Abstract
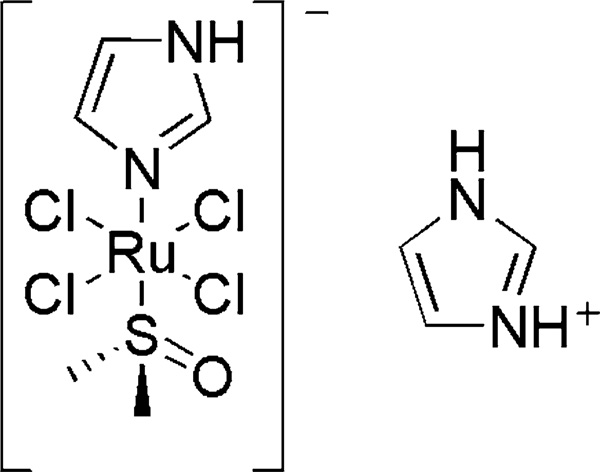
[ImH][trans-RuIIICl4(DMSO)(Im)] (where DMSO is dimethyl sulfoxide and Im is imidazole) (NAMI-A) is an antimetastatic prodrug currently in phase II clinical trials. The mechanisms of action of this and related Ru-based anticancer agents are not well understood, but several cellular targets have been suggested. Although Ru has been observed to bind to DNA following in vitro NAMI-A exposure, little is known about Ru–DNA interactions in vivo and even less is known about how this or related metallodrugs might influence cellular RNA. In this study, Ru accumulation in cellular RNA was measured following treatment of Saccharomyces cerevisiae with NAMI-A. Drug-dependent growth and cell viability indicate relatively high tolerance, with approximately 40% cell death occurring at 6 h for 450 µM NAMI-A. Significant dose-dependent accumulation of Ru in cellular RNA was observed by inductively coupled plasma mass spectrometry measurements on RNA extracted from yeast treated with NAMI-A. In vitro, binding of Ru species to drug-treated model DNA and RNA oligonucleotides at pH 6.0 and 7.4 was characterized by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in the presence and absence of the reductant ascorbate. The extent of Ru–nucleotide interactions increases slightly with lower pH and significantly in the presence of ascorbate, with differences in observed species distribution. Taken together, these studies demonstrate the accumulation of aquated and reduced derivatives of NAMI-A on RNA in vitro and in cellulo, and enhanced binding with nucleic acid targets in a tumorlike acidic, reducing environment. To our knowledge, this is also the first study to characterize NAMI-A treatment of S. cerevisiae, a genetically tractable model organism.
Keywords: NAMI-A, RNA, DNA, Anticancer drug, Ruthenium
Introduction
Current metal-based anticancer therapeutics such as cisplatin are effective against primary tumors, but suffer from several drawbacks: severe side effects, the development of resistance, and only minimal antimetastatic activity [1]. Research efforts to find more successful treatments have led to a number of promising new drugs. One Ru-based complex has received particular attention and is currently in phase II of clinical trials: [ImH][trans-RuIIICl4(DMSO) (Im)] (where DMSO is dimethyl sulfoxide and Im is imidazole), or NAMI-A (Structure 1) [2, 3]. This complex has shown low toxicity and remarkable efficacy against metastases, especially among lung carcinomas. Multiple effects may be involved in this antimetastatic activity, including interactions with the extracellular matrix and the cell surface, interference with NO metabolism, effects on tumor metalloproteinases, and transport into tumors through binding to serum proteins such as transferrin [4, 5]. NAMI-A also causes transient tumor cell cycle arrest in the premitotic G2/M phase [6]. In addition, the highly related [Na][trans-RuIIICl4(DMSO)(Im)] (NAMI) has been shown to cause a marked reduction of tumor nucleic acid content [7]. This effect and its consequences have not yet been explored for NAMI-A. In short, NAMI-A is an extremely attractive drug candidate owing to both its antimetastatic activity and its low toxicity to healthy cells, but much remains to be determined regarding its mechanism of action.
Structure 1.
Redox and ligand-exchange chemistry are also considered important to the mechanism of action of NAMI-A, and the in vivo effectiveness of aquated and reduced drug products has led to the conceptualization of NAMI-A as a prodrug [8]. The reduction potential of NAMI-A is +0.016 V versus the standard calomel electrode in pH 7.4 phosphate buffer [9], and as such, reduction from Ru(III) to Ru(II) is electrochemically attainable by the plasma and cellular reducing agents ascorbate and glutathione. In addition to the presence of ubiquitous reducing agents, rapidly dividing cancer cells provide a hypoxic environment [10, 11] accompanied by low pH levels [12] that affect reduction and subsequent reactivity. Furthermore, several pH-dependent equilibria are possible in vivo and, as such, there are expected to be multiple Ru species present, many of which have the potential to be therapeutically active [8].
Ru anticancer therapeutics are known to bind to DNA in vitro and in cellulo [13, 14]. Indeed, treatment with NAMI-A results in Ru–DNA adducts. Similar to the case of cisplatin, the cytotoxic activity of NAMI-A has been correlated with Ru–DNA accumulation in several cancer cell lines [15]. Unlike the case of cisplatin, however, in the case of NAMI-A significant Ru–DNA accumulation and cytotoxicity occur at much higher drug concentrations than those needed for anticancer activity. In addition, although both drugs result in metal–DNA adducts, the types and potential consequences of those adducts differ significantly. Results from in vitro studies using calf thymus or plasmid DNA to investigate the consequences of NAMI-A treatment on global measures of DNA structure such as thermal denaturation or circular dichroism spectroscopic signatures suggest less overall structural effects for NAMI-A than with cisplatin, which could be due to the types of adducts formed or less accumulation for a given treatment method [16, 17]. The “soft,” nucleophilic N7 site of guanine is the main site of coordination for cisplatin-derived Pt(II), with coordination to the N7 of adenine also observed [18]. Similarly, a hydrolysis product of NAMI-A, [trans-RuCl4(H2O)(Im)]−, has been observed binding to N7 of 9-methyladenine by NMR spectroscopy [19]. Ru binding to GMP following NAMI-A treatment has been documented by capillary electrophoresis coupled with inductively coupled plasma mass spectrometry (ICP-MS), and a strong preference for guanine over adenine bases was confirmed by electrospray ionization mass spectrometry [20, 21]. Because of their different propensities for monoadduct and diadduct formation, the in vivo adduct profiles produced by NAMI-A and cisplatin are expected to be different. This has been shown for NAMI in vitro, where transcriptional mapping found that the Ru drug produced fewer intrastrand GG adducts on double-stranded DNA than did cisplatin [22]. A minor amount of interstrand cross-links were formed (also fewer than for cisplatin), along with monofunctional adducts. To our knowledge, the specific geometries and percent occurrences of the different DNA adducts formed following NAMI-A treatment are not known.
