rm, Law M, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis for patients with oligodendroglioma. Cancer Res. 2006;66:9852–9861. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 34.Jenkins RB, Wrensch MR, Johnson D, Fridley BL, Decker PA, Xiao Y, et al. Distinct germ line polymorphisms underlie glioma morphologic heterogeneity. Cancer Genet. 2011;204:13–18. doi: 10.1016/j.cancergencyto.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenkins RB, Xiao Y, Sicotte H, Decker PA, Kollmeyer TM, Hansen, et al. A low-frequency variant at 8q24.21 is strongly associated with risk of oligodendroglial tumors and astrocytomas with IDH1 or IDH2 mutation. Nat Genet. 2012;44:1122–1125. doi: 10.1038/ng.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiao Y, Killela PJ, Reitman ZJ, Rasheed BA, Heaphy CM, de Wilde RF, et al. Frequent ATRX, CIC, FUBP1 and IDH mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:710–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaloshi G, Benouaich-Amiel A, Diakite F, Taillibert S, Lejeune J, Laigle-Donadey F, et al. Temozolamide for low-grade gliomas. Predictive impact of 1p/19q loss on response and outcome. Neurology. 2007;68:1831–1836. doi: 10.1212/01.wnl.0000262034.26310.a2. [DOI] [PubMed] [Google Scholar]
- 38.Karim AB, Maat B, Hatlevoll R, Menten J, Rutten EH, Thomas DG, et al. A randomized trial on dose-response in radiation therapy of low-grade cerebral glioma: European Organization for Research and Treatment of Cancer (EORTC) Study 22844. Int J Radiat Oncol Biol Phys. 1996;36:549–556. doi: 10.1016/s0360-3016(96)00352-5. [DOI] [PubMed] [Google Scholar]
- 39.Kesari S, Schiff D, Drappatz J, LaFrankie D, Doherty L, Macklin EA, et al. Phase II study of protracted daily temozolomide for low-grade gliomas in adults. Clin Cancer Res. 2009;15:330–337. doi: 10.1158/1078-0432.CCR-08-0888. [DOI] [PubMed] [Google Scholar]
- 40.Kiebert GM, Curran D, Aaronson NK, Bolla M, Menten J, Rutten EH, et al. Quality of life after radiation therapy of cerebral low-grade gliomas of the adult: results of a randomised phase III trial on dose response (EORTC trial 22844) Eur J Cancer. 1998;34:1902–1909. doi: 10.1016/s0959-8049(98)00268-8. [DOI] [PubMed] [Google Scholar]
- 41.Levin N, Lavon I, Zelikovitsh B, Fuchs D, Bokstein F, Fellig Y, et al. Progressive low-grade oligodendrogliomas. Response to temozolamide and correlation between genetic profile and O6-methylguanine-DNA methyltransferase protein expression. Cancer. 2006;106:1759–1765. doi: 10.1002/cncr.21809. [DOI] [PubMed] [Google Scholar]
- 42.Lima-Ramos V, Pacheco-Figueiredo L, Costa S, Pardal F, Silva A, Amorim J, et al. TP53 codon 72 polymorphism in susceptibility, overall survival, and adjuvant therapy response of gliomas. Cancer Genet Cytogenet. 2008;180:14–19. doi: 10.1016/j.cancergencyto.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Shete S, Etzel CJ, Scheurer M, Alexiou G, Armstrong G, et al. Polymorphisms of LIG4, BTBD2, HMGA2, and RTEL1 genes involved in the double-strand break repair pathway predict glioblastoma survival. J Clin Oncol. 2010;28:2467–2474. doi: 10.1200/JCO.2009.26.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu R, Solheim K, Polley MY, Lamborn KR, Page M, Fedoroff A, et al. Quality of life in low-grade glioma patients receiving temozolomide. Neuro Oncol. 2009;11:59–68. doi: 10.1215/15228517-2008-063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Louis DN, Ohgaki H, Wiestler OD, Cavanee WK, Burger PC, Jouvet A, et al. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mainio A, Tuunanen S, Hakko H, Niemela A, Koivukangas J, Rasanen P. Decreased quality of life and depression as predictors for shorter survival among patients with low-grade gliomas: a follow-up from 1990 to 2003. Eur Arch Psychiatr Clin Neurosci. 2006;256:516–521. doi: 10.1007/s00406-006-0674-2. [DOI] [PubMed] [Google Scholar]
