Abstract
Within the past few decades, drug combination therapy has been intensively studied in oncology and other complex disease areas, especially during the early drug discovery stage, as drug combinations have the potential to improve treatment response, minimize development of resistance or minimize adverse events. In the present, designing combination trials relies mainly on clinical and empirical experience. While empirical experience has indeed crafted efficacious combination therapy clinical trials (combination trials), however, garnering experience with patients can take a lifetime. The preliminary step to eliminating this barrier of time, then, is to understand the current state of combination trials. Thus, we present the first large-scale study of clinical trials (2008–2013) from ClinicalTrials.gov to compare combination trials to non-combination trials, with a focus on oncology. In this work, we developed a classifier to identify combination trials and oncology trials through natural language processing techniques. After clustering trials, we categorized them based on selected characteristics and observed trends present. Among the characteristics studied were primary purpose, funding source, endpoint measurement, allocation, and trial phase. We observe a higher prevalence of combination therapy in oncology (25.6% use combination trials) in comparison to other disease trials (6.9%). However, surprisingly the prevalence of combinations does not increase over the years. In addition, the trials supported by the NIH are significantly more likely to use combinations of drugs than those supported by industry. Our preliminary study of current combination trials may facilitate future trial design and move more preclinical combination studies to the clinical trial stage.
1. Introduction
Since many diseases including cancer are driven by complex molecular and environmental interactions, targeting a single component may not be sufficient to disrupt those mechanisms [1, 2]. Interest in early drug discovery stages has increasingly evolved to target multiple molecules, pathways, or networks [3–6].
Due to the myriad of potential targets and causes of disease, the rate-limiting step to making a meaningful difference with personalized and precision medicine will be the availability of therapeutic options, which are only validated in the context of clinical trials. Increasingly, panomics technologies are being used to identify novel therapeutic options (e.g. target discovery, repositioned drugs) among existing conventional sets of treatments. Of particular note, the visionary document of The American Society of Clinical Oncology stated that combination therapy is critical to developing prevention strategies and curative therapies [7]. Successful combination therapies in oncology include trastuzumab in combination with paclitaxel for breast cancer, and cetuximab in combination with irinotecan for metastatic colorectal cancer. In addition, combinational antiretroviral therapy has become an effective treatment for HIV infections. Despite the promises of combination therapy, however, its success requires the precise optimization of effective doses, the prevention of adverse drug-drug interactions, and many other factors [8, 9]. Currently, combination design is still based primarily on empirical clinical studies [10]. Thus, systematic examination of current combination trials could facilitate more rational design of clinical trials and guide preclinical tests in the early drug discovery stage. To our knowledge, the characteristics of drug combinations in clinical trials remain elusive.
ClinicalTrials.gov, the most robust of the international clinical trial registries, provides a unique opportunity to take a snapshot of all drug combination trial therapy. In September 2007, the federal law required sponsors or designees to register trials and record key elements in this registry. In addition, many journals require the registration of clinical trials before publication. This registry currently (as of July 2014) contains 171,527 studies in 187 countries and increases at a rate of approximately 350 studies per week. A recent effort on the creation of the database for Aggregate Analysis of ClincalTrials.gov (AACT) facilitates systematic analysis of clinical trials in this registry [11]. Several recent studies have been conducted to examine the characteristics of all the trials [12] or of individual disease areas, including oncology [13–16]. However, neither AACT nor the ClinicalTrials.gov website explicitly annotates combination trials, as their free text data delimits only between individual treatments; combinations, on the other hand, are reported in various ways, including multiple delimited drugs or strings of natural language drug combinations. This inconsistency renders identification and analysis of these trials to be impossible without mining the free text.
In this work, we aimed to learn more about combination trial design in the United States, focusing on basic characteristics such as funding source, primary purpose, trial phase, and prevalence of such trials over time. We first developed a classifier to identify combination trials and oncology trials. By leveraging the information from AACT, we present an initial view of combination trials in oncology. By systematically identifying combinatorial studies within the database, we make it possible to answer future questions regarding combination therapy and further guide the design of combination trials and preclinical studies.