RNA is chemically similar to DNA, but plays diverse roles in cell regulation and gene expression. In addition to well-known functional roles in translation (messenger RNA, transfer RNA, and ribosomes) a great deal is now being learned about the regulatory roles performed by RNAs such as small interfering RNA, microRNA, piwi-interacting RNA, and long noncoding RNA in both transcription and translation [23]. Consistent with these regulatory roles, the integrity of cellular RNA is monitored. RNA damage is linked to disease and can trigger programmed cell death [24–26]. Examples of “ribotoxic” responses include those produced by sarcin [25] and ricin [26], toxic proteins that induce apoptosis through their interaction with the sarcin/ricin loop of the large subunit of the eukaryotic ribosome. In addition, there is evidence that both ribosomes and transfer RNA may play other specific roles in programmed cell death pathways [27, 28].
Drug binding to RNA can impact cell fate via downstream effects on a wide range of RNA and regulatory pathways. Studies with cisplatin have shown significant drug accumulation in RNA as well as inhibition of RNA function in extracts [29–31]. This could be true for other DNA binding drugs such as NAMI-A. To our knowledge, the effect on RNA of treatment with NAMI-A has not been previously studied. In the work reported here, we used Saccharomyces cerevisiae for in cellulo analysis of drug accumulation in RNA, and we performed in vitro analysis of binding to RNA and DNA oligonucleotides. S. cerevisiae was chosen as a model system because it has been used to study a range of anticancer drugs [32] and RNA pathways [33, 34], and is a genetically tractable system for future studies [35]. When S. cerevisiae is treated with NAMI-A, growth inhibition and accumulation of Ru in whole cells and cellular RNA is observed. The extent of Ru–nucleotide binding to model DNA and RNA oligonucleotides depends on both pH and reduction by ascorbate, where enhanced binding occurs under acidic, reducing conditions similar to those found in solid tumors. Taken together, these studies demonstrate significant binding of Ru from aquated and/or reduced derivatives of NAMI-A to RNA in vitro and in cellulo, and enhanced binding with nucleic acid targets in a tumorlike environment.
Materials and methods
Cell cultures and treatments
The S. cerevisiae strain used in this study was BY4741, a gift from the Stevens laboratory at the University of Oregon. All liquid cultures were grown on synthetic complete medium consisting of 0.67% yeast nitrogen base and 2% glucose as a carbon source supplemented with amino acids and nucleotide bases. Plated cells were grown on yeast extract–peptone–glucose (YEPD) agar plates (1% yeast extract, 2% peptone, 2% glucose, and 2% agar). Liquid culture growth was maintained in the dark at 30 °C with shaking at 200 rpm. For each yeast treatment, a 10 mM NAMI-A stock solution was prepared fresh in 50 mM NaHPO4 (pH 7.4) with 0.1 M NaCl exactly 10 min before the start of treatment. Yeast cultures were pregrown to an optical density at 600 nm (OD600) of 5, medium at 30 °C was then inoculated with them to an OD600 of 0.075, and the cultures were treated with the given concentrations of NAMI-A.
Synthesis and characterization of NAMI-A
The two-step procedure described in the international patent held by Alessio et al. [36] was followed with slight modifications, as follows. In the second step, 0.782 g of [(DMSO)2H][trans-RuCl4(DMSO)2] was dissolved in 15 mL of acetone and the mixture was stirred for 15 min to allow the compound to dissolve before adding 0.391 g of Im. The suspension was then heated and stirred at 30 °C for 21 h. The product was collected via vacuum filtration, washing with acetone and ethyl ether, and drying under a vacuum. The final product was recrystallized by dissolving crude NAMI-A in DMSO to form a saturated paste, and then acetone was added to precipitate the product. NAMI-A was characterized using Fourier transform IR, Raman, and UV–vis spectroscopy. The spectra were in agreement with results from samples that were characterized via X-ray diffraction [37].
Measurement of culture growth and cell survival
Culture growth was measured as OD600 (conversion factor OD600, 1 absorbance unit at 600 nm is equivalent to 2.0 × 107 cells/mL). Cell viability was measured by plating serial dilutions of treated and untreated yeast onto drug-free YEPD agar plates (approximately 200 cells/plate) and counting the number of colonies formed after 3 days at 30 °C. The number of colony-forming units (cfu) was determined by dividing the cfu counts of treated cultures by those of untreated cultures, which were assumed to be 100%.
Measurement of Ru content in whole cells and extracted RNA
Yeast cultures treated with NAMI-A were pelleted (6 × 107 cells for whole-cell measurements and 1.2 × 108 cells for RNA extraction) at 4 °C and washed three times with Milli-Q water. RNA was extracted using a MasterPure RNA purification kit (Epicentre) according to the manufacturer’s specifications. The RNA was then desalted on in-house-prepared G-25 Sephadex spin columns (Bio-Rad) and quantified by absorbance at 260 nm. Mass was converted to moles of ribonucleotides using the average molecular weight of a ribonucleotide monophosphate. Both RNA and whole-cell samples were digested in 70% nitric acid (TraceSELECT, Fluka) for 2 h at 65 °C and were then diluted to 2% (v/v) nitric acid with Milli-Q water. Ru content was determined by ICP-MS using a Thermo VG PQExcell quadrupole ICP-MS instrument equipped with a Gilson 222 autosampler at the W. M. Keck Collaboratory for Plasma Spectrometry (Oregon State University, Corvallis, OR, USA). The instrument was calibrated for 99Ru, 101Ru, and 102Ru by developing standard curves from a Ru standard (High Purity Standards). All measurements were done in triplicate using 115In as an internal standard. Intracellular Ru content was estimated using an average volume of 36.6 fL calculated for a wild-type haploid strain of S. cerevisiae grown on YEPD medium [38], whose cell size matches estimates for control samples from this study (Fig. S1, and data not shown).
