Abstract
Translation initiation in the hepatitis C virus (HCV) occurs through a cap-independent mechanism that involves an internal ribosome entry site (IRES) capable of interacting with and utilizing the eukaryotic translational machinery. In this review, we focus on the structural configuration of the different HCV IRES domains and the impact of IRES primary sequence variations on secondary structure conservation and function. In some cases, multiple mutations, even those scattered across different domains, led to restoration of the translational activity of the HCV IRES, although the individual occurrences of these mutations were found to be deleterious. We propose that such observation may be attributed to probable long-range inter- and/or intra-domain functional interactions. The precise functioning of the HCV IRES requires the specific interaction of its domains with ribosomal subunits and a subset of eukaryotic translation initiation factors (eIFs). The structural conformation, sequence preservation and variability, and translational machinery association with the HCV IRES regions are also thoroughly discussed, along with other factors that can affect and influence the formation of translation initiation complexes. WIREs RNA 2015, 6:211–224. doi: 10.1002/wrna.1268
INTRODUCTION
Hepatitis C virus (HCV) is a blood-borne pathogen with an estimated global prevalence of 130–200 million cases of chronic infection,1 increasing these patients' risk of developing liver cirrhosis and hepatocellular carcinoma. The standard chemotherapeutic treatment for HCV is the combination of pegylated interferon α (IFN-α) with guanosine analogue ribavirin, which is linked to side effects and can often lead to a relapse in many patients.2,3 Only recently, other treatments based on the inhibition of either the NS5B viral polymerase (sofosbuvir)4,5 or the NS3/4A viral protease (simeprevir,6 boceprevir,7 and telaprevir8) have been approved.
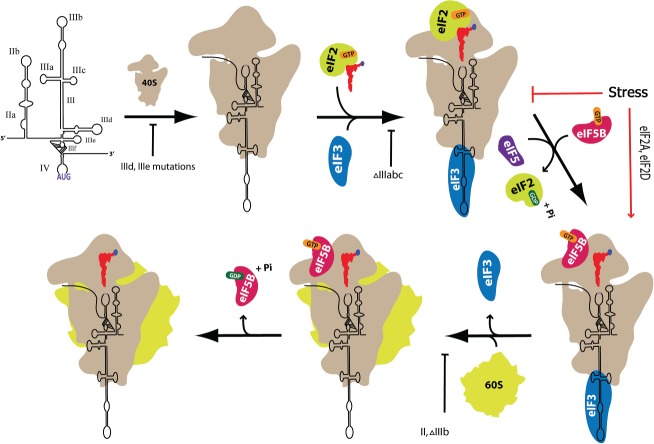
The viral genome comprises a 9.6 kb long, single-stranded, positive sense RNA molecule9,10 that possesses a highly conserved 5′ untranslated region (5′ UTR), a single open reading frame (ORF) and a conserved 3′ UTR. The ORF encodes a single large polyprotein of >3000 amino acids, which is co- and post-translationally cleaved by cellular and viral proteases to yield the mature viral structural (core, E1, and E2) and nonstructural (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins. Translation of the polyprotein is driven by an internal ribosomal entry site (IRES) that occupies most of the 5′ UTR of the viral RNA.11 Translation initiation mediated by the HCV IRES is distinct from canonical eukaryotic mRNA translation initiation in that it does not require the presence of the 7mG cap moiety at the 5′ mRNA end along with all of the eukaryotic translation initiation factors (eIFs) that are normally responsible for the recruitment of a ribosome to mRNA and its scanning for the correct AUG codon. Instead, the HCV IRES directly recruits and positions the small ribosomal subunit at the start AUG codon without the need for any known initiation factor.12 This positioning is followed by the binding of eukaryotic translation initiation factor 3 (eIF3) and eIF2-GTP-Met-tRNAi, the ternary complex (eIF2-TC) that stabilizes the pre-initiation translation assembly.12,13 The release of eIFs via GTP hydrolysis further allows the binding of a 60S subunit to assemble an 80S ribosome and initiate protein synthesis14 (Figure 1). The establishment of 48S and 80S pre-initiation complexes requires many other intermediate processes that, if not performed properly, can result in the blockage of efficient translation.
Figure 1.
A stepwise demonstration of the hepatitis C virus internal ribosome entry site (HCV IRES) translation initiation pathway. The specified arrangement of the HCV IRES domains and ribosomal subunits along with the particular placement of a subset of canonical eukaryotic translation initiation factors (eIFs) required to assemble the pre-initiation 48S and 80S complexes are depicted. The red arrows indicate eIF2-GTP-independent translation under stress (upon eIF2α phosphorylation), when protein synthesis continues with eIF5B or other reported factors (eIF2A, eIF2D). Mutations in different HCV IRES domains that may inhibit particular stages of translation complex formation are shown under the arrows.
HCV IRES RNA has a conserved primary sequence and secondary structure that spans ∼341 nucleotides composing domains I, II, III, and IV15–17 (Figure 2). The HCV IRES adopts a characteristic 3D conformation or specific tertiary structure under physiological concentrations of metal ions.25 The conservation of the IRES structure determines the efficacy of viral protein synthesis. Various structural,19,26,20 in vitro mutagenesis,27,28 biochemical, and enzymatic studies29,30 have demonstrated the importance of the conservation and specificity of domains II-IV, which are required to interact with translational machinery and perform the HCV IRES-mediated translation. Whereas the deletion of domain I results in an increased level of translational response,31,32 domains II and III have been found to interact with the 40S subunit, which is the first step directed toward protein synthesis. A pancreatic RNase protection assay has been utilized to locate sequences that occur in close proximity to the 40S subunit but are not necessarily in direct contact.33,34 In addition to making contact with 40S, the basal region of domain III also helps in the placement of the HCV IRES start codon, which is present in domain IV, in the ribosomal decoding groove.35 Domain III also regulates the binding of eIF3 through its own apical half,30,36 while domain II modulates GTP hydrolysis of the TC, mediated by eukaryotic translation initiation factor 5 (eIF5).14 The HCV IRES can also initiate translation through an alternative eIF2-independent pathway under stress conditions and increased eIF2-α phosphorylation.37,38
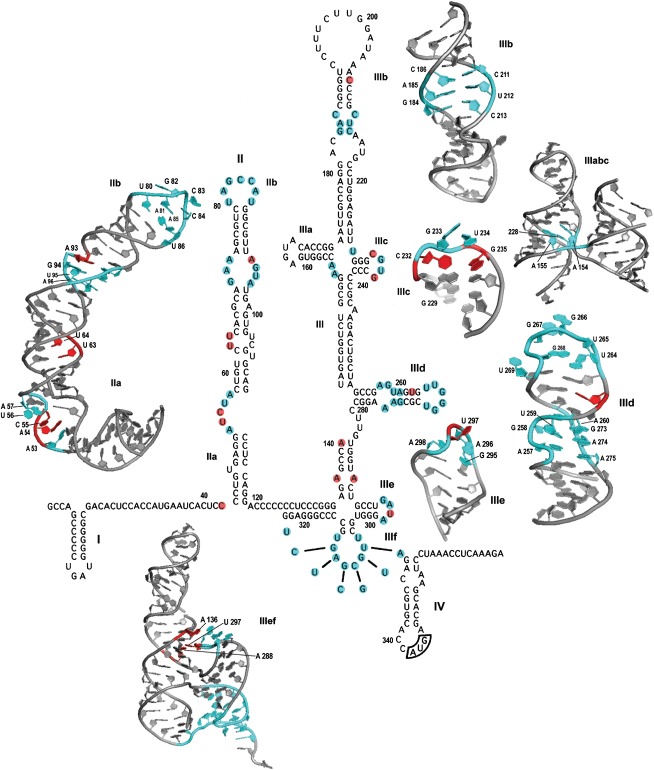
Figure 2.