2. Methods
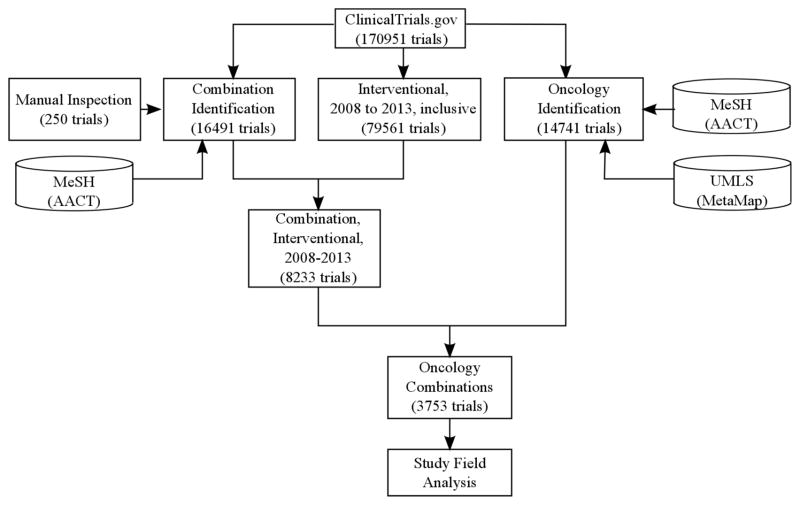
The clinical trial data used in this study were downloaded from ClinicalTrials.gov on July 15, 2014. The AACT database referenced here reflects ClinicalTrials.gov as of March 27, 2014. We restricted our analysis of clinical trials to interventional trials between 2008 and 2013, as those trials are a complete and unbiased sample following the legislation passed in 2007 requiring all ongoing clinical trials to be registered. The overall workflow is shown in Figure 1.
Figure 1.
Overview of study workflow. This flowchart documents our process of standardizing and subsetting clinical trial data based on type and content of study.
2.1 Identifying drug combination trials
Though neither ClinicalTrials.gov nor AACT expressly annotates drug combination trials, some study fields may indicate if a combination is used. We developed a scoring system to identify combination trials based on the information extracted from these fields. A combination trial is defined as a trial in which at least two drugs are administered to a group of patients. Without specific note, a drug can be a small molecule or a biological agent in our study. We summarized discriminating features and assigned different scores to each feature, depending on how confidently that feature could identify combinations. The scores were first assigned based on empirical experience and later adjusted using a training set consisting of 300 manually annotated trials. For example, if the word, “combination,” appeared in the title, then the trial was highly probable to contain combinations, so we considered the occurrence of “combination” in the title a highly weighted feature. On the other hand, if the title included the word “and,” it may indicate that two drugs are used together, but it also may refer to unrelated words, so we weighed this feature lower. Such features were also extracted manually from a list of interventions that contained non-standard, free text descriptions of combinations. These strings often did not map to the standardized vocabulary for drugs as defined by PubChem and DrugBank. For example, trial NCT01121575 specifies its intervention as “PF-00299804 followed by combined PF-02341066 and PF-00299804.” A complete list of our features and study fields can be found in Table 1. Sub-scores were added together to generate the final score for the trial. Trials with scores greater than 2 were classified as combination therapy trials. We validated the scoring system using independent 250 manually annotated trials, with which the system could achieve an impressive performance (with precision 0.94 and recall 0.85). Due to the small feature set, other classifiers such as random forest did not produce a better performance (data not shown), so we decided to choose this simpler scoring system for the classification.
Table 1.
Features for combination detection. Each feature is labeled with its field in AACT, the exact words that were looked for, and the sub-score assigned to it for each occurrence. Sub-scores were added together to generate the final score for the trial. Trials with scores greater than 2 were classified as combination therapy trials.