In vitro oligonucleotide treatment with NAMI-A
RNA (Dharmacon) and DNA (IDT) oligonucleotides (T6GGT5, U6GGU5, and A5CCA6) were heated to 90 °C for 90 s, cooled to room temperature, and then incubated with 0, 150, and 450 µM NAMI-A (freshly dissolved in the appropriate buffer immediately before use) in 25 mM phosphate buffer (pH 6.0 or 7.4) at 37 °C in the dark. All reactions were conducted for 6 or 24 h in 100 mM NaNO3 and 2 mM Mg(NO3)2. Ascorbate was freshly dissolved in water immediately before use and added to the buffered oligonucleotide samples (final concentration 2.5 mM) immediately before NAMI-A. Samples were desalted using in-house-prepared G-25 Sephadex spin columns (Bio-Rad) to stop the reaction.
Matrix-assisted laser desorption/ionization time-of-flight analysis
RNA and DNA samples were purified using C18 ZipTips (Millipore) with a procedure modified by Chapman and DeRose [39] from the manufacturer’s protocol for RNA [40]. ZipTips were washed by aspiration three times with 1:1 MeCN/H2O, and were equilibrated by washing them three times with 0.1% trifluoroacetic acid. RNAs were bound to the tip by repeated aspiration of the analyte solution. Bound RNA was washed three times by aspiration with 0.1% trifluoroacetic acid and three times with Milli-Q water, and was then eluted from the column using two washes of 1:1 MeCN/H2O. The eluent was dried to completion by a SpeedVac and resuspended in a matrix consisting of 375 mM 2′,4′,6′-trihydroxyacetophenone [THAP; Sigma-Aldrich, puriss. p.a., matrix substance for matrix-assisted laser desorption/ionization (MALDI) mass spectrometry], 30 mM diammonium citrate in 3:1 EtOH/H2O, with added NH4+-loaded Dowex cation-exchange beads (Aldrich) and applied to the sample plate. MALDI time-of-flight (TOF) mass spectrometry analysis was performed with a Waters QToF Premier mass spectrometer in positive-ion mode using V-mode optics. THAP is known as a “hot” matrix, which results in higher rates of depurination [41]. It was chosen as the matrix for this study because it gave a better signal-to-noise ratio for the short oligonucleotide sequences used. In addition, it is well known that the extent of depurination observed in MALDI is greater for DNA than for RNA [42]. These factors are the most likely contributors to the higher-than-typical extent of depurination of the DNA that was observed.
Results
Growth inhibition and cell viability following NAMI-A treatment
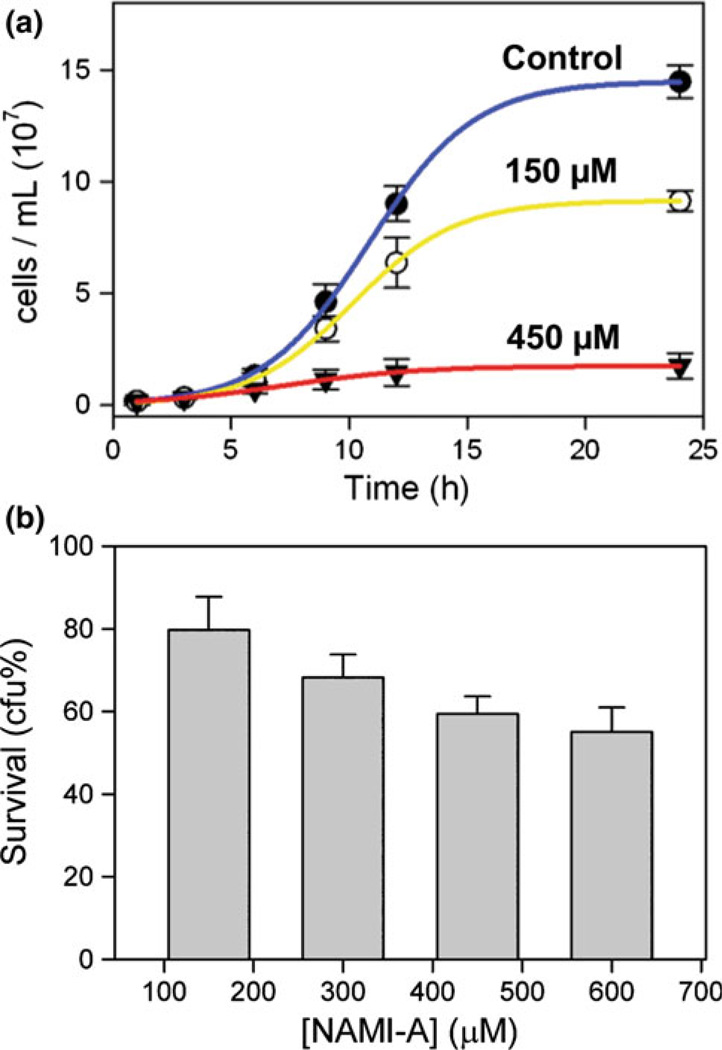
Controlled conditions were first established for subsequent measurements of Ru accumulation in S. cerevisiae following treatment with NAMI-A. Yeast in synthetic complete medium continuously treated with freshly dissolved NAMI-A shows high tolerance for this drug. Concentrations below 100 µM NAMI-A did not result in significant change in culture density for 6 h treatment, whereas treatments with 100–300 µM showed a gradual decrease in cell density, making 100 µM a threshold concentration above which decreases in culture density are clearly observable (Fig. S1a). Full growth curves for 0, 150, and 450 µM NAMI-A are shown in Fig. 1a, demonstrating a moderate (63 ± 7% of the control at 24 h) and severe (12 ± 3% of the control at 24 h) reduction in culture density for 150 and 450 µM, respectively.