The proposed secondary structure of the hepatitis C virus internal ribosome entry site (HCV IRES),11,18 with three-dimensional structures of domains II-IV resolved by NMR and X-ray crystallography.19–24 The domains of the HCV IRES are labeled. Nucleotides colored in cyan indicate the highly conserved regions of the IRES, whereas nucleotides in red denote mutations in the domains involved in a proposed long-range inter or intra-domain interaction or an unusual translational activity. The colors in the secondary structure correspond to the 3D structure. The PDB entries for the HCV IRES domains are domains II (1P5P), IIIb (1KP7), IIIc (1IDV), IIIabc (1KH6), IIId (1F84), IIIe (1 F85), and IIIef (3T4B).
In this review, we attempt to describe the current understanding of the HCV IRES, encompassing the importance of the HCV IRES structural elements in the mediation of translation initiation. We examined the influence of mutations in the context of possible alterations of the IRES configuration and its interaction with translational machinery that may disrupt the HCV IRES function. We also highlight combinations of mutations collectively expressing restored translational activity, the individual occurrence of which otherwise displayed reduced HCV IRES efficiency.
DOMAIN II STRUCTURE CONSERVATION IS VITAL FOR HCV IRES FUNCTION
L-Shaped Domain IIa may Serve as a Target for New Antivirals
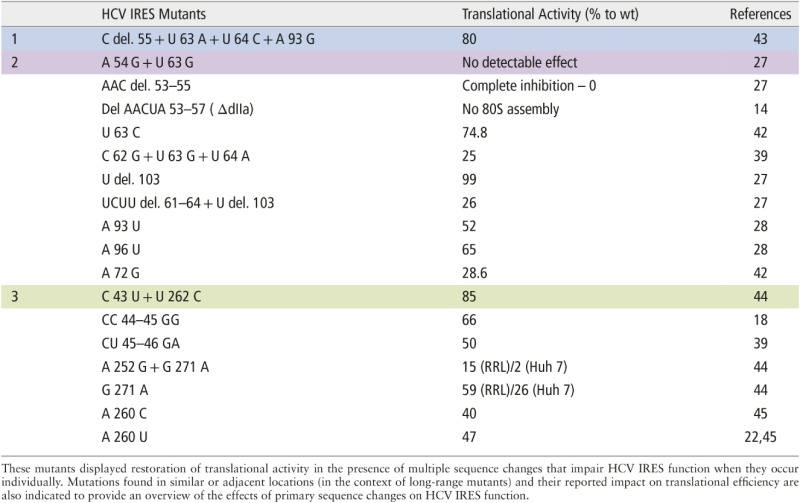
On the basis of phylogenetic and mutational analyses, domain II was found to be highly conserved among the IRES RNA of HCV, pestivirus, and GB virus B isolates.39,18 The NMR structure of HCV IRES domain II exhibited an L-shaped conformation that retains an identical configuration in both its free and 40S-bound forms.19 The structure of domain IIa (nucleotides 49–69 and 100–115) was also investigated using X-ray crystallography.40,41 Domain IIa is an independently folded domain of the HCV IRES that adopts a 90° helical bent structure including nucleotides A53, A54, C55, U56, and A57. Continuous base stacking and hydrogen bonding between the nucleotides determine the characteristic structural features of this region comprising an extended conformation and a curve in the RNA backbone with a looped out U56 (Figure 2). The divalent metal ions present in the internal bulge of domain IIa interact with different bases, contributing to the stability of the helical bend.19,40,41 The sequence and structural integrity of this asymmetrical loop are important for IRES function, and the deletion of the loop has deleterious effects on translation efficiency.27 The regions adjacent to the helical bend forming the base and upper stem of domain IIa are also susceptible to substitutions that display lower translation activity.27,39,42 Surprisingly, some mutants with multiple mutations in these regions do not display appreciably low translation activity despite the lower translation response observed upon the individual occurrence of these mutations. For example, the double mutant A54G, U63G shows a translation response closer to that of the wild type.27 Likewise, the deletion of nucleotide C55, with three additional mutations at locations U63, U64, and A93 in domain II, results in an activity of 80% relative to the wild type43 (Table1, color: purple, blue). These phenotypes might be results of the long-range functional interaction of nucleotides that even if localized spatially in different regions have a structural impact upon specific mutations in a way that restores the translational activity (Figure 2).
Table 1.
Examples of the Hepatitis C Virus Internal Ribosome Entry Site (HCV IRES) Compensatory Mutations Indicating Probable Long-Range Inter- and Intra-Domain Interactions Described in the Literature (colored: blue, purple, green)
 |
A subdomain IIa helical bend has also been identified as a vital target site for a new class of benzimidazole molecule inhibitors that exhibit binding affinity with this region.46 Binding of the inhibitory compound changes the structural conformation by widening the interhelical angle in the domain IIa bend in HCV-infected cells. The interference in the structure of the bend helps undock domain IIb from the 40S subunit, eventually restricting the formation of the 80S pre-initiation translational complex to inhibit protein synthesis.47 NMR was employed to study the conformational changes of domain IIa induced by the ligand inhibitor molecule that straightens the helical bend, consequently disturbing 80S assembly.48 Furthermore, X-ray crystallography data give much deeper insight into the binding interactions of domain IIa in complex with benzimidazole translation initiation inhibitors.49
Variations in Domain II Impact the Assembly of Translation Pre-Initiation Complexes
Subdomain IIb contains an apical loop with a looped-out uracil (U86) at its 3′ side and a loop E motif constituting an S-turn. Both the apical loop and the loop E motif are highly conserved among HCV isolates.19 These structural features are somewhat similar to those of domain IIId. However, the location of the looped-out uracil (U269) in the IIId apical loop and the S-turn from the loop E motif in the same domain are both on the 3′ side in contrast to domain IIb, where they reside on opposite sides. These features constitute binding surfaces that are characteristic of both domains IIb and IIId for the interaction with 40S21 (Figure 2). Conservation of the domain II loop E motif and the apical loop has been shown to be of some significance, with deletions and substitutions abrogating HCV IRES function in translation initiation.27,28 The substitutions of nucleotides G82 and U86 at the apical loop somehow do not affect the IRES activity, contrary to substitutions of the other nucleotides in this region. However, the deletion of nucleotides in the apical loop generates low translation feedback, suggesting the importance of structural conservation.28 Cross-linking data have shown that the domain II apical loop interacts with ribosomal proteins S14 and S16 of the 40S subunit, while G87 in the IIb stem was not cross-linked to any of the ribosomal proteins or rRNA.50,51 The bend conformation allows domain II to reach the E site upon binding to the 40S subunit. The interaction of domain II with 40S also occurs through ribosomal protein S5 in the head region, which allows a conformational change and movement of the head relative to the 40S body.26,52 The apical loop of domain II lies in close proximity to the loop regions of helices 23 and 24 of the 40S subunit.53
Through chemical probing, it was shown that mutations in domain IIb affect the domain IV structure in the HCV IRES-40S complex, disrupting its arrangement at the AUG start codon and leading to a decreased translational response. It has been proposed that a long-range structural interaction between two domains that lie in close proximity within the IRES-40S complex can have important implications.54 Mutations in the apical loop of domain IIb result in decreased HCV IRES functionality, but translation is not aborted, and the formation of all translation complexes occurs with equal efficiency to that with the wild type IRES. However, the interaction of domain IIb apical loop mutants with the β-hairpin of S5 in the E site is altered. The disruptive contact with the β-hairpin of protein S5 caused by local structural perturbation of the mutants is suggested to induce a global change in the IRES-40S conformation that may stall the first event of ribosomal translocation, as observed through toe-printing, after 80S assembly.55 While the HCV IRES is still able to bind the 40S ribosomal subunit and recruit eIF2-TC and eIF3 to form a 48S pre-initiation complex when domain II is deleted entirely, it is inefficient at 60S subunit joining and thus 80S ribosome assembly.13,56 This inefficient 80S formation is mainly due to the fact that the processes facilitated by domain II that are downstream of 48S assembly are also affected upon domain II deletion. The conformational change induced by domain II in the head region may assist eIF5, a GTPase-activating protein, and eIF2-TC interaction on the 40S subunit. This contact would initiate GTP hydrolysis and subsequent release/dissociation of eIF2-GDP, after the establishment of AUG codon recognition by initiating Met-tRNAi. A reduction in 80S assembly compared to the wild type was also encountered when conserved structural motifs of domain II were deleted individually.14 The release of inorganic phosphate (Pi) in cap-dependent translation is dependent on AUG codon recognition in eIF5-promoted eIF2-GTP hydrolysis, with eIF1 being a negative regulator of this Pi release.57 Once eIF2-GDP is released from the complex, the catalysis of second GTP hydrolysis for the displacement of eIF3 is driven by eIF5B, thereby promoting the formation of the 80S pre-initiation complex by allowing subunit joining58 (Figure 1).