| Field | Term | Score |
|---|---|---|
| title | “combine”, “combination”, “combining” | 2 |
| title | “with/without”, “and/or” | 2 |
| title | “plus” | 2 |
| title | “interaction”, “co-administered” | 1 |
| title | “and”, “with” | 0.5 |
|
| ||
| intervention | “plus”, “and”, “+”, “/” | 1 |
| intervention | “placebo”, “vehicle” | −1 |
|
| ||
| summary | “combine”, “combination”, “combining” | 2 |
| summary | “together with”, “interaction with”, “alone or with”, “co-administered”, “parallel assignment” | 1 |
|
| ||
| arm | “combine”, “combination”, “combining” | 2 |
| arm | “together with”, “interaction with”, “alone or with”, “parallel assignment”, “plus” | 1 |
| arm | “Drug:” (frequency per individual arm) | 1 |
|
| ||
| keyword | “combination” | 1 |
2.2 Identifying oncology trials
Oncology trials were inferred using disease condition terms (including both Medical Subject Heading [MeSH] and non-MeSH terms) provided by the data submitters and additional condition MeSH terms annotated by a National Library of Medicine (NLM) algorithm [11]. The average number of MeSH terms for the interventional trials between 2008 and 2013 was 2.3. If the trial had at least one MeSH term that started with C04 (Neoplasms), it was considered an oncology trial; otherwise, it was considered a non-oncology trial. For example, C04.557.337 represents leukemia, while C02.839.040 represents acquired immunodeficiency disorder and does not start with “C04”. The trials were also grouped into other disease categories (e.g., Cardiovascular Diseases with MeSH ID starting with C14, Nervous System Diseases with MeSH ID starting with C10, and others) based on their MeSH terms. The MeSH IDs associated with each trial were provided by AACT.
Unfortunately, 15,306 out of 79,561 trials were not associated with any MeSH terms, but provided conditions using free text. To include these trials in our analysis, we used MetaMap [17] to annotate the conditions that they studied. MetaMap uses a knowledge-intensive approach based on symbolic, NLP and computational-linguistic techniques to map free text into Unified Medical Language System (UMLS) concepts. This tool also provides confidence scores and concept categories. Trials with at least one term categorized as “Neoplastic Process” with the maximum confidence score of 1000 were considered oncology trials. Trials categorized otherwise, or with lower confidence scores, were considered non-oncology trials. For example, trial NCT00546247 is not associated any MeSH terms, even though it studied “Advanced Solid Tumors” according to the condition description; this trial was as a Neoplastic Process successfully recognized by MetaMap.
Both the MeSH-based approach and the UMLS-based approach used text mining to process clinical trial conditions. We further compared their predictions using the trials where MeSH and non-MeSH terms were provided. MeSH identified 35,144 oncology trials, MetaMap identified 21,292 oncology trials, and 18,960 of them were common. This shows that 89% of oncology trials identified by the UMLS-based approach were corroborated by the MeSH-based approach.
2.3 Characterization of drug combination clinical trials in oncology
We examined combination trials in oncology in the context of: (1) start year, (2) primary purpose, (3) endpoint measurements (type of study), (4) phases, (5) allocation (randomization), and (6) funding sources. These were extracted from the trials’ study design, start date, phase, and funded by fields. Trials with missing features were ignored for analysis of that particular feature. For funded sources, some trials may have multiple sources. The results were compared across combination trails in oncology, combination trials in non-oncology, and non-combination trials in oncology.
2.4 Statistical analysis
Pearson’s Chi-Squared test was used to compare frequencies of trials associated with different features in Tables 2 and 3, for oncology combinations vs. oncology non-combinations and oncology combinations vs. non-oncology combinations, respectively. Linear regression and analysis of variance (ANOVA) were used to identify significant factors associated with trial start year. P-value < 0.001 was the default significance cutoff.
Table 2.
Combination trials across different disease types
| Disease Type | Oncology | Viral Diseases | Digestive Diseases | Cardiovascular Diseases | Pathological Conditions | Neurological Diseases |
|---|---|---|---|---|---|---|
| Combination (number of trials) | 3753 | 797 | 1295 | 722 | 963 | 463 |
| Non-Combination (number of trials) | 10899 | 2771 | 5662 | 7782 | 16056 | 8056 |
| % Combination | 25.6 | 22.3 | 18.6 | 8.5 | 5.7 | 5.4 |
Table 3.
