Abstract
Several single nucleotide variants (SNVs) in low-density lipoprotein receptor-related protein 6 (Lrp6) cause neural tube defects (NTDs) in mice. We therefore examined LRP6 in 192 unrelated infants from California with the NTD, spina bifida, and found four heterozygous missense SNVs, three of which were predicted to be deleterious, among NTD cases and not in 190 ethnically matched non-malformed controls. Parents and siblings could not be tested because of the study design. Like Cd and rs mouse variants, the p.Tyr544Cys Lrp6 protein failed to bind the chaperone protein MESD and impaired Lrp6 subcellular localization to the plasma membrane of MDCK II cells. Only the p.Tyr544Cys Lrp6 variant down-regulated canonical Wnt signaling in a TopFlash luciferase reporter in vitro assay. In contrast, three Lrp6 mutants (p.Ala3Val, p.Tyr544Cys and p.Arg1574Leu) increased non-canonical Wnt/PCP signaling in an Ap1-luciferase assay. Thus, LRP6 variants outside of YWTD-repeats could potentially predispose embryos to NTDs, while Lrp6 modulation of Wnt/PCP signaling would be more essential than its canonical pathway role in neural tube closure.
Keywords: LRP6, SNV, Spina bifida, Wnt signaling, PCP signaling
INTRODUCTION
Neural tube defects (NTDs) are severe central nervous system (CNS) congenital malformations affecting approximately 1 per 2000 livebirths annually in the US [Mitchell, 2005]. They remain the second most common human structural birth defect in the world today. Common NTDs include anencephaly, spina bifida, and craniorachischisis. NTDs occur in developing human embryos when the neural tube improperly closes during the third and fourth weeks of gestation [Wallingford, et al., 2013]. The complicated mechanism of the events of neural tube closure, along with dietary/nutritional, genetic and environmental factors makes the study of the etiology of NTDs very complex.
Significant advancement has been made in the NTD field in recent decades, with efforts towards their prevention and treatment remaining the areas of greatest research activity. It has been over 3 decades since the discovery that dietary folic acid greatly reduces the risk of having a child afflicted with an NTD [Smithells, et al., 1980], and still the mechanism by which folate responsive and folate non-responsive NTDs arise is little understood. In recent years, the development of genetically modified mouse models have fueled research and increased understanding of potential genetic causes of NTDs [Harris and Juriloff, 2010]. Despite the definition of hundreds of genetically modified mouse models, translation to humans is not always successful. In this study, we draw comparisons between a folate sensitive NTD genetic mouse model and rare SNVs found in a cohort of spina bifida patients, thus broadening the potential of an already available tool, the Lrp6 Crooked Tail (Cd) mouse mutant.
An important approach to mechanistic investigation into the causes of NTDs examines the major bioactive signaling pathways in animals. The Wnt signaling pathways regulate multiple genes, and are considered essential during embryonic neural tube development. In the canonical Wnt pathway, LRP6 (or Lrp5) is the obligate co-receptor that, together with Frizzled, responds to Wnt ligand binding to increase β-catenin entry to the nucleus and stimulate TCF/Lef-dependent transcription [Carter, et al., 2005]. Non-canonical Wnt signaling is also activated through Frizzled as well as other receptors like Vangl and ROR that do not enhance nuclear shuttling of β-catenin, but rather triggers a Rho family GTPase-dependent cascade. Non-canonical Wnt signaling has been found to influence the planar cell polarity (PCP) pathway. Defects in PCP genes have been associated with the severe NTD, craniorachischisis [Wang, et al., 2006], as well as spina bifida [Lei, et al., 2013].