IRES DOMAIN III CONSERVED REGIONS ARE IMPORTANT FOR 40S AND eIF3 BINDING
The Domain III Apical Region Is Crucial for Recruitment of eIF3 and for Efficient Translation
Domain III is a principal domain of the HCV IRES and is further sub-divided into domains IIIa–IIIf. The structural elements of domain III are involved in making most of the direct contacts with the host translational machinery, 40S and eIF3, as observed through a variety of experimental approaches, including biochemical, mutational and structural analyses.36,21,59
The apical half of domain III comprises domains IIIa, IIIb, IIIc, and a four-way junction (jIIIabc). Mutations in domain IIIa were found to affect the stability of the binding affinity with eIF3. Substituting the IIIa loop causes a >6-fold reduction in the eIF3 binding affinity, with loss of IRES function.13,36 It was demonstrated through toe-printing, mutagenesis, and chemical and enzymatic footprinting that for eIF3-HCV IRES binding, domain IIIb and junction IIIabc (jIIIabc) are vital determinants of this interaction (Figure 1).12,30,36,60 Domain IIIb comprises an apical loop and an internal loop variable among HCV isolates that adopts a conserved three-dimensional secondary structure, observed through NMR. The interhelical region, consisting of a C186•C211 mismatch and adjacent base pairs G184-C213 and A185-U212, is conserved (Figure 2) and mutations in this region lead to low levels of translation. Variations at nucleotides 182 and 217 result in a range of activities, from as low as 33% to activity equivalent to the wild type, which also suggests that sequence preservation in this region might be important for eIF3 recognition. Among the domain IIIb most common variants that were structurally analyzed, C183•A214 and A183-U214 adopt a similar structural conformation with a signature S-turn and display an efficient IRES response, contrary to mutations at A183-G214 pair that alter the geometry of the structure within the loop. This finding may suggest that the interaction of eIF3 with this region is more dependent on the geometry and characteristic of its backbone rather than the primary sequence.22
The primary sequence of the domain IIIb apical loop may not be vital for IRES function as revealed by mutagenesis.61 Deletion of domain IIIb causes a reduced affinity for eIF3, which has been shown to affect the stable association of eIF2 with the 48S complex and the deposition of Met-tRNAi to the AUG binding site. Domain IIIb deletion mutants may assemble an IRES-80S complex but do not do so as efficiently as the wild type.13,56 Similarly, a low translation response upon deletion of the domain IIIb apical loop has also been reported.62 Any nucleotide insertion between nucleotides 206 and 207, adjacent to the 3′ end of the IIIb apical loop has been shown to increase IRES activity in both monocistronic and bicistronic luciferase expression systems compared to the wild type. The inserted nucleotide at 207 is speculated to increase the eIF3 binding affinity for the IRES, which might be responsible for the increase in cap-independent translation. The stimulatory effect of nucleotide insertion in position 207 remains preserved even with combination of other mutations. Point substitution A119C, which individually retains the HCV IRES activity to a near wild type, appeared to had no negative effect on translation with simultaneous insertion at the 207 position.63,44 Likewise, with an additional substitution of G82A in one sample (G82A + A insertion 207, 108%) and U248G in another (A insertion 207 + U248G, 109%), insertions at 207 displayed an increased IRES response compared to the wild type.44
The helical junctions in HCV IRES interact with 40S and eIF3 by providing specific recognition sites, as observed using various biochemical assays. Junction IIIabc (jIIIabc) plays an important role in the interaction with eIF3 and 40S.36 Structural analysis using X-ray crystallography determined the arrangement of junction nucleotides and the adjacent helices that provide the specific binding site for eIF3. JIIIabc adopts a parallel orientation in which helix IIIc is stacked on the rest of stem III in an almost perfectly coaxial manner. Helixes IIIa and IIIb have disrupted stacking due to the insertion of junction residues (A154, A155, and U228) into the minor groove (Figure 2). These features of the junction suggest that the distortion of the helical structure is important for providing a recognition site for eIF3.20 The introduction of mutations at the junction has been shown to be devastating for IRES efficiency, as it apparently destroys the fine intricacy of the hydrogen bonding between the junction fold and the adjacent nucleotides. The mutant U228C achieves assembly of the 48S complex with a >15-fold reduced eIF3 binding affinity and a significant reduction in 80S formation, leading to a decrease in IRES activity.13,25,20 JIIIabc interacts with the 40S subunit body near expansion segment 6 in IRES-40S and IRES-80S complexes. A shift from a parallel to antiparallel orientation of coaxially stacked helices between domain III and IIIb in jIIIabc was suggested for HCV IRES-80S.53 The presence of two different parallel and antiparallel conformations of jIIIabc has also been shown to exist in solution using time-resolved fluorescence measurements.64
Domain IIIc is a conserved region with a stem-loop structure composed of 10 nucleotides. The stem constitutes three G-C base pairs, along with a tetraloop (CGUG). The interaction of domain IIIc with eIF3 was established through chemical and enzymatic footprinting. The deletion of stem-loop IIIc affects the eIF3 binding stability to this region.30 Using NMR, the first and fourth nucleotides of the tetraloop, C232 and G235, were revealed to be a Watson-Crick base pair. The center nucleotides of the loop, G233 and U234, are the only residues in the structure that adopt a C2′-endo conformation. The substitutions of G233 and U234 in the apical loop are well-tolerated. An examination of the mutations at the base pair C232-G235 showed that conservation of the structure through usual Watson-Crick base-pairing maintains the translation response similar to that of the wild type, while other mutations have deleterious effects.23 However, Tang et al.42 showed that the compensatory base pair mutations C232G/G235C and C232U/G235A restore only 25 and 40% of the IRES activity, respectively, which suggests that the primary sequence is equally important to the maintenance of the secondary structure. The stacking of the bases and hydrogen bonding at C232-G235 provide stability for this domain.42,23
Conservation of Both Structure and Sequence Is Required for Multiple Interactions of the Domain III Basal Region with the 40S Subunit
Data obtained by chemical probing of domains IIId and IIIe demonstrated the importance of the stem-loop residues involved in the 40S subunit interaction. Structural analysis using NMR further revealed the configurations of domains IIId and IIIe. A helical stem, an internal loop and an apical hexanucleotide loop constitute domain IIId. The internal loop is conserved among HCV isolates, and it folds into an E loop motif. The inversion of backbone direction at A257 and G258 leads to an S-turn formation, characteristic of an E loop motif, that further allows the parallel hydrogen bonding of A257•A275.21 Transition and transversion at A276 causes a reduction in the IRES response and substituting CUC for AAG at nucleotides 275–277 generates only 25% activity relative to the wild type. Moreover, substituting A257G and A275G of the loop E motif in combination distorts translation almost completely, with no 48S and 80S formation.56