Fig. 1.
a Exponential growth curves of yeast treated with 0, 150, and 450 µM [ImH][trans-RuIIICl4(DMSO)(Im)] (where DMSO is dimethyl sulfoxide and Im is imidazole) (NAMI-A). b Survival of yeast treated for 6 h with NAMI-A measured as percentage colony-forming units (cfu) after 3 days in drug-free medium. The results were averaged from three independent experiments presented as the mean ± the standard deviation
To discriminate between slow and/or reversibly inhibited cell division and full loss of cell viability due to NAMI-A treatment, cell survival was measured by clonogenic assay. After 6 h of drug treatment, NAMI-A-treated yeast was plated on drug-free YEPD medium and allowed to grow for 3 days, at which time the number of resultant colonies was compared with that of the control. The results for four concentrations of NAMI-A ranging from 150 to 600 µM show a moderate decrease in cell viability that does not scale linearly with treatment concentration (Fig. 1b). Tripling the NAMI-A dose from 150 to 450 µM results in a doubling of the number of nonviable cells (viable cells going down from 80 ± 8 to 59 ± 4%) and a somewhat higher loss of cell culture density (from 88 ± 20 to 53 ± 14%, Table 1). This relatively low loss of viability contrasts with the properties of the more cytotoxic cisplatin, which after 6 h at a 200 µM treatment concentration results in a culture density of 72 ± 14%, but only 27 ± 8% viable cells in comparison with controls (Hostetter, Osborn, and DeRose, unpublished results). Taken together, these results indicate that the reduced culture density of NAMI-A-treated yeast is due to a combination of cell death and slowed growth. Typically, yeast that is undergoing slower cell division owing to general metabolic factors has a smaller average cell size [38]. Yeast treated with NAMI-A, however, exhibits an increased average cell size (Fig. S1b), which might be indicative of cell cycle arrest.
Table 1.
Effects of [ImH][trans-RuIIICl4(DMSO)(Im)] (where DMSO is dimethyl sulfoxide and Im is imidazole) (NAMI-A) on yeast at 6 h
| NAMI-A concentration |
||
|---|---|---|
| 150 µM | 450 µM | |
| Culture density (%) | 88 ± 20 | 53 ± 14 |
| Cell viability (%) | 80 ± 8 | 59 ± 4 |
Accumulation of Ru in yeast cells and cellular RNA
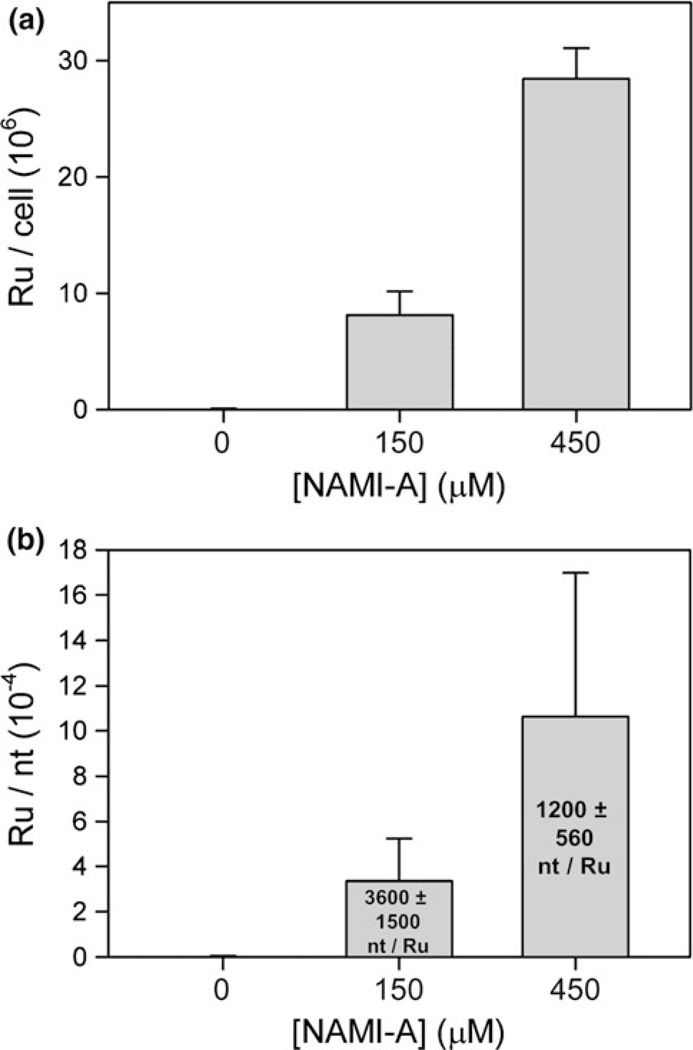
The Ru content of whole cells and that of total RNA isolated from yeast treated for 6 h with 0, 150, and 450 µM NAMI-A were quantified by ICP-MS. The results for both whole cells and total RNA show significant Ru accumulation that is proportional to the treatment concentration, roughly tripling when the treatment concentration is tripled (Fig. 2). Very approximate average cellular Ru concentrations of 370 and 1,200 µM (for 150 and 450 µM NAMI-A treatment, respectively) can be calculated on the basis of these results and estimates for the average volume of growing yeast (see “Materials and methods”); these values are approximate upper limits, since as noted already, yeast cell sizes increase with treatment. Although cells were washed in triplicate before analysis, an influence from extracellular accumulation of Ru on cell walls cannot be excluded. Nonetheless, the proportional increase in Ru content for both whole cells and extracted RNA indicates that under these conditions cellular uptake of NAMI-A is not saturated and that Ru accumulation in RNA is proportional to the cellular Ru accumulation.
Fig. 2.
a Accumulation of Ru atoms per cell in yeast treated with NAMI-A for 6 h. b Accumulation of Ru atoms per nucleotide (nt) in total RNA from yeast treated with NAMI-A for 6 h. The average number of nucleotides per Ru atom is given in the text. Each result was averaged from three independent experiments and presented as the mean ± the standard deviation
Ru adduct formation on RNA and DNA oligonucleotides following in vitro treatment with NAMI-A
Measurements from the in cellulo studies described already indicate significant accumulation of Ru in cellular RNA following 6 h of NAMI-A exposure in a yeast cell environment. To provide context for these results, the reactivity of NAMI-A with RNA and DNA oligonucleotides was examined in vitro. NAMI-A was incubated with single-stranded RNA and DNA oligonucleotides in an ionic background similar to biological conditions [100 mM NaNO3 and 2 mM Mg(NO3)2]. Comparative studies were performed at pH 6.0, chosen to simulate the lower pH often found in tumor tissue [12], and at pH 7.4 (similar to the pH of blood and healthy cells). A strong preference for Ru coordination to G has been previously observed for DNA [21], and so a simple model containing a single GG site embedded in a sequence of U or T nucleotides (U6GGU5 and T6GGT5) was chosen for this study.
