Abstract
We investigated cross-talk between the membrane-associated, myosin II-regulatory protein supervillin and the actin-regulatory small GTPases Rac1, RhoA, and Cdc42. Supervillin knockdown reduced Rac1-GTP loading, but not the GTP loading of RhoA or Cdc42, in HeLa cells with normal levels of the Rac1-activating protein Trio. No reduction in Rac1-GTP loading was observed when supervillin levels were reduced in Trio-depleted cells. Conversely, overexpression of supervillin isoform 1 (SV1) or, especially, isoform 4 (SV4) increased Rac1 activation. Inhibition of the Trio-mediated Rac1 guanine nucleotide exchange (GEF) activity with ITX3 partially blocked the SV4-mediated increase in Rac1-GTP. Both SV4 and SV1 co-localized with Trio at or near the plasma membrane in ruffles and cell surface projections. Two sequences within supervillin bound directly to Trio spectrin repeats 4–7: SV1-171, which contains N-terminal residues found in both SV1 and SV4 and the SV4-specific differentially spliced coding exons 3, 4, and 5 within SV4 (SV4-E345; SV4 amino acids 276 – 669). In addition, SV4-E345 interacted with the homologous sequence in rat kalirin (repeats 4–7, amino acids 531 – 1101). Overexpressed SV1-174 and SV4-E345 affected Rac1-GTP loading, but only in cells with endogenous levels of Trio. Trio residues 771 – 1057, which contain both supervillin-interaction sites, exerted a dominant-negative effect on cell spreading. Supervillin and Trio knockdowns, separately or together, inhibited cell spreading, suggesting that supervillin regulates the Rac1 guanine nucleotide exchange activity of Trio, and potentially also kalirin, during cell spreading and lamellipodia extension.
Keywords: Lamellipodia, cell spreading, spectrin repeats, kalirin, RhoA, Cdc42
INTRODUCTION
Cell translocation requires activation of actin-based cell surface extension and adhesion to the extracellular matrix, processes that are regulated by small GTPases in the Rho family (reviewed in (Sit and Manser, 2011; Blanchoin et al., 2014; Lawson and Burridge, 2014)). Rac1, RhoA, and Cdc42 are the best characterized of the cytoskeleton-reorganizing GTPases (Heasman and Ridley, 2008). Extracellular stimuli increase the activities of guanine nucleotide exchange factors (GEFs), which activate target GTPases by facilitating the exchange of GTP for GDP at their nucleotide binding sites (Rossman et al., 2005). Activation times are limited by the rate of GTP hydrolysis, which is regulated by GTPase activating proteins (GAPs) (Scheffzek et al., 1998). Downstream effects are cell type-specific but can be generalized as increased formation of cell processes by GTP-loaded Rac1 and Cdc42, increased cell polarity promoted by Cdc42, and RhoA-mediated increases in stress fibers and myosin II-based contractility (Jaffe and Hall, 2005).
During lamellipodial extension and associated cell migration, the cytoskeletal machinery is regulated by cyclical cross-talk between Rac1 and RhoA signalling pathways (Wertheimer et al., 2012; Miller et al., 2013; Nayak et al., 2013; Lawson and Burridge, 2014). In murine fibroblasts, RhoA is activated within 2 μm of the cell edge (Machacek et al., 2009), where RhoA-GTP promotes the assembly of parallel actin filaments by the formin mDia1 (Li and Higgs, 2003); mDia1 in turn activates Rac1 (Tsuji et al., 2002). Rac1 activation reaches a peak at ~1.8 μm behind the advancing edge ~40-s after the peak of RhoA activation (Li and Higgs, 2003) and leads to the assembly of Arp2/3-nucleated, branched actin filaments (Ridley et al., 2003). Other avenues of RhoA/Rac1 cross-talk involve direct and indirect regulation of each others GEFs and GAPs (Guilluy et al., 2011; Lawson and Burridge, 2014). In general, Rac1 activation and suppression of RhoA facilitate the formation of early cell-substrate adhesions, whereas RhoA activation and Rac1 inhibition promote adhesion maturation and the formation of myosin II-associated contractile actin bundles (reviewed in Lawson and Burridge, 2014).
One category of GEFs implicated in promoting cell spreading (van Rijssel and van Buul, 2012; Miller et al., 2013; Schmidt and Debant, 2014) includes the nearly ubiquitous, triple-function protein Trio (Debant et al., 1996) and kalirin, a related protein most concentrated in brain (Alam et al., 1997). Trio and kalirin are structurally similar GTPase-activating proteins; their longest isoforms contain a Sec14 lipid-binding domain, nine spectrin repeats, two GEF domains (GEFD1 and GEFD2), an immunoglobulin (Ig) domain, and a C-terminal serine kinase domain (van Rijssel and van Buul, 2012; Miller et al., 2013). The Trio GEFD1 activates Rac1 and RhoG, whereas Trio GEFD2 activates RhoA (Debant et al., 1996; Bellanger et al., 1998; Blangy et al., 2000; Medley et al., 2000; Chhatriwala et al., 2007). Best characterized in the brain, kalirin is required for normal formation of dendritic spines and neuronal synapses; kalirin variants are associated with schizophrenia, Alzheimer’s disease, and other disorders (Remmers et al., 2014). Trio/UNC-73 also regulates neuronal development (recently reviewed by (van Rijssel and van Buul, 2012; Miller et al., 2013)). In addition, Trio functions in cell growth, cytokinesis, spreading, migration, and matrix invasion (Bellanger et al., 1998; Bellanger et al., 2000; Bellanger et al., 2003; Debreceni et al., 2004; Skowronek et al., 2004; Salhia et al., 2008; Fortin et al., 2012; van Rijssel et al., 2012a; Vaqué et al., 2013; Moshfegh et al., 2014) and is linked to poor outcome in bladder and breast cancer (Zheng et al., 2004; Lane et al., 2008).
Supervillin is a myosin II- and actin-binding peripheral membrane protein that regulates cell-substrate adhesion, cell polarization, and the rates of cell spreading and translocation (Pestonjamasp et al., 1997; Chen et al., 2003; Takizawa et al., 2006; Takizawa et al., 2007; Bhuwania et al., 2012; Edelstein et al., 2012). A hub protein with > 80 interaction partners, supervillin also functions during cytokinetic furrowing, invadopodia-mediated degradation of extracellular matrix, survival signalling, and rapid membrane recycling of integrins in a Rab5/Arf6-associated pathway (Kim et al., 2001; Sampson et al., 2001; Nebl et al., 2002; Ting et al., 2002; Gangopadhyay et al., 2004; Jin et al., 2004; Crowley et al., 2009; Fang et al., 2010; Smith et al., 2010; Fang and Luna, 2013; Hasegawa et al., 2013; Smith et al., 2013). Arf6 is a small GTPase that cross-talks with Rac1 through interactions with both Rac1 GEFs and GAPs (Radhakrishna et al., 1999; Zhang et al., 1999; Santy and Casanova, 2001; Koo et al., 2007; Myers and Casanova, 2008; Kawaguchi et al., 2014). Supervillin-mediated regulation of cell spreading, cytokinesis, and cell polarization is effected through increases in myosin II contractility (Takizawa et al., 2007; Bhuwania et al., 2012; Smith et al., 2013), which also may underlie supervillin’s effects on matrix degradation (Alexander et al., 2008; Crowley et al., 2009). However, other cytoskeletal effects are not easily explained by this mechanism. For instance, supervillin overexpression decreases the extent of lamellipodia and enhances the appearance of narrow cell surface extensions (Wulfkuhle et al., 1999; Crowley et al., 2009).
We show here that supervillin cross-talks with the Rac1 signaling pathway through interactions with Trio. Supervillin isoform 4 (SV4) interacts with spectrin repeats 4–7 from both Trio and kalirin, activates Rac1 GTP-loading by Trio, and coordinates with Trio to regulate the initial spreading of HeLa cells on fibronectin.
RESULTS
Supervillin increases Rac1 activation
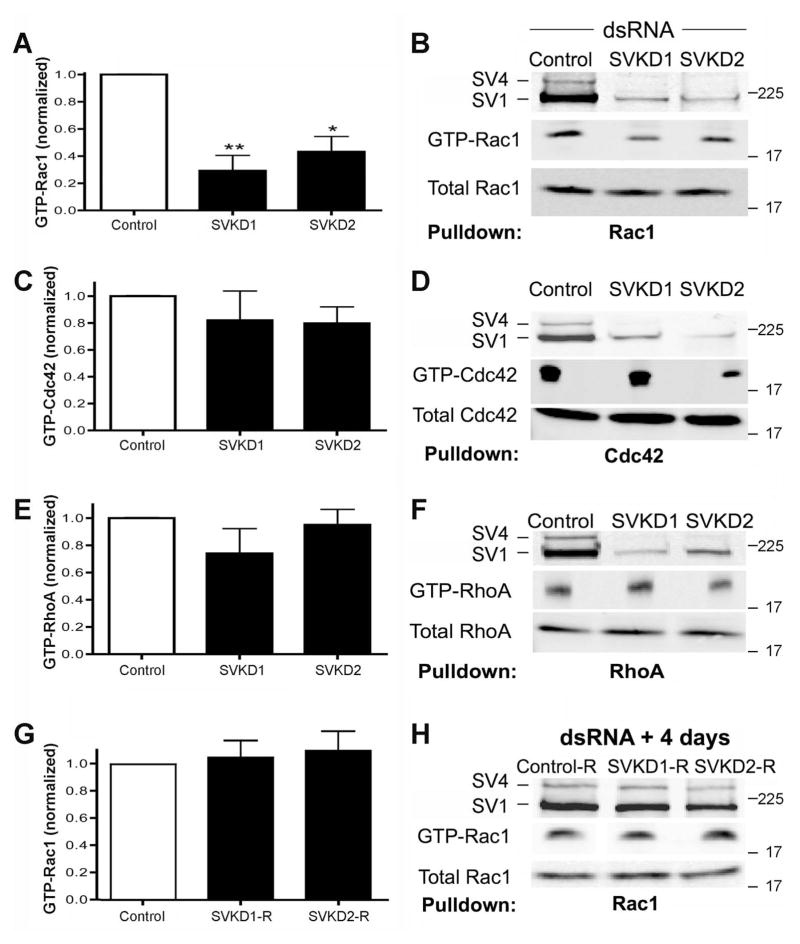
We screened for functional cross-talk between supervillin and the small GTPases Rac1, Cdc42 and RhoA in HeLa cells (Figure 1). Unlike many other cell lines, HeLa cells survive after supervillin knockdown because they contain a supervillin-independent pathway for maintaining low levels of p53 (Smith et al., 2010; Fang and Luna, 2013; Smith et al., 2013). As described previously (Fang and Luna, 2013), each of two double-stranded RNAs (dsRNAs) that target both HeLa cell supervillin splice forms (SV1, SV4) reduced the level of each isoform by ≥90% (Figure 1B, D, F). The knockdown of supervillin was accompanied by a significant (~3-fold) reduction in the amount of GTP-loaded (activated) Rac1 (Figure 1A), as assessed in pulldown assays with GST-tagged Pak1-binding domain (PBD) (Ren and Schwartz, 2000; Benard and Bokoch, 2002). By contrast, no significant changes were observed in GTP loading of Cdc42 (Figure 1C–D) or RhoA (Figure 1E–F) in similar experiments. When supervillin expression was allowed to recover to initial levels, GTP-Rac1 levels returned to control values (Figure 1G–H).
Figure 1. Supervillin knockdown decreases GTP loading of Rac1, but not Cdc42 or RhoA.
HeLa CCL-2 cervical adenocarcinoma cells were transfected with control or supervillin-specific dsRNAs for 2 days, and lysates were assayed for the relative amounts of GTP-loaded (activated) (A, B) Rac1, (C, D) Cdc42, and (E, F) RhoA. (G, H) HeLa cells that had been treated with dsRNAs to control or supervillin dsRNAs were allowed to recover expression of supervillin isoforms 1 (SV1) and 4 (SV4) before assay for GTP-Rac1 levels. (A, C, E, G) Means ± s.e.m., N = 4. *, P < 0.05; ** P < 0.01, as assessed using a one-way analysis of variance assay (ANOVA). (B, D, F, H) Representative immunoblots of active (GTP-bound) and total Rac1, Cdc42, and RhoA.