The terminal loop of domain IIId (5′-UUGGGU-3′) is one of the most important conserved regions of the HCV IRES. The GGG (266–268) is strongly conserved and is essential for HCV IRES translation in all HCV genotypes and HCV-like IRESs. The nucleotide U269 is looped out into the solution, favoring the positioning of G268 toward the major groove and an inversion in the backbone, which causes the formation of another S-turn between G267 and C270 (Figure 2). The mutation of GGG (266–268) to AAA to preserve the structure still results in a 50% decrease in translation. Any disruption of the IIId terminal loop causes a low translation response.21,45 The functional capacity of HCV IRES translation has been shown to be seriously challenged upon mutations in this domain.13,25,59 A molecular dynamics simulation followed by circular dichroism spectroscopy on a G266A/G268U mutant showed a similar structural conformation in domain IIId as that of the wild type and at different magnesium concentrations. The activity of this mutant in translation assays, however, was abrogated, which highlighted the importance of conservation of the primary sequence of the domain IIId hexaloop.44 With an exception of a double mutant (Table1; green), which, having one of the mutation in domain IIId (U262C) and the other apparently stabilizing the basal stem of domain IIa, has exhibited only a mild decrease in IRES response.44 This can probably be associated with a long-range inter-domain functional interaction for compensation of HCV IRES translation since the region of occurrence of the additional mutation at C43 has also shown a reduced level of activity upon mutations.39,18
The HCV IRES contacts the 40S ribosomal subunit at multiple interaction sites that are specific and important. The basal portion of domain III, particularly domain IIId, and domain IV have been observed to be the interaction sites for the 40S subunit. The majority of nucleotides in domain IIId have been shown to be protected from RNase V1, RNase T1, and iodine cleavage upon 40S binding.29,36 Domain IIId is proposed to contact the 40S subunit near helix 26, which is extended to expansion segment 7. The cross-linking data also suggest that there are contact points at ribosomal proteins S14 and S16 (cross-linked to A275 or G263). This interaction of IIId-40S is the most extensive and provides stability to the structure.50,53 The G triplet (266–268) in domain IIId has been shown to contact 18S rRNA through a CCC sequence in the apical loop of expansion segment 7 with complementary base pairing, as analyzed through DMS modification.65
Domain IIIe consists of a tetraloop (5′-GA[U/C]A-3′) that is conserved among HCV isolates and HCV-like IRES RNAs. The NMR and crystal structures of this domain display base pairing of the bottom nucleotides, G295 and A298, of the tetraloop. The conformation of the IIIe hairpin alone observed in NMR is distinct from the X-ray crystallographic IIIe tetraloop structure, shown with a pseudoknot and stem III junction, most likely due to its tertiary interaction (Figure 2). The overall configuration of this tetraloop is different (in the positioning and stacking of the bases) from the standard GNRA tetraloop.21,24 The central nucleotide, U297, flips out of the tetraloop, and it base pairs with A288 from stem III, which is also bulged outward (Figure 2). This tertiary interaction constitutes a second pseudoknot in the HCV IRES.24 The mutation analysis demonstrates that the conservation of a canonical base pair at 288–297 is important for IRES function in translation initiation.66 However, it was also demonstrated that the maintenance of purine–pyrimidine—not necessarily the Watson-Crick base pairing—interaction between A288 and U297, generates IRES activity similar to that of wild type and that altering the orientation of this identity disrupts translation. In the context of an existing destabilized IIIf pseudoknot with 40% activity, the compensatory mutations restore the translation activity to 45% of that of the wild type.35,24 Another bulging nucleotide, A136, that is flipped out from stem III is stacked over A296 and A298 of the IIIe tetraloop. Substituting this nucleotide causes no hindrance to HCV IRES function.24 Interestingly, another study reported that in HCV genotype 1b, the presence of nucleotide substitution A136G along with another mutation, A140U, decreases the translation activity to merely 35% relative to that of the wild type.44
Deletions and substitutions in the IIIe tetraloop result in a severe deterioration of the translation response from 60 to 90% in all types of transitions and transversions, demonstrating the importance of the conservation of the sequence of each of the four nucleotides.67 Using a site-specific crosslinking method, hairpin IIIe has been identified in proximity to ribosomal proteins S3a, S5, and S16 on the solvent side of the 40S subunit.51
The domain III basal region is composed of another helical junction that comprises a pseudoknot structure. The pseudoknot in domain IIIf, located upstream of the AUG codon, is highly conserved, and maintenance of its secondary structure is necessary for HCV IRES function.61,68 The pseudoknot domain configuration was revealed using computational modeling and X-ray crystallography.24,69 The junction arranges itself into a unique, complex double-pseudoknot fold that further allows the formation of two helices, coaxially stacked and nonparallel with a tilt of approximately 40° between the helical stacks. The significance of the pseudoknot has been determined through mutational and functional analyses that revealed the pseudoknot's contribution to the translational activity and its importance in properly positioning the AUG codon at the 40S binding site.35,36,24,68 The pseudoknot contacts the helices 28, 37, and 40 at the back of the 40S subunit head region.53
The characterization of the HCV IRES structures was also performed with the development and combination of small-angle X-ray scattering (SAXS) data and molecular modeling that gave rise to the ensemble of conformers for the full-length HCV IRES RNA.70
CHANGES IN SEQUENCE COMPOSITION DOWNSTREAM OF AUG CAN ALTER THE HCV IRES ACTIVITY
The initiator AUG codon of the HCV IRES is localized in a stem-loop IV71,72; a conserved domain among the HCV genotypes and distant GB virus B. The chemical and enzymatic probing data have demonstrated the interaction of domain IV with the 40S ribosome.29,36 The nucleotide changes in domain IV that resulted in an increased stabilization of this domain exhibit translation with decreased efficiency.11 As it binds to the mRNA binding cleft of the ribosome, domain IV is unwound, due to its increased flexibility, to support the correct positioning of the initiation codon and the subsequent binding of eIF2-TC.54 Evidence for a possible long-range structural and functional interaction between domain IIb and domain IV has been discussed earlier in the text.