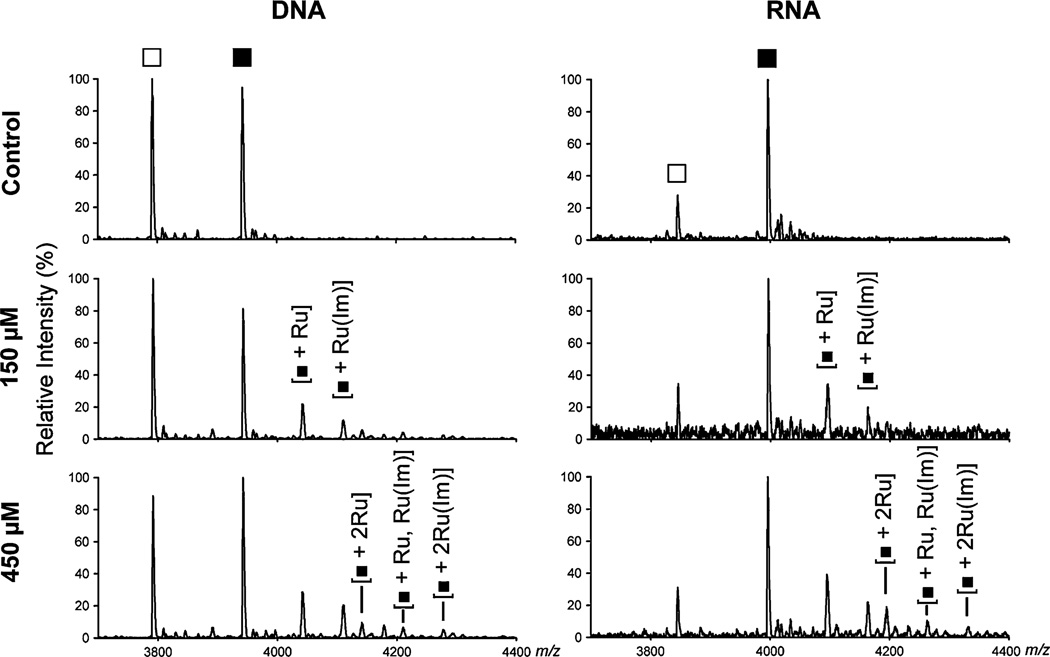
Figure 3 shows the MALDI-TOF mass spectra following oligonucleotide treatment with NAMI-A for 24 h at pH 6.0. Owing to the natural abundances of Ru isotopes, of which there are six isotopes that have more than 5% natural abundance (with 101Ru having the highest abundance at 31.6%) [43], the product peaks appear broad and of low intensity. In addition to the parent DNA or RNA oligonucleotide ion, the major +1 Ru products correspond to the mass of the oligonucleotide bound to either a single Ru or a single Ru bearing an Im ligand (assigned as [oligo + 1Ru] and [oligo + 1Ru(Im)]). Minor peaks for all three +2 Ru product species are also present ([oligo + 2Ru], [oligo + 1Ru, Ru(Im)], [oligo + 2Ru(Im)]), corresponding to binding at either both Gs or to one G and one U/T. As expected, the relative amount of these three different +2 Ru products increases with an increase in NAMI-A concentration from 150 to 450 µM. No chloride, aqua, hydroxo, or DMSO ligands are observed in these mass spectra, indicating loss of all but the least exchangeable ligands. This matches previous observations of the loss of labile ligands in both electrospray ionization and MALDI-TOF mass spectra for other metallodrugs bound to oligonucleotides [21, 39, 44]. Because these labile ligands are not observed in MALDI-TOF mass spectra, it is not possible with these data to identify which of these ligands were displaced during aquation or upon binding to the oligonucleotides.
Fig. 3.
Representative matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectra of the products of the incubation of NAMI-A with 20 µM DNA T6GGT5 or RNA U6GGU5 for 24 h at 37 °C and pH 6.0 in 100 mM NaNO3 and 2 mM Mg(NO3)2. Filled squares denote unmodified, full-length T6GGT5 and U6GGU5 and open squares denote singly depurinated oligonucleotides
In these mass spectra the overall amount of Ru adduct formation appears to be similar with RNA and DNA oligonucleotides. MALDI-TOF mass spectra are not quantitative owing to differing ionization efficiencies, but qualitative conclusions can be drawn from similar experiments performed with the same oligonucleotides, ionic conditions, and matrix protocols as were used for the data in Fig. 3. A more exact comparison between RNA and DNA products is not possible because the absolute ratio of product to starting material may be influenced by differential ionization, and is additionally difficult owing to the high amount of depurination observed for the DNA oligonucleotide strand.
Interestingly, the apparent increase in the amount of the two different +1 Ru product species with treatments with 150–450 µM NAMI-A is modest, and a comparison of reaction products at 6 and 24 h shows that the reaction has run to completion by 6 h (Figs. 3, 4, Fig. S2a). This is surprising as it appears that a significant amount of unreacted oligonucleotides remain despite the 7.5-fold to 22.5-fold excess of NAMI-A. Less reaction is observed for A5CCA6 when compared with T6GGT5 in a side-by-side study, suggesting that the preference for G previously reported in the literature is observed under these conditions as well (Fig. S2b) [21].
Fig. 4.
