Abstract
The composition of the gut microbiome represents a very important environmental factor that influences the development of type 1 diabetes (T1D). We have previously shown that MyD88-deficient non-obese diabetic (MyD88−/−NOD) mice, that were protected from T1D development, had a different composition of gut microbiota compared to wild type NOD mice. The aim of our study was to investigate whether this protection could be transferred. We demonstrate that transfer of gut microbiota from diabetes-protected MyD88-deficient NOD mice, reduced insulitis and significantly delayed the onset of diabetes. Gut bacteria from MyD88-deficient mice, administered over a 3-week period, starting at 4 weeks of age, stably altered the family composition of the gut microbiome, with principally Lachnospiraceae and Clostridiaceae increased and Lactobacillaceae decreased. The transferred mice had a higher concentration of IgA and TGFβ in the lumen that was accompanied by an increase in CD8+CD103+ and CD8αβ T cells in the lamina propria of the large intestine. These data indicate not only that gut bacterial composition can be altered after the neonatal/weaning period, but that the composition of the microbiome affects the mucosal immune system and can delay the development of autoimmune diabetes. This result has important implications for the development of probiotic treatment for T1D.
1. Introduction
Development of type 1 diabetes (T1D) requires a genetic predisposition that interacts with environmental factors [1]. The exact nature of these environmental factors has not been clearly understood, although infection has long been thought to play a role [2]. Recent evidence suggests that gut bacteria play a role in Non Obese Diabetic (NOD) mouse and the BioBreeding (BB) rat models of T1D and this role is also true for humans [3].
The incidence of T1D has increased over the last 40 years, in common with allergic diseases [4–6]. To account for these changes in incidence and prevalence, the “Hygiene hypothesis” or a refinement of this, the “Old Friends hypothesis” has been suggested [5, 7]. This postulates that a reduction in exposure to microorganisms in the environment can lead to a failure of immunoregulation [8–10]. These “Old Friends” could either be non-pathogenic organisms, as in saprophytic mycobacteria [11] or lactobacilli [12, 13], or parasitic infections, such as with helminths [14–16] that are more common in developing countries. The idea is that these organisms influence the maturation of dendritic cells, stimulating regulatory T cells and reducing pathogenic effector cells [10]. In addition to the possible effect of increasing tolerance and/or bystander suppression, there may also be other mechanisms of importance.
It is interesting that the BB rat, the main rat model of T1D, was originally derived in germ-free (GF) conditions [17]. It was later reported that the BB rat has an abnormal intestinal barrier, [18]. There are numerous studies, in both humans and animal models of human diseases, which strongly support the role of gut microbiota as an important factor in balancing health and disease. Development of inflammatory bowel disease (IBD) is influenced by gut microbiota as most, if not all, of the experimental IBD animal models are disease free if they are housed in GF conditions. There is an increasing public interest in probiotic compounds as an alternative medicine. Probiotics are cultures of beneficial bacteria from the healthy gut microbiota that improve the balance of the intestinal milieu by modifying the gut microbiota and suppressing inflammatory responses caused by the host immune cells in response to harmful microbes in the intestine. Recent studies have shown that oral probiotic administration prevents diabetes development in NOD mice [19]. This suggests that normal commensal microbes and their balance in the gut are extremely important for maintenance of health. In this study, we investigated the effect of gut microbiota transfer on diabetes development in NOD mice and our results suggested that transient gut microbiota transfer at a young age could have long- lasting effects on diabetes development in adulthood in the NOD mouse model of human T1D.
2. Materials and Methods
2.1. Mice
NOD/LtJ mice purchased from the Jackson Laboratory were used for studying diabetes development. NOD/Caj mice were originally obtained from the Jackson Laboratory (NOD/LtJ) and have been maintained at Yale University for over 25 years. MyD88−/−NOD mice were generated as described previously [20] and have been maintained at Yale University for over 7 years. MyD88−/−B6 mice were kindly provided by Dr. Akira [21] and have been maintained at Yale University for over 10 years. B6g7 breeders were kindly provided by Drs. Mathis and Benoist (Harvard University) and have been bred at Yale University for over 10 years. MyD88−/−B6g7 mice were generated by breeding B6g7 with MyD88−/−B6 mice. C57BL/6J (B6) mice were originally obtained form the Jackson Laboratory and have been maintained at Yale University for over 5 years. All mice used in this study were kept in the same room, in specific pathogen–free conditions, in a 12-hour dark/light cycle and housed in individually-ventilated filter cages with autoclaved food at the Yale University animal facility. The use of the animals in this study was approved by the Yale University Institutional Animal Care and Use Committee.
2.2. Antibodies and reagents
All fluorochrome-conjugated monoclonal antibodies (mAbs) were purchased from Biolegend Inc. All the reagents for detection of mouse immunogobulins were purchased from Southern Biotech Inc. The reagents for detection of TGFβ were from R&D Systems. The reagents for isolation of bacterial DNA and pyrosequencing were from Qiagen and Roche, respectively.
2.3. Gut microbiota transfer
Fresh feces (10 fecal pellets ~150mg) were collected from each of the following female donor mice (n=3–4/strain): MyD88−/−NOD, MyD88−/−B6, MyD88−/−B6g7 and wild type B6 mice (all at 12–15 wks of age) and resuspended in 250 ml of sterile water, containing approximately 6×105/ml bacteria. The treated water was given to wild type female NOD/LtJ mice for 3 weeks (at ~4 wk of age, n=15/group) and the freshly treated water was changed twice a week.
2.4. Bacterial DNA isolation
Total bacterial DNA was extracted from 0.25 g fecal sample using the repeated bead beating method described by Favier [22] with modifications. Briefly, 250 mg of fresh mouse fecal samples from colon were first loosened by vortex in TE buffer before Proteinase K (200 μg/ml) digestion. Repeated bead beating was done in 50% PCI solution (phenol/chloroform/isoamyl alcohol: 25/24/1) and spun after bead beating. DNA was precipitated and washed sequentially using isopropanol and 70% (v/v) alcohol, respectively.
2.5. 16S rRNA gene sequencing
The V2 region of the 16S rRNA gene was amplified from each DNA sample using a composite broadly conserved bacterial forward primer (5′-CATGCTGCCTCCCGTAGGAGT-3′) and bar-coded broad-range bacterial reverse primer (5′-TCAGAGTTTGATCCTGGCTCAG-3′) as described by Vaishnava et al [23]. The PCR products were purified with a Qiagen gel extraction kit (Qiagen, CA). After quantification of DNA concentration by NanoDrop, each sample was diluted to a concentration of 1×109 molecules/μl in TE buffer and pooled. 20μl of the pooled sample was used for pyrosequencing with GS Junior Titanium Series 454 sequencing system according to the manufacturer’s instructions (Roche 454, Life Sciences Corp. Branford, CT, USA).