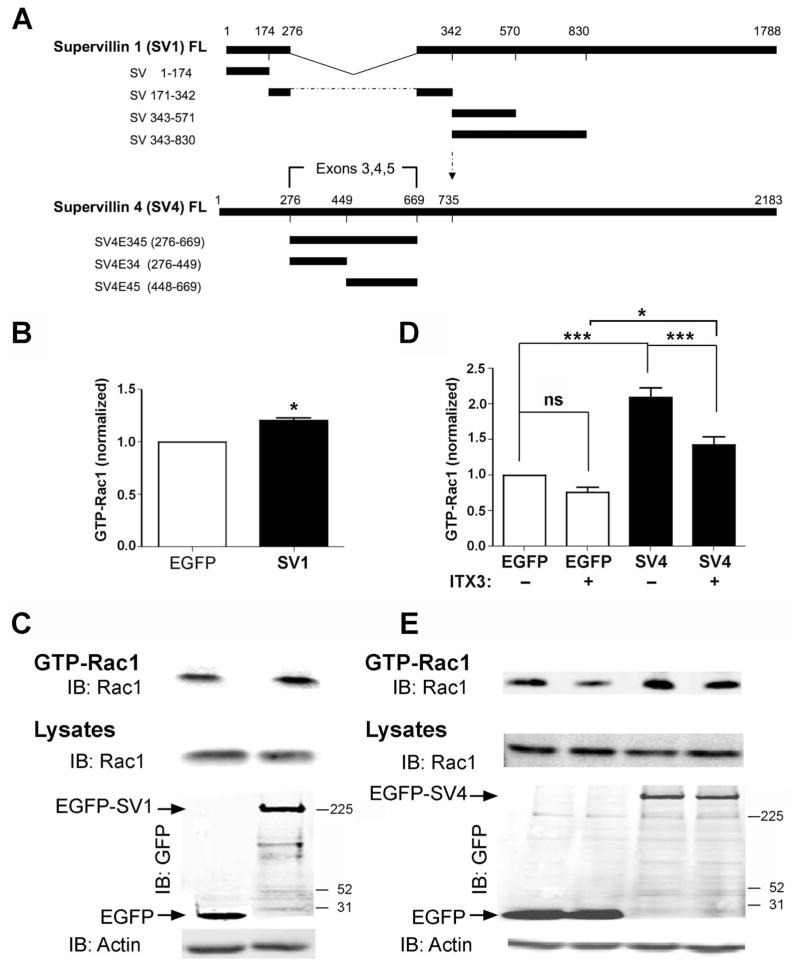
To determine which major supervillin splice-forms are involved, we analyzed GTP-Rac1 levels in HeLa cells expressing EGFP-tagged human SV1 or SV4 (Figure 2). As described previously (Pope et al., 1998; Fang and Luna, 2013), these proteins are products from a single gene, with SV4 containing an additional 393 amino acids encoded by the differentially spliced coding exons 3, 4, and 5 (Oh et al., 2003) (Figure 2A). Overexpression of EGFP-SV1 resulted in a small, but significant, increase in the amount of GTP-Rac1 (~25%), relative to cells expressing EGFP alone (Figure 2B, 2C). By contrast, overexpression of EGFP-SV4 resulted in a larger, ≥ 2-fold increase in the relative amounts of GTP-Rac1 (Figure 2D, 2E). These results suggested that, while the SV1 isoform had an effect on Rac1 activation, the sequence encoded by the SV4-specific exons (SV4-E345; SV4 amino acids 276 – 669) was quantitatively more important for the GTP loading of Rac1.
Figure 2. GTP loading (activation) of Rac1 increases after overexpression of supervillin, especially isoform 4 (SV4).
(A) Schematics of the differentially spliced full-length (FL) SV1 and SV4 proteins and protein fragments. SV4 (2183 amino acids, ~245 kDa) contains an additional 393 amino acids (E345, residues 276–669), as compared with SV1 (1788 amino acids, ~201 kDa) (Fang et al., 2010). (B) Quantification of values normalized to those for cells expressing EGFP alone and (C) representative immunoblots of GTP-Rac1 from HeLa cells expressing EGFP or EGFP-SV1 for 2 days. Means ± SEM; N = 3; *, P = 0.019 (two-tailed Student t-test). (D) Quantification of normalized values and (E) representative immunoblots of GTP-Rac1 from HeLa cells expressing EGFP or EGFP-SV4. Transfected cells were incubated with either the carrier (DMSO) or 50 μM ITX3 for 1 h before lysis and GST-PBD pulldowns. Levels in each experiment were normalized to those for DMSO-treated cells expressing EGFP. Means ± SEM; N = 5; ns, not significant; P: * < 0.05; *** < 0.001 (ANOVA).
Because a sequence within the N-terminus of the Rac1-GEF Trio emerged as a candidate interactor for SV4-E345 in a yeast two-hybrid screen (Spinazzola et al., 2015), we carried out GTP-Rac1 loading assays in the presence of ITX3 (Figure 2D, 2E). ITX3 is a specific inhibitor of the membrane ruffling and Rac1 GEF activity attributable to the Trio N-terminal GEF-D1 domain (Bouquier et al., 2009). Consistent with cross-talk between SV4 and Trio, ITX3 treatment partially inhibited the SV4-induced increase in GTP loading of Rac1 (Figure 2D, 2E), suggesting a potential involvement of Trio in supervillin-mediated Rac1 activation.
Supervillin interacts with the Rac1-GEF Trio
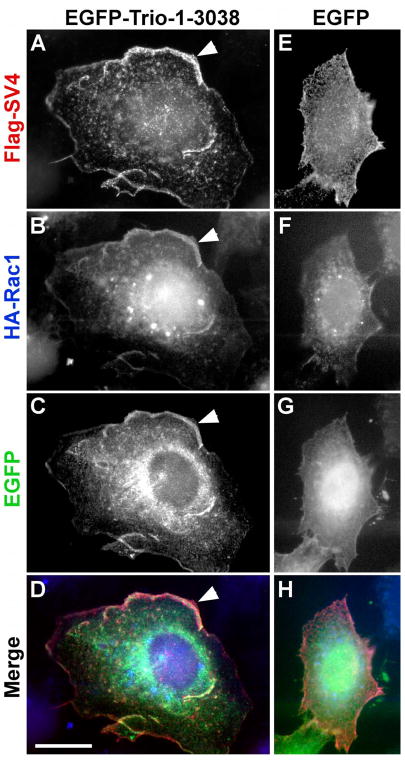
To test for interactions of supervillin with Trio and Rac1, we first looked for co-localization by immunofluorescence microscopy (Figures 3, 4). Antibodies against the endogenous proteins were insufficient for immunofluorescence, requiring co-expression of tagged proteins. In wide-field images of HeLa cells that co-expressed tagged SV4, Rac1 and full-length Trio (Figure 3), all three proteins co-localized at areas of ruffling membranes (Figure 3A–D). As originally described (Seipel et al., 1999; Debreceni et al., 2004; van Rijssel et al., 2012a), exogenous expression of EGFP-Trio amplified the formation of these lamellipodia (Figure 3A–D, arrowheads) although limited areas of overlap near the membrane also were observed in the less well-spread cells that overexpressed SV4 and Rac1 with the EGFP tag alone (Figure 3E–H).
Figure 3. Co-localization of Trio, SV4, and Rac1 at lamellipodia.
HeLa cells were transfected with Flag-tagged SV4 (A, E), HA-Rac1 (B, F) and either EGFP-Trio-1-3038 (C) or EGFP (G). Flag-SV4 is red, HA-Rac1 is blue, and EGFP-Trio and EGFP appear in green in the merged images (D, H). Cells were fixed before detergent permeabilization. Cells expressing EGFP-Trio-1-3038 are larger, with extensive lamellipodia containing all three proteins (E-H, arrowheads). Bar, 20 μm.
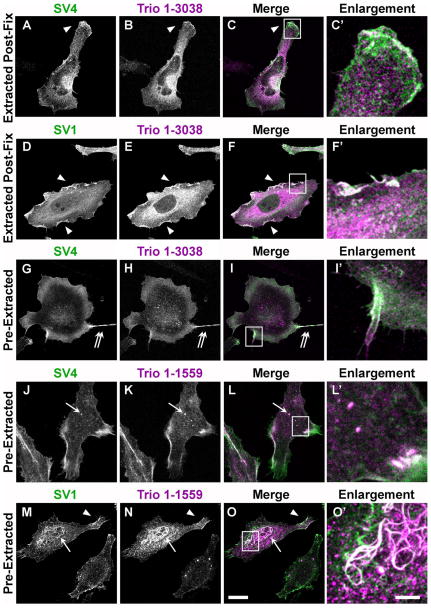
Figure 4. Supervillin co-localization with Trio in HeLa cells fixed before and after detergent permeabilization.
HeLa cells co-expressing Flag-tagged SV4 (A–C, C′, G–L, L′) or SV1 (D–F, F′, M–O, O′) with HA-tagged Trio (Trio 1-3038) (A–I′) or the Trio N-terminus (Trio 1-1559) (J–O′) were fixed with 4% paraformaldehyde. (A–F′) Cells fixed before detergent permeabilization; (G–O′) cells permeabilized prior to fixation, as described in Methods. Supervillin isoforms appear green, Trio signals appear magenta, and areas of co-localization appear white in merged images (C, F, I, L, O). Areas of signal overlap include membrane ruffles (arrowheads), cell surface extensions (G–I′, double arrows), cytoplasmic punctae (J–L′, arrow) and supervillin-induced nuclear bundles (J–L′, arrow); bar, 20 μm. Outlined areas are shown as five-fold enlargements (C′, F′, I′, L′, O′); bar, 5 μm.
Due to the potential for artifactual localizations caused by crosslinking of EGFP to F-actin (Schmitz and Bereiter-Hahn, 2001), which binds directly to supervillin (Chen et al., 2003), we used confocal microscopy and HA-tagged Trio constructs to determine the co-localization of Flag-tagged SV4 and SV1 with Trio or the Trio N-terminus (Figure 4). As described previously (Bellanger et al., 1998; Seipel et al., 1999; van Rijssel et al., 2012a), the Trio N-terminus is sufficient for the Trio-mediated formation of lamellipodia and is more readily transfectable due to its smaller size. Signals from both SV4 and SV1 overlapped with Trio in peripheral membrane ruffles, especially in HeLa cells fixed before permeabilization with detergent (Figure 4A – 4F, arrowheads). A similar SV1 localization was seen previously with F-actin and Arf6 pathway components at membrane ruffles and rapidly recycling endosomes (Fang et al., 2010). As observed for SV1 (Wulfkuhle et al., 1999), much of the signals associated with SV4, Trio, and the Trio N-terminus (Trio1-1559) remained associated with membranes and the cytoskeleton in cells extracted with Triton X-100 prior to fixation (Figure 4G – 4O′). Prominent areas of co-localization in these pre-extracted cells included peripheral membranes (Figure 4G – 4I′, double arrows), cytoplasmic punctae reminiscent of recycling endosomes (Fang et al., 2010) (Figure 4J – 4L′, arrows), filamentous nuclear bundles induced by SV1 overexpression (Wulfkuhle et al., 1999) (Figure 4M – 4O′, arrows), and lamellipodial ruffles (Figure 4M – 4O′, arrowheads). These results suggest lamellipodia, plasma membranes, and recycling endosomes as sites for functional cross-talk between supervillin and Trio and support the possibility of an interaction between these proteins.
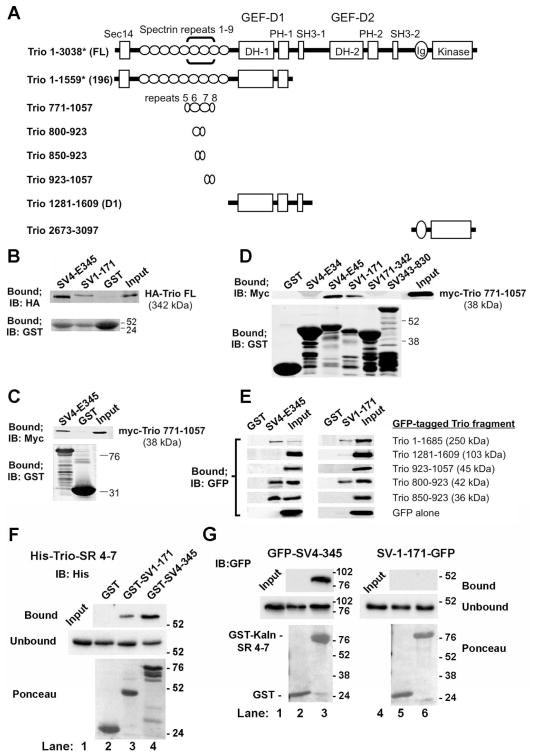
We used GST-pulldown assays to confirm interactions between supervillin sequences and Trio protein fragments (Figure 5). The domain organization and contents of each Trio construct are shown in Figure 5A. A single prey that started with amino acid 751 in Trio (NP_009049) emerged as one of 59 candidate interactors in a yeast two-hybrid screen with SV4-E345 as bait. Although no 3′ DNA sequence was obtainable for this clone, prey protein fragments averaged ~270 residues, suggesting a binding site for SV4-E345 within Trio 751-1021. This sequence encodes spectrin repeats 6 and 7 within Trio and flanking sequences (Figure 5A, brackets). Consistent with an interaction, full-length HA-Trio (1-3038) co-sedimented with GST-SV4-E345, but not with GST alone (Figure 5B). Surprisingly, HA-Trio also co-sedimented with beads containing the myosin II-binding region of supervillin GST-SV1-171 (Figure 5B). In addition, myc-Trio 771-1057 co-sedimented with GST-SV4-E345 and GST-SV1-171, but not with GST alone (Figure 5C). Other GST-tagged supervillin protein fragments (SV1-171-342, SV1-343-830) were not pulled down with myc-Trio 771-1057 (Figure 5D). These results suggested the presence of two Trio-interacting sites, one within SV4 amino acids 448 – 669 (SV4-E45) and one within the N-terminal 171 residues found in both SV1 and SV4 (SV1-171). We then generated GFP-tagged truncated Trio proteins containing deletions around and within spectrin repeats 6–7. We found that GST-SV1-171 did not interact with Trio residues 850–923 but did pull down Trio residues 800–923, indicating that Trio residues 800–850 are required for the interaction with SV1-171. By contrast, the SV4-E345 interaction site was located within Trio residues 850 – 923 (Figure 5E), indicating the presence of distinguishable interaction sites within or near Trio spectrin repeats 6–7. In direct binding experiments, purified recombinant His-tagged Trio spectrin repeats 4–7 (amino acids 565–1011) co-sedimented with GST-SV1-171 and GST-SV4-E345, but not with GST alone (Figure 5F). Interestingly, an interaction with GFP-SV4-E345, but not GFP-SV1-171, also was observed using GST-tagged spectrin repeats 4–7 from the Trio-like protein kalirin (Figure 5G). Thus, SV4-E345 and SV1-171 each binds directly to sequences containing Trio spectrin repeats 6–7 while only SV4-E345 interacts with the similar region in kalirin, presumably at a conserved site in spectrin repeats 6–7.