Representative MALDI-TOF mass spectra of the products of the incubation of 450 µM NAMI-A with 20 µM DNA T6GGT5 or RNA U6GGU5 at pH 6.0 and 7.4, with and without 2.5 mM ascorbate. Reactions were run for 6 h at 37 °C in 100 mM NaNO3 and 2 mM Mg(NO3)2. Filled squares denote the unmodified, full-length T6GGT5 and U6GGU5 strands and open squares denote singly depurinated oligonucleotides
Effects of pH and ascorbate on Ru–oligonucleotide formation following in vitro treatment with NAMI-A
Oligonucleotide reactivities at pH 6.0 (simulation of tumor tissue pH) and pH 7.4 (simulation of the pH of blood and healthy cells) were compared and only a slight increase of reactivity at lower pH was observed (Fig. 4). By contrast, the addition of 2.5 mM ascorbate to the reaction greatly enhanced the reactivity at both pH values, leading to a nearly complete loss of unreacted oligonucleotides, in particular at pH 6.0. Treatment with ascorbate also causes a change in the observed product species; product species bearing a DMSO ligand are now observed ([oligo + 1Ru(DMSO)] and [oligo + 1Ru(DMSO)(Im)]), especially at pH 6.0 (Fig. 4). The changes observed upon treatment with ascorbate are consistent with reduction of Ru(III) to Ru(II), leading to greater oligonucleotide binding and retention of the DMSO ligand.
Discussion
Ru accumulation following NAMI-A treatment in cellulo
Growth, viability, and accumulation of Ru in cells and cellular RNA were characterized following treatment of S. cerevisiae with 150 and 450 µM NAMI-A, concentrations that produce a moderate and severe reduction in culture density, respectively. On the basis of whole-cell ICP-MS measurements, growth for 6 h in these conditions resulted in estimated intracellular Ru concentrations of approximately 370 and 1,200 µM, indicating significant uptake of Ru from the medium. As a basis of comparison, in a phase I clinical trial the mean maximum Ru concentration observed in patient blood was approximately 1,800 µM (183 ± 17.3 µg/mL) for the maximum recommended dosage (300 mg/m2/day, on day 5, course 1) [3]. Both the 150 µM and the 450 µM NAMI-A treatments caused a decrease in the number of viable yeast cells, but this loss is modest in comparison with the more cytotoxic cisplatin, indicating that in the case of NAMI-A a significant portion of the decreased culture density is due to reversible growth inhibition. Indeed, transient cell cycle arrest in human tumor cell lines in the G2/M phase has been seen following treatment with NAMI-A [6]. The relatively high threshold of 100 µM NAMI-A that is required to clearly observe decreases in budding yeast culture density is consistent with previous observations. For example, a threshold of 100 µM NAMI-A was necessary to cause cell cycle effects in the KB tumor cell line, in which a “significant increase of cells in the S and G2/M phases” was observed 24 h after exposure [45]. The observed increases in yeast cell size also indicate a halting of the cell cycle. These observations taken together, it is likely that a portion of the reduced culture growth in NAMI-A-treated budding yeast is due to transient cell cycle arrest [38].
In both total RNA and whole yeast cells Ru accumulated in a dose-dependent manner, indicating that drug uptake is not saturated at 450 µM and that drug accumulation in RNA is proportional to cellular accumulation. This result is consistent with the lack of saturation of Ru accumulation observed in human tumor cell lines treated with NAMI-A at concentrations up to 600 µM [15]. In treated yeast, a significant amount of Ru accumulated in RNA: one Ru every 3,500 ± 1,500 and 1,100 ± 550 nucleotides for 150 and 450 µM NAMI-A, respectively. Assuming that yeast ribosomes, which are 80% of total cellular RNA in log-phase growth, accumulate the same amount of Ru as total RNA, there would be on average two or five Ru bound per ribosome [46]. RNA binding of this scale has the potential to cause significant downstream effects and may be involved in the mechanism of cell cycle arrest. The elucidation of the nonfunctional ribosomal RNA decay pathway in yeast is revealing connections between cellular responses to RNA damage and DNA damage [47]. In addition, in humans, a growing body of knowledge connects ribosomal stress (such as the disruption of ribosome biogenesis) to p53 activation and cell cycle arrest [48]. To our knowledge, this is the first study of the effect of NAMI-A on the genetically tractable S. cerevisiae, and also the first to report results of NAMI-A treatment regarding Ru interactions with cellular RNA. The results establish yeast as a model system for studying the effects of NAMI-A on RNA, observing a significant level of drug accumulation that has the potential to cause disruption of RNA function.
Ru–oligonucleotide adduct formation following NAMI-A treatment in vitro
When NAMI-A is dissolved in water it undergoes a series of aquation reactions resulting in loss of CI−, Im, and/or DMSO ligands [19, 49]. The aqua derivatives of the complex can also be in equilibrium with the hydroxo species, producing a wide range of possible active species in solution. The first aquation reaction was measured to take place with a half-life of 20 min at pH 7.4 and of 2 h at pH 6.0 (25 °C) [19]. MALDI-TOF mass spectra showed that both RNA and DNA oligonucleotides with a single GG site demonstrated a greater extent of reaction at pH 6.0 than at pH 7.4 when reacted for 6 h in a physiological salt background. The higher yield of products at pH 6.0 could be due to the greater exchangeability of Ru-aqua ligands as compared with hydroxo ligands, which would be expected to form more readily at higher pH, or due to formation of unreactive Ru–hydroxo polymers at higher pH values [8, 19].
The reduction of NAMI-A or its aqua derivatives by ascorbate or other biological reductants prior to treatment has been reported to enhance subsequent in vivo antimetastatic activity [8]. In vitro, NAMI-A reduction by ascorbate has been measured to take place in seconds at pH 7.4 (37 °C) and significantly increases the rate of subsequent aquation reactions [49, 50]. For the DNA and RNA oligonucleotides examined here, the presence of ascorbate caused a significant increase in the extent of reaction along with changes in the observed products in the MALDI-TOF mass spectra. Without ascorbate, the main observed products had a single bound Ru or Ru(Im). At pH 6.0, the ascorbate-treated samples showed mainly a single bound Ru or Ru(DMSO). At pH 7.4, both Ru(Im)- and Ru(DMSO)-bound products were present. The increased resiliency of the Ru–DMSO bond following reduction to Ru(II) is likely due to the greater extent of dπ–S backbonding that tends to occur in Ru(II)–DMSO complexes as compared with Ru(III) [51]. This increased retention of the DMSO ligand as compared with Im is consistent with the findings of previous studies by Bacac et al. [19] and Brindell et al. [50], both of which observed Im hydrolysis following reduction of NAMI-A, but to different extents. Our observations are consistent with ascorbate-induced reduction, and indicate that the greater yield of Ru adducts is due to the binding of reduced Ru(II) species to the oligonucleotides [51].