The Crooked tail mouse (Cd) is one of the over 240 distinct mouse genetic models that are prone to NTDs and has been found to closely mimic human clinical response to folic acid [Carter, et al., 1999]. The heterozygous mice display a crooked tail, while homozygous mutants reveal severe NTDs. The homozygous mutants are usually non-viable, but they can be rescued by folate supplementation. Carter et al [Carter, et al., 2005] found that NTDs in the Cd mouse are due to a point mutation of G494D in a highly conserved amino acid in the second YWTD-EGF-like repeat in the LRP6 extracellular domain [Carter, et al., 2005]. This point mutation was observed to interfere with localization of the protein within cells and therefore interfere with Wnt canonical and non-canonical signaling [Gray, et al., 2013]. The mesoderm development (MESD) protein in mice is a chaperone protein in the endoplasmic reticulum that serves the LDL family of proteins [Herz and Marschang, 2003]. MESD is required for the appropriate trafficking of LRP5/6 to the plasma membrane [Hsieh, et al., 2003]. The Cd variant of Lrp6 has been shown to ablate the binding of MESD and LRP6, affecting Lrp6 protein plasma membrane localization [Gray, et al., 2013]. An additional recent report has also indicated human SNVs in the LRP6 (MIM# 603507) to be associated with NTDs in a cohort of patients [Allache, et al., 2014], underlining the potentially great role that LRP6 plays during development.
The present study examines the localization and function of LRP6 when different human SNPs, found by Sanger sequencing in a cohort of spina bifida patients, are introduced into the protein. Wildtype Lrp6 and Cd Lrp6, a known variant, is included along-side the rare variants. Thus, specific patient DNA sequence variants are evaluated using in vitro assays compared to known mouse NTD model systems to identify functionally significant human SNPs that can illuminate mechanisms and contribute to therapeutic strategies.
MATERIALS AND METHODS
Human subjects
Spina bifida cases and non-affected controls were obtained the California Birth Defects Monitoring Program (CBDMP) [Croen, et al., 1991]. Included were 192 infants with isolated spina bifida and without other major birth defects (cases), along with 190 non-malformed infants (controls) randomly selected from among all livebirths in the same time period as cases. Among the 192 spina bifida cases, 82 are Non-Hispanic White, 54 are native US born Hispanics and 56 are foreign born Hispanics. Among the 190 controls, 81 are White Non-Hispanic, 54 are US born Hispanics and 55 are foreign born Hispanics. All of the 192 spina bifida phenotypes were myelomeningocele [Lei, et al., 2013; Lei, et al., 2014]. The approval process includes detailed review by the State of California Committee for the Protection of Human Subjects (the primary IRB). All newborn bloodspot samples (source of DNA) were obtained with approval from the State of California Health and Welfare Agency Committee for the Protection of Human Subjects.
DNA Sequencing and Data Analysis
Genomic DNA was prepared and amplified as previously described [Lei, et al., 2013; Lei, et al., 2014]. Coding exons and flanking exon-intron regions of the human LRP6 gene (NM_002336.2 and NP_002327.2) were amplified by polymerase chain reactions (PCR) from the whole genome amplification (WGA) product. Primer sequences and PCR conditions are available upon request. The PCR products were sequenced using the BigDye® Terminator v3.1 Cycle Sequencing Kit. DNA was sequenced with either a specific forward or reverse primer. Samples were run on ABI 3700 automated sequencer and analyzed using the Mutation Surveyor software V4.0.7 (Softgenetics, Stage College, PA). Human LRP6 variants were verified for absence in 190 ethnically matched controls by sequence analysis and in 4 public databases: dbSNP (http://www.ncbi.nlm.nih.gov/snp), the 1000 genome project (http://www.1000genomes.org), the NHLBI GO Exome Sequencing Project (http://snp.gs.washington.edu/EVS/), and the Exome Aggregation Consortium (ExAC) Browser (http://exac.broadinstitute.org/). Rare variants (<1%) were subsequently confirmed by a second round of whole genome amplification, PCR and sequencing analysis. The Fisher Exact Test was used to compare the frequency of the rare variants between NTDs and controls in public database. A two-tailed p value <0.05 was considered as statistically significant. The effect of rare missense variants on the protein of the coded protein was predicted in silico using the PolyPhen-2 (Polymorphism Phenotyping version 2.1.0; http://genetics.bwh.harvard.edu/pph/) and SIFT (Sorting Intolerant From Tolerant; http://sift.jcvi.org/) programs. In the PolyPhen-2 program, HumDiv-trained model was used for evaluating the damaging effect of the identified LRP6 variants. This model was used because NTDs are complex phenotypes and the four identified variants are rare alleles. The PolyPhen-2 false positive rate (FPR) 5% (0.05)/10%(0.10) was used as the threshold for the ternary classification. Mutations and their Naïve Bayes posterior probability scores associated with estimated FPR below 0.05 are predicted to be probably damaging. Mutations with the Naïve Bayes posterior probability scores associated with FPR between 0.05–0.10 are predicted to be possibly damaging. Mutations with estimated FPR values above 0.10 are classified as benign. SIFT is a sequence homology-based tool which takes a query and uses multiple alignment information to predict tolerated and deleterious substitutions for every position of the query sequence. SIFT searches and compares similar sequences and calculates normalized probabilities for the every possible substitution. Variants with normalized probabilities less than 0.05 are predicted to be deleterious, others are predicted to be tolerated. Multiple alignments of orthologous Lrp6 proteins were done using ClustalW program in Mega 5 software package (http://www.megasoftware.net/).