- 48.Malmer B, Barnholtz-Sloan J, Bernstein J, Claus EB, Houlston R, Jenkins R, et al. Gliogene: An international effort for delineating the underlying causes of familial glioma. Cancer Epid Biomarkers Prev. 2007;16:1730–1734. doi: 10.1158/1055-9965.EPI-07-0081. [DOI] [PubMed] [Google Scholar]
- 49.Malmer B, Feychting M, Lonn S, Ahlbom A, Henriksson R. p53 Genotypes and risk of glioma and meningioma. Cancer Epidemiol Biomarkers Prev. 2005;14:2220–2223. doi: 10.1158/1055-9965.EPI-05-0234. [DOI] [PubMed] [Google Scholar]
- 50.Malmer BS, Feychting M, Lonn S, Lindstrom S, Gronberg H, Ahlbom A, et al. Genetic variation in p53 and ATM haplotypes and risk of glioma and meningioma. J Neurooncol. 2007;82:229–237. doi: 10.1007/s11060-006-9275-1. [DOI] [PubMed] [Google Scholar]
- 51.Nigro JM, Misra A, Zhang L, Smirnov I, Colman H, Griffin C, et al. Integrated array-comparative genomic hybridization and expression array profiles identify clinically relevant molecular subtypes of glioblastoma. Cancer Res. 2005;65:1678–1686. doi: 10.1158/0008-5472.CAN-04-2921. [DOI] [PubMed] [Google Scholar]
- 52.Okcu MF, Selvan M, Wang LE, Stout L, Erana R, Airewele G, et al. Glutathione S-transferase polymorphisms and survival in primary malignant glioma. Clinical Cancer Research. 2004;10:2618–2625. doi: 10.1158/1078-0432.ccr-03-0053. [DOI] [PubMed] [Google Scholar]
- 53.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 55.Pignatti F, et al. Prognostic factors for survival in adult patients in cerebral low grade glioma. J Clin Oncol. 2002;20:2076–2084. doi: 10.1200/JCO.2002.08.121. [DOI] [PubMed] [Google Scholar]
- 56.Quinn JA, Reardon DA, Friedman AH, Rich JN, Sampson JH, Provenzale JM, et al. Phase II trial of temozolamide in patients with progressive low-grade glioma. J Clin Oncol. 2003;21:646–651. doi: 10.1200/JCO.2003.01.009. [DOI] [PubMed] [Google Scholar]
- 57.Rajaraman P, Wang SW, Rothman N, Brown MM, Black PM, Fine HA, et al. Polymorphisms in apoptosis and cell cycle control genes and risk of brain tumors in adults. Cancer Epidemiol Biomarkers Prev. 2007;16:1655–1661. doi: 10.1158/1055-9965.EPI-07-0314. [DOI] [PubMed] [Google Scholar]
- 58.Rajaraman P, Stewart PA, Samet JM, Schwartz BS, Linet MS, Zahm SH, et al. Lead, genetic susceptibility, and risk of adult brain tumors. Cancer Epidemiol Biomarkers Prev. 2006;15:2514–2522. doi: 10.1158/1055-9965.EPI-06-0482. [DOI] [PubMed] [Google Scholar]
- 59.Rajaraman P, Hutchinson A, Wichner S, Black PM, Fine HA, Loeffler JS, et al. DNA repair gene polymorphisms and risk of adult meningioma, glioma, and acoustic neuroma. Neuro Oncol. 2010;12:37–48. doi: 10.1093/neuonc/nop012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rice T, Zheng S, Decker PA, Walsh KM, Bracci P, Xiao Y, et al. Inherited variant on chromosome 11q23 increases susceptibility to IDH mutated but not IDH normal gliomas regardless of grade or histology. Neuro-Oncology. 2012 doi: 10.1093/neuonc/nos324. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanai N, Berger MS. Glioma Extent of Resection and Its Impact on Patient Outcome. Neurosurgery. 2008;62:753–766. doi: 10.1227/01.neu.0000318159.21731.cf. [DOI] [PubMed] [Google Scholar]