Figure 5. The Trio and kalirin N-termini both interact with the SV4-specific sequence, SV4-E345; Trio contains a second binding site for N-terminal residues common to both SV1 and SV4.
(A) Domain structure of Trio constructs used in this study. Full-length Trio (Trio FL, ~342 kDa) contains a Sec14 homology domain (aa 8-145), followed by nine spectrin repeats (aa 161–1187), tandem DH-PH-SH3 domain combinations with GEF activity for Rac1 (aa 1234–1533) and RhoA (aa 1911–2213), an immunoglobulin (Ig)-like domain (aa 2626–2717) and a catalytic kinase domain (aa 2737–2979) (Debant et al., 1996; Medley et al., 2000). Brackets in the first line indicate the location of the Trio prey sequence recovered in a yeast two-hybrid screen, using SV4-E345 (SV4 residues 276 – 669) as bait. *The numbering for Trio amino acids is that used in the literature; Trio 771-1057, Trio 1281-1609, Trio 2673-3097 and smaller fragments correspond to the current NCBI Reference Sequence (NP_990949.2) (Liu et al., 2012), whereas the larger constructs begin at Met-60 in this sequence. (B) Immunoblots of pull down assays with HA-tagged Trio FL and GST-tagged SV4-E345 and SV1-171. (C, D) Immunoblots showing the interactions between myc-Trio 771-1057 and GST-tagged sequences unique to SV4 (SV4-E345; SV4-E34, amino acids 276–449; SV4-E45, amino acids 448 – 669) and GST-tagged SV1 sequences (SV1-174, SV171-342, SV343-830). (E) Immunoblots of GFP-tagged Trio N-terminal protein fragments from (A) and GST-tagged SV4-E345 and SV1-174, the N-terminus of both SV1 and SV4. Immunoblots showing the corresponding amounts of column-bound GST proteins are shown in Supplemental Figure S1A. (F) Immunoblots of purified recombinant His-tagged Trio spectrin repeats 4–7 (His-Trio-SR 4-7; amino acids 565–1011) show direct binding to GST-SV1-171 and GST-SV4-345, but not to GST alone. (G) Immunoblots of pull down assays with GFP-tagged SV4-E345 or SV1-171 and GST-tagged rat kalirin spectrin repeats 4–7 (GST-Kaln-SR 4-7; amino acids 517 – 976) or GST alone. Locations of molecular mass markers, in kDa, are indicated to the right in panels in B–G.
Trio is required for supervillin-mediated increases in GTP loading of Rac1
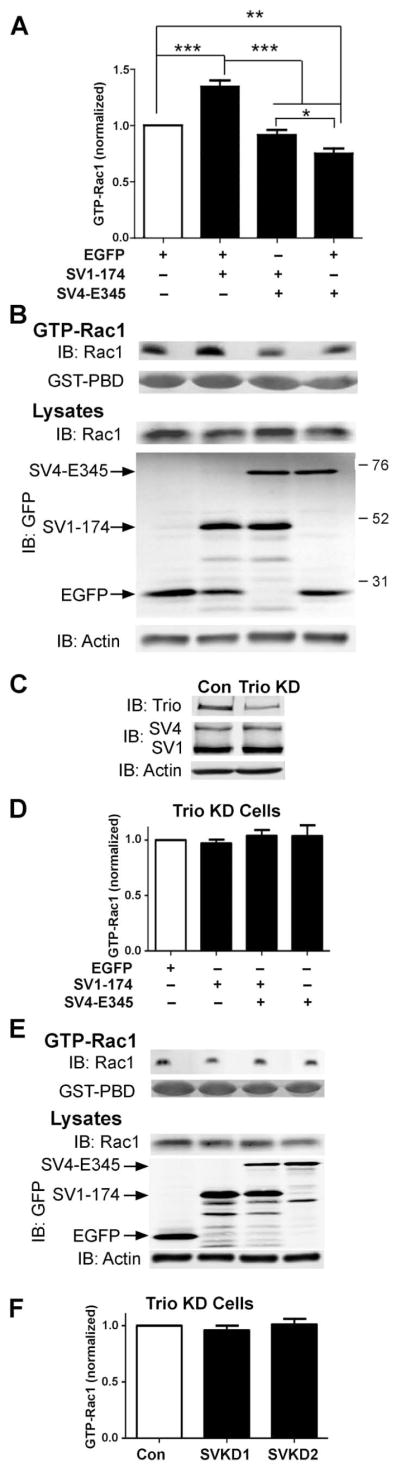
We first explored the functional implications of the interactions between supervillin and Trio by overexpressing the two Trio-binding supervillin protein fragments in control and stably Trio-deficient HeLa cells (Figure 6). GTP loading of Rac1 is increased in control cells expressing EGFP-SV1-174 and decreased in cells expressing EGFP-SV4-E345, as compared with cells expressing EGFP alone (Figure 6A, 6B). GTP-Rac1 loading in cells expressing both supervillin protein fragments was not significantly different from that of controls (Figure 6A, 6B). No interaction between SV1-174 and SV4-E345 was detected in reciprocal GST-pulldown experiments (not shown), suggesting different mechanisms of action for these two protein fragments on Rac1 activation. To determine whether these effects require normal levels of endogenous Trio, we repeated GTP-Rac1 loading experiments in HeLa cells stably expressing a Trio shRNA (Despras et al., 2007). These Trio-knockdown cells contain only ~30% as much Trio as a similarly generated control HeLa cell line stably transfected with a pEBV-based vector encoding a nonspecific shRNA (Figure 6C). SV4 and SV1 levels were not detectably affected by the long-term Trio knockdown (Figure 6C). No significant changes were observed in GTP-Rac1 levels after overexpression of either EGFP-SV1-174 or EGFP-SV4-E345 in Trio-knockdown cells (Figure 6D, 6E). In addition, reduction of supervillin levels in Trio-knockdown cells did not further reduce the extent of GTP-Rac1 loading (Figure 6F), in contrast to the results observed in HeLa cells with endogenous levels of Trio (Figure 1A). These results strongly suggest that Trio is required for supervillin-mediated effects on GTP loading of Rac1.
Figure 6. Trio is required for supervillin-dependent effects on Rac1 activation.
(A)Quantification and (B) representative immunoblots of GTP-Rac1 from control HeLa SilenciX cells after a 24-h transfection with EGFP or EGFP-tagged Trio-binding sequences in supervillin, as shown. Activated Rac1 levels were normalized to those for cells expressing EGFP alone. Means ± s.d.; N = 6. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (ANOVA). (C) Immunoblots showing the ~70% reduction in Trio levels in TRIO HeLa SilenciX cells (Trio KD) and unchanged supervillin (SV) levels; actin used as loading control. (D) Quantification and (E) representative immunoblots of GTP-Rac1 from Trio KD cells after a 24-h transfection with EGFP, EGFP-SV1-174, and/or EGFP-SV4-E345 constructs. Levels were normalized to those for cells expressing EGFP alone. Means ± s.d.; N = 4. (F) Quantification of GTP-Rac1 loading in Trio KD cells treated for 2d with control (Con) or supervillin-specific dsRNAs (SVKD1, SVKD2). Means + s.d.; N = 3.
Supervillin and Trio coordinate to promote initial spreading of HeLa cells
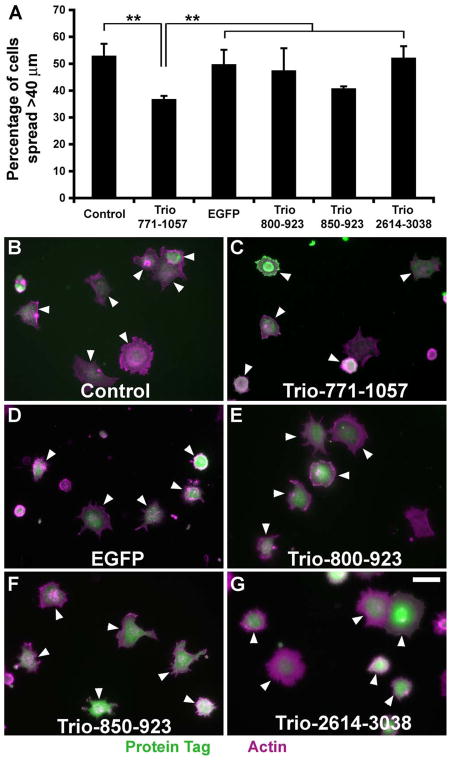
To determine whether the supervillin-Trio interaction functions in vivo, we first looked for dominant-negative effects of supervillin-interacting Trio protein fragments on early cell spreading (Figure 7). Based on our prior observations that 30 min after replating is a point of maximal difference in the diameters of spreading cells (Takizawa et al., 2007; Smith et al., 2010), we quantified the percentages of spread HeLa cells, defined as cells with lengths >2 times that of the mean diameter of initially plated cells. Overexpression of myc-Trio 771-1057, but not other Trio protein fragments assayed, significantly delayed cell spreading (Figure 7A, 7B). By contrast, no differences in cell dimensions were observed after 60 minutes of spreading (not shown). These results are consistent with an in vivo effect of the Trio-supervillin interaction.
Figure 7. Inhibition of cell spreading by Trio 771-1057.
(A) Percentages of transfected cells that were spread 30 minutes after plating onto fibronectin-coated coverslips. HeLa cells were transfected with plasmids encoding either myc (Control), myc-Trio-771-1057, EGFP, EGFP-Trio-800-923, EGFP-Trio-850-923, or EGFP-Trio-2673-3097. “Spread” cells were defined as cells with longest dimensions > 40 μm, which is twice the mean diameter of the initially plated cells (Smith et al., 2010). Means ± s.d. of three experiments; N >150 cells counted per condition in each experiment. **, P < 0.01 (ANOVA). (B–F) Representative fields of cells counted for panel A. Cells expressing tagged proteins, shown in green, are indicated with arrowheads. F-actin stained with phalloidin shown in magenta, with overlaps appearing white. Bar, 40 μm.