Conclusion
In summary, our investigations of the effect of NAMI-A treatment on RNA in cellulo and in vitro add to the growing understanding of Ru-based anticancer drugs. Firstly, we observed a nonlinear decrease in cell viability in S. cerevisiae with increasing NAMI-A concentration that corresponds with a moderate decrease in the number of viable cells, consistent with previous reports of high tolerance for this drug. We quantified accumulation of Ru within the cells and found that hydrolysis products of NAMI-A bind to intracellular RNA in numbers large enough to potentially result in an average of two to five Ru bound per ribosome. Secondly, to support our in cellulo studies, we conducted in vitro treatment of DNA and RNA oligonucleotides with NAMI-A both at physiological (7.4) and at lower (6.0) pH values, and in normal and reducing environments. The greatest extent of Ru–RNA adduct formation was observed in the acidic, reducing environment, which is analogous to that of tumor tissue [10–12], giving a strong indication that NAMI-A can become activated to react with nucleic acids in the environment of a tumor.
Supplementary Material
Acknowledgements
We thank Andy Ungerer for assistance with the ICP-MS experiments, the W. M. Keck Collaboratory for Plasma Spectrometry at Oregon State University, J. David Sumega for synthesizing and characterizing NAMI-A, and Laurie Graham for assistance with protocols and imaging. The Stevens laboratory at the University of Oregon is gratefully acknowledged for the use of a Carl Zeiss Axioplan 2 fluorescence microscope and for a gift of the BY4741 strain. This work was supported by a Willamette University Atkinson Grant (K.L.M.H.), the NIH (GM058096, V.J.D.), and the University of Oregon (V.J.D.).
Abbreviations
- cfu
Colony-forming units
- DMSO
Dimethy sulfoxide
- ICP-MS
Inductively coupled plasma mass spectrometry
- Im
Imidazole
- MALDI
Matrix-assisted laser desorption/ionization
- OD600
Optical density at 600 nm
- NAMI
[Na][trans-RuIIICl4(DMSO)(Im)]
- NAMI-A
[ImH][trans-RuIIICl4(DMSO)(Im)]
- THAP
2′,4′,6′-Trihydroxyacetophenone
- TOF
Time-of-flight
- YEPD
Yeast extract–peptone–glucose
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00775-011-0806-7) contains supplementary material, which is available to authorized users.
Contributor Information
Alethia A. Hostetter, Department of Chemistry, University of Oregon, 1253 University of Oregon, Eugene, OR 97403, USA
Michelle L. Miranda, Department of Chemistry, University of Oregon, 1253 University of Oregon, Eugene, OR 97403, USA Department of Chemistry, Willamette University, 900 State Street, Salem, OR 97301, USA.
Victoria J. DeRose, Department of Chemistry, University of Oregon, 1253 University of Oregon, Eugene, OR 97403, USA, derose@uoregon.edu
Karen L. McFarlane Holman, Department of Chemistry, Willamette University, 900 State Street, Salem, OR 97301, USA, kholman@willamette.edu.
References
- 1.Hannon MJ. Pure Appl Chem. 2007;79:2243–2261. [Google Scholar]
- 2.Dyson PJ, Sava G. Dalton Trans. 2006:1929–1933. doi: 10.1039/b601840h. [DOI] [PubMed] [Google Scholar]
- 3.Rademaker-Lakhai JM, van den Bongard D, Pluim D, Beijnen JH, Schellens JHM. Clin Cancer Res. 2004;10:3717–3727. doi: 10.1158/1078-0432.CCR-03-0746. [DOI] [PubMed] [Google Scholar]
- 4.Levina A, Mitra A, Lay PA. Metallomics. 2009;1:458–470. doi: 10.1039/b904071d. [DOI] [PubMed] [Google Scholar]
- 5.Kostova I. Curr Med Chem. 2006;13:1085–1107. doi: 10.2174/092986706776360941. [DOI] [PubMed] [Google Scholar]
- 6.Bergamo A, Gagliardi R, Scarcia V, Furlani A, Alessio E, Mestroni G, Sava G. J Pharmacol Exp Ther. 1999;289:559–564. [PubMed] [Google Scholar]
- 7.Sava G, Capozzi I, Bergamo A, Gagliardi R, Cocchietto M, Masiero L, Onisto M, Alessio E, Mestroni G, Garbisa S. Int J Cancer. 1996;68:60–66. doi: 10.1002/(SICI)1097-0215(19960927)68:1<60::AID-IJC12>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 8.Sava G, Bergamo A, Zorzet S, Gava B, Casarsa C, Cocchietto M, Furlani A, Scarcia V, Serli B, Iengo E, Alessio E, Mestroni G. Eur J Cancer. 2002;38:427–435. doi: 10.1016/s0959-8049(01)00389-6. [DOI] [PubMed] [Google Scholar]
- 9.Ravera M, Baracco S, Cassino C, Zanello P, Osella D. Dalton Trans. 2004:2347–2351. doi: 10.1039/b400952E. [DOI] [PubMed] [Google Scholar]
- 10.Gullino PM. Adv Exp Biol Med. 1976;75:521–536. doi: 10.1007/978-1-4684-3273-2_62. [DOI] [PubMed] [Google Scholar]
- 11.Richard DE, Berra E, Pouyssegur J. Biochem Biophys Res Commun. 1999;266:718–722. doi: 10.1006/bbrc.1999.1889. [DOI] [PubMed] [Google Scholar]
- 12.Gerweck LE, Vijayappa S, Kozin S. Mol Cancer Ther. 2006;5:1275–1279. doi: 10.1158/1535-7163.MCT-06-0024. [DOI] [PubMed] [Google Scholar]
- 13.Brabec V, Novakova O. Drug Resist Update. 2006;9:111–122. doi: 10.1016/j.drup.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Pizarro AM, Sadler PJ. Biochimie. 2009;91:1198–1211. doi: 10.1016/j.biochi.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pluim D, van Waardenburg RCAM, Beijnen JH, Schellens JHM. Cancer Chemother Pharmacol. 2004;54:71–78. doi: 10.1007/s00280-004-0773-6. [DOI] [PubMed] [Google Scholar]