Comparison of combination trials and non-combination trials in oncology and non-oncology
|
|
|||
|---|---|---|---|
| Non-Oncology Combination | Oncology Combination | Oncology Non-Combination | |
| Funded By(a) | n=4480 | n=3753 | n=10899 |
|
| |||
| Industry | 3086 (68.9%) | 1839 (49.0%) | 3946 (36.2%) |
| NIH | 230 (5.1%) | 853 (22.7%) | 1205 (11.1%) |
| Other | 1654 (36.9%) | 2318 (61.8%) | 8195 (75.2%) |
| U.S. Fed | 39 (0.9%) | 14 (0.4%) | 90 (0.8%) |
|
| |||
| Primary Purpose | n=4217 | n=3732 | n=10545 |
|
| |||
| Basic Science | 236 (5.6%) | 13 (0.3%) | 148 (1.4%) |
| Diagnostic | 44 (1.0%) | 12 (0.3%) | 858 (8.1%) |
| Health Services Research | 9 (0.2%) | 1 (0.03%) | 97 (0.9%) |
| Prevention | 458 (10.9%) | 34 (0.9%) | 508 (4.8%) |
| Screening | 6 (0.1%) | 3 (0.08%) | 145 (1.4%) |
| Supportive Care | 43 (1.0%) | 30 (0.8%) | 597 (5.7%) |
| Treatment | 3421 (81.1%) | 3639 (97.5%) | 8192 (77.7%) |
|
| |||
| Endpoint Measures | n=4179 | n=3311 | n=9202 |
|
| |||
| Bio-availability Study | 63 (1.5%) | 4 (0.1%) | 24 (0.3%) |
| Bio-equivalence Study | 95 (2.3%) | 0 (0%) | 52 (0.6%) |
| Efficacy Study | 809 (19.4%) | 795 (24.0%) | 3051 (33.2%) |
| Pharmacodynamics | 78 (1.9%) | 7 (0.2%) | 86 (0.9%) |
| Pharmacokinetics | 425 (10.2%) | 24 (0.7%) | 103 (1.1%) |
| Pharmacokinetics/Dynamics | 137 (3.3%) | 12 (0.4%) | 71 (0.8%) |
| Safety Study | 437 (10.7%) | 492 (14.9%) | 947 (10.3%) |
| Safety/Efficacy Study | 2135 (51.1%) | 1977 (59.7%) | 4868 (52.9%) |
|
| |||
| Allocation | n=4176 | n=2413 | n=6142 |
|
| |||
| Non-Randomized | 563 (13.5%) | 795 (33.0%) | 2189 (35.6%) |
| Randomized | 3613 (86.5%) | 1618 (67.0%) | 3953 (64.4%) |
|
| |||
| Phase | n=4134 | n=3634 | n=8530 |
|
| |||
| Phase 0 | 12 (0.3%) | 15 (0.4%) | 180 (2.1%) |
| Phase 1 | 1114 (27.0%) | 778 (21.4%) | 1657 (19.4%) |
| Phase 1/Phase 2 | 133 (3.2%) | 582 (16.0%) | 896 (10.5%) |
| Phase 2 | 851 (20.6%) | 1655 (45.5%) | 3950 (46.3%) |
| Phase 2/Phase 3 | 88 (2.1%) | 52 (1.4%) | 201 (2.4%) |
| Phase 3 | 1136 (27.5%) | 499 (13.7%) | 1183 (13.9%) |
| Phase 4 | 800 (19.4%) | 53 (1.5%) | 463 (5.4%) |
The sum of percentages exceed 100 because trials may be funded by more than agencies
3. Results
We examined 170,951 clinical trials available through ClinicalTrials.gov. We further identified 16,491 combination trials and 36,430 oncology trials. After restricting the trials to interventional studies from 2008 to 2013 to obtain the most comprehensive and unbiased set of trials for our analysis, 8,233 combination trials and 14,652 oncology trials remained. Among the 8,233 combination trials, 45.6% (3,753 trials) are related to oncology. In addition, among the 14,652 oncology trials, 25.6% contain combinations, while only 6.9% of non-oncology trials contain combinations, indicating that oncology trials have significantly more combination trials than non-oncology trials (Chi-Squared test, P-value < 0.001).
When looking at other specific disease types, we found that viral diseases and digestive diseases were also more likely to contain combinations (22.3% and 18.6%, respectively), while cardiovascular conditions, pathological conditions, and nervous systems diseases were less likely to contain combinations (8.5%, 5.6%, and 5.4%, respectively).