2.6. Microbiota classification
The sequencing data were analyzed with QIIME software [24] package (version 1.6) to assign operational taxonomic units (OTUs). After quality filtering based on the characteristics of each sequence, any low quality or ambiguous reads will be removed. Taxonomy assignment was performed at various levels using representative sequences of each OTU. Beta-diversity was calculated to compare differences between microbial communities and the data was shown as Principal Coordinate Analysis (PCoA) [24].
2.7. Gut lumen IgA and TGFβ measurement
Intestine (small and large) was harvested from the mice and flushed with 10 ml of sterile PBS. The total material was centrifuged for 5 min at 2,000 rpm. The supernatant was collected and IgA (Southern Biotech) or TGFβ (eBioscience) was measured by ELISA. The results were expressed as the total IgA or TGFβ content, which was calculated by concentration per milliliter x 10 (ml).
2.8. Lamina propria lymphocyte isolation
Mouse intestine was divided into small intestine (SI) and colon. Luminal contents were washed off with sterile PBS. After removing Peyer’s patches (PP), the intestine was cut longitudinally into 0.5cm lengths. Mucus in the gut segments was washed off by gently shaking the tube. The gut segments were then transferred to a new 50 ml tube into pre-warmed HBSS and 1 mM EDTA and shaken for 20 min at 250 rpm at 37°C, followed by further vigorous shaking for 30 seconds. The sample was then filtered through nylon mesh. The remaining gut tissue was further cut into small pieces and digested with 1 mg/ml collagenase D and 500 U DNAse1 in RPMI medium and incubated for 1 hr at 250 rpm, 37°C. Lamina propria (LP) lymphocytes were isolated using 40% (w/v) Percoll.
2.9. Intracellular staining
Foxp3 staining was performed using a Foxp3 staining kit (eBioscience) following the manufacturer’s instructions. For intracellular cytokine staining, 106 cells were cultured for 5 hours in the presence of 50 ng/ml PMA (Sigma), 500 ng/ml of ionomycin (Sigma) and 1 μl/ml of Golgi plug (BD Bioscience). After staining of surface markers, cells were fixed in IC fixation buffer (eBioscience) for 20 minutes at room temperature. After 2 washes with permeabilization buffer (eBioscience), cells were stained with anti-cytokine antibodies.
2.10. Histopathology and insulitis score
Pancreata were fixed in 10% buffered formalin and paraffin-embedded when the mice were terminated either at the time of diabetes development or at the end of study (7 month old). Tissues were sectioned and stained with H&E. Insulitis was scored under light microscopy using the following grading: 0, no insulitis; I, insulitis affecting less than 25% of the islet; II, insulitis affecting 25~50% of the islet; III, more than 50~75% islet and IV, >75% islet was infiltrated. Fifty-six to 120 islets were scored for insulitis in each group (n=9 to 15 mice) by an individual blinded to the experimental design. The statistical analysis was performed with χ2 analysis.
2.11. Statistics
PCoA based on multivariate statistical analysis that maps the samples in different dimensions, was performed with QIIME software [24] for finding clusters of similar groups of microbial species (qiime.org). All other statistical tests were performed using GraphPad Prism software (V4). For incidence of diabetes, a log rank test was used and for the comparison between the groups, Student t test was used. P values of less than 0.05 were considered significant.
3. Results
3.1. Transient gut microbiota transfer induces a delay in onset of diabetes and reduced insulitis in NOD mice
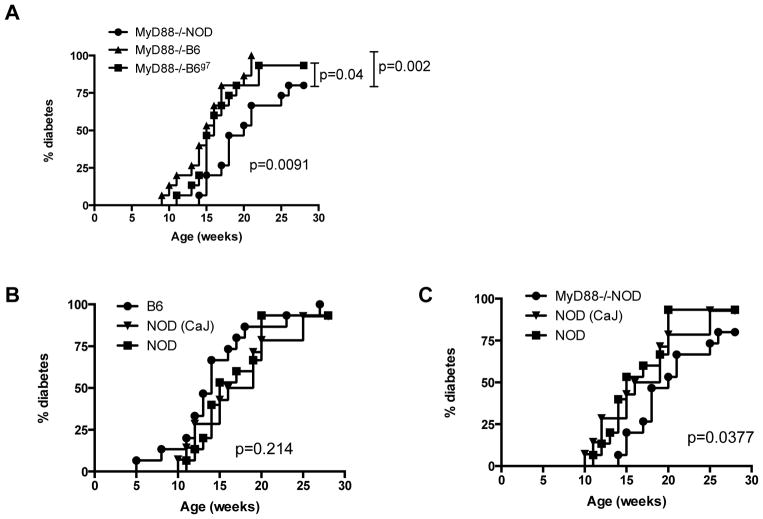
Our previous study suggested that gut microbiota play an important role in diabetes development [20] and the dysbiosis of gut microbiota in MyD88−/−NOD mice resulted in complete protection from diabetes development [20]. We hypothesized that gut bacteria from diabetes resistant mice could transfer diabetes protection to otherwise highly susceptible (to diabetes) hosts. To test our hypothesis, we transferred fecal bacteria from diabetes-resistant female MyD88−/−NOD to female NOD/LtJ mice (~4 wks, n=15) through drinking water. The incidence of diabetes in recipient mice was observed after exposure to the “bacterially-contaminated” water, which was freshly prepared twice a week for 3 weeks. To ascertain whether the composition of gut microbiota of MyD88−/−NOD mice was related to lack of MyD88, we treated another group of female NOD/LtJ mice (n=15) with water similarly prepared using fresh MyD88−/−B6 (females) fecal pellets for 3 weeks. To test whether the MHC plays a role in the composition of gut microbiota, in addition to lack of MyD88, we treated a second control group where female NOD/LtJ mice (n=15) were given water treated with fresh MyD88−/−B6g7 (females) fecal pellets for 3 weeks. We also set up three more control groups (n=15/group), in which female NOD/LtJ mice were given wild type B6 or NOD/Caj fecally-treated or normal clean water. Table 1 shows a summary of the mice used in the study. Figure 1 shows diabetes development in NOD mice that received orally-transferred gut microbiota from different mouse strains. Our data show that transient gut microbiota transfer at a young age, for a limited period, can affect the outcome of diabetes development in adulthood. Strikingly, transfer of the gut bacteria of the diabetes-resistant MyD88−/−NOD strain conferred diabetes protection to wild type NOD mice. In contrast, transfer of microbiota from the MyD88−/−B6 and MyD88−/−B6g7 donors did not provide diabetes protection (Figure 1A, p=0.002 and p=0.04, respectively). It is interesting that the gut microbiota from wild type B6 donors appeared to have a diabetes promoting effect as the recipient NOD/LtJ mice developed earliest onset of diabetes at 5 wks of age although this did not reach statistical significance when compared with the NOD/LtJ mice that received gut microbiota from NOD/Caj donors (Figure 1B). We had a third control group, in which NOD/LtJ mice were not given “exogenous” gut bacteria but clean water, although these mice can transmit their gut microbiota naturally through the fecal-oral route. The incidence of diabetes in this control group was similar to the incidence of disease in mice that ingested fecal materials from NOD/Caj or B6 mice (p=0.214, Figure 1B). Thus, in this large cohort of bacterial transfer experiment, only gut microbiota from MyD88−/−NOD mice delayed and reduced diabetes development (Figures 1A+C). In addition, in line with the delay in diabetes seen in the mice receiving MyD88−/−NOD bacteria, there was also a reduction in insulitis in these mice compared with the mice receiving gut bacteria from the control mouse strains (Figure 2A+B).