Plasmid preparation
Mesdc2-Flag plasmid was kindly provided by Dr. Bernadette Holdener (Stony Brook University, New York, USA) [Hsieh, et al., 2003].. Human LRP6 and mouse Lrp6 have 98% identity and all four identified variants described here are predicted to affect the same amino acid residue in both human and mice. Therefore, mouse Lrp6 plasmid, as previously described [Gray, et al., 2013], was used to make the mutant constructs. Mutations to Lrp6-GFP plasmids were made using the GeneArt® Site-Directed Mutagenesis System (Life Technologies). The mutations made were Alanine 3 to Valine, Tyrosine 544 to Cysteine, Proline 1482 to Lysine, and Argine 1574 to Lysine. All mutants were validated by sequencing analyses. M50 Super 8× TOPFlash [Veeman, et al., 2003], human beta-catenin pcDNA3, and pcDNA3-S33Y Beta-catenin [Kolligs, et al., 1999] were purchased from Addgene (https://www.addgene.org/). The AP1 reporter construct was purchased from Agilent. Plasmid DNA was purified from the E. coli using HiSpeed® Plasmid Maxi Kit (QIAGEN) per manufacturer’s instructions. Concentrations were determined using NanoDrop 2000.
Co-Immunoprecipitaiton
HEK293T cells were transfected with Lrp6 cDNA constructs and MESD-Flag using Lipofectamine ® 2000 reagent (Life Technologies) per manufacturer’s instructions with a 0.5:1 ratio of reagent to DNA. Cells were allowed to incubate with transfection media for 24 hours then were washed with PBS and harvested in TBS with 1% Triton × 100 with protease inhibitors (Roche). Cells were lysed by rocking at 4°C for 2 hours. Total protein concentrations of lysate were determined using a Bradford assay. After normalization to total protein, lysate was incubated with Flag antibody (Sigma) for 2 hours. Protein A/G agarose beads (Pierce) were added to mixture and incubated at 4°C overnight while rocking. Beads were washed 5 times with TBS with 0.5% Tween 20, then boiled in SDS loading buffer and supernatant loaded onto a 10% PAGE gel. Nitrocellulose membranes were used for western blotting. Beta Tubulin (Developmental Studies Hybridoma Bank, 1:500), Lrp6 (Cell Signaling, 1:2000), and Flag (Sigma, 1:500) primary antibodies were used. LiCor secondary antibodies were used at 1:10000 and protein bands were analyzed on a LiCor Odyssey CLx.