In our previous work, we studied the necessity of the AUG codon at position 342 in yeast and mammalian systems by introducing mutations. The substitution of UUG for the AUG codon resulted in a substantial inhibition of the IRES-mediated response in both mammals and yeast.73 The sequence conservation of nucleotides located downstream of the AUG codon has been proposed to be very crucial for the maintenance and modulation of HCV IRES activity.32,74 Our results suggested that a high AT content within the first 15 codons of the HCV polyprotein is important in maintaining the HCV IRES activity. The study reflects the evolutionary conservation of basic translation processes by displaying similarities of HCV IRES translation between two unrelated organisms, such as yeast and humans.73 However, no specific requirement has also been reported for either the nucleotides or amino acid sequences downstream of the IRES for efficient translation in HCV and other flaviviruses.75 The core gene stem loops SL47 and SL87 are highly structured elements, which have been concluded to contribute to HCV IRES translation in JFH-1 viral strain. The introduction of silent mutations in SL47 and SL87 separately and simultaneously led to a decreased translation of the viral RNA and pointed to the importance of these structural elements for robust viral production both in vitro and in vivo.76 The stability of the long-range RNA–RNA interaction between the IRES (nt. 24–38) and the core gene (nt. 428–442) was shown to be involved in modulating viral gene expression as well. Mutational analysis revealed that disruption in the stability of this interaction increases the IRES translation. In contrast, compensatory mutations that restore stability led to slightly reduced translation activity compared to the wild type, both in vivo and in vitro. This reduced translation may be important for viral persistence during chronic infections.77,78 A region between domain I and domain II (nt. 22–28) was also shown to be the site of attachment for the liver miRNA-122, which may cause interference with the long-range interaction between the HCV IRES and the core gene, and stimulate the HCV translation, although under different conditions, in vitro and in vivo.79,80 Annealing of miRNA-122 induces a switch from close to an open conformation of HCV RNA, as observed in vitro.81
STABILIZATION OF THE TRANSLATION COMPLEX WITH CHARACTERISTIC BINDING OF eIF3 SUBUNITS TO THE IRES
Function of the HCV IRES has been repeatedly reported to depend on the specific interaction with the eukaryotic translation initiation factor 3 (eIF3). Mammalian eIF3 is a 13-subunit complex (subunits eIF3a-m) of approximately 800 kDa.82 Cryo-electron microscopy (EM) revealed the structure of eIF3 and its arrangement on the IRES-40S subunit. The interaction occurs through the front face of eIF3, a 5-lobed structure consisting of a head, arms, and legs. The left arm is composed of an extended portion located near the HCV IRES domain II toward the E site, while the left leg covers S15/rpS13 below the 40S platform blocking its contact with helix 34 of the 60S in order to prevent premature ribosomal subunit association. The domains IIIdef and IIIabc are located near the center and right leg of eIF3.83
A number of eIF3 subunits interact with the HCV IRES, as observed using limited proteolysis and mass spectrometry (MS). The subunits eIF3a, eIF3c, eIF3e, eIF3f, eIF3h, and eIF3l reside mostly in the right and left legs of eIF3 and are responsible for high-affinity binding with the IIIabc domain of the HCV IRES.84 However, other studies suggest that the eIF3b and eIF3g subunits are also involved in a direct interaction, whereas the eIF3i and eIF3l:k dimer is proposed to have no immediate contact with the HCV IRES.85,86
The association of eIF3 with the HCV IRES places the C-terminus of eIF3j, a subunit of eIF3, in the 40S mRNA entry channel. The dissociation of eIF3j is necessary for the proper placement of HCV mRNA in the decoding groove. Directed hydroxyl radical probing and toe-printing showed that the dissociation of eIf3j is promoted by a conformational change induced by domain II in the head region of the 40S subunit. Moreover, the recruitment of eIF2-TC is needed to displace the eIF3j, providing more stability to the HCV mRNA in the 40S mRNA binding channel.87 Using cryo-EM, it has recently been proposed that in the HCV-like CSFV IRES (classical swine fever virus IRES), the eIF3 is displaced from the 40S subunit observed in the 43S pre-initiation complex. Instead, eIF3 interacts with the apical region of domain III, presenting its ribosome-binding surface, eIF3a and eIF3c, thus forming a 40S-IRES-eIF3 complex.88 Another study using low-resolution EM along with biochemical analysis revealed that conserved regions in eIF3a and eIF3c directly bind with domain IIIabc of the HCV IRES.89
eIF2-INDEPENDENT TRANSLATION MECHANISM (STRESS-INDUCED HCV PROTEIN SYNTHESIS)
The protein kinase R (PKR) undergoes activation during HCV infection which leads to eIF2 inhibition and a consequential decrease in protein synthesis, including that of the antiviral interferon-stimulated genes (ISG), in the host cells. While suppressing host protein synthesis by activating PKR to its own advantage, the HCV IRES persists in translation90 through an eIF2α-independent mechanism. The HCV IRES domains III–IV have been found to be critical for PKR activation.91 Domain II has also been reported to act as a potent PKR activator.92 The ability of the HCV IRES to undergo an alternate bacteria-like mode of translation upon eIF2 inactivation only requires two initiation factors, eIF3 and eIF5B-GTP, which are necessary for 80S pre-initiation assembly. The initiation factor eIF5B was shown to be required to direct Met-tRNAi to the IRES-40S complex before 60S subunit attachment and provides stability without any direct binding (Figure 1).37 The role of eIF5B, in addition to the displacement of eIF3 and other initiation factors in cap-dependent translation, has also been implied in assisting the subunit joining of 60S to 40S, forming an 80S complex.93,94 GTP is required because it provides an active conformation for eIF5B.95 Another factor, eIF2A, is also proposed to help with the recruitment of Met-tRNAi to the P-site by binding to domain IIId. By using a filter-binding assay, eIF2A was shown to have a binding affinity for Met-tRNAi. Moreover, the deletion of domain IIId or mutations in the IIId hexaloop causes a low binding capability with eIF2A, thus affecting translation activity under stress conditions. eIF2A knockdown using siRNA has an inhibitory effect on HCV IRES translation under conditions where the eIF2α subunit is phosphorylated.96 eIF2D is also one of the factors that have been reported to facilitate Met-tRNAi assembly with the 40S under stress conditions (Figure 1).97 The alternate mode of translation occurs only when the eIF2α subunit is phosphorylated under stress conditions to avoid the severe inhibition of viral protein synthesis. Both the HCV and related CSFV IRES-mediated translation were shown to be resistant to the reduced levels of eIF2-TC.37,38,96,98
The HCV IRES can also constitute a pre-initiation 80S translation complex without the requirement of any of the initiation factors at a higher Mg+2 concentration of 5 mM and continue protein synthesis with the aid of elongation factors and tRNAs.99
CONCLUSION
There has been considerable advancement into the insights of HCV IRES RNA structure and function over the past few years. The atomic resolution structures and cryo-EM reconstructions have allowed to visualize and to study the characteristic structural conformation of the IRES domains and their interactions with the ribosome and the eIFs in a specific manner. In this review, we aimed to show how sequence and structural features of the HCV IRES are vitally interlinked with its function in the initiation of the viral protein synthesis. Structural and/or sequence changes in the conserved domains can prove to be devastating in the context of IRES function. Interestingly, some HCV IRES mutants containing multiple mutations restored the IRES activity to levels near that of the wild type, although the occurrence of these mutations individually significantly reduced the translational activity. A careful analysis of published mutational data also revealed an increase in IRES activity upon the insertion of nucleotides at a specific location. Further study is required to understand the structural adaptability of the HCV IRES with regard to the probable long-range inter- and intra-domain interactions that restore a functional response in translation initiation. Progress in better understanding of the possible protein–protein and protein–RNA interactions that may play a role in stabilizing functional complexes in a course of translation initiation in the context of long-range interaction of and/or within the IRES domains is also required. Successful development of benzimidazole inhibitors suggested that an extensive analysis of HCV IRES structure and structure variations including their impact on translation mediation can further be utilized in the development of more effective HCV IRES-targeted antivirals.