- 16.Gallori E, Vettori C, Alessio E, Vilchez FG, Vilaplana R, Orioli P, Casini A, Messori L. Arch Biochem Biophys. 2000;376:156–162. doi: 10.1006/abbi.1999.1654. [DOI] [PubMed] [Google Scholar]
- 17.Messori L, Casini A, Vullo D, Haroutiunian SG, Dalian EB, Orioli P. Inorg Chim Acta. 2000;303:283–286. [Google Scholar]
- 18.Jung Y, Lippard SJ. Chem Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 19.Bacac M, Hotze ACG, van der Schilden K, Haasnoot JG, Pacor S, Alessio E, Sava G, Reedijk J. J Inorg Biochem. 2004;98:402–412. doi: 10.1016/j.jinorgbio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Schluga P, Hartinger CG, Egger A, Reisner E, Galanski M, Jakupec MA, Keppler BK. Dalton Trans. 2006:1796–1802. doi: 10.1039/b511792e. [DOI] [PubMed] [Google Scholar]
- 21.Groessl M, Tsybin YO, Hartinger CG, Keppler BK, Dyson PJ. J Biol Inorg Chem. 2010;15:677–688. doi: 10.1007/s00775-010-0635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malina J, Novakova O, Keppler BK, Alessio E, Brabec V. J Biol Inorg Chem. 2001;6:435–445. doi: 10.1007/s007750100223. [DOI] [PubMed] [Google Scholar]
- 23.Mattick JS. Nat Rev Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 24.Kong QM, Lin CLG. Cell Mol Life Sci. 2010;67:1817–1829. doi: 10.1007/s00018-010-0277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olmo N, Turnay J, González de Buitrago G, López de Silanes I, Gavilanes JG, Lizarbe MA. Eur J Biochem. 2001;268:2113–2123. doi: 10.1046/j.1432-1327.2001.02086.x. [DOI] [PubMed] [Google Scholar]
- 26.Jetzt AE, Cheng JS, Tumer NE, Cohick WS. Int J Biochem Cell B. 2009;41:2503–2510. doi: 10.1016/j.biocel.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mroczek S, Kufel J. Nucleic Acids Res. 2008;36:2874–2888. doi: 10.1093/nar/gkm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mei Y, Yong J, Liu H, Shi Y, Meinkoth J, Dreyfuss G, Yang X. Mol Cell. 2010;37:668–678. doi: 10.1016/j.molcel.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akaboshi M, Kawai K, Maki H, Akuta K, Ujeno Y, Miyahara T. Jpn J Cancer Res. 1992;83:522–526. doi: 10.1111/j.1349-7006.1992.tb01959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmittgen TD, Ju J-F, Danenberg KD, Danenberg PV. Int J Oncol. 2003;23:785–789. [PubMed] [Google Scholar]
- 31.Heminger KA, Hartson SD, Rogers J, Matts RL. Arch Biochem Biophys. 1997;344:200–207. doi: 10.1006/abbi.1997.0198. [DOI] [PubMed] [Google Scholar]
- 32.Menacho-Marquez M, Murguia JR. Clin Trans Oncol. 2007;9:221–228. doi: 10.1007/s12094-007-0043-2. [DOI] [PubMed] [Google Scholar]
- 33.McCarthy JEG. Microbiol Mol Biol Rev. 1998;62:1492–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phizicky EM, Hopper AK. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barr MM. Physiol Genomics. 2003;13:15–24. doi: 10.1152/physiolgenomics.00075.2002. [DOI] [PubMed] [Google Scholar]
- 36.Alessio E, Balducci G, Calligaris M, Costa G, Attia WM, Mestroni G. Inorg Chem. 1991;30:609–618. [Google Scholar]
- 37.Witkowsky L. Senior thesis. Willamette University; 2006. [Google Scholar]
- 38.Tyson CB, Lord PG, Wheals AE. J Bacteriol. 1979;138:92–98. doi: 10.1128/jb.138.1.92-98.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chapman EG, DeRose VJ. J Am Chem Soc. 2010;132:1946–1952. doi: 10.1021/ja908419j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ragas JA, Simmons TA, Limbach PA. Analyst. 2000;125:575–581. doi: 10.1039/a909709k. [DOI] [PubMed] [Google Scholar]
- 41.Sauer S. J Biochem Biophys Methods. 2007;70:311–318. doi: 10.1016/j.jbbm.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 42.Christian NP, Reilly JP, Mokler VR, Wincott FE, Ellington AD. J Am Soc Mass Spectrom. 2001;12:744–753. doi: 10.1016/S1044-0305(01)00228-8. [DOI] [PubMed] [Google Scholar]
- 43.National Institute of Standards and Technology. [Accessed 5 Jan 2011];Physical measurement laboratory: basic atomic spectroscopic data. 2010 http://physics.nist.gov/PhysRefData/Handbook/Tables/rutheniumtable1.htm.
- 44.Ang WH, Daldini E, Scolaro C, Scopelliti R, Juillerat-Jeannerat L, Dyson PJ. Inorg Chem. 2006;45:9006–9013. doi: 10.1021/ic061008y. [DOI] [PubMed] [Google Scholar]
- 45.Bergamo A, Messori L, Piccoli F, Cocchietto M, Sava G. Invest New Drugs. 2003;21:401–411. doi: 10.1023/a:1026243000320. [DOI] [PubMed] [Google Scholar]
- 46.Warner JR. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 47.Hinnebusch AG. Genes Dev. 2009;23:891–895. doi: 10.1101/gad.1800509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horn HF, Vousden KH. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- 49.Brindell M, Stawoska I, Supel J, Skoczowski A, Stochel G, van Eldik R. J Biol Inorg Chem. 2008;13:909–918. doi: 10.1007/s00775-008-0378-3. [DOI] [PubMed] [Google Scholar]
- 50.Brindell M, Piotrowska D, Shoukry AA, Stochel G, van Eldik R. J Biol Inorg Chem. 2007;12:809–818. doi: 10.1007/s00775-007-0234-x. [DOI] [PubMed] [Google Scholar]
- 51.Calligaris M, Carugo O. Coord Chem Rev. 1996;153:83–154. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.