Cellular Localization
MDCK II cells were used for transient transfection with Lipfectamine® 2000 Reagent. MDCK II cells were plated in a 12 well plate on cover slips (Corning) in MEM media and grown up to 70-90% confluence. Mesdc2 and Lrp6 wildtype or its mutant plasmids were co-transfected using Lipofectamine® 2000 reagent in a 1:1 DNA: reagent ratio. Cells were incubated for 24 hours at 37°C in transfection media. Cells were fixed with paraformaldehyde on cover slips. After rinsing with PBS, cell membranes were permeablized with 1% Triton × 100. Cells were blocked with 5% BSA then incubated with anti-SCRIB antibody (1:1000, Santa Cruz Biotechnology). Secondary anti-goat antibody was used (1:2000, Alexa Fluor® 647). Cells were finally placed in PBS with DAPI (300 ng/mL) for imaging. Using a laser scanning confocal microscope (LSM710, Leica), images were taken with a 10× objective so that at least 100 cells could be counted per field. Cell images were viewed and overlaid in Image J. Chi-squared analysis was performed in Microsoft Excel to compare the localization difference between wildtype and different Lrp6 mutants.
Assessment of Canonical and Non-Canonical Wnt Signaling
Luciferase Reporter Assays were used in order to determine the effects that the various mouse Lrp6 constructs had on both canonical and non-canonical Wnt signaling. TopFlash is a TCF/LEF reporter plasmid that will indicate the activation of the canonical Wnt signaling pathway, whereas AP1 is a reporter plasmid for the downstream activation by Jun-kinase, a measure of the flux through the non-canonical Wnt signaling pathway. HEK293T cells were transfected using Lipofectamine ® 2000 with TopFlash, and various Lrp6 constructs or human beta-catenin pcDNA3 (positive control), or pcDNA3-S33Y Beta-catenin (negative control) at a ratio of 1µg DNA to 0.5µL Lipofectamine ® 2000. Cells were harvested 24 hours post transfection and the Promega Dual Luciferase Assay kit was used per manufacturer’s instructions. AP1 luciferase reporter plasmid (Agilent) was used to evaluate PCP signaling. Cells were transfected with AP1 and various Lrp6 constructs with VANGL2. After 24 hours in transfection media, cells were overlaid with Wnt5a conditioned media, or DMEM-complete media for negative controls as previously described [Allache, et al., 2014]. Conditioned medium was harvested from Wnt-5a secreting L cells per manufacturer’s instructions (ATCC). Cells were harvested after a 24-hour incubation and the Promega Dual Luciferase Assay kit was used per manufacturer’s instructions. A Biotek-2 plate reader was used for luminescence readings and data were analyzed using a 2-tailed student’s T-test in Microsoft Excel.
RESULTS
Rare variants identified in LRP6 among spina bifida patients
The open reading frame and the exon–intron junction of LRP6 were sequenced in 192 spina bifida cases. We identified four rare missense SNPs, c.8C>T ((p.Ala3Val) RNA not analysed), c.1631A>G ((p.Tyr544Cys) RNA not analysed), c.4445C>T ((p.Pro1482Leu) RNA not analysed) and c.4721G>T ((p.Arg1574Let) RNA not analysed), in the LRP6 coding sequence of four cases that were not present in 192 ethnically matched controls, 1000 genome project, nor NHLBI GO Exome Sequencing Project databases (Table 1). Three of the four rare variants were found in the ExAC exome variant browser. The p.Ala3Val variant was found in 1/10932 alleles in Latino populations, the p.Tyr544Cys variant was found in 2/67702 alleles in European populations, and the p.Pro1482Leu variant was found in 3/11586 alleles in Latino populations. Use of a Fisher Exact Test showed that the p.Ala3Val (p=0.011<0.05) and p.Tyr544Cys (p<0.001) variants have a significant difference between the NTD cohort presented here and the compared control populations. Each of these variants was detected in a heterozygous state and was identified in only one case. The ORF and exon–intron junctions of LRP6 were also sequenced in 96 controls, and no rare variants were detected in this gene. Novel LRP6 variants were submitted to Leiden Open Variation Database (http://LRP6.lovd.nl)
Table 1.