Table 1.
Fecal donors used in the study
| Fecal donor | MHC class II gene | Non-MHC genes | MyD88 | Diabetes |
|---|---|---|---|---|
|
| ||||
| MyD88−/−NOD | I-Ag7 | NOD | −/− | − |
| MyD88−/−B6 | I-Ab | B6 | −/− | − |
| MyD88−/−B6g7 | I-Ag7 | B6 | −/− | − |
| B6 | I-Ab | B6 | +/+ | − |
| NOD (Caj) | I-Ag7 | NOD | +/+ | + |
Fecal samples from the mice listed above (all females) were collected and transfterred to the NOD/LtJ female mice for diabetes study.
Figure 1.
Incidence of diabetes after ingestion of “exogenous” gut microbiota. (A) Female NOD/LtJ mice were given water containing female MyD88−/−NOD gut microbiota; MyD88−/−B6 gut microbiota or MyD88−/−B6g7 gut microbiota (n=15/group; n=3–4 gut microbiota donor mice/strain). The water was administered for 3 weeks (at 4wk to 7wk of age) and mice were screened for diabetes by testing for glycosuria weekly and diabetes was confirmed by blood glucose >250mg/dl (13.9mmol/l). (B) Female NOD/LtJ mice were given water containing female wild type C57BL/6 (B6) or NOD/Caj gut microbiota as described above or normal clean water (n=15/group; n=3–4 gut microbiota donor mice/strain). (C) Comparison of diabetes development in NOD/LtJ mice that received MyD88−/−NOD gut microbiota in (A) with NOD/LtJ mice that received NOD/Caj gut microbiota or normal clean water in (B).
Figure 2.
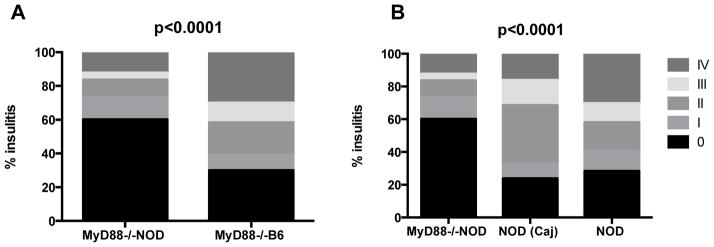
Insulitis score. Pancreata were fixed in 10% buffered formalin and paraffin-embedded when the mice were terminated either at the time of diabetes development or at the end of study (7 month). Tissues were sectioned and stained with H&E. Insulitis was scored under light microscopy using the following grading: 0, no insulitis; I, insulitis affecting less than 25% of the islet; II, insulitis affecting 25~50% of the islet; III, more than 50~75% islet and IV, >75% islet was infiltrated. Fifty-six to 120 islets were scored for insulitis in each group (n=9 to 15 mice) by an individual blinded to the experimental design. The statistical analysis was performed with χ2 analysis.
3.2. Gut microbiota transfer resulted in a long-term change of host intestinal microbiota
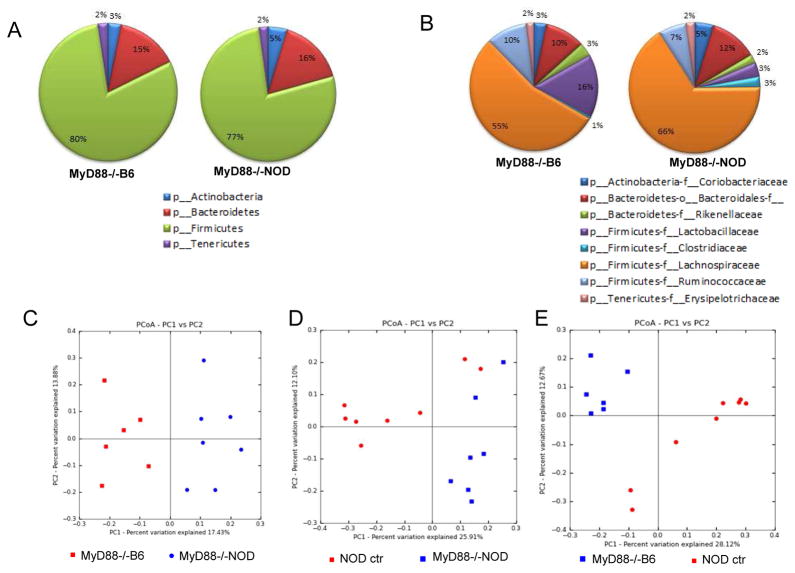
To investigate whether the difference in diabetes development was associated with the recipients’ gut microbiota, we collected fecal samples from the recipient NOD/LtJ mice 4–6 months after transient gut microbial transfer and extracted bacterial DNA. The fecal samples were taken from colon, instead of excrement, in order to better represent the gut microbiota in vivo. We focused on 2 groups of recipient mice that were given either MyD88−/−NOD or MyD88−/−B6 gut microbiota, as the different incidence of diabetes between these 2 groups was highly significant (p=0.002). We compared the gut microbiota in the recipients with that from the donor MyD88−/−NOD or MyD88−/−B6 mice. We also analyzed the gut microbiota of NOD/LtJ mice that had not received “exogenous” gut microbiota as baseline controls. We performed 16S rRNA gene sequencing and analyzed the composition of gut microbiota in these groups. After filtering low quality reads from each sample, analysis at the phylum level showed that NOD mice that received gut microbiota from MyD88−/−NOD mice carried more Bacteroidetes (16% of total microbiome identified) and Actinobacteria (5%) but less Firmicutes (77%) than those that received gut microbiota from MyD88−/−B6 mice (15%, 3% and 80%, respectively) (Figure 3A). Although these differences at the phylum level are small, the difference in the gut microbiome between the two groups was greater at the family level. NOD/LtJ mice that were given gut microbiota from MyD88−/−NOD mice had less Ruminococcaceae and Lactobacillaceae (7% and 3%, respectively) compared to the NOD/LtJ mice that received MyD88−/−B6 gut bacteria (10 and 16%, respectively) (Figure 3B). However, NOD/LtJ mice treated with MyD88−/−NOD gut bacteria showed a higher abundance of Lachnospiraceae family (66%) compared with MyD88−/−B6 treated NOD/LtJ mice (55%). Principal component analysis (PCA) at family level revealed that the composition of gut microbiota was very different in NOD/LtJ mice that had different donor microbiota (Figure 3C). The composition of gut microbiota of untreated control NOD/LtJ mice was also very different from that in treated NOD/LtJ mice (Figure 3D+E). This demonstrates that the change of gut microbiota in treated mice was due to the exposure to exogenous gut bacteria. It is noteworthy that although the NOD/LtJ mice received donor microbiota at a young age, the gut microbiota was distinct between the groups several months after bacterial transfer. It is interesting that the composition of the gut microbiome in the recipient mice does not faithfully mirror the composition of microbiota of the donor mice (Supplementary Fig. 1A). This is not surprising since our recipient mice are housed in SPF conditions and have a full community of endogenous gut microbiota, unlike germ-free mice that often faithfully present bacteria introduced exogenously. However, one family of gut bacteria, F. Lachnospiraceae, remained in high abundance in both MyD88−/−NOD donor and NOD recipients (Supplementary Fig. 1A and Table 2) whereas the same family showed a 13.4 fold reduction in NOD recipients after transferring MyD88−/−B6 gut bacteria (Table 2). Interestingly, there was a 22.6 fold increase of Erysipelotrichaceae in the same recipients (Table 2). Our results indicate that gut microbiota transfer can have a considerable effect on the composition of gut microbiota in the hosts, long after the transfers had been performed and this would most likely be responsible for the different diabetes incidences as all the recipient mice originated from the same source.
Figure 3.
Taxonomic analysis of gut microbiota by 16S rRNA sequencing. (A) Composition of gut microbiota (Phylum) of the NOD/LtJ mice received gut bacteria from MyD88−/−NOD or MyD88−/−B6 mice. NOD/LtJ mice without gut bacterial transfer were controls. (B) Composition of gut microbiota (family) of the NOD/LtJ mice used in (A). (C) Principal component analysis (PCA, unweighted) of taxonomic families of gut microbiota from the NOD/LtJ mice that received gut bacteria from MyD88−/− NOD or MyD88−/−B6 mice. (D) Principal component analysis (PCA, unweighted) of taxonomic families of gut microbiota from the control NOD/LtJ mice (that did not receive exogenous gut bacteria) compared with those that received gut bacteria from MyD88−/−NOD mice. (E) Principal component analysis (PCA, unweighted) of taxonomic families of gut microbiota from the control NOD/LtJ mice (without receiving exogenous gut bacteria) or received gut bacteria from MyD88−/−B6 mice.
Table 2.
Comparison of gut microbiota of controls, recipients and donors
| Phylum – family | % of total bacterial sequences | |
|---|---|---|
| Controls : Recipients : MyD88−/−B6 donors | Controls : Recipients : MyD88−/−NOD donors | |
|
| ||
| Actinobacteria – Coriobacteriaceae | 1 : 2.1 : 2 | 1 : 1.5 : 5 |
| Bacteroidetes – Bacteroidales | 19 : 7.4 : 10 | 19 : 1.1 : 12 |
| Bacteroidetes – Rikenellaceae | 2 : 0.5 : 3 | 2 : 0.2 : 2 |
| Firmicutes – Lactobacillaceae | 9 : 30.9 : 16 | 9 : 48.1 : 3 |
| Firmicutes – Clostridiaceae | 1 : <0.2 : 3 | 1 : <0.2 : 5 |
| Firmicutes – Lachnospiraceae | 61 : 4.1 : 55 | 61 : 35 : 66 |
| Firmicutes – Ruminococcaceae | 6 : <0.2 : 10 | 6 : 3.5 : 7 |
| Firmicutes – Turicibacteraceae | <0.2 : 9.4 : <0.2 | <0.2 : 10.3 : <0.2 |
| Tenericutes – Erysipelotrichaceae | 1 : 45.2 : 2 | 1 : <0.2 : 2 |
NOD/LtJ mice (females), as the recipients, were given no exogenous gut bacteria (controls) or gut bacteria (pooled) from MyD88−/−B6 or MyD88−/−NOD donor mice (all females) as described in the Materials and Methods. The composition of gut microbiota of control, donor and recipient mice was analyzed by 16S rRNA sequencing (Materials and Methods). The numbers in the table are the % of total bacterial sequences in the controls (n=8), recipients (n=7–8/group), and the donors (n=3–4) respectively.