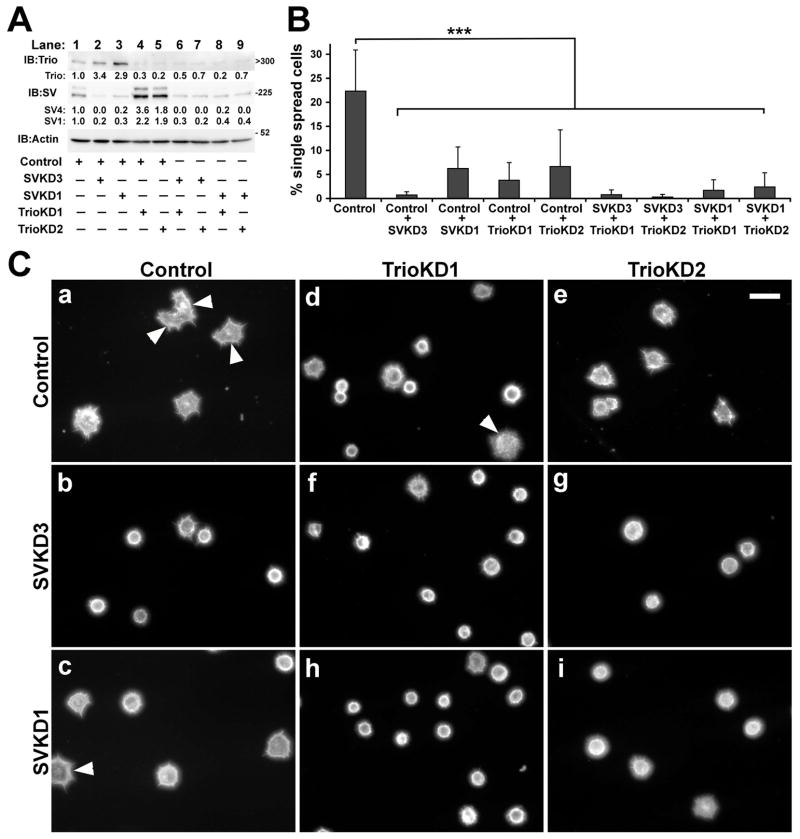
We further explored functional cross-talk between supervillin and Trio with a series of transient single and double protein knockdowns in HeLa cells (Figure 8). When levels of SV4 and SV1 were reduced with each of two dsRNAs directed against separate sequences, we found a significant 3.1 ± 1.4 (mean ± s.d., P < 0.01, N = 7) fold increase in Trio levels (Figure 8A). Similarly, SV4 and SV1 levels tended to increase ~2 fold after a transient Trio knockdown (Figure 8A) although the variability in supervillin levels precluded statistical significance (SV4: 2.7 ± 2.5; SV1: 2.1 ± 1.2; P > 0.05; N = 6) and was not apparent in the stable Trio-deficient cell line (Figure 6C). The knockdown of supervillin alone, Trio alone, or both proteins together significantly reduced the percentage of single HeLa cells scored as spread 30 min after re-plating (Figure 8B, 8C). These results are consistent with functional cross-talk at the level of protein expression or stability and suggest that supervillin and Trio function together in vivo to promote the spreading of HeLa cells.
Figure 8. Inhibition of cell spreading by knockdown of supervillin and Trio.
(A) Immunoblots showing knockdown of Trio and the supervillin isoforms SV4 and SV1. Actin was used as loading control. Locations of molecular mass markers, in kDa, are indicated to the right for supervillin and actin blots; the estimated kDa is shown for Trio. Levels of Trio, SV4 and SV1 were normalized to the amounts in cells treated with control dsRNA (lane 1) for each experiment (N ≥ 3); the mean fold increase or decrease is shown for each experimental condition. Lane 2, Control + SVKD3; lane 3, Control + SVKD1; lane 4, Control + TrioKD1; lane 5, Control + TrioKD2; lane 6, SVKD3 + TrioKD1; lane 7,SVKD3 + TrioKD2; lane 8, SVKD1 + TrioKD1; lane 9, SVKD1 + TrioKD2. (B) Percentages of cells treated with dsRNAs, as shown in panel A, that were spread 30 minutes after plating onto fibronectin-coated coverslips. Cells were treated with dsRNAs for 48 hours before being lifted for assay; only single cells unattached to others were scored. Quantification of the percentage of spread transfected cells assayed as described in Figure 6. Means ± s.d. of three to five experiments; N > 95 cells counted per condition in each experiment. ***, P < 0.001 (ANOVA). (C) Representative fields of phalloidin-stained cells counted for panel B. Control (a), Control + SVKD3 (b), Control + SVKD1 (c), Control + TrioKD1 (d), Control + TrioKD2 (e), SVKD3 + TrioKD1 (f), SVKD3 + TrioKD2 (g), SVKD1 + TrioKD1 (h), SVKD1 + TrioKD2 (i). Cells considered to be spread are indicated with arrowheads. Bar, 40 μm.
DISCUSSION
We show here that the myosin II-regulatory protein supervillin promotes GTP loading of Rac1 by the Rac1-GEF Trio. Rac1-GTP loading is decreased by supervillin knockdown (Figure 1A–B), even though Trio levels are increased (Figure 8A). Rac1, Trio and both supervillin isoforms localize near the plasma membrane in cell surface protrusions and intracellular punctae (Figure 3, 4), sites reminiscent of the locations of early endosomes associated with Arf6, supervillin and the Trio isoform Solo/Trio8 (Song et al., 1998; Sun et al., 2006; Fang et al., 2010). Sites within or near Trio spectrin repeats 6 – 7 bind directly to SV4-specific amino acids 448 – 669 (SV4-E45) and to the N-terminal 171 residues common to both SV1 and SV4 (SV1-171); SV4-E345 also interacts with spectrin repeats 4 – 7 from kalirin (Figure 5). Rac1-GTP loading is increased by overexpression of either SV1, which contains only one Trio-interaction site, or especially SV4, which contains both sites (Figure 2). The Trio GEF-D1 inhibitor ITX3 reverses SV4-mediated Rac1 activation (Figure 2D–E). Furthermore, overexpression of SV4-E345 inhibits Rac1 GTP loading, as expected for a dominant-negative disruption of the SV4-Trio interaction (Figure 6A–B). Similarly, overexpression of the supervillin-interaction site within Trio (Trio 771-1057) inhibits cell spreading (Figure 7), consistent with dominant-negative inhibition of Trio-, Arf6- and Rac1-mediated promotion of lamellipodial extension (Song et al., 1998; Guo et al., 2006; van Rijssel et al., 2012a). Supervillin and Trio cross-talk also is evinced by alterations in each other’s protein levels during transient RNAi knockdowns (Figure 8A). Trio is required for supervillin-mediated Rac1-GTP loading because no effects are seen for either supervillin knockdown (Figure 6F) or overexpression of the individual SV4 Trio-interacting domains on Rac1 activation (Figure 6D–E) in stable Trio-knockdown cells. Finally, the conclusion that supervillin and Trio function in the same pathway is supported by the absence of significant differences between single and double transient knockdowns of these two proteins during cell spreading (Figure 8B–C).
The simplest hypothesis to explain the effect of SV1 on Trio-mediated Rac1 GTP loading is that SV1-174 binding to the spectrin repeats in Trio promotes increased accessibility or activity of the Trio GEF-D1 domain for Rac1. Overexpression of full-length SV1 or of SV1-174, each of which contains only a single Trio-interaction site, results in a similar 20% - 30% increase in Rac1 GTP loading (Figures 2B–C, 6A–B). About 4-fold increases on Trio binding to Rac1 or on Trio GEF-D1 activity have been described after binding of Trio spectrin repeats 1–4 to the Kidins220/ARMS protein (Neubrand et al., 2010) or to Disrupted in Schizophrenia 1 (DISC1) (Chen et al., 2011). However, only Trio spectrin repeats 1–4 interact with the Trio GEF-D1 in a GST pulldown assay (Chen et al., 2011). Thus, SV1-174 binding to Trio spectrin repeats 6–7 may be modestly disrupting the auto-inhibition of GEF-D1 by Trio spectrin repeats 1–4 through steric hindrance or by indirect effects on the conformation of the GEF-D1 interaction site in the Trio N-terminus.
A more complicated hypothesis is necessary to explain the effects of SV4 and SV4-E345 on Trio-mediated Rac1 activation. Overexpression of SV4 increases Trio-mediated Rac1 GTP loading by >2-fold, whereas overexpression of SV4-E345 decreases Rac1 activation by ~25% (Figures 2D–E, 6A–B). These results suggest that SV4-E345 acts as a dominant-negative inhibitor of Trio GEF-D1 activity, perhaps by competing with full-length SV4 for binding to Trio. In the presence of co-expressed SV1-174, the opposite effects of these two protein fragments evidently cancel each other out because Rac1-GTP levels are indistinguishable from controls. By contrast, SV1-174 and SV4-E345 act synergistically to increase Rac1 activation when both fragments are present within full-length SV4. This synergy could be due to increased binding avidity due to the proximity of SV1-171 and SV4-E345 in the context of the full-length protein, or to the presence of other supervillin sequences capable of recruiting additional interactors into a complex that enhances Trio activation.
Based on the strong interaction of SV4-E345 with kalirin spectrin repeats 4–7, SV4-E345 also may bind to the Trio-like protein kalirin. Primarily investigated for its role in synapse formation and neuropsychiatric diseases (Mandela and Ma, 2012; Miller et al., 2013; Remmers et al., 2014), kalirin also is important for normal muscle function (Mandela et al., 2012). Kalirin-deficient mice exhibit abnormal ultrastructure in skeletal muscles (Mandela et al., 2012), a tissue with abundant expression of supervillin isoform 2 (archvillin), which contains the SV4-E345 sequence (Oh et al., 2003). Supervillin isoform expression has not been investigated in the brain, largely because overall expression levels there are low (Pope et al., 1998). However, in situ hybridizations for supervillin (SVIL) and kalirin (KALRN) messages in mouse brains show overlapping, high expression in Purkinje cells within the cerebellum (Allen Brain Atlas, http://mouse.brain-map.org), suggesting a potential role for the supervillin-kalirin interaction in motor control.
In addition to the direct binding shown here between supervillin and Trio, additional, intermediary proteins are likely to be important for in vivo function. Supervillin and Trio are each hub proteins with numerous interactors, including myosin II and the lamellipodial protein filamin, each of which is also a hub for cytoskeletal regulation (Bellanger et al., 2000; Chen et al., 2003; Lee et al., 2010; Smith et al., 2010; Nakamura et al., 2011; Razinia et al., 2012; van Rijssel and van Buul, 2012; Miller et al., 2013; Smith et al., 2013). Myosin II interactions with GEF domains, such as those in Trio and kalirin, inhibit GEF activity (Lee et al., 2010; Shin et al., 2014). Therefore, the direct binding of SV1-171 to the myosin II heavy chain (Chen et al., 2003; Takizawa et al., 2007) could facilitate Rac1 GTP loading by alleviating myosin II inhibition of Trio GEF-D1 activity.
SV1 and SV4 also could regulate the interactions between Trio and the downstream Rac1 effector filamin (Bellanger et al., 2000). Both supervillin isoforms contain sequences that interact with filamin immunoglobulin-like (IgFLN) domains 8–10 and 20–22 (Smith et al., 2010). The Trio GEF-D1 binds to the nearby IgFLN domains 23–24 (Bellanger et al., 2000). Filamin is required for Trio-mediated formation of lamellipodia and adhesive leukocyte docking structures (Bellanger et al., 2000; van Rijssel et al., 2012b). On the other hand, the kalirin GEF-D1 can regulate lamellipodia formation through Rac1-independent protein-protein interactions (Schiller et al., 2005). The lack of interaction of kalirin with SV1-174, which predicts a lack of interaction with SV1, may contribute to differential regulatory requirements for Trio vs. kalirin, especially in cell types that lack SV4-E345-containing supervillin isoforms.
Cell type-specific differences in supervillin isoforms or their interactors are necessary to explain the differences observed here on initial spreading behavior in HeLa cells vs. previous work. Genetic ablation of SV1, the only isoform present, from murine platelets (Edelstein et al., 2012) or ~90% reduction of all supervillin isoforms in human adenocarcinoma A549 cells (Takizawa et al., 2007) increased early cell spreading. Because similar effects were observed after inhibition of nonmuscle myosin II ATPase activity or inhibition of myosin light chain kinase and because all three of these proteins interacted directly (Chen et al., 2003; Takizawa et al., 2007), the previous conclusion was that supervillin’s primary function during cell spreading was regulation of myosin II contractility. Our results here with HeLa cells agree with the prior results showing the importance of Trio for early cell spreading (van Rijssel et al., 2012a) and suggest that supervillin’s role in cell spreading is more nuanced than previously realized. In some cellular contexts, effects on Trio GEF-D1 activation take precedence over myosin II activation. Consistent with this possibility, overexpression of myosin II isoforms has been shown to have opposite effects in HeLa cells vs. monkey fibroblastic COS-7 cells (Betapudi, 2010).
We speculate that the molecular ratios and localizations of supervillin, Trio vs. myosin II and their interactions with other direct and indirect regulators are important for full mechanistic understanding. Supervillin cross-talk with Rac1, Trio and filamin during lamellipodia formation is supported by the effects on lamellipodia observed after overexpression of EGFP-tagged SV1 in COS-7 cells (Crowley et al., 2009). In these cells, filamin is displaced away from the plasma membrane, lamellipodia formation is suppressed, and the SV1 interactor cortactin is redistributed with exogenous SV1 to other membrane sites (Crowley et al., 2009). The loss of lamellipodia in the presence of excess supervillin may be a dominant-negative effect because lamellipodia are retained in cells that co-overexpress Trio with supervillin (Figure 3).