Acknowledgments
This work was supported by projects GCP305/10/J026 and GBP305/12/G034 awarded to MP by the Czech Science Foundation and project No. 68609 awarded to AK by the Charles University Grant Agency. The work was also supported by Charles University institutional projects UNCE 204013 and SVV-2014-260081.
Conflict of interest:
The authors have declared no conflicts of interest for this article.
Further Reading/Resources
http://HCVIVdb.org http://www.hcvivdb.org/ ), a database of mutations within the IRES of the hepatitis C virus
REFERENCES
- 1.Gravitz L. A smouldering public-health crisis. Nature. 2011;474:S2–S4. doi: 10.1038/474S2a. [DOI] [PubMed] [Google Scholar]
- 2.Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967–972. doi: 10.1038/nature04082. [DOI] [PubMed] [Google Scholar]
- 3.Dienstag JL, McHutchison JG. American Gastroenterological Association technical review on the management of hepatitis C. Gastroenterology. 2006;130:231–264. doi: 10.1053/j.gastro.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Sofia MJ, Bao D, Chang W, Du JF, Nagarathnam D, Rachakonda S, Reddy PG, Ross BS, Wang PY, Zhang HR. Discovery of a β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J Med Chem. 2010;53:7202–7218. doi: 10.1021/jm100863x. et al. [DOI] [PubMed] [Google Scholar]
- 5.Lawitz E, Mangia A, Wyles D, Rodriguez-Torres M, Hassanein T, Gordon SC, Schultz M, Davis MN, Kayali Z, Reddy KR. Sofosbuvir for previously untreated chronic hepatitis C infection. New Engl J Med. 2013;368:1878–1887. doi: 10.1056/NEJMoa1214853. et al. [DOI] [PubMed] [Google Scholar]
- 6.Fried MW, Buti M, Dore GJ, Flisiak R, Ferenci P, Jacobson I, Marcellin P, Manns M, Nikitin I, Poordad F. Once-daily simeprevir (TMC435) with pegylated interferon and ribavirin in treatment-naive genotype 1 hepatitis C: the randomized PILLAR study. Hepatology. 2013;58:1918–1929. doi: 10.1002/hep.26641. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malcolm BA, Arassappan A, Bennett F, Bogen S, Chase R, Chen K, Chen T, Ingravallo P, Jao E, Kong S. SCH 503034, a mechanism-based inhibitor of hepatitis C virus (HCV) NS3 protease suppresses polyprotein maturation and enhances the antiviral activity of interferona-211 (INF) Hepatology. 2005;42:535a–536a. doi: 10.1128/AAC.50.3.1013-1020.2006. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perni RB, Almquist SJ, Byrn RA, Chandorkar G, Chaturvedi PR, Courtney LF, Decker CJ, Dinehart K, Gates CA, Harbeson SL. Preclinical profile of VX-950, a potent, selective, and orally bioavailable inhibitor of hepatitis C virus NS3-4A serine protease. Antimicrob Agents Chemother. 2006;50:899–909. doi: 10.1128/AAC.50.3.899-909.2006. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choo QL, Richman KH, Han JH, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby R, Barr PJ. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honda M, Brown EA, Lemon SM. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 12.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji H, Fraser CS, Yu YH, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci USA. 2004;101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Locker N, Easton LE, Lukavsky PJ. HCV and CSFVIRES domain II mediate eIF2 release during 80S ribosome assembly. Embo J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsukiyamakohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis-C virus-Rna. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown EA, Zhang HC, Ping LH, Lemon SM. Secondary structure of the 5′ nontranslated regions of hepatitis-C virus and pestivirus genomic Rnas. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao WD, Wimmer E. Genetic analysis of a poliovirus/hepatitis C virus chimera: new structure for domain II of the internal ribosomal entry site of hepatitis C virus. J Virol. 2001;75:3719–3730. doi: 10.1128/JVI.75.8.3719-3730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCVIRES domain II determined by NMR. Nat Struct Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 20.Kieft JS, Zhou KH, Grech A, Jubin R, Doudna JA. Crystal structure of an RNA tertiary domain essential to HCVIRES-mediated translation initiation. Nat Struct Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- 21.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat Struct Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 22.Collier AJ, Gallego J, Klinck R, Cole PT, Harris SJ, Harrison GP, Aboul-ela F, Varani G, Walker S. A conserved RNA structure within the HCVIRES eIF3-binding site. Nat Struct Biol. 2002;9:375–380. doi: 10.1038/nsb785. [DOI] [PubMed] [Google Scholar]
- 23.Rijnbrand R, Thiviyanathan V, Kaluarachchi K, Lemon SM, Gorenstein DG. Mutational and structural analysis of stem-loop IIIc of the hepatitis C virus and GB virus B internal ribosome entry sites. J Mol Biol. 2004;343:805–817. doi: 10.1016/j.jmb.2004.08.095. [DOI] [PubMed] [Google Scholar]
- 24.Berry KE, Waghray S, Mortimer SA, Bai Y, Doudna JA. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure. 2011;19:1456–1466. doi: 10.1016/j.str.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kieft JS, Zhou KH, Jubin R, Murray MG, Lau JYN, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 26.Spahn CMT, Kieft JS, Grassucci RA, Penczek PA, Zhou KH, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 27.Odreman-Macchioli F, Baralle FE, Buratti E. Mutational analysis of the different bulge regions of hepatitis C virus domain II and their influence on internal ribosome entry site translational ability. J Biol Chem. 2001;276:41648–41655. doi: 10.1074/jbc.M104128200. [DOI] [PubMed] [Google Scholar]
- 28.Kalliampakou KI, Psaridi-Linardaki L, Mavromara P. Mutational analysis of the apical region of domain II of the HCVIRES. Febs Lett. 2002;511:79–84. doi: 10.1016/s0014-5793(01)03300-2. [DOI] [PubMed] [Google Scholar]
- 29.Kolupaeva VG, Pestova TV, Hellen CUT. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J Virol. 2000;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rijnbrand R, Bredenbeek P, Vanderstraaten T, Whetter L, Inchauspe G, Lemon S, Spaan W. Almost the Entire 5′ Non-Translated Region of Hepatitis-C Virus Is Required for Cap-Independent Translation. Febs Lett. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- 32.Honda M, Ping LH, Rijnbrand RCA, Amphlett E, Clarke B, Rowlands D, Lemon SM. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 33.Lytle JR, Wu L, Robertson HD. The ribosome binding site of hepatitis C virus mRNA. J Virol. 2001;75:7629–7636. doi: 10.1128/JVI.75.16.7629-7636.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lytle JR, Wu L, Robertson HD. Domains on the hepatitis C virus internal ribosome entry site for 40s subunit binding. RNA. 2002;8:1045–1055. doi: 10.1017/s1355838202029965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berry KE, Waghray S, Doudna JA. The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA. 2010;16:1559–1569. doi: 10.1261/rna.2197210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kieft JS, Zhou KH, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis CIRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol. 2008;15:836–841. doi: 10.1038/nsmb.1445. [DOI] [PubMed] [Google Scholar]