3.3. Higher IgA and TGFβ in NOD mice with lower diabetes incidence
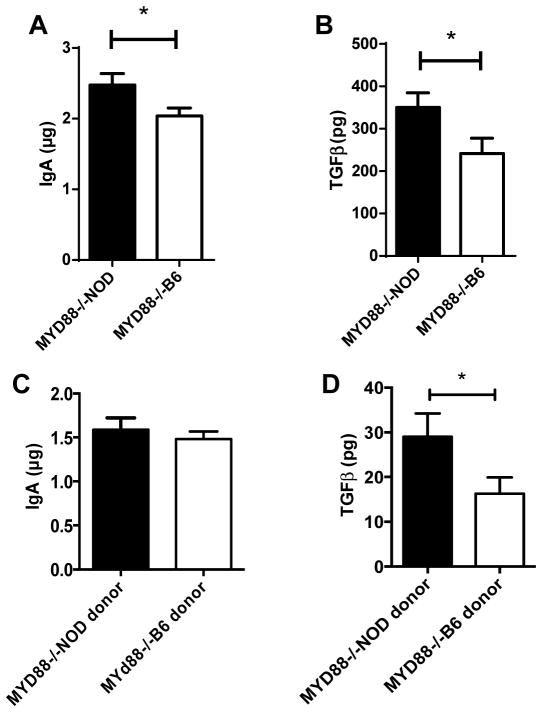
We hypothesized that different gut microbial communities would have different effects on host mucosal immunity, which in turn affects diabetes development. To test our hypothesis, we measured the content of IgA and TGFβ in the gut lumen of the NOD/LtJ mice that had been transferred with gut microbiota from MyD88−/−NOD or MyD88−/−B6 mice. As shown in Figure 4, NOD/LtJ mice that had delayed diabetes onset when MyD88−/−NOD gut microbiota were introduced compared to their counterparts that were treated with MyD88−/−B6 gut microbiota had higher levels of IgA (Fig. 4A) and TGFβ (Fig. 4B) in the gut lumen. There was no difference in any of the other isotypes of mucosal immunoglobulin between the two groups (data not shown). We also measured IgA and TGFβ in the gut lumen of the donor mice and found that whereas the level of IgA in the gut lumen was comparable (Fig. 4C), TGFβ was higher in the MyD88−/−NOD donor mice compared to that in the MyD88−/−B6 mice (Fig. 4D). It is noteworthy that the scale of TGFβ in the donor mice was approximately 10 times lower than that in the recipients, which indicates that gut bacteria transfer induced an active immune response in the intestine of the recipient mice.
Figure 4.
IgA and TGFβ in gut lumen. Total IgA (A) and TGFβ (B) content in gut lumen from NOD/LtJ mice, that were transferred with gut microbiota from MyD88−/−NOD or MyD88−/−B6 mice, was measured by ELISA as described in Materials & Methods. N= 8–10/group. Total IgA (C) and TGFβ (D) content in gut lumen of donor MyD88−/−NOD or MyD88−/−B6 mice were also measured. N= 3–4/group.
3.4. Higher inflammatory cytokine producing T cells in NOD mice with lower diabetes
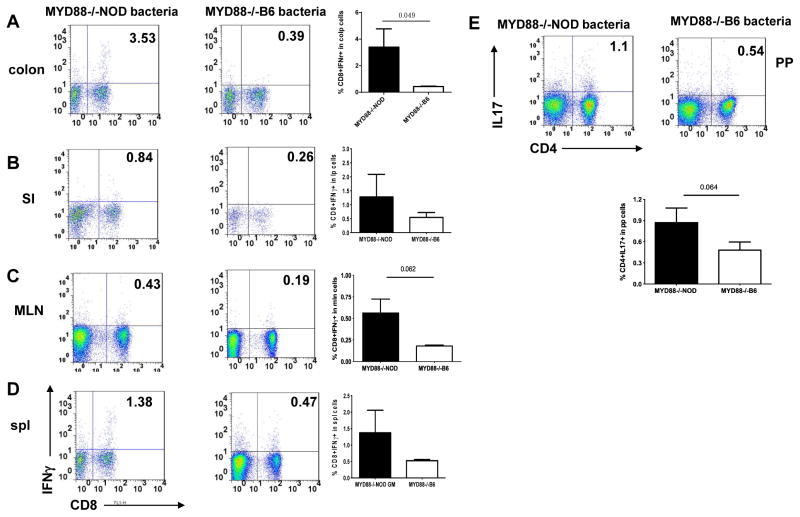
We next investigated the cytokine profile of T cells from gut associated lymphoid tissue (GALT) in the two groups of mice. We isolated lymphocytes from mesenteric lymph nodes (MLN), Peyer’s patches (PP), small intestine (SI) and colon. Unexpectedly, we found that although T cells from the GALT in NOD/LtJ mice that had a later onset of diabetes had higher levels of mucosal IgA and TGFβ, the CD8 T cells also expressed more inflammatory cytokine IFNγ. We also observed an increased percentage of IL-17 producing CD4 T cells from PP (Figure 5A–E). It is interesting that we did not find any difference in IFNγ or IL-17 producing T cells in the pancreatic lymph nodes (PLN, data not shown).
Figure 5.
IFNγ and IL-17A producing T cells in NOD/LtJ mice that were transferred with gut microbiota from MyD88−/−NOD or MyD88−/−B6 donors. Lymphocytes were isolated from different lymphoid tissues as indicated and intracellular IFNγ and IL-17A producing T cells were examined by flow cytometry as described in Materials & Methods. Examples of FACS plots from each lymphoid tissue in each experimental group are shown. Bar charts represent the summary percentage of IFNγ and IL-17A producing T cells in NOD/LtJ mice that were transferred with gut microbiota from MyD88−/−NOD or MyD88−/−B6 donors. N=5–7/group.
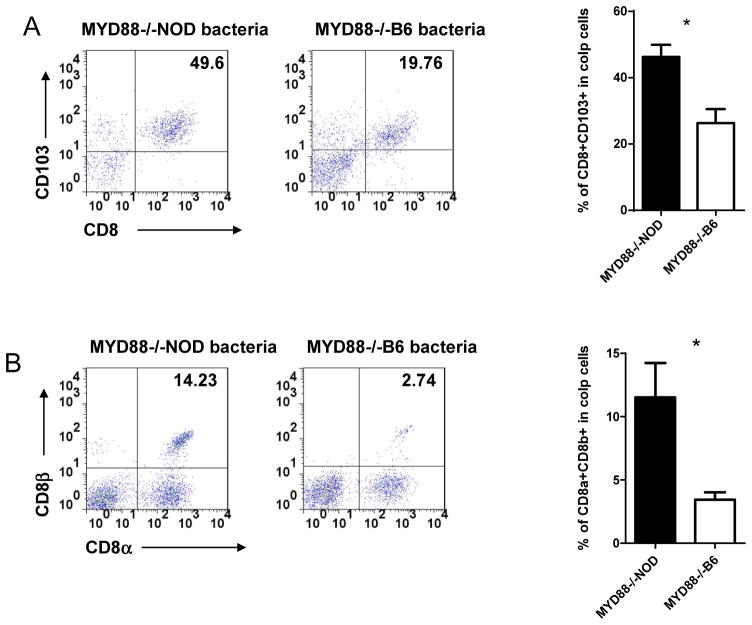
3.5. Increased CD103+ gut homing T cells and CD8αβ T cells in NOD mice with lower diabetes
CD103 is an intergrin protein and expressed on most T cells from GALT [25]. To investigate whether transient transfer of gut microbiota would affect gut-homing T cells in NOD recipient mice used in this study, we examined CD103+ T cells from spleen and GALT. Figure 6A showed a significant increase in the percentage of CD103+ T cells, both TCRαβ and TCRγδ, only in the large intestines of the NOD/LtJ recipient mice that received MyD88−/−NOD gut microbiota, which led to a later onset of diabetes. It is interesting that we did not find this increase in T cells harvested from MLN, PP and small intestine (data not shown).