- 62.Sanson M, Hosking FJ, Shete S, Zelenika D, Dobbins SE, Ma Y, et al. Chromosome 7p11.2 (EGFR) variation influences glioma risk. Hum Mol Genet. 2011;20:2897–2904. doi: 10.1093/hmg/ddr192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.SAS Institute Inc. SAS/STAT User's Guide. Vol. 1. Cary, NC: SAS Institute Inc.; 2006. [Google Scholar]
- 64.Schwartzbaum JA, Ahlbom A, Lonn S, Warholm M, Rannug A, Auvinen A, et al. An international case-control study of glutathione transferase and functionally related polymorphisms and risk of primary adult brain tumors. Cancer Epidemiol Biomarkers Prev. 2007;16:559–565. doi: 10.1158/1055-9965.EPI-06-0918. [DOI] [PubMed] [Google Scholar]
- 65.Surveillance, Epidemiology, and End Results (SEER) Program( www.seer.cancer.gov) Research Data (1973–2011), National Cancer Institute, DCCPS, Surveillance Research Program, Surveillance Systems Branch. [based on the November 2013]; released April 2014, submission.
- 66.Shaw E, Arusell R, Scheithauer B, O’Fallon J, O’Neill B, Dinapoli R, et al. Prospective randomized trial of low- versus high-dose radiation therapy in adults with supratentorial low-grade glioma: initial report of a North Central Cancer Treatment Group/Radiation Therapy Oncology Group/Eastern Cooperative Oncology Group study. J Clin Oncol. 2002;120:2267–2276. doi: 10.1200/JCO.2002.09.126. [DOI] [PubMed] [Google Scholar]
- 67.Shaw EG, Wang M, Coons SW, Brachman DG, Buckner JC, Stelzer KJ, et al. Randomized trial of radiation therapy plus procarbazine, lomustine, and vincristine chemotherapy for supratentorial adult low-grade glioma: initial results of RTOG 9802. J Clin Oncol. 2012;30:3065–3070. doi: 10.1200/JCO.2011.35.8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shete S, Hosking F, Robertson LB, Dobbins SE, Sanson M, Malmer B, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nature Genetics. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shete S, Lau C, Houlston R, Claus EB, Barnholtz-Sloan J, Lai R, et al. A High-density SNP Genome-wide Linkage Search identifies Susceptibility Loci for Glioma: Results from the GLIOGENE Consortium. Cancer Res. 2011;71:7568–7575. doi: 10.1158/0008-5472.CAN-11-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simon M, Ludwig M, Fimmers R, Mahlberg R, Müller-Erkwoh A, Köster G, et al. Variant of the CHEK2 gene as a prognostic marker in glioblastoma multiforme. Neurosurgery. 2006;59:1078–1085. doi: 10.1227/01.NEU.0000245590.08463.5B. [DOI] [PubMed] [Google Scholar]
- 71.Simon M, Hosking FJ, Marie Y, Gousias K, Boisselier B, Carpentier C, et al. Genetic risk profiles identify different molecular etiologies for glioma. Clin Cancer Res. 2010;16:5252–5259. doi: 10.1158/1078-0432.CCR-10-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang J, Shao W, Dorak MT, Li Y, Miike R, Lobashevsky E, et al. Positive and negative associations of human leukocyte antigen variants with the onsent and prognosis of adult glioblastoma mulitforme. Cancer Epidemiol Biosmarkers Prev. 2005;14:2040–2044. doi: 10.1158/1055-9965.EPI-05-0136. [DOI] [PubMed] [Google Scholar]
- 73.Tosoni A, Franceschi E, Ermani M, Bertorelle R, Bonaldi L, Blatt V, et al. Temozolomide three weeks on and one week off as first line therapy for patients with recurrent or progressive low grade gliomas. J Neurooncol. 2008;89:179–185. doi: 10.1007/s11060-008-9600-y. [DOI] [PubMed] [Google Scholar]