In addition to potential effects on the Trio-filamin interaction, high levels of supervillin may alter the association between filamin and its binding partners (Nakamura et al., 2011). For example, the supervillin interaction with IgFLN domains 8–10, which decrease HeLa cell spreading (Smith et al., 2010), could affect the binding between IgFLN domains 10–13 and the regulatory protein migfilin/FBLP-1 (Takafuta et al., 2003). The interaction of supervillin with IgFLN domains 20 – 22 could influence filamin interactions with migfilin/FBLP-1 at IgFLN domain 21 (Lad et al., 2008) or with the Rac1 GAP FilGAP at IgFLN domain 23 (Nakamura et al., 2009). In conjunction with the Rac1-activation activity described here, localized effects on Rac1-GAP activity could contribute to the spatiotemporal coordination of Rac activation within lamellipodia (Machacek et al., 2009; Hinde et al., 2013).
Localized, potentially cyclic, control of Trio’s Rac1-activating GEF-D1 and RhoA-activating GEF-D2 activities could contribute to the oscillations observed in lamellipodial protrusion and at podosome-type cell-substrate adhesions (Machacek et al., 2009; Deakin et al., 2012; Hinde et al., 2013; van den Dries et al., 2013; Labernadie et al., 2014). Trio, supervillin, myosin II, filamin and cortactin are all implicated in the regulation of dynamic cell-substrate interactions at focal adhesions, podosomes and invadopodia (Medley et al., 2003; Sandquist et al., 2006; Takizawa et al., 2006; Zhou et al., 2006; Clark et al., 2007; Alexander et al., 2008; Crowley et al., 2009; Bhuwania et al., 2012; Guiet et al., 2012; Tomar et al., 2012; van den Dries et al., 2013; Moshfegh et al., 2014). Trio binds to focal adhesion kinase and the LAR transmembrane protein tyrosine phosphatase (PTPRF) at focal adhesions (Debant et al., 1996; Medley et al., 2003). Supervillin binds to the focal adhesion proteins TRIP6 and LPP (Takizawa et al., 2006) and is recruited to podosomes and focal adhesions by myosin II activation (Kuo et al., 2011; Bhuwania et al., 2012). Increased myosin II activation promoted by supervillin leads to disassembly of cell-substrate adhesions (Takizawa et al., 2006; Bhuwania et al., 2012), and Trio-mediated increases in Rac1 activity drive invadopodia disassembly (Moshfegh et al., 2014). Thus, RhoA-mediated myosin II assembly could recruit a supervillin-Trio complex, followed by supervillin stimulation of Trio GEF-D1 activity and locally high Rac1-GTP levels that could facilitate local actin reorganization. To complete Rho/Rac cycling during oscillatory force generation (van den Dries et al., 2013; Labernadie et al., 2014), it has recently been shown that Abl tyrosine kinase in dorsal ruffles phosphorylates Trio and activates RhoA-GTP loading by the Trio GEF-D2 (Jin and Wang, 2007; Sonoshita et al., 2014). Therefore, our discovery of supervillin-Trio-Rac1 cross-talk contributes to the evolving understanding of Rac/Rho regulatory pathways during cell spreading.
MATERIALS AND METHODS
Antibodies and Reagents
Rabbit anti-Trio polyclonal antibodies (raised against spectrin repeats or Ig/kinase domain) were generous gifts from Dr. Betty A. Eipper, University of Connecticut (Farmington, CT) (McPherson et al., 2005). Rabbit polyclonal anti-supervillin (H340) has been described previously (Nebl et al., 2002; Oh et al., 2003). The following mouse primary antibodies were used: anti-Rac1 (Cytoskeleton, Denver, CO); anti-actin monoclonal (clone C4) and anti-Rac1 (clone 23A8) (EMD-Millipore, Billerica, MA); monoclonal (6E2) anti-HA (Cell Signaling Technology, Beverly, MA); anti-GST (Santa Cruz Biotechnology, Dallas, TX); a mix of monoclonals (clones 7.1 and 13.1) to GFP (Roche Life Science, Indianapolis, IN); and anti-Flag M2 monoclonal antibody (Sigma-Aldrich, St. Louis, MO). The following rabbit primary antibodies were used: anti-RhoA monoclonal (Cytoskeleton); monoclonal anti-Cdc42 (11A11), -Myc (clone 71D10), -GFP (D5.1 XP), and -Flag M2, and polyclonal anti-His (Cell Signaling Technology). A C-terminally targeted goat anti-Trio polyclonal antibody (C20) was from Santa Cruz Biotechnology. Secondary antibodies conjugated to AlexaFluor 488 and 568, as well as Texas Red-X phalloidin, were from Life Technologies (Grand Island, NY). Secondary goat anti-rabbit IR 680 and IR 800, goat anti-mouse IR 680 and IR 800, and donkey anti-goat IR 800 antibodies were purchased from Li-Cor (Lincoln, NE). Secondary donkey anti-mouse, anti-rabbit, and anti-goat antibodies conjugated to HRP were from Jackson ImmunoResearch (West Grove, PA). Glutathione-Sepharose 4B was from GE Healthcare BioSciences (Piscataway, NJ), and the Trio-specific inhibitor ITX3 was from Chembridge (San Diego, CA). Restriction enzymes were from New England Biolabs (Beverly, MA), oligonucleotides were from Integrated DNA Technologies (Coralville, IA) and other chemicals were from Sigma-Aldrich.
Yeast Two-Hybrid Screen
The yeast two-hybrid screen was performed by Hybrigenics Services, S.A.S. (Paris, France), as described previously (Spinazzola et al., 2015). Briefly, SV4-E345 was PCR-amplified and cloned into pB27 as a C-terminal fusion to LexA and into pB66 as a C-terminal fusion to Gal4 DNA-binding domain, and used as bait to screen for interacting sequences in a random-primed human adult and fetal skeletal muscle cDNA library in pP6. Mating approaches were used to generate and confirm candidate prey colonies (Fromont-Racine et al., 1997).
Plasmids
Plasmids encoding GFP-tagged human Trio residues 1 – 3038 (Trio “full length”), 1281 – 1609 (Trio-D1) and 1854 – 3038 (Trio C) were generously provided by Dr. J. D. van Buul (University of Amsterdam, The Netherlands) (van Rijssel et al., 2012a). Plasmids encoding HA-tagged Trio residues 1–3038 and 1–1559 (Trio 196) were gifts from Dr. Quintus G. Medley (Pfizer, Boston, MA) (Medley et al., 2000). The pcDNA3.1-myc-Trio 771-1057 (Trio spectrin repeats 5–8) was kindly provided by Dr. Ira Pastan (NIH, Bethesda, MD) (Liu et al., 2012). The plasmid encoding GST-tagged rat kalirin spectrin repeats 4–7 (amino acids 517–976) was generously supplied by Dr. Betty A. Eipper (Vishwanatha et al., 2012). The GST-human Pak1-binding domain (GST-PBD) construct was a generous gift from Dr. Gary Bokoch (The Scripps Research Institute, CA) (Benard and Bokoch, 2002). GST-rhotekin binding domain (GST-RBD, plasmid 15247) was purchased from Addgene (Cambridge, MA) (Ren et al., 1999).
We generated the plasmid encoding EGFP-tagged Trio 800-923 (spectrin repeats 6–7) by digesting pcDNA3.1-myc-Trio 771-1057 with XhoI and BamHI and ligating the fragment into the corresponding sites in pEGFP-C3 (Clontech Laboratories, Mountain View, CA). We produced the plasmid encoding EGFP-Trio 850-923 by deleting a BglII-digested fragment from pEGFP-Trio 800-923. We generated pEGFP-Trio 923-1057 (spectrin repeats 7–8) by digesting pcDNA3.1-myc-Trio 771-1057 with BamHI and cloning the fragment into BamHI-digested pEGFP-C2 (Clontech Laboratories). EGFP-Trio 2673-3097 was generated by deleting N-terminal sequences from EGFPC3-Trio C with HindIII. The His-Trio-SR4-7 (6xHis-tagged Trio spectrin repeats 4–7, amino acids 565–1011) used for direct binding was created through PCR using the primers listed in Table I. The resulting fragment was cloned in-frame to the EcoRI and HindIII sites in pET30a (EMD-Millipore) as described (Takizawa et al., 2007), and verified by sequencing. The numbering for Trio amino acids shown here is that used originally; older sequences begin with what is now accepted as Met-60. For the Trio 771-1057, Trio 565-1011, and derived protein fragments, we use the numbering in the more recent NCBI Reference Sequence (NP_990949.2).
Table I.
Primers and dsRNAs.
| Name | Sequence |
|---|---|
| PCR primers: | |
| SV4-345-F | 5′-TACAGGGAAACCCAAACATG |
| SV4-345-R | 5′-GTCGACCTAATCCGATTCCTTTCGTTC |
| EcoRI-Trio565F | 5′-GAATTCAGGCTGCAGCTGTGTGTTTTCCAGCAG |
| HindIII-Trio1101R | 5′-AAGCTTGTTGACGAGCTTGAGGCGATCTTCCATC |
| dsRNAs: | |
| Control | 5′-GAACUAUGAAGGACCACCAGAGAUA |
| SVKD1 | 5′-GCGAAUCAACCUUUCUACCUUAAUA |
| SVKD2 | 5′-CCCCUGGAAGAUAUCGAAGCCAGAC |
| SVKD3 | 5′-GAAGAUAUCGAAGCCAGACCAGAUA |
| TrioKD1 | 5′-GCUUAGACUUUAGCUUCCCAGAUGA |
| TrioKD2 | 5′-AAGAACUUCUGGAUAGGUCAAACUC |
PCR primers and Stealth dsRNAs used. Underlined bases indicate embedded restriction sites.
The cDNAs encoding EGFP- and Flag-tagged human supervillin isoforms 1 and 4 were generated by PCR and ligated into pEGFP-C2 between EcoRI and XbaI (Pope et al., 1998; Fang and Luna, 2013). GST-SV4-E345 (aa 277–669) was a gift from Dr. Zhiyou Fang, and generated by PCR with the primers listed in Table I. The PCR product was cloned into the pCR2.1 TOPO-TA vector (Life Technologies), and the insert was transferred with EcoRI and SalI into both pGEX-5X-1 (GE Healthcare Bio-Sciences) and pEGFP-C2 (Clontech Laboratories) for bacterial and mammalian expression, respectively. PCR and subcloning added a two-residue linker (AL) and resulted in an L517P point mutation, which was subsequently repaired using the QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). GST-SV4-E34 (aa 277–449) was generated by deleting an SV450-669 fragment from GST-SV4-E345 with XhoI. We made GST-SV4-E45 (aa 448–669) by digesting GST-SV4-E345 with XhoI and ligating the SV448-669 fragment into XhoI-digested pGEX-5X-1. GST-SV1-171, GST-SV171-342, GST-SV343-830, EGFP-SV343-571, and both GFP tagged SV-1-174 constructs were described previously (Chen et al., 2003; Smith et al., 2013).
Fusion proteins and GST pulldown assays
GST fusion proteins were produced and purified as described previously (Ren and Schwartz, 2000; Benard and Bokoch, 2002; Chen et al., 2003; Smith et al., 2010; Vishwanatha et al., 2012). The GST-PBD and GST-RBD peptides were stored bound to the glutathione Sepharose 4B (GE Healthcare Life Sciences) in 10% (v/v) glycerol at −80°C until use. HeLa cells were transfected with myc- or GFP-tagged Trio fragments for 24 hours, washed twice with ice-cold PBS, and extracted in ice-cold lysis buffer (25 mM Tris, 150 mM NaCl, 10 mM MgCl2, 2 mM EDTA, 0.02% SDS, 0.2% deoxycholate, 1% Igepal CA-630, 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml aprotinin, pH 7.5). Lysates were mixed with GST or GST-fusions for 2 h at 4°C, and bound proteins were recovered by sedimentation of the glutathione-Sepharose beads. The beads were rinsed 4 times with ice-cold wash buffer A (25mM Tris, pH 7.5, 40 mM NaCl, 30 mM MgCl2), and the bound proteins were eluted by 2x Laemmli sample buffer (Laemmli, 1970) before being resolved by SDS-PAGE.
For binding experiments with His-tagged Trio spectrin repeats 4–7 (His-Trio-SR 4-7), the recombinant protein was produced in Rosetta BL21 cells (EMD-Millipore Biosciences) and purified as described previously (Takizawa et al., 2003). Briefly, the cleared bacterial lysate was incubated with 1 ml slurry volume of NiNTA agarose (Qiagen, Valencia, CA) for 2 h at 4°C and transferred to a 10ml poly-prep chromatography column (BioRad, Hercules, CA). The beads were washed five times with 10 ml of wash buffer B (10 mM phosphate buffer, pH 7.5, 60 mM NaCl, 2% glycerol, 0.01% DTT) with 20 mM imidazole. Five 1-ml fractions were eluted using wash buffer B containing 250 mM imidazole and stored at −80°C. Previously frozen GST and GST-supervillin fusion proteins were thawed and approximate molar equivalents were diluted to 200 μl in MOPS buffer (20 mM MOPS, 60 mM KCl, 1mM DTT, 0.1 mM EGTA). The GST proteins were then pre-bound to 50 μl (slurry volume) of MOPS buffer-rinsed glutathione Sepharose 4B beads by incubation at 4°C for 1 hour before collection and removal of the buffer by aspiration. The thawed recombinant His-Trio-4-7 solution was modified to contain 90 mM NaCl, 2 mM DTT, 10 mg/ml BSA, and 1% Tween-20 and clarified by centrifugation at 15,000 x g for 15 minutes. The supernatant was transferred to a fresh tube and 100 μl aliquots were added to the GST or GST-supervillin Sepharose beads, and incubated for 1.5 hours at 4°C with rotation. The beads were collected by centrifugation, and the supernatants saved as the unbound fractions. Beads were washed five times with 500 μl of 0.5x TBST (83.5 mM NaCl, 5 mM Tris, 0.025% Tween-20, pH 7.5); at the second wash, the bead slurry was moved to a fresh tube. Bound fractions were eluted with 100 μl of 1x Laemmli sample buffer (Laemmli, 1970).