Figure 6.
Phenotype of CD8 T cells from the large intestine of NOD/LtJ mice that were transferred gut microbiota from MyD88−/−NOD or MyD88−/−B6 donors. Lymphocytes from lamina propria (LP) in the large intestine of NOD/LtJ mice was isolated as described in Materials & Methods. LP lymphocytes were examined by flow cytometry after staining with different markers. (A) Examples of FACS plots of CD103+ CD8 T cells are shown on the left and the summary of % of CD103+ CD8 T cells in total LP lymphocytes is shown on the right (n=6–8/group). (B) Examples of FACS plots of CD8αβ and CD8αα T cells are shown on the left and the summary percentage of CD8αβ T cells in total LP lymphocytes is shown on the right (n=6–8/group).
It is known that most CD8 T cells from GALT express CD8αα homo-dimer co-receptor whereas most CD8 T cells from non-GALT, including spleen and lymph nodes, express the CD8αβ hetero-dimer co-receptor [26, 27]. We postulated that the change of gut microbiota attracted peripheral T cells, especially CD8 T cells from other peripheral lymphoid tissue to GALT. To investigate this possibility, we examined expression of the CD8 co-receptor from T cells in spleen and intestine. As predicted, we found a significant increase of CD8αβ expressing CD8 T cells in the large intestine of the NOD mice that received MyD88−/−NOD gut microbiota (Figure 6B), whereas no difference was found in spleen (data not shown). Consistent with the CD103+ T cells, the increase of CD8αβ expressing CD8 T cells also appeared to be only in large intestine.
4. Discussion
Our studies have shown that the composition of gut bacteria can have a significant effect on development of diabetes in the genetically diabetes-predisposed NOD mouse. The innate immune response, together with mouse genetic background also has an influence on the composition of the gut microbiota. We have shown that mice of different genetic backgrounds, housed in similar environments, and fed with the same food and water have differences in their gut microbiota. This interaction between gut bacteria and the host is clearly important as transfer of gut bacteria from diabetes resistant MyD88−/−NOD mice significantly reduced insulitis and delayed disease onset in recipient NOD mice, whereas neither MyD88−/−B6 nor MyD88−/−B6g7 gut bacteria had obvious effect. Interestingly, we found that the NOD mice that were given gut bacteria from wild-type mice on a B6 genetic background had a trend towards promoting earlier diabetes onset, although this did not reach statistical significance. Thus, any host effect of MyD88 deficiency on the gut microbiome is also modified by other genetic factors.
In this report, we have also shown that short-term oral transfer of fecal bacteria, from different diabetes resistant mouse strains on NOD or B6 genetic backgrounds, into diabetes-prone NOD mice can have a long-term effect on the incidence of autoimmune diabetes. The composition of the gut bacteria in the mice that received MyD88−/−NOD gut microbiota or the bacteria from the various control strains was altered compared with untreated recipient NOD mice, even months after the initial feeding of bacteria. However, the mice in which onset of diabetes was delayed, had gut bacterial composition of increased diversity, which concords with the observation that greater diversity of gut microbiota is present in non-diabetic individuals who are age and sex matched with T1D patients [28]. Conversely, it is interesting that there was a trend to accelerated diabetes in mice that received B6 microbiota, whether wild type or MyD88 deficient. This indicates that the MyD88 deficiency per se does not necessarily predispose to more diverse gut microbiota, but rather that this deficiency, together with the NOD genetic background leads to this change. This is not due to influences of the MHC, as the effects of transfer on diabetes incidence of MyD88−/−B6g7 microbiota was similar to the transfer of MyD88−/−B6 microbiota.
Comparing the composition of the flora from mice transferred with faecal bacteria from MyD88−/−B6 and the MyD88−/−NOD mice in more detail, it is clear from the PCA that while the overall proportion of the phyla is not different, the individual families are quite different. It was notable that the main differences in the families was seen in the Firmicutes, principally in Lachnospiraceae and Clostridiaceae that were increased and the Lactobacillaceae were decreased in the mice that received the MyD88−/−NOD bacteria. Unlike germ-free mice, the composition of the gut bacteria from the NOD recipients, that are housed in SPF conditions and have the full spectrum of gut microbiota, does not faithfully mirror the composition of gut bacteria from the donor mice. However, the increase of Lachnospiraceae remains consistent between donors and recipients. There was also a sharp reduction (11-fold) of Bacteroidales between MyD88−/−NOD donors and the NOD recipients. It is interesting that Lachnospiraceae had over 13-fold decrease between the MyD88−/−B6 donors and the NOD recipients whereas Erysipelotrichaceae showed approximately 23-fold increase in NOD recipients compared to the MyD88−/−B6 donors (45.2 vs 2), but over 226 times higher than the NOD mice received MyD88−/−NOD gut bacteria (45.2 vs <0.2). We also found changes in other families of gut bacteria, which indicates that introducing exogenous gut bacteria possibly promotes competition and interaction among gut commensals. Furthermore, the content of TGFβ in gut lumen remains consistent between donors and recipients albeit on a lower scale. Our results suggest that transferring exogenous gut bacteria not only alters the community of gut bacteria in the hosts but also immune responses. Although individual bacterial families have not been identified that have a particularly pathogenic role in diabetes, the different composition is likely to have an influence on development of diabetes. These effects were also not limited to individual cages [29] but rather was seen in the groups of mice as a whole, spread over several separate cages.
The second generation of high throughput sequencing has advanced rapidly over the past 5 years and the Roche 454 platform is one of the technologies among the second generation sequencing. We have used sequencing of 16SrRNA with the 454 platform as a quantitative means of phylogenetic identification, which also allows for identification of unknown bacteria. For more in depth analysis, it has been reported that whole genome sequencing may give better resolution of bacterial nucleic acid sequences and possibly identify single base changes in the whole genome [30–32]. The whole genome sequencing approach has recently been used for identifying bacterial or viral variants in infectious diseases [33–36]. It is possible that one could find individual bacterial families or even strains that have a particularly pathogenic or protective role in diabetes, although in this context, we feel that it is unlikely that there would be a single strain responsible for the phenotype. However, this has important implications for future development of probiotic treatment that could be used to delay or prevent diabetes.