3.1 Trend of oncology combination trials across years
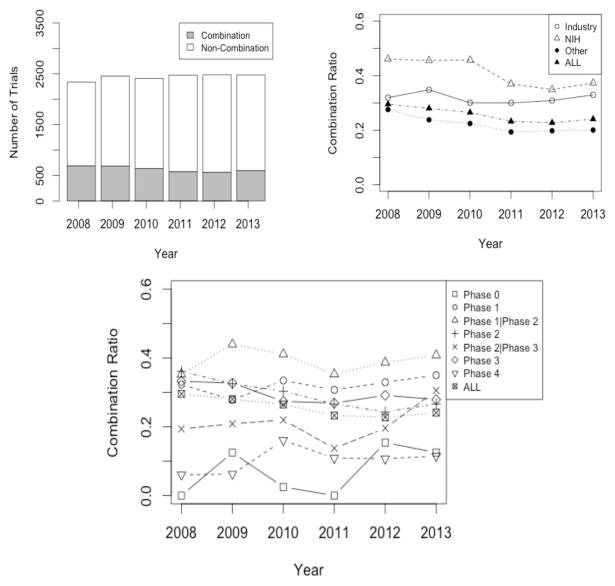
The number of total oncology trials and combination oncology trials stayed constant from 2008 to 2013 (Figure 2a). Surprisingly, the ratio of oncology combination trials to all oncology trials decreased significantly (P value < 0.05; Figure 2b). In 2008, there are 2,339 oncology trials in total, and 691 of these trials contained combinations (29.5%). The ratio decreases to 22.7% in 2012. As shown in Figure 2c, the primary decrease is caused by the steady decrease in phase 2 trials. In addition, over the years, industry has consistently shown less interest in combinations than the NIH, but the NIH has recently decreased involvement in combinations as well (Figure 2b, P value < 0.05 by ANOVA).
Figure 2.
a) Total oncology trials and oncology combination trials over the years, b) The ratio of oncology combination trials vs. all oncology trials over the years, grouped by funding sources, c) The ratio of oncology combination trials vs. all oncology trials over the years, grouped by phases.
3.2 Comparison of combination and non-combination trials in oncology
Table 3 lists the comparison across combination trials in non-oncology, combination trials in oncology, and non-combination trials in oncology. Oncology combinations are more likely to be funded by the NIH than non-oncology combinations (22.7% vs. 5.1%; P-value < 0.001). Combinations in oncology are less likely to be used for prevention (0.9% in oncology vs. 10.9% in non-oncology; P-value < 0.001) and basic science (0.4% in oncology vs. 5.6% in non-oncology; P value < 0.001). Combination trials in oncology are less likely to be reported as phase 4 than those in non-oncology (1.5% vs. 19.4%; P value < 0.001) and are more likely to be non-randomized (33.0% vs. 13.5%; P value < 0.001).
Then within oncology trials, the primary funding source of combination trials is “Other” (61.7%), which includes universities, independent institutes, etc., followed by industry (49.0%), the NIH (22.7%), and the U.S. Federal Government (0.4%). Of the 2,058 oncology trials supported by the NIH, 853 trials are combinations (22.7% of all combination trials) and 1,205 trials are non-combinations (11.1% of all non-combination trials), indicating that combination trials are more likely to be supported by the NIH than non-combination trials (P-value < 0.001).
As expected, the primary purpose of combination trials is to test treatments (97.5%). Very few combination trials are conducted for prevention (0.9%), screening (0.08%), supportive care (0.8%), basic science (0.3%), diagnosis (0.3%), or health services research (0.03%). On the other hand, larger portions of non-combination trials are conducted for diagnosis (8.1%), supportive care (5.7%) and prevention (4.8%). Prevention is less likely to appear as a primary purpose in combination trials (4.8% in non-combination trials vs. 0.9% in combination trials; P value < 0.001).
Nearly half of the combination trials are reported as phase 2 (45.5%), and very few are reported as phase 0 (0.4%) or phase 4 (1.5%). Of the 526 phase 4 trials in oncology, only 10.1% (53 trials) include combinations, while 26.5% of all oncology trials include combinations.