- 74.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van den Bent MJ, Afra D, de Witte O, Ben Hassel M, Schraub S, Hoang-Xuan K, et al. EORTC Radiotherapy and Brain Tumor Groups and the UK Medical Research Council. Long-term efficacy of early versus delayed radiotherapy for low-grade astrocytoma and oligodendroglioma in adults: the EORTC 22845 randomised trial. Lancet. 2005;366:985–990. doi: 10.1016/S0140-6736(05)67070-5. [DOI] [PubMed] [Google Scholar]
- 76.Vauleon E, Auger N, Benouaich-Amiel A, Laigle-Donadey F, Kaloshi G, Lejeune J, et al. The 61 A/G EGF polymorphism is functional but is neither a prognostic marker nor a risk factor for glioblastoma. Cancer Genet Cytogenet. 2007;172:33–37. doi: 10.1016/j.cancergencyto.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 77.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Verhaak RGW, et al. San Diego: 2014. AACR 2014: Presentation Abstract: Comprehensive and Integrative Genomic Characterization of Diffuse Lower Grade Gliomas. ( http://cancergenome.nih.gov/newsevents/events/AACR2014_Abstract_LGGs) [Google Scholar]
- 79.Walsh KM, Anderson E, Hansen HM, Decker PA, Kosel ML, Kollmeyer T, et al. Analysis of 60 reported glioma risk SNPs replicates published GWAS findings but fails to replicate associations from published candidate-gene studies. Genetic Epidemiology. 2012;37:222–228. doi: 10.1002/gepi.21707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Walsh KM, Codd V, Smirnov IV, Rice T, Decker PA, Hansen HM, et al. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet. 2014;46:731–735. doi: 10.1038/ng.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wiencke JK, Aldape K, McMillan A, Wiemels J, Maghadassi M, Miike R, et al. Molecular features of adult glioma associated with patient race/ethnicity, age, and a polymorphism in O6-methylguanine-DNA methyltransferase. Cancer Epidemiol Biomarkers Prev. 2005;14:1774–1783. doi: 10.1158/1055-9965.EPI-05-0089. [DOI] [PubMed] [Google Scholar]
- 82.Wrensch M, Jenkins RB, Chang JS, Yeh R-F, Xiao Y, Decker PA, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nature Genetics. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wrensch M, Wiencke JK, Wiemels J, Miike R, Patoka J, Moghadassi M, et al. Serum IgE, tumor epidermal growth factor receptor expression, and inherited polymorphisms associated with glioma survival. Cancer Res. 2006;66:4531–4541. doi: 10.1158/0008-5472.CAN-05-4032. [DOI] [PubMed] [Google Scholar]
- 84.Wrensch M, McMillan A, Wiencke J, Wiemels J, Kelsey K, Patoka J, et al. Non synonymous coding single-nucleotide polymorphisms spanning the genome in relation to glioblastoma survival and age at diagnosis. Clin Cancer Res. 2007;13:197–205. doi: 10.1158/1078-0432.CCR-06-1199. [DOI] [PubMed] [Google Scholar]
- 85.Xiao Y, Decker PA, Rice T, McCoy LS, Smirnov I, Patoka JS, et al. SSBP2 variants impact survival in glioblastoma patients. Clin Cancer Res. 2012;18:3154–3162. doi: 10.1158/1078-0432.CCR-11-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in Gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang P, Kollmeyer TM, Buckner K, Bamlet W, Ballman KV, Jenkins RB, et al. Polymorphisms in GLTSCR1 and ERCC2 are associated with the development of oligodendrogliomas. Cancer. 2005;103:2363–2372. doi: 10.1002/cncr.21028. [DOI] [PubMed] [Google Scholar]