In addition to the possible influence of bacteria themselves on the autoimmune response, the effect of these bacteria on the mucosal immune system is likely to play a central role in the observed diabetes-protective effects. Gut bacteria shape immune homeostasis and influence the generation of regulatory T cells [37]. Antigen-presenting cells within the gut are directly affected [38], with recent evidence suggesting that particular metabolites such as short-chain fatty acids are key players [39, 40]. We have shown that the mice transferred with the MyD88−/−NOD microbiota expressed regulatory immune responses in the gut, with increased IgA and TGFβ as well as a considerable increase in a population of CD8+CD103+ T cells which have been identified to be a regulatory CD8 population [41]. It is not clear which cells produce the increased TGFβ, but recent evidence suggests that TGFβ is important for upregulation of CD103 and necessary for the maintenance of T resident memory cells of the gut [42]. These CD8+CD103+ T cells may play an immunoregulatory role in the mice that induces delay in the onset of diabetes, although their regulatory function is yet to be determined. Not only are regulatory cells stimulated in situ by the interaction with the gut bacteria, but we have shown that there is also an increase in CD4+IL17+ T cells as well, that may occur also as a result of increased TGFβ production within the gut environment. Whether these cells have a local function to maintain homeostasis or recirculate to other lymphoid tissue is not known. There is also a general increase in IFNγ producing cells in the GALT as well as the spleen. Interestingly, although IFNγ producing cells are generally associated with a Th1 inflammatory response, it is clear that IFNγ is a necessary cytokine for regulatory function of cells [43]. A similar finding was also observed in autoimmune thyroiditis [44]. Thus, the altered composition of bacterial families in the NOD mice transferred with MyD88−/−NOD mice induces changes in the GALT that could contribute to the delayed onset of diabetes. However, this protection may be counterbalanced by effects of potentially pathogenic cells, leading to delay in onset of disease rather than complete protection. Further experiments will help clarify the roles of the expanded populations.
In conclusion, we have shown that altering the gut bacteria, in this case by feeding the bacteria over a short period at young age can have a lasting effect on alteration of the gut microbiome, gut mucosal immunity and ameliorate autoimmunity. By altering the bacterial composition, the interactions of these gut bacteria with the mucosal immune system can induce regulatory CD8 T cells that may influence the development of autoimmune diabetes. The challenge remains to determine how and why the different bacterial families induce these mucosal responses. If these can be understood, this may form a rational basis for selection of specific probiotic agents to boost mucosal regulatory responses.
Supplementary Material
S Figure 1: Taxonomic analysis of gut microbiota by 16S rRNA sequencing. (A) Composition of gut microbiota (family) of the donor MyD88−/−NOD or MyD88−/−B6 mice. (B) Principal component analysis (PCA, unweighted) of taxonomic families of gut microbiota from the donor MyD88−/−NOD or MyD88−/−B6 mice.
Highlights.
Gut microbiota transfer has a long-term effect on diabetes development.
Gut microbiota transfer alters the communities within the host gut microbiome.
Diabetes resistant B6 mice harbor diabetes promoting gut microbiota.
Diabetes protective gut microbiota increase IgA and TGF-β mucosal immune responses in the hosts.
This work supports the concept of a probiotic approach for prevention of type 1 diabetes.
Acknowledgments
This study was supported by JDRF (5-2010-664), NIH (RC1DK087699 and RO1 DK088181).
Footnotes
Author contribution:
JP performed the experiments and analyzed the data. SN helped with 16S rRNA gene sequencing and edited the manuscript. JRM and AB analyzed the data and edited the mauscripts. FSW analyzed the data and wrote the manuscript. WL designed the experiments, analyzed the data and wrote the manuscript.
The authors declare no conflict interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Atkinson MA, Eisenbarth GS, Michels AW. Type 1 diabetes. Lancet. 2013 doi: 10.1016/S0140-6736(13)60591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stene LC, Rewers M. Immunology in the clinic review series; focus on type 1 diabetes and viruses: the enterovirus link to type 1 diabetes: critical review of human studies. Clinical and experimental immunology. 2012;168:12–23. doi: 10.1111/j.1365-2249.2011.04555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson MA, Chervonsky A. Does the gut microbiota have a role in type 1 diabetes? Early evidence from humans and animal models of the disease. Diabetologia. 2012;55:2868–77. doi: 10.1007/s00125-012-2672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gale EA. A missing link in the hygiene hypothesis? Diabetologia. 2002;45:588–94. doi: 10.1007/s00125-002-0801-1. [DOI] [PubMed] [Google Scholar]
- 5.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. The New England journal of medicine. 2002;347:911–20. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 6.Gale EA. The rise of childhood type 1 diabetes in the 20th century. Diabetes. 2002;51:3353–61. doi: 10.2337/diabetes.51.12.3353. [DOI] [PubMed] [Google Scholar]
- 7.Rook GA. The hygiene hypothesis and the increasing prevalence of chronic inflammatory disorders. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2007;101:1072–4. doi: 10.1016/j.trstmh.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Zuany-Amorim C, Sawicka E, Manlius C, Le Moine A, Brunet LR, Kemeny DM, et al. Suppression of airway eosinophilia by killed Mycobacterium vaccae-induced allergen-specific regulatory T-cells. Nature medicine. 2002;8:625–9. doi: 10.1038/nm0602-625. [DOI] [PubMed] [Google Scholar]
- 9.Rook GA. Hygiene and other early childhood influences on the subsequent function of the immune system. Digestive diseases. 2011;29:144–53. doi: 10.1159/000323877. [DOI] [PubMed] [Google Scholar]
- 10.Rook GA. Hygiene hypothesis and autoimmune diseases. Clinical reviews in allergy & immunology. 2012;42:5–15. doi: 10.1007/s12016-011-8285-8. [DOI] [PubMed] [Google Scholar]
- 11.Barlan IB, Bahceciler N, Akdis M, Akdis CA. Role of bacillus Calmette-Guerin as an immunomodulator for the prevention and treatment of allergy and asthma. Current opinion in allergy and clinical immunology. 2005;5:552–7. doi: 10.1097/01.all.0000191238.20632.e2. [DOI] [PubMed] [Google Scholar]
- 12.Bjorksten B, Naaber P, Sepp E, Mikelsaar M. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 1999;29:342–6. doi: 10.1046/j.1365-2222.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- 13.Penders J, Thijs C, Mommers M, Stobberingh EE, Dompeling E, Reijmerink NE, et al. Intestinal lactobacilli and the DC-SIGN gene for their recognition by dendritic cells play a role in the aetiology of allergic manifestations. Microbiology. 2010;156:3298–305. doi: 10.1099/mic.0.042069-0. [DOI] [PubMed] [Google Scholar]