Cell Culture and Transfection
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM-HG with sodium pyruvate, Life Technologies) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS), 300 μg/ml L-glutamine, and 100 U/ml penicillin and streptomycin at 37°C and 5% CO2. Transient transfections were performed using Lipofectamine 2000 (Life Technologies) according to the manufacturers’ instructions. Control and stable Trio knockdown HeLa SilenciX cells (tebu-bio, Peterborough, United Kingdom) were kindly provided by Dr. J. D. van Buul (University of Amsterdam, The Netherlands). These cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM, Life Technologies) supplemented with 10% (v/v) heat-inactivated fetal calf serum, 1% glutamine and and 100 U/ml penicillin and streptomycin (van Rijssel et al., 2012b). For transient knock down of supervillin and Trio, HeLa cells were transfected for 2 days with Stealth dsRNAs and Lipofectamine RNAiMAX (Life Technologies) as described previously (Smith et al., 2010; Fang and Luna, 2013; Smith et al., 2013). All Stealth dsRNA (Life Technologies) sequences are listed in Table I. The first supervillin dsRNA (SVKD1) targeted a 3′-UTR sequence, beginning with nucleotide 6016 (Smith et al., 2010). The second and third supervillin dsRNAs (SVKD2 and SVKD3) were designed against coding exon 16, starting with nucleotides 2468 and 2473, respectively (Smith et al., 2010; Fang and Luna, 2013). The two Trio dsRNAs (TrioKD1 and TrioKD2) were targeted to separate sequences in the C-terminus, and a scrambled sequence was used as Control.
RhoA, Cdc42 and Rac1 Activation Assay
GTP loading assays were carried out as described (Ren and Schwartz, 2000; Benard and Bokoch, 2002). Briefly, HeLa cells were extracted in lysis buffer (25 mM Tris, 150 mM, NaCl, 10 mM MgCl2, 1 % Igepal CA-630, 5% glycerol, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride, 1 mM Na3VO4, 20 mM NaF, pH 7.5) at 4°C and centrifuged for 10 min at 14,000 x g. Supernatants were immediately flash frozen in aliquots and stored at −80°C until assay. Aliquots were thawed at room temperature and incubated with GST- RBD or GST-PBD pre-bound to glutathione–Sepharose beads for 1 h at 4°C. GST-peptide beads were centrifuged, washed three times in washing buffer A (25 mM Tris, 40 mM NaCl, 30 mM MgCl2, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride, pH 7.5), boiled in 1× Laemmli sample buffer, and resolved by SDS-PAGE.
Immunoblot Analyses
Full-length Trio was assayed using 4–8% gradient SDS-PAGE; 10 or 12% gels were used for lower molecular mass proteins. The Odyssey Infrared Imaging system (Li-Cor Inc.) was used for detection and analysis of signals in GTP loading and the Trio pull down assays. Resolved proteins were transferred to 0.25 μm nitrocellulose membranes and blocked with Odyssey blocking buffer (both from Li-Cor Inc.), before overnight incubation with primary antibodies. Protein sizes were estimated using Full-Range Rainbow Molecular Weight Markers (GE Healthcare Life Sciences). Band intensities were quantified using the Odyssey Software after subtracting the calculated average background value and multiplying by the area of the band. The resulting active G-protein values for each experiment were first ratioed to the amount of total G-protein in the sample, and then normalized to the control value. Statistics were calculated using InStat 3 software (GraphPad Software, Inc., La Jolla, CA).
Direct binding assays and co-sedimentations of EGFP-tagged supervillin constructs with GST-kalirin were resolved on 12% SDS-PAGE gels, transferred to 0.45 μm nitrocellulose (Whatman GmBH, Dassel, Germany), and probed with rabbit anti-His (1:1000) or rabbit-anti GFP (1:1000), respectively. The blots were developed by chemiluminescence with either SuperSignal West-Pico or -Femto reagents (Thermo Scientific, Rockford, IL) and imaged using a BioRad Gel Doc Molecular Imager and Image Lab 4.1 software (BioRad).
Cell-spreading Assay
The cell-spreading assays were performed as described previously (Smith et al., 2010). Briefly, HeLa cells were transfected for 24 hours, trypsinized for passaging and plated onto fibronectin-coated coverslips. Cells were allowed to settle and spread for 30 minutes before being fixed with 4% paraformaldehyde and stained for tagged protein and phalloidin.
For the spreading experiments requiring knockdown of both supervillin and Trio, HeLa cells in antibiotic-free medium were reverse transfected in 6 well dishes with 10 nM each of two dsRNAs (see Table I), for a total of 20 nM dsRNA per well, or 20 nM total for Control only. After incubation for 48 hours, cells were either lifted with trypsin and plated on fibronectin-coated coverslips for 30 minutes as above, or harvested using RIPA buffer (150 mM NaCl, 50 mM Tris pH 7.5, 1.0 % Igepal CA-630, 0.5% deoxycholic acid, 0.1% SDS) and analyzed on 5–15% SDS-PAGE gels (Laemmli, 1970) to assess knockdown efficiencies as described previously (Smith et al., 2013). Cells were also allowed to spread for > 1 hour to assess assay viability. Immunoblots were separated below the 76 kDa molecular weight marker and probed for supervillin (1:1000) and actin (1:2000), followed by probing for Trio (C20, 1:500). Immunoblots were imaged and analyzed on a BioRad Gel Doc Molecular Imager using Image Lab 4.1 software (BioRad), and relative protein amounts were calculated using the Volume Tools with local background subtraction and Microsoft Excel (Redmond, WA) software.
Immunofluorescence Microscopy
Cells were plated, fixed, and stained as previously described (Chen et al., 2003; Smith et al., 2010,Takizawa, 2006 #40). Briefly, cells were plated on glass cover slips overnight, transfected for 24–48 hours, rinsed in 1x PBS, fixed in 4% formaldehyde in PBS for 10 min, and permeabilized with 0.1% Triton X-100 in PBS for 5 min before staining. In some experiments, cells were permeabilized for 5 min at 0°C with 0.1% Triton X-100 in a cytoskeleton stabilizing buffer (50 mM NaCl, 3 mM MgCl2, 30 mM sucrose, 10 mM PIPES, pH 6.8) (Adams et al., 1996) before washing with PBS and fixation with 4% para-formaldehyde. Cells were incubated with anti-Flag (1:800) and anti-HA (1:100) antibodies, and subsequently stained with fluorescently labeled secondary antibodies. Wide field images for Figure 3 were obtained on a Axioskop fluorescent microscope using a 100X (NA 1.3) Plan-NeoFluor oil immersion objective (Carl Zeiss Microscopy, LLC, Thornwood, NY), a RETIGA 1300 CCD camera (QImaging, Surrey, BC, Canada), and OpenLab 3.5.2 software (Improvision, Waltham, MA). Cells in Figure 4 were imaged in the UMASS Cell & Developmental Biology Three-Dimensional Microscopy Laboratory using a 63X (NA 1.4) HCX PL APO CS oil immersion objective with a 1.4X zoom on a Leica SP5 (II) AOBS laser scanning confocal microscope (Leica Microsystems, Exton, PA). Image stacks were obtained sequentially using laser lines at 405 nm and 488 nm. Optical sections of 0.25 nm were acquired for the basal ~2 μm of each cell, and maximum point projection images were generated using Leica Confocal Software (Leica Microsystems). For the spreading experiments, micrographs of 15 random fields were taken from each of 2 coverslips per condition per experiment on a Leica DMI 6000B using Leica Application Suite Advanced Fluorescence 3.2.0.9652 software and a HCX PL Fluotar 40X/0.60 corrected lens. Cells were scored as spread if they were more than twice the width of an unspread cell in any linear direction (> 40 μm), as visualized by Texas Red-X phalloidin stain (Life Technologies). Means, standard deviations, and ANOVA results were all calculated using InStat 3 (GraphPad Software, Inc.). Images were assembled and labeled using Adobe Photoshop software (Adobe Systems Inc., San Jose, CA).
Supplementary Material
Immunoblots stained with anti-GST to show the relative amounts of column-bound GST proteins corresponding to the experiments shown in Figure 4D.
Acknowledgments
We thank Dr. Zhiyou Fang for the plasmids encoding GST-tagged SV4-E345 and EGFP- and Flag-tagged human SV1 and SV4; Dr. Gary Bokoch for the GST-PBD construct; Dr. Martin A. Schwartz for the HA-Rac1 plasmid; and Drs. Ira Pastan, Quintus Medley, and Betty Eipper for plasmids encoding full-length Trio proteins and Trio and kalirin protein fragments. We gratefully acknowledge Dr. Eipper for generously supplying us with anti-Trio antibodies and Dr. Jap van Buul for plasmids, experimental advice, and HeLa cells deficient in Trio expression. We are also grateful to Dr. Marcela Nunez and her colleagues at Hybrigenics Services, S.A.S. for their assistance and expertise in yeast two-hybrid screening. We thank Dr. Jeffrey Nickerson and Jean Underwood for training and assistance in the Cell Biology Confocal Core Facility. We also thank Dr. Sebastian Mana-Capelli for experimental input and helpful discussions. This work was supported by NIH grant R01 GM033048-26S1 (EJL) and by the Department of Cell and Developmental Biology at the University of Massachusetts Medical School.