- 38.Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU. eIF2-dependent and eIF2-independent modes of initiation on the CSFVIRES: a common role of domain II. Embo J. 2008;27:1060–1072. doi: 10.1038/emboj.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Honda M, Beard MR, Ping LH, Lemon SM. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dibrov SM, Johnston-Cox H, Weng YH, Hermann T. Functional architecture of HCVIRES domain II stabilized by divalent metal ions in the crystal and in solution. Angew Chem Int Ed. 2007;46:226–229. doi: 10.1002/anie.200603807. [DOI] [PubMed] [Google Scholar]
- 41.Zhao Q, Han Q, Kissinger CR, Hermann T, Thompson PA. Structure of hepatitis C virus IRES subdomain IIa. Acta Crystallogr Sect D Biol Crystallogr. 2008;64:436–443. doi: 10.1107/S0907444908002011. [DOI] [PubMed] [Google Scholar]
- 42.Tang SX, Collier AJ, Elliott RM. Alterations to both the primary and predicted secondary structure of stem-loop IIIc of the hepatitis C virus ib 5′ untranslated region (5′ UTR) lead to mutants severely defective in translation which cannot be complemented in trans by the wild-type 5′ UTR sequence. J Virol. 1999;73:2359–2364. doi: 10.1128/jvi.73.3.2359-2364.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laporte J, Malet I, Andrieu T, Thibault V, Toulme JJ, Wychowski C, Pawlotsky JM, Huraux JM, Agut H, Cahour A. Comparative analysis of translation efficiencies of hepatitis C virus 5′ untranslated regions among intraindividual quasispecies present in chronic infection: Opposite behaviors depending on cell type. J Virol. 2000;74:10827–10833. doi: 10.1128/jvi.74.22.10827-10833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barria MI, Gonzalez A, Vera-Otarola J, Leon U, Vollrath V, Marsac D, Monasterio O, Perez-Acle T, Soza A, Lopez-Lastra M. Analysis of natural variants of the hepatitis C virus internal ribosome entry site reveals that primary sequence plays a key role in cap-independent translation. Nucleic Acids Res. 2009;37:957–971. doi: 10.1093/nar/gkn1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klinck R, Westhof E, Walker S, Afshar M, Collier A, Aboul-Ela F. A potential RNA drug target in the hepatitis C virus internal ribosomal entry site. RNA. 2000;6:1423–1431. doi: 10.1017/s1355838200000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seth PP, Miyaji A, Jefferson EA, Sannes-Lowery KA, Osgood SA, Propp SS, Ranken R, Massire C, Sampath R, Ecker DJ. SAR by MS: discovery of a new class of RNA-binding small molecules for the hepatitis C virus: internal ribosome entry site IIA subdomain. J Med Chem. 2005;48:7099–7102. doi: 10.1021/jm050815o. et al. [DOI] [PubMed] [Google Scholar]
- 47.Parsons J, Castaldi MP, Dutta S, Dibrov SM, Wyles DL, Hermann T. Conformational inhibition of the hepatitis C virus internal ribosome entry site RNA. Nat Chem Biol. 2009;5:823–825. doi: 10.1038/nchembio.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paulsen RB, Seth PP, Swayze EE, Griffey RH, Skalicky JJ, Cheatham TE, Davis DR. Inhibitor-induced structural change in the HCV IRES domain IIa RNA. Proc Natl Acad Sci USA. 2010;107:7263–7268. doi: 10.1073/pnas.0911896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dibrov SM, Ding K, Brunn ND, Parker MA, Bergdahl BM, Wyles DL, Hermann T. Structure of a hepatitis C virus RNA domain in complex with a translation inhibitor reveals a binding mode reminiscent of riboswitches. Proc Natl Acad Sci USA. 2012;109:5223–5228. doi: 10.1073/pnas.1118699109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Babaylova E, Graifer D, Malygin A, Stahl J, Shatsky I, Karpova G. Positioning of subdomain IIId and apical loop of domain II of the hepatitis C IRES on the human 40S ribosome. Nucleic Acids Res. 2009;37:1141–1151. doi: 10.1093/nar/gkn1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laletina E, Graifer D, Malygin A, Ivanov A, Shatsky I, Karpova G. Proteins surrounding hairpin IIIe of the hepatitis C virus internal ribosome entry site on the human 40S ribosomal subunit. Nucleic Acids Res. 2006;34:2027–2036. doi: 10.1093/nar/gkl155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fukushi S, Okada M, Stahl J, Kageyama T, Hoshino FB, Katayama K. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J Biol Chem. 2001;276:20824–20826. doi: 10.1074/jbc.C100206200. [DOI] [PubMed] [Google Scholar]
- 53.Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCVIRES. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 54.Filbin ME, Kieft JS. HCV IRES domain IIb affects the configuration of coding RNA in the 40S subunit′s decoding groove. RNA. 2011;17:1258–1273. doi: 10.1261/rna.2594011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Filbin ME, Vollmar BS, Shi D, Gonen T, Kieft JS. HCV IRES manipulates the ribosome to promote the switch from translation initiation to elongation. Nat Struct Mol Biol. 2013;20:150–158. doi: 10.1038/nsmb.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Otto GA, Puglisi JD. The pathway of HCVIRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 57.Algire MA, Maag D, Lorsch JR. P-i release from elF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 58.Lee JH, Pestova TV, Shin BS, Cao C, Choi SK, Dever TE. Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation. Proc Natl Acad Sci USA. 2002;99:16689–16694. doi: 10.1073/pnas.262569399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jubin R, Vantuno NE, Kieft JS, Murray MG, Doudna JA, Lau JYN, Baroudy BM. Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding. J Virol. 2000;74:10430–10437. doi: 10.1128/jvi.74.22.10430-10437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buratti E, Tisminetzky S, Zotti M, Baralle FE. Functional analysis of the interaction between HCV 5′ UTR and putative subunits of eukaryotic translation initiation factor elF3. Nucleic Acids Res. 1998;26:3179–3187. doi: 10.1093/nar/26.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang CY, Sarnow P, Siddiqui A. A conserved helical element is essential for internal initiation of translation of hepatitis-C virus-Rna. J Virol. 1994;68:7301–7307. doi: 10.1128/jvi.68.11.7301-7307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buratti E, Gerotto M, Pontisso P, Alberti A, Tisminetzky SG, Baralle FE. In vivo translational efficiency of different hepatitis C virus 5′-UTRs. Febs Lett. 1997;411:275–280. doi: 10.1016/s0014-5793(97)00715-1. [DOI] [PubMed] [Google Scholar]
- 63.Zhang J, Yamada O, Ito T, Akiyama M, Hashimoto Y, Yoshida H, Makino R, Masago A, Uemura H, Araki H. A single nucleotide insertion in the 5′-untranslated region of hepatitis C virus leads to enhanced cap-independent translation. Virology. 1999;261:263–270. doi: 10.1006/viro.1999.9879. [DOI] [PubMed] [Google Scholar]
- 64.Melcher SE, Wilson TJ, Lilley DMJ. The dynamic nature of the four-way junction of the hepatitis C virus IRES. RNA. 2003;9:809–820. doi: 10.1261/rna.5130703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malygin AA, Kossinova OA, Shatsky IN, Karpova GG. HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res. 2013;41:8706–8714. doi: 10.1093/nar/gkt632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Easton LE, Locker N, Lukavsky PJ. Conserved functional domains and a novel tertiary interaction near the pseudoknot drive translational activity of hepatitis C virus and hepatitis C virus-like internal ribosome entry sites. Nucleic Acids Res. 2009;37:5537–5549. doi: 10.1093/nar/gkp588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Psaridi L, Georgopoulou U, Varaklioti A, Mavromara P. Mutational analysis of a conserved tetraloop in the 5′ untranslated region of hepatitis C virus identifies a novel RNA element essential for the internal ribosome entry site function. Febs Lett. 1999;453:49–53. doi: 10.1016/s0014-5793(99)00662-6. [DOI] [PubMed] [Google Scholar]