- 14.Scrivener S, Yemaneberhan H, Zebenigus M, Tilahun D, Girma S, Ali S, et al. Independent effects of intestinal parasite infection and domestic allergen exposure on risk of wheeze in Ethiopia: a nested case-control study. Lancet. 2001;358:1493–9. doi: 10.1016/S0140-6736(01)06579-5. [DOI] [PubMed] [Google Scholar]
- 15.Cooper PJ, Chico ME, Rodrigues LC, Ordonez M, Strachan D, Griffin GE, et al. Reduced risk of atopy among school-age children infected with geohelminth parasites in a rural area of the tropics. The Journal of allergy and clinical immunology. 2003;111:995–1000. doi: 10.1067/mai.2003.1348. [DOI] [PubMed] [Google Scholar]
- 16.Rook GA. Review series on helminths, immune modulation and the hygiene hypothesis: the broader implications of the hygiene hypothesis. Immunology. 2009;126:3–11. doi: 10.1111/j.1365-2567.2008.03007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chappel CI, Chappel WR. The discovery and development of the BB rat colony: an animal model of spontaneous diabetes mellitus. Metabolism: clinical and experimental. 1983;32:8–10. doi: 10.1016/s0026-0495(83)80004-3. [DOI] [PubMed] [Google Scholar]
- 18.Meddings JB, Jarand J, Urbanski SJ, Hardin J, Gall DG. Increased gastrointestinal permeability is an early lesion in the spontaneously diabetic BB rat. The American journal of physiology. 1999;276:G951–7. doi: 10.1152/ajpgi.1999.276.4.G951. [DOI] [PubMed] [Google Scholar]
- 19.Calcinaro F, Dionisi S, Marinaro M, Candeloro P, Bonato V, Marzotti S, et al. Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse. Diabetologia. 2005;48:1565–75. doi: 10.1007/s00125-005-1831-2. [DOI] [PubMed] [Google Scholar]
- 20.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–13. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–50. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 22.Favier CF, Vaughan EE, De Vos WM, Akkermans AD. Molecular monitoring of succession of bacterial communities in human neonates. Applied and environmental microbiology. 2002;68:219–26. doi: 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–8. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Annacker O, Coombes JL, Malmstrom V, Uhlig HH, Bourne T, Johansson-Lindbom B, et al. Essential role for CD103 in the T cell-mediated regulation of experimental colitis. The Journal of experimental medicine. 2005;202:1051–61. doi: 10.1084/jem.20040662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das G, Gould DS, Augustine MM, Fragoso G, Sciutto E, Stroynowski I, et al. Qa-2-dependent selection of CD8alpha/alpha T cell receptor alpha/beta(+) cells in murine intestinal intraepithelial lymphocytes. The Journal of experimental medicine. 2000;192:1521–8. doi: 10.1084/jem.192.10.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konkel JE, Maruyama T, Carpenter AC, Xiong Y, Zamarron BF, Hall BE, et al. Control of the development of CD8alphaalpha+ intestinal intraepithelial lymphocytes by TGF-beta. Nature immunology. 2011;12:312–9. doi: 10.1038/ni.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, et al. Toward defining the autoimmune microbiome for type 1 diabetes. The ISME journal. 2011;5:82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, Equinda M, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. The Journal of experimental medicine. 2012;209:1445–56. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niedringhaus TP, Milanova D, Kerby MB, Snyder MP, Barron AE. Landscape of next-generation sequencing technologies. Analytical chemistry. 2011;83:4327–41. doi: 10.1021/ac2010857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkhill J, Wren BW. Bacterial epidemiology and biology--lessons from genome sequencing. Genome biology. 2011;12:230. doi: 10.1186/gb-2011-12-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koser CU, Fraser LJ, Ioannou A, Becq J, Ellington MJ, Holden MT, et al. Rapid single-colony whole-genome sequencing of bacterial pathogens. The Journal of antimicrobial chemotherapy. 2013 doi: 10.1093/jac/dkt494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koser CU, Bryant JM, Becq J, Torok ME, Ellington MJ, Marti-Renom MA, et al. Whole-genome sequencing for rapid susceptibility testing of M. tuberculosis. The New England journal of medicine. 2013;369:290–2. doi: 10.1056/NEJMc1215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reuter S, Ellington MJ, Cartwright EJ, Koser CU, Torok ME, Gouliouris T, et al. Rapid bacterial whole-genome sequencing to enhance diagnostic and public health microbiology. JAMA internal medicine. 2013;173:1397–404. doi: 10.1001/jamainternmed.2013.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reuter S, Harrison TG, Koser CU, Ellington MJ, Smith GP, Parkhill J, et al. A pilot study of rapid whole-genome sequencing for the investigation of a Legionella outbreak. BMJ open. 2013:3. doi: 10.1136/bmjopen-2012-002175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torok ME, Reuter S, Bryant J, Koser CU, Stinchcombe SV, Nazareth B, et al. Rapid whole-genome sequencing for investigation of a suspected tuberculosis outbreak. Journal of clinical microbiology. 2013;51:611–4. doi: 10.1128/JCM.02279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 38.Ng SC, Kamm MA, Stagg AJ, Knight SC. Intestinal dendritic cells: their role in bacterial recognition, lymphocyte homing, and intestinal inflammation. Inflammatory bowel diseases. 2010;16:1787–807. doi: 10.1002/ibd.21247. [DOI] [PubMed] [Google Scholar]
- 39.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–5. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Lan Q, Lu L, Chen M, Xia Z, Ma J, et al. Phenotypic and functional characteristic of a newly identified CD8+Foxp3-CD103+ regulatory T cells. Journal of molecular cell biology. 2013 doi: 10.1093/jmcb/mjt026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang N, Bevan MJ. Transforming growth factor-beta signaling controls the formation and maintenance of gut-resident memory T cells by regulating migration and retention. Immunity. 2013;39:687–96. doi: 10.1016/j.immuni.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–12. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barin JG, Afanasyeva M, Talor MV, Rose NR, Burek CL, Caturegli P. Thyroid-specific expression of IFN-gamma limits experimental autoimmune thyroiditis by suppressing lymphocyte activation in cervical lymph nodes. Journal of immunology. 2003;170:5523–9. doi: 10.4049/jimmunol.170.11.5523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S Figure 1: Taxonomic analysis of gut microbiota by 16S rRNA sequencing. (A) Composition of gut microbiota (family) of the donor MyD88−/−NOD or MyD88−/−B6 mice. (B) Principal component analysis (PCA, unweighted) of taxonomic families of gut microbiota from the donor MyD88−/−NOD or MyD88−/−B6 mice.