Abbreviations
- GEF
guanine nucleotide exchange factor
- SV1
human supervillin isoform 1
- SV4
human supervillin isoform 4
- SVKD
supervillin knockdown by specific dsRNA
References
- Adams CL, Nelson WJ, Smith SJ. Quantitative analysis of cadherin-catenin-actin reorganization during development of cell-cell adhesion. J Cell Biol. 1996;135:1899–1911. doi: 10.1083/jcb.135.6.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam MR, Johnson RC, Darlington DN, Hand TA, Mains RE, Eipper BA. Kalirin, a cytosolic protein with spectrin-like and GDP/GTP exchange factor-like domains that interacts with peptidylglycine alpha-amidating monooxygenase, an integral membrane peptide-processing enzyme. J Biol Chem. 1997;272:12667–12675. doi: 10.1074/jbc.272.19.12667. [DOI] [PubMed] [Google Scholar]
- Alexander NR, Branch KM, Parekh A, Clark ES, Iwueke IC, Guelcher SA, Weaver AM. Extracellular matrix rigidity promotes invadopodia activity. Curr Biol. 2008;18:1295–1299. doi: 10.1016/j.cub.2008.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger JM, Astier C, Sardet C, Ohta Y, Stossel TP, Debant A. The Rac1- and RhoG-specific GEF domain of Trio targets filamin to remodel cytoskeletal actin. Nat Cell Biol. 2000;2:888–892. doi: 10.1038/35046533. [DOI] [PubMed] [Google Scholar]
- Bellanger JM, Estrach S, Schmidt S, Briancon-Marjollet A, Zugasti O, Fromont S, Debant A. Different regulation of the Trio Dbl-Homology domains by their associated PH domains. Biol Cell. 2003;95:625–634. doi: 10.1016/j.biolcel.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Bellanger JM, Lazaro JB, Diriong S, Fernandez A, Lamb N, Debant A. The two guanine nucleotide exchange factor domains of Trio link the Rac1 and the RhoA pathways in vivo. Oncogene. 1998;16:147–152. doi: 10.1038/sj.onc.1201532. [DOI] [PubMed] [Google Scholar]
- Benard V, Bokoch GM. Assay of Cdc42, Rac, and Rho GTPase activation by affinity methods. Methods Enzymol. 2002;345:349–359. doi: 10.1016/s0076-6879(02)45028-8. [DOI] [PubMed] [Google Scholar]
- Betapudi V. Myosin II motor proteins with different functions determine the fate of lamellipodia extension during cell spreading. PLoS One. 2010;5:e8560. doi: 10.1371/journal.pone.0008560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuwania R, Cornfine S, Fang Z, Kruger M, Luna EJ, Linder S. Supervillin couples myosin-dependent contractility to podosomes and enables their turnover. J Cell Sci. 2012;125:2300–2314. doi: 10.1242/jcs.100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- Blangy A, Vignal E, Schmidt S, Debant A, Gauthier-Rouviere C, Fort P. TrioGEF1 controls Rac- and Cdc42-dependent cell structures through the direct activation of RhoG. J Cell Sci. 2000;113(Pt 4):729–739. doi: 10.1242/jcs.113.4.729. [DOI] [PubMed] [Google Scholar]
- Bouquier N, Fromont S, Zeeh JC, Auziol C, Larrousse P, Robert B, Zeghouf M, Cherfils J, Debant A, Schmidt S. Aptamer-derived peptides as potent inhibitors of the oncogenic RhoGEF Tgat. Chem Biol. 2009;16:391–400. doi: 10.1016/j.chembiol.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Chen SY, Huang PH, Cheng HJ. Disrupted-in-Schizophrenia 1-mediated axon guidance involves TRIO-RAC-PAK small GTPase pathway signaling. Proc Natl Acad Sci U S A. 2011;108:5861–5866. doi: 10.1073/pnas.1018128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Takizawa N, Crowley JL, Oh SW, Gatto CL, Kambara T, Sato O, Li X, Ikebe M, Luna EJ. F-actin and myosin II binding domains in supervillin. J Biol Chem. 2003;278:46094–46106. doi: 10.1074/jbc.M305311200. [DOI] [PubMed] [Google Scholar]
- Chhatriwala MK, Betts L, Worthylake DK, Sondek J. The DH and PH domains of Trio coordinately engage Rho GTPases for their efficient activation. J Mol Biol. 2007;368:1307–1320. doi: 10.1016/j.jmb.2007.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark ES, Whigham AS, Yarbrough WG, Weaver AM. Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 2007;67:4227–4235. doi: 10.1158/0008-5472.CAN-06-3928. [DOI] [PubMed] [Google Scholar]
- Crowley JL, Smith TC, Fang Z, Takizawa N, Luna EJ. Supervillin reorganizes the actin cytoskeleton and increases invadopodial efficiency. Mol Biol Cell. 2009;20:948–962. doi: 10.1091/mbc.E08-08-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin NO, Ballestrem C, Turner CE. Paxillin and Hic-5 interaction with vinculin is differentially regulated by Rac1 and RhoA. PLoS One. 2012;7:e37990. doi: 10.1371/journal.pone.0037990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debant A, Serra-Pages C, Seipel K, O’Brien S, Tang M, Park SH, Streuli M. The multidomain protein Trio binds the LAR transmembrane tyrosine phosphatase, contains a protein kinase domain, and has separate rac- specific and rho-specific guanine nucleotide exchange factor domains. Proc Natl Acad Sci U S A. 1996;93:5466–5471. doi: 10.1073/pnas.93.11.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debreceni B, Gao Y, Guo F, Zhu K, Jia B, Zheng Y. Mechanisms of guanine nucleotide exchange and Rac-mediated signaling revealed by a dominant negative trio mutant. J Biol Chem. 2004;279:3777–3786. doi: 10.1074/jbc.M308282200. [DOI] [PubMed] [Google Scholar]
- Despras E, Pfeiffer P, Salles B, Calsou P, Kuhfittig-Kulle S, Angulo JF, Biard DS. Long-term XPC silencing reduces DNA double-strand break repair. Cancer Res. 2007;67:2526–2534. doi: 10.1158/0008-5472.CAN-06-3371. [DOI] [PubMed] [Google Scholar]
- Edelstein LC, Luna EJ, Gibson IB, Bray M, Jin Y, Kondkar A, Nagalla S, Hadjout-Rabi N, Smith TC, Covarrubias D, Jones SN, Ahmad F, Stolla M, Kong X, Fang Z, Bergmeier W, Shaw C, Leal SM, Bray PF. Human genome-wide association and mouse knockout approaches identify platelet supervillin as an inhibitor of thrombus formation under shear stress. Circulation. 2012;125:2762–2771. doi: 10.1161/CIRCULATIONAHA.112.091462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z, Takizawa N, Wilson KA, Smith TC, Delprato A, Davidson MW, Lambright DG, Luna EJ. The membrane-associated protein, supervillin, accelerates F-actin-dependent rapid integrin recycling and cell motility. Traffic. 2010;11:782–799. doi: 10.1111/j.1600-0854.2010.01062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang ZY, Luna EJ. Supervillin-mediated suppression of p53 protein enhances cell survival. J Biol Chem. 2013;288:7918–7929. doi: 10.1074/jbc.M112.416842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin SP, Ennis MJ, Schumacher CA, Zylstra-Diegel CR, Williams BO, Ross JT, Winkles JA, Loftus JC, Symons MH, Tran NL. Cdc42 and the guanine nucleotide exchange factors Ect2 and trio mediate Fn14-induced migration and invasion of glioblastoma cells. Mol Cancer Res. 2012;10:958–968. doi: 10.1158/1541-7786.MCR-11-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain JC, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- Gangopadhyay SS, Takizawa N, Gallant C, Barber AL, Je HD, Smith TC, Luna EJ, Morgan KG. Smooth muscle archvillin: a novel regulator of signaling and contractility in vascular smooth muscle. J Cell Sci. 2004;117:5043–5057. doi: 10.1242/jcs.01378. [DOI] [PubMed] [Google Scholar]
- Guiet R, Verollet C, Lamsoul I, Cougoule C, Poincloux R, Labrousse A, Calderwood DA, Glogauer M, Lutz PG, Maridonneau-Parini I. Macrophage mesenchymal migration requires podosome stabilization by filamin A. J Biol Chem. 2012;287:13051–13062. doi: 10.1074/jbc.M111.307124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilluy C, Garcia-Mata R, Burridge K. Rho protein crosstalk: another social network? Trends Cell Biol. 2011;21:718–726. doi: 10.1016/j.tcb.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Debidda M, Yang L, Williams DA, Zheng Y. Genetic deletion of Rac1 GTPase reveals its critical role in actin stress fiber formation and focal adhesion complex assembly. J Biol Chem. 2006;281:18652–18659. doi: 10.1074/jbc.M603508200. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Hyodo T, Asano E, Ito S, Maeda M, Kuribayashi H, Natsume A, Wakabayashi T, Hamaguchi M, Senga T. The role of PLK1-phosphorylated SVIL in myosin II activation and cytokinetic furrowing. J Cell Sci. 2013;126:3627–3637. doi: 10.1242/jcs.124818. [DOI] [PubMed] [Google Scholar]
- Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- Hinde E, Digman MA, Hahn KM, Gratton E. Millisecond spatiotemporal dynamics of FRET biosensors by the pair correlation function and the phasor approach to FLIM. Proc Natl Acad Sci U S A. 2013;110:135–140. doi: 10.1073/pnas.1211882110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Jin H, Wang JY. Abl tyrosine kinase promotes dorsal ruffles but restrains lamellipodia extension during cell spreading on fibronectin. Mol Biol Cell. 2007;18:4143–4154. doi: 10.1091/mbc.E07-01-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, Metalnikov P, O’Donnell P, Taylor P, Taylor L, Zougman A, Woodgett JR, Langeberg LK, Scott JD, Pawson T. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Kawaguchi K, Saito K, Asami H, Ohta Y. ADP ribosylation factor 6 (Arf6) acts through FilGAP protein to down-regulate Rac protein and regulates plasma membrane blebbing. J Biol Chem. 2014;289:9675–9682. doi: 10.1074/jbc.M113.546051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Jiang LH, Wilson HL, North RA, Surprenant A. Proteomic and functional evidence for a P2X7 receptor signalling complex. EMBO J. 2001;20:6347–6358. doi: 10.1093/emboj/20.22.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo TH, Eipper BA, Donaldson JG. Arf6 recruits the Rac GEF Kalirin to the plasma membrane facilitating Rac activation. BMC Cell Biol. 2007;8:29. doi: 10.1186/1471-2121-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo JC, Han X, Hsiao CT, Yates JR, 3rd, Waterman CM. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for beta-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol. 2011;13:383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labernadie A, Bouissou A, Delobelle P, Balor S, Voituriez R, Proag A, Fourquaux I, Thibault C, Vieu C, Poincloux R, Charriere GM, Maridonneau-Parini I. Protrusion force microscopy reveals oscillatory force generation and mechanosensing activity of human macrophage podosomes. Nat Commun. 2014;5:5343. doi: 10.1038/ncomms6343. [DOI] [PubMed] [Google Scholar]
- Lad Y, Jiang P, Ruskamo S, Harburger DS, Ylanne J, Campbell ID, Calderwood DA. Structural basis of the migfilin-filamin interaction and competition with integrin beta tails. J Biol Chem. 2008;283:35154–35163. doi: 10.1074/jbc.M802592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (Lond) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lane J, Martin TA, Mansel RE, Jiang WG. The expression and prognostic value of the guanine nucleotide exchange factors (GEFs) Trio, Vav1 and TIAM-1 in human breast cancer. Int Semin Surg Oncol. 2008;5:23. doi: 10.1186/1477-7800-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson CD, Burridge K. The on-off relationship of Rho and Rac during integrin-mediated adhesion and cell migration. Small GTPases. 2014;5:e27958. doi: 10.4161/sgtp.27958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Choi CK, Shin EY, Schwartz MA, Kim EG. Myosin II directly binds and inhibits Dbl family guanine nucleotide exchange factors: a possible link to Rho family GTPases. J Cell Biol. 2010;190:663–674. doi: 10.1083/jcb.201003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Higgs HN. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr Biol. 2003;13:1335–1340. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- Liu XF, Bera TK, Kahue C, Escobar T, Fei Z, Raciti GA, Pastan I. ANKRD26 and its interacting partners TRIO, GPS2, HMMR and DIPA regulate adipogenesis in 3T3-L1 cells. PLoS One. 2012;7:e38130. doi: 10.1371/journal.pone.0038130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandela P, Ma XM. Kalirin, a key player in synapse formation, is implicated in human diseases. Neural Plast. 2012;2012:728161. doi: 10.1155/2012/728161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandela P, Yankova M, Conti LH, Ma XM, Grady J, Eipper BA, Mains RE. Kalrn plays key roles within and outside of the nervous system. BMC Neurosci. 2012;13:136. doi: 10.1186/1471-2202-13-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson CE, Eipper BA, Mains RE. Multiple novel isoforms of Trio are expressed in the developing rat brain. Gene. 2005;347:125–135. doi: 10.1016/j.gene.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Medley QG, Buchbinder EG, Tachibana K, Ngo H, Serra-Pages C, Streuli M. Signaling between focal adhesion kinase and trio. J Biol Chem. 2003;278:13265–13270. doi: 10.1074/jbc.M300277200. [DOI] [PubMed] [Google Scholar]
- Medley QG, Serra-Pages C, Iannotti E, Seipel K, Tang M, O’Brien SP, Streuli M. The Trio guanine nucleotide exchange factor is a RhoA target. Binding of RhoA to the trio immunoglobulin-like domain. J Biol Chem. 2000;275:36116–36123. doi: 10.1074/jbc.M003775200. [DOI] [PubMed] [Google Scholar]