4. Discussion
The advances of genomics and high-throughput technologies enable identifying molecular aberrations of individual tumors and other molecular features that could guide individualized treatment. As tumors are consequences of defects in a complex network comprising a multitude of environmental factors, genetic mutations, and polymorphisms, increasingly more preclinical combinatory studies suggest that targeting multiple components in the tumors may be necessary [4–6]. However, the parameters in clinical trials are very different with those in preclinical settings, while in the present, designing combination trials relies mainly on clinical and empirical experience. Therefore, understanding existing combination trials is of critical importance for future design of clinical trials and preclinical studies.
To our knowledge, there has not yet been any systematic study of combination clinical trials conducted. One critical barrier is that no combination clinical trial dataset is publicly available. Hence, we took the first step towards collecting combination trials and extracting useful quantitative data about these combination trial characteristics from a massive data repository. We developed a simple, yet precise, classifier to identify combination trials, and we also leveraged public natural language processing tools (e.g., MetaMap) and datasets (e.g., MeSH) to identify oncology trials. The dataset we collected paves the way for future drug combination studies.
Our analysis shows that nearly half of all combination trials are conducted in oncology, and a quarter of oncology trials use combination therapies, indicating drug combination is indeed, prevalent in oncology. According to FDA guidance, combinations are intended to treat serious diseases or conditions associated with morbidity that have substantial impact on day-to-day functioning. Oncology and infectious diseases are among the most popular severe diseases for which combination therapy is highly desired, while less severe diseases such as pathological conditions and neurological diseases do not demand combinatory therapy [18].
We found that the trials supported by the NIH are significantly more likely to use combinations of drugs than those supported by industry in the last few years. This phenomenon may be caused by companies’ tendencies to focus on developing their own specific drugs rather than testing drugs from possible outside sources; conversely, academic labs tend to focus less on the drug vendors and more on finding efficacious combinations. Over the years, industry’s interest in combination usage remains constant. Surprisingly, academic interest in combinations appears to be declining from 2010 to 2012 and then starts increasing in 2012. Notably, the interest drops significantly from 2010 to 2011; this coincides the release of the draft guidance on combination therapy codevelopment issued by Food and Drug Administration (FDA) in December 2010 [18]. In the guidance, FDA suggests all of the following criteria should be met for the consideration of co-development of combination therapy: 1) the combination is intended to treat a serious disease or condition, 2) there is a strong biological rationale for use of the combination, 3) a full nonclinical or a short-term clinical study suggests the combination is superior to the individual agents, and 4) there is a compelling reason why the new investigational drugs cannot be developed independently. This requirement may lend increases in costs and time to combination trials, leading a deceasing interest to initiate combination trials. Another possible explanation is that industry and academia are increasingly linked, and conflicts of interest may exist [19–21]. The success of many single-target drugs in oncology may partially explain why industry still prefers to single agent therapy [22,23], and why academia has followed this trend in recent years. In addition, this requirement explains why we found more safety and safety/efficacy studies within oncology combination trials, and why the main primary purpose is treatment.
We also found that combination trials are more likely conducted for treatment and happen in phase1/phase2 and phase 2, signifying that most combinations are still being tested for safety and/or efficacy. FDA suggests whenever possible, the safety profile and dose response of individual new drugs should be characterized in phase 1, resulting in a fewer number of phase 1 combination trials in phase 1 than phase 2 trials [18].
Our current work has several limitations. First, ClinicalTrials.gov does not capture all the studies performed in the USA, especially phase 0–1 trials, which are not required to be registered in the repository. Nonetheless, it still covers over 80% of all studies [12]. Second, some information is missing or misrepresented in the database. For example, 3,872 trials in our data set do not specify a start date. As we only retained trials between 2008 and 2013 for final analysis, the trials without start dates were unable to be used. Third, although our classifier has good performance, some trials are still misclassified. Much effort is required for manual inspection of trials, but even human error in annotating trials cannot be avoided due to the ambiguous information provided. In the future, we will incorporate a greater number of features and keywords to improve classifier performance and potentially introduce more sophisticated algorithms to identify combination trials. For this study, however, due to the low number of falsely identified trials and our classifier’s satisfactory precision, the overall trend is unlikely to change.