- 68.Wang CY, Le SY, Ali N, Siddiqui A. An Rna pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis-C virus 5′-noncoding region. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]
- 69.Lavender CA, Ding F, Dokholyan NV, Weeks KM. Robust and generic RNA modeling using inferred constraints: a structure for the hepatitis C virus IRES pseudoknot domain. Biochemistry. 2010;49:4931–4933. doi: 10.1021/bi100142y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perard J, Leyrat C, Baudin F, Drouet E, Jamin M. Structure of the full-length HCV IRES in solution. Nat Commun. 2013;4:1612. doi: 10.1038/ncomms2611. [DOI] [PubMed] [Google Scholar]
- 71.Rijnbrand RCA, Abbink TEM, Haasnoot PCJ, Spaan WJM, Bredenbeek PJ. The influence of AUG codons in the hepatitis C virus 5′ nontranslated region on translation and mapping of the translation initiation window. Virology. 1996;226:47–56. doi: 10.1006/viro.1996.0626. [DOI] [PubMed] [Google Scholar]
- 72.Reynolds JE, Kaminski A, Carroll AR, Clarke BE, Rowlands DJ, Jackson RJ. Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA. 1996;2:867–878. [PMC free article] [PubMed] [Google Scholar]
- 73.Masek T, Vopalensky V, Horvath O, Vortelova L, Feketova Z, Pospisek M. Hepatitis C virus internal ribosome entry site initiates protein synthesis at the authentic initiation codon in yeast. J Gen Virol. 2007;88:1992–2002. doi: 10.1099/vir.0.82782-0. [DOI] [PubMed] [Google Scholar]
- 74.Reynolds JE, Kaminski A, Kettinen HJ, Grace K, Clarke BE, Carroll AR, Rowlands DJ, Jackson RJ. Unique features of internal initiation of hepatitis-C virus-Rna translation. Embo J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rijnbrand R, Bredenbeek PJ, Haasnoot PC, Keift JS, Spaan WJM, Lemon SM. The influence of downstream protein-coding sequence on internal ribosome entry on hepatitis C virus and other flavivirus RNAs. RNA. 2001;7:585–597. doi: 10.1017/s1355838201000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vassilaki N, Friebe P, Meuleman P, Kallis S, Kaul A, Paranhos-Baccala G, Leroux-Roels G, Mavromara P, Bartenschlager R. Role of the hepatitis C virus core + 1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J Virol. 2008;82:11503–11515. doi: 10.1128/JVI.01640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim YK, Lee SH, Kim CS, Seol SK, Jang SK. Long-range RNA-RNA interaction between the 5′ nontranslated region and the core-coding sequences of hepatitis C virus modulates the IRES-dependent translation. RNA. 2003;9:599–606. doi: 10.1261/rna.2185603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang TH, Rijnbrand RCA, Lemon SM. Core protein-coding sequence, but not core protein, modulates the efficiency of cap-independent translation directed by the internal ribosome entry site of hepatitis c virus. J Virol. 2000;74:11347–11358. doi: 10.1128/jvi.74.23.11347-11358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Henke JI, Goergen D, Zheng JF, Song YT, Schuttler CG, Fehr C, Junemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. Embo J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goergen D, Niepmann M. Stimulation of Hepatitis C Virus RNA translation by microRNA-122 occurs under different conditions in vivo and in vitro. Virus Res. 2012;167:343–352. doi: 10.1016/j.virusres.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 81.Diaz-Toledano R, Ariza-Mateos A, Birk A, Martinez-Garcia B, Gomez J. In vitro characterization of a miR-122-sensitive double-helical switch element in the 5′ region of hepatitis C virus RNA. Nucleic Acids Res. 2009;37:5498–5510. doi: 10.1093/nar/gkp553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Damoc E, Fraser CS, Zhou M, Videler H, Mayeur GL, Hershey JWB, Doudna JA, Robinson CV, Leary JA. Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Mol Cell Proteomics. 2007;6:1135–1146. doi: 10.1074/mcp.M600399-MCP200. [DOI] [PubMed] [Google Scholar]
- 83.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 84.Cai Q, Todorovic A, Andaya A, Gao JY, Leary JA, Cate JHD. Distinct regions of human eIF3 are sufficient for binding to the HCV IRES and the 40S ribosomal subunit. J Mol Biol. 2010;403:185–196. doi: 10.1016/j.jmb.2010.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou M, Sandercock AM, Fraser CS, Ridlova G, Stephens E, Schenauer MR, Yokoi-Fong T, Barsky D, Leary JA, Hershey JW. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc Natl Acad Sci USA. 2008;105:18139–18144. doi: 10.1073/pnas.0801313105. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Perard J, Rasia R, Medenbach J, Ayala I, Boisbouvier J, Drouet E, Baudin F. Human initiation factor eIF3 subunit b interacts with HCV IRES RNA through its N-terminal RNA recognition motif. Febs Lett. 2009;583:70–74. doi: 10.1016/j.febslet.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 87.Fraser CS, Hershey JWB, Doudna JA. The pathway of hepatitis C virus mRNA recruitment to the human ribosome. Nat Struct Mol Biol. 2009;16:397–404. doi: 10.1038/nsmb.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CUT, Frank J. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013;503:539–543. doi: 10.1038/nature12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sun C, Querol-Audi J, Mortimer SA, Arias-Palomo E, Doudna JA, Nogales E, Cate JH. Two RNA-binding motifs in eIF3 direct HCV IRES-dependent translation. Nucleic Acids Res. 41:7512–7521. doi: 10.1093/nar/gkt510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Garaigorta U, Chisari FV. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe. 2013;6:513–522. doi: 10.1016/j.chom.2009.11.004. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shimoike T, McKenna SA, Lindhout DA, Puglisi JD. Translational insensitivity to potent activation of PKR by HCV IRES RNA. Antiviral Res. 2009;83:228–237. doi: 10.1016/j.antiviral.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 92.Toroney R, Nallagatla SR, Boyer JA, Cameron CE, Bevilacqua PC. Regulation of PKR by HCV IRES RNA: importance of domain II and NS5A. J Mol Biol. 2010;400:393–412. doi: 10.1016/j.jmb.2010.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CUT. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- 94.Unbehaun A, Borukhov SI, Hellen CUT, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roll-Mecak A, Cao C, Dever TE, Burley SK. X-ray structures of the universal translation initiation factor IF2/elF5B: conformational changes on GDP and GTP binding. Cell. 2000;103:781–792. doi: 10.1016/s0092-8674(00)00181-1. [DOI] [PubMed] [Google Scholar]
- 96.Kim JH, Park SM, Park JH, Keum SJ, Jang SK. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. Embo J. 2011;30:2454–2464. doi: 10.1038/emboj.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem. 2010;285:26779–26787. doi: 10.1074/jbc.M110.119693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Robert F, Kapp LD, Khan SN, Acker MG, Kolitz S, Kazemi S, Kaufman RJ, Merrick WC, Koromilas AE, Lorsch JR. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2 center dot GTP center dot Met-tRNA(i)(Met) ternary complex availability. Mol Biol Cell. 2006;17:4632–4644. doi: 10.1091/mbc.E06-06-0478. , et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lancaster AM, Jan E, Sarnow P. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA. 2006;12:894–902. doi: 10.1261/rna.2342306. [DOI] [PMC free article] [PubMed] [Google Scholar]