- Miller MB, Yan Y, Eipper BA, Mains RE. Neuronal Rho GEFs in synaptic physiology and behavior. Neuroscientist. 2013;19:255–273. doi: 10.1177/1073858413475486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshfegh Y, Bravo-Cordero JJ, Miskolci V, Condeelis J, Hodgson L. A Trio-Rac1-Pak1 signalling axis drives invadopodia disassembly. Nat Cell Biol. 2014;16:574–586. doi: 10.1038/ncb2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers KR, Casanova JE. Regulation of actin cytoskeleton dynamics by Arf-family GTPases. Trends Cell Biol. 2008;18:184–192. doi: 10.1016/j.tcb.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F, Heikkinen O, Pentikainen OT, Osborn TM, Kasza KE, Weitz DA, Kupiainen O, Permi P, Kilpelainen I, Ylanne J, Hartwig JH, Stossel TP. Molecular basis of filamin A-FilGAP interaction and its impairment in congenital disorders associated with filamin A mutations. PLoS One. 2009;4:e4928. doi: 10.1371/journal.pone.0004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F, Stossel TP, Hartwig JH. The filamins: organizers of cell structure and function. Cell Adh Migr. 2011;5:160–169. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak RC, Chang KH, Vaitinadin NS, Cancelas JA. Rho GTPases control specific cytoskeleton-dependent functions of hematopoietic stem cells. Immunol Rev. 2013;256:255–268. doi: 10.1111/imr.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebl T, Pestonjamasp KN, Leszyk JD, Crowley JL, Oh SW, Luna EJ. Proteomic analysis of a detergent-resistant membrane skeleton from neutrophil plasma membranes. J Biol Chem. 2002;277:43399–43409. doi: 10.1074/jbc.M205386200. [DOI] [PubMed] [Google Scholar]
- Neubrand VE, Thomas C, Schmidt S, Debant A, Schiavo G. Kidins220/ARMS regulates Rac1-dependent neurite outgrowth by direct interaction with the RhoGEF Trio. J Cell Sci. 2010;123:2111–2123. doi: 10.1242/jcs.064055. [DOI] [PubMed] [Google Scholar]
- Oh SW, Pope RK, Smith KP, Crowley JL, Nebl T, Lawrence JB, Luna EJ. Archvillin, a muscle-specific isoform of supervillin, is an early expressed component of the costameric membrane skeleton. J Cell Sci. 2003;116:2261–2275. doi: 10.1242/jcs.00422. [DOI] [PubMed] [Google Scholar]
- Pestonjamasp KN, Pope RK, Wulfkuhle JD, Luna EJ. Supervillin (p205): A novel membrane-associated, F-actin-binding protein in the villin/gelsolin superfamily. J Cell Biol. 1997;139:1255–1269. doi: 10.1083/jcb.139.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope RK, Pestonjamasp KN, Smith KP, Wulfkuhle JD, Strassel CP, Lawrence JB, Luna EJ. Cloning, characterization, and chromosomal localization of human supervillin (SVIL) Genomics. 1998;52:342–351. doi: 10.1006/geno.1998.5466. [DOI] [PubMed] [Google Scholar]
- Radhakrishna H, Al-Awar O, Khachikian Z, Donaldson JG. ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J Cell Sci. 1999;112(Pt 6):855–866. doi: 10.1242/jcs.112.6.855. [DOI] [PubMed] [Google Scholar]
- Razinia Z, Makela T, Ylanne J, Calderwood DA. Filamins in mechanosensing and signaling. Annu Rev Biophys. 2012;41:227–246. doi: 10.1146/annurev-biophys-050511-102252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmers C, Sweet RA, Penzes P. Abnormal kalirin signaling in neuropsychiatric disorders. Brain Res Bull. 2014;103:29–38. doi: 10.1016/j.brainresbull.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XD, Kiosses WB, Schwartz MA. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 1999;18:578–585. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XD, Schwartz MA. Determination of GTP loading on Rho. Methods Enzymol. 2000;325:264–272. doi: 10.1016/s0076-6879(00)25448-7. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Salhia B, Tran NL, Chan A, Wolf A, Nakada M, Rutka F, Ennis M, McDonough WS, Berens ME, Symons M, Rutka JT. The guanine nucleotide exchange factors trio, Ect2, and Vav3 mediate the invasive behavior of glioblastoma. Am J Pathol. 2008;173:1828–1838. doi: 10.2353/ajpath.2008.080043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson ER, Yeh SY, Chang HC, Tsai MY, Wang X, Ting HJ, Chang C. Identification and characterization of androgen receptor associated coregulators in prostate cancer cells. J Biol Regul Homeost Agents. 2001;15:123–129. [PubMed] [Google Scholar]
- Sandquist JC, Swenson KI, Demali KA, Burridge K, Means AR. Rho kinase differentially regulates phosphorylation of nonmuscle myosin II isoforms A and B during cell rounding and migration. J Biol Chem. 2006;281:35873–35883. doi: 10.1074/jbc.M605343200. [DOI] [PubMed] [Google Scholar]
- Santy LC, Casanova JE. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J Cell Biol. 2001;154:599–610. doi: 10.1083/jcb.200104019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Wittinghofer A. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem Sci. 1998;23:257–262. doi: 10.1016/s0968-0004(98)01224-9. [DOI] [PubMed] [Google Scholar]
- Schiller MR, Blangy A, Huang J, Mains RE, Eipper BA. Induction of lamellipodia by Kalirin does not require its guanine nucleotide exchange factor activity. Exp Cell Res. 2005;307:402–417. doi: 10.1016/j.yexcr.2005.03.024. [DOI] [PubMed] [Google Scholar]
- Schmidt S, Debant A. Function and regulation of the Rho guanine nucleotide exchange factor Trio. Small GTPases. 2014;5:e29769. doi: 10.4161/sgtp.29769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz HD, Bereiter-Hahn J. GFP associates with microfilaments in fixed cells. Histochem Cell Biol. 2001;116:89–94. doi: 10.1007/s004180100293. [DOI] [PubMed] [Google Scholar]
- Seipel K, Medley QG, Kedersha NL, Zhang XA, O’Brien SP, Serra-Pages C, Hemler ME, Streuli M. Trio amino-terminal guanine nucleotide exchange factor domain expression promotes actin cytoskeleton reorganization, cell migration and anchorage-independent cell growth. J Cell Sci. 1999;112(Pt 12):1825–1834. doi: 10.1242/jcs.112.12.1825. [DOI] [PubMed] [Google Scholar]
- Shin EY, Lee CS, Yun CY, Won SY, Kim HK, Lee YH, Kwak SJ, Kim EG. Non-muscle myosin II regulates neuronal actin dynamics by interacting with guanine nucleotide exchange Factors. PLoS One. 2014;9:e95212. doi: 10.1371/journal.pone.0095212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit ST, Manser E. Rho GTPases and their role in organizing the actin cytoskeleton. J Cell Sci. 2011;124:679–683. doi: 10.1242/jcs.064964. [DOI] [PubMed] [Google Scholar]
- Skowronek KR, Guo F, Zheng Y, Nassar N. The C-terminal basic tail of RhoG assists the guanine nucleotide exchange factor trio in binding to phospholipids. J Biol Chem. 2004;279:37895–37907. doi: 10.1074/jbc.M312677200. [DOI] [PubMed] [Google Scholar]
- Smith TC, Fang Z, Luna EJ. Novel interactors and a role for supervillin in early cytokinesis. Cytoskeleton (Hoboken) 2010;67:346–364. doi: 10.1002/cm.20449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TC, Fridy PC, Li Y, Basil S, Arjun S, Friesen RM, Leszyk J, Chait BT, Rout MP, Luna EJ. Supervillin binding to myosin II and synergism with anillin are required for cytokinesis. Mol Biol Cell. 2013;24:3603–3619. doi: 10.1091/mbc.E12-10-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Khachikian Z, Radhakrishna H, Donaldson JG. Localization of endogenous ARF6 to sites of cortical actin rearrangement and involvement of ARF6 in cell spreading. J Cell Sci. 1998;111(Pt 15):2257–2267. doi: 10.1242/jcs.111.15.2257. [DOI] [PubMed] [Google Scholar]
- Sonoshita M, Itatani Y, Kakizaki F, Sakimura K, Terashima T, Katsuyama Y, Sakai Y, Taketo MM. Promotion of Colorectal Cancer Invasion and Metastasis Through Activation of Notch-Dab1-Abl-RhoGEF Protein Trio. Cancer discovery. 2014 doi: 10.1158/2159-8290.CD-14-0595. In press. [DOI] [PubMed] [Google Scholar]
- Spinazzola JM, Smith TC, Liu M, Luna EJ, Barton ER. Gamma-sarcoglycan is required for the response of archvillin to mechanical stimulation in skeletal muscle. Hum Mol Gen. 2015 doi: 10.1093/hmg/ddv008. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YJ, Nishikawa K, Yuda H, Wang YL, Osaka H, Fukazawa N, Naito A, Kudo Y, Wada K, Aoki S. Solo/Trio8, a membrane-associated short isoform of Trio, modulates endosome dynamics and neurite elongation. Mol Cell Biol. 2006;26:6923–6935. doi: 10.1128/MCB.02474-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takafuta T, Saeki M, Fujimoto TT, Fujimura K, Shapiro SS. A new member of the LIM protein family binds to filamin B and localizes at stress fibers. J Biol Chem. 2003;278:12175–12181. doi: 10.1074/jbc.M209339200. [DOI] [PubMed] [Google Scholar]
- Takizawa N, Ikebe R, Ikebe M, Luna EJ. Supervillin slows cell spreading by facilitating myosin II activation at the cell periphery. J Cell Sci. 2007;120:3792–3803. doi: 10.1242/jcs.008219. [DOI] [PubMed] [Google Scholar]
- Takizawa N, Schmidt DJ, Mabuchi K, Villa-Moruzzi E, Tuft RA, Ikebe M. M20, the small subunit of PP1M, binds to microtubules. Am J Physiol Cell Physiol. 2003;284:C250–262. doi: 10.1152/ajpcell.00153.2002. [DOI] [PubMed] [Google Scholar]
- Takizawa N, Smith TC, Nebl T, Crowley JL, Palmieri SJ, Lifshitz LM, Ehrhardt AG, Hoffman LM, Beckerle MC, Luna EJ. Supervillin modulation of focal adhesions involving TRIP6/ZRP-1. J Cell Biol. 2006;174:447–458. doi: 10.1083/jcb.200512051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting HJ, Yeh S, Nishimura K, Chang C. Supervillin associates with androgen receptor and modulates its transcriptional activity. Proc Natl Acad Sci U S A. 2002;99:661–666. doi: 10.1073/pnas.022469899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar A, Lawson C, Ghassemian M, Schlaepfer DD. Cortactin as a target for FAK in the regulation of focal adhesion dynamics. PLoS One. 2012;7:e44041. doi: 10.1371/journal.pone.0044041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji T, Ishizaki T, Okamoto M, Higashida C, Kimura K, Furuyashiki T, Arakawa Y, Birge RB, Nakamoto T, Hirai H, Narumiya S. ROCK and mDia1 antagonize in Rho-dependent Rac activation in Swiss 3T3 fibroblasts. J Cell Biol. 2002;157:819–830. doi: 10.1083/jcb.200112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Dries K, Meddens MB, de Keijzer S, Shekhar S, Subramaniam V, Figdor CG, Cambi A. Interplay between myosin IIA-mediated contractility and actin network integrity orchestrates podosome composition and oscillations. Nat Commun. 2013;4:1412. doi: 10.1038/ncomms2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijssel J, Hoogenboezem M, Wester L, Hordijk PL, Van Buul JD. The N-terminal DH-PH domain of Trio induces cell spreading and migration by regulating lamellipodia dynamics in a Rac1-dependent fashion. PLoS One. 2012a;7:e29912. doi: 10.1371/journal.pone.0029912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijssel J, Kroon J, Hoogenboezem M, van Alphen FP, de Jong RJ, Kostadinova E, Geerts D, Hordijk PL, van Buul JD. The Rho-guanine nucleotide exchange factor Trio controls leukocyte transendothelial migration by promoting docking structure formation. Mol Biol Cell. 2012b;23:2831–2844. doi: 10.1091/mbc.E11-11-0907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijssel J, van Buul JD. The many faces of the guanine-nucleotide exchange factor trio. Cell Adh Migr. 2012;6:482–487. doi: 10.4161/cam.21418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaqué JP, Dorsam RT, Feng X, Iglesias-Bartolome R, Forsthoefel DJ, Chen Q, Debant A, Seeger MA, Ksander BR, Teramoto H, Gutkind JS. A genome-wide RNAi screen reveals a Trio-regulated Rho GTPase circuitry transducing mitogenic signals initiated by G protein-coupled receptors. Mol Cell. 2013;49:94–108. doi: 10.1016/j.molcel.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwanatha KS, Wang YP, Keutmann HT, Mains RE, Eipper BA. Structural organization of the nine spectrin repeats of Kalirin. Biochemistry. 2012;51:5663–5673. doi: 10.1021/bi300583s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheimer E, Gutierrez-Uzquiza A, Rosemblit C, Lopez-Haber C, Sosa MS, Kazanietz MG. Rac signaling in breast cancer: a tale of GEFs and GAPs. Cell Signal. 2012;24:353–362. doi: 10.1016/j.cellsig.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulfkuhle JD, Donina IE, Stark NH, Pope RK, Pestonjamasp KN, Niswonger ML, Luna EJ. Domain analysis of supervillin, an F-actin bundling plasma membrane protein with functional nuclear localization signals. J Cell Sci. 1999;112:2125–2136. doi: 10.1242/jcs.112.13.2125. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Calafat J, Janssen H, Greenberg S. ARF6 is required for growth factor- and rac-mediated membrane ruffling in macrophages at a stage distal to rac membrane targeting. Mol Cell Biol. 1999;19:8158–8168. doi: 10.1128/mcb.19.12.8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Simon R, Mirlacher M, Maurer R, Gasser T, Forster T, Diener PA, Mihatsch MJ, Sauter G, Schraml P. TRIO amplification and abundant mRNA expression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Am J Pathol. 2004;165:63–69. doi: 10.1016/S0002-9440(10)63275-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Webb BA, Eves R, Mak AS. Effects of tyrosine phosphorylation of cortactin on podosome formation in A7r5 vascular smooth muscle cells. Am J Physiol Cell Physiol. 2006;290:C463–471. doi: 10.1152/ajpcell.00350.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunoblots stained with anti-GST to show the relative amounts of column-bound GST proteins corresponding to the experiments shown in Figure 4D.