Finally, we only assessed the fundamental characteristics of combination trials in this work. In order to facilitate the design of combination trials, it would be important to assess other features as specific disease types and interventions applied. The data contained within ClinicalTrials.gov is free text submitted without strict guidelines for sponsors, so much manual effort is required to process these data. In the future, we will stratify our data into different cancer types (e.g., lung cancer, leukemia, etc.) and seek characteristics within the different types. In addition, we will identify patterns within specific drug combinations (e.g., which two drugs are used together frequently, which drugs reach further stages of clinical trials) by analyzing various drug features. We will aim to extract more information from our datasets regarding the interactions between drugs listed in clinical trials, especially regarding the exact combinations formed from the individual drugs. Many drugs that are tested in clinical trials are unapproved compounds or biological agents, which renders them hard to standardize and extract from the clinical trial data. In addition, many combination trials only list all the interventions applied without specifying the exact relationships between them. We believe by analyzing other fields and integrating with other sources (e.g., drug-target relationships [24]), we can better understand the nature of drug combinations in these trials.
5. Conclusion
Cancer is a heterogeneous disease that involves a multitude of genetic and environment factors. Its complexity also accounts for its resistance against many current targeted therapies. Increasing numbers of studies aim to target multiple factors and have proven successful in many cases, especially in preclinical models. However, moving from preclinical models to clinical trials requires consideration of many more clinical factors. Thus, understanding the characteristics of current combination trials may facilitate the design of future trials. In this work, we developed methods to identify combination trials and oncology trials from ClinicalTrials.gov, and we took the first step to exploring the fundamental characteristic of drug combination trials. Surprisingly, we found that interest in drug combinations does not increase. Understanding the barriers that prevent combinations from reaching the clinical stage may help advance more combinational therapy studies. In the future, we will integrate our clinical data with other molecular features to extract more patterns regarding drug combinations.
Acknowledgments
This research was funded in part by the Lucile Packard Foundation for Children’s Health, the Stanford Child Health Research Institute, the Hewlett Packard Foundation, and the National Institute of General Medical Sciences (R01 GM079719). Menghua Wu was supported by Stanford Institutes of Medicine Summer Research Program (SIMR). We thank Jenna Bollyky for providing valuable comments.
Contributor Information
MENGHUA WU, Email: rachel.wu@gmail.com, Division of Systems Medicine, Department of Pediatrics, Stanford University School of Medicine, 1265 Welch Road, Stanford, CA, 94305, USA. The Harker High School, 500 Saratoga Ave, San Jose, CA, 95129, USA.
MARINA SIROTA, Email: msirota@stanford.edu, Division of Systems Medicine, Department of Pediatrics, Stanford University School of Medicine, 1265 Welch Road, Stanford, CA, 94305, USA.
ATUL J. BUTTE, Email: abutte@stanford.edu, Division of Systems Medicine, Department of Pediatrics, Stanford University School of Medicine, 1265 Welch Road, Stanford, CA, 94305, USA
BIN CHEN, Email: binchen1@stanford.edu, Division of Systems Medicine, Department of Pediatrics, Stanford University School of Medicine, 1265 Welch Road, Stanford, CA, 94305, USA.
References
- 1.Chen B, Butte AJ. Network medicine in disease analysis and therapeutics. Clin Pharmacol Ther. 2013 Dec;94(6):627–9. doi: 10.1038/clpt.2013.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nature reviews Genetics. 2011 Jan;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nature chemical biology. 2008 Nov;4(11):682–90. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 4.Lee MJ, Ye AS, Gardino AK, et al. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012 May 11;149(4):780–94. doi: 10.1016/j.cell.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller ML, Molinelli EJ, Nair JS, et al. Drug synergy screen and network modeling in dedifferentiated liposarcoma identifies CDK4 and IGF1R as synergistic drug targets. Science signaling. 2013 Sep 24;6(294):ra85. doi: 10.1126/scisignal.2004014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathews Griner LA, Guha R, Shinn P, et al. High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proceedings of the National Academy of Sciences of the United States of America. 2014 Feb 11;111(6):2349–54. doi: 10.1073/pnas.1311846111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oncology ASoC. Shaping the Future of Oncology: Envisioning Cancer Care in 2030. 2014 Available from: http://www.asco.org/about-asco/asco-vision.
- 8.Verweij J, Disis ML, Cannistra SA. Phase I studies of drug combinations. J Clin Oncol. 2010 Oct 20;28(30):4545–6. doi: 10.1200/JCO.2010.30.6282. [DOI] [PubMed] [Google Scholar]
- 9.Ananthakrishnan R, Menon S. Design of oncology clinical trials: a review. Critical reviews in oncology/hematology. 2013 Oct;88(1):144–53. doi: 10.1016/j.critrevonc.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol. 2012 Jul;30(7):679–92. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- 11.Tasneem A, Aberle L, Ananth H, et al. The database for aggregate analysis of ClinicalTrials.gov (AACT) and subsequent regrouping by clinical specialty. PloS one. 2012;7(3):e33677. doi: 10.1371/journal.pone.0033677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Califf RM, Zarin DA, Kramer JM, Sherman RE, Aberle LH, Tasneem A. Characteristics of clinical trials registered in ClinicalTrials.gov, 2007–2010. JAMA. 2012 May 2;307(17):1838–47. doi: 10.1001/jama.2012.3424. [DOI] [PubMed] [Google Scholar]
- 13.Goswami ND, Pfeiffer CD, Horton JR, Chiswell K, Tasneem A, Tsalik EL. The state of infectious diseases clinical trials: a systematic review of ClinicalTrials.gov. PloS one. 2013;8(10):e77086. doi: 10.1371/journal.pone.0077086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill KD, Chiswell K, Califf RM, Pearson G, Li JS. Characteristics of pediatric cardiovascular clinical trials registered on ClinicalTrials.gov. American heart journal. 2014 Jun;167(6):921–9. e2. doi: 10.1016/j.ahj.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirsch BR, Califf RM, Cheng SK, et al. Characteristics of oncology clinical trials: insights from a systematic analysis of ClinicalTrials.gov. JAMA internal medicine. 2013 Jun 10;173(11):972–9. doi: 10.1001/jamainternmed.2013.627. [DOI] [PubMed] [Google Scholar]
- 16.Stockmann C, Sherwin CM, Ampofo K, et al. Characteristics of antimicrobial studies registered in the USA through ClinicalTrials.Gov. International journal of antimicrobial agents. 2013 Aug;42(2):161–6. doi: 10.1016/j.ijantimicag.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. Journal of the American Medical Informatics Association : JAMIA. 2010 May-Jun;17(3):229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.U.S. Department of Health and Human Services. Guidance for Industry: Codevelopment of Two or More New Investigational Drugs for Use in Combination. Clinical Medical. 2013 Jun [Google Scholar]
- 19.Lexchin J, Bero LA, Benjamin Djulbegovic, et al. Pharmaceutical industry sponsorship and research outcome and quality: systematic review. BMJ. 2003;326:1167. doi: 10.1136/bmj.326.7400.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bodenheimer T. Uneasy alliance: Clinical investigators and the Pharmaceutical Industry. Health Policy Report. 2000 May;342(20):1539–1544. doi: 10.1056/NEJM200005183422024. [DOI] [PubMed] [Google Scholar]
- 21.Angell M. Industry-sponsored clinical research: A broken system. JAMA. 2008;300(9):1069–1071. doi: 10.1001/jama.300.9.1069. [DOI] [PubMed] [Google Scholar]
- 22.DiMasi JA, Grabowski HG. Economics of new oncology drug development. Journal of Clinical Oncology. 2007 Jan;25(2):209–216. doi: 10.1200/JCO.2006.09.0803. [DOI] [PubMed] [Google Scholar]
- 23.Kamb A, Wee S, Lengauer C. Why is cancer drug discovery so difficult? Nature Reviews Drug Discovery. 2007 Feb;6:115–120. doi: 10.1038/nrd2155. [DOI] [PubMed] [Google Scholar]
- 24.Rask-Andersen M, Masuram S, Schioth HB. The druggable genome: Evaluation of drug targets in clinical trials suggests major shifts in molecular class and indication. Annual review of pharmacology and toxicology. 2014;54:9–26. doi: 10.1146/annurev-pharmtox-011613-135943. [DOI] [PubMed] [Google Scholar]