Novel rare mutations (<1%) identified in LRP6 in human spina bifida but not in controls
| Nucleotide changea | NCBI/1k/EVSb | ExACd(p valuee) | Protein change | PolyPhen-2 predict (FPR score)f |
SIFT predict (SIFT Score)g |
Conservation |
|---|---|---|---|---|---|---|
| c.8C>T | NAc | 1/10932 (0.011) | p.Ala3Val | benign (1.00) | DAMAGING (0.04) | Yes |
| c.1631A>G | NA | 2/67702 (<0.001) | p.Tyr544Cys | probably damaging (0.0387) | TOLERATED (0.09) | Yes |
| c.4445C>T | NA | 3/11586 (0.086) | p.Pro1482Leu | benign (0.158) | TOLERATED (0.16) | Yes |
| c.4721G>T | NA | NA | p.Arg1574Leu | possibly damaging (0.0556) | DAMAGING (0.01) | Yes |
The DNA mutation and amino acid numbering systems are based on LRP6 cDNA sequence with the accession NM_002336.2 and LRP6 protein sequence with the accession NP_002327.2 respectively. Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence.
Frequency in NCBI, 1000 genome and Exome variants server.
NA not availiable
Data from Exome Aggregation Consortium (ExAC).Data presented as (allele count)/(tested allele number in relative population)
p value represented Fisher's exact test.
FPR (false positive rate) score <0.05: probably damaging; 0.5=< FPR score<0.10: possibly damaging; FRP score>=0.10: benign;
SIFT (sorts intolerant from tolerant) score. Damaging L: SIFT socre<0.05; Tolerated: SIFT socre>=0.05;
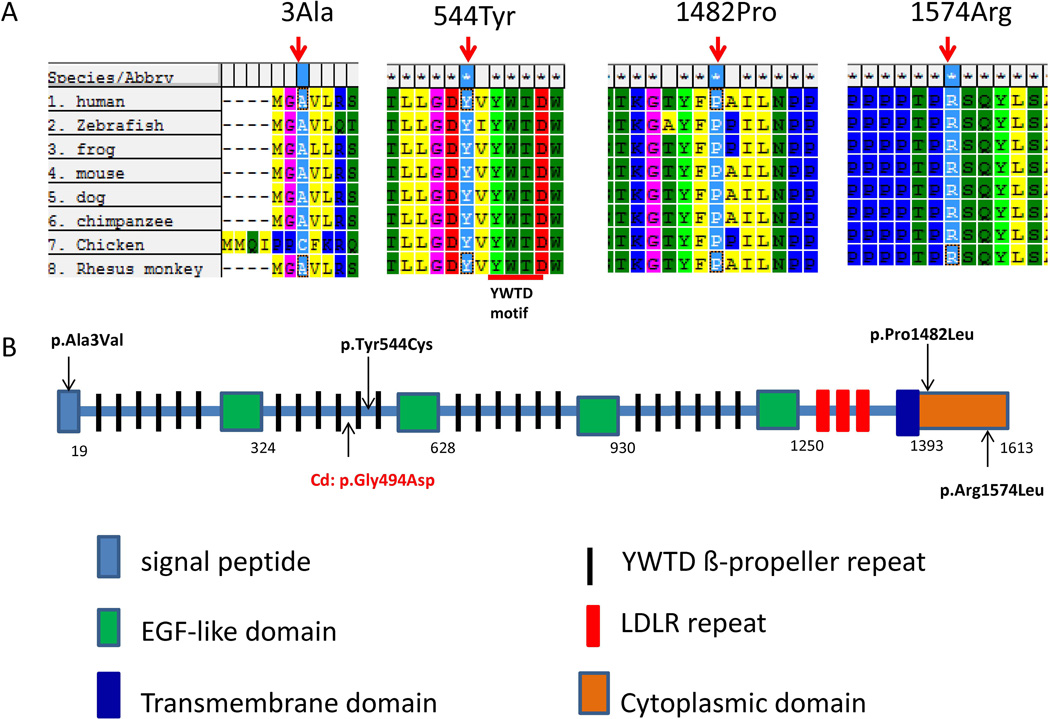
The variant c.8C>T changes a highly conserved alanine residue in the signal peptide region to a valine residue (p.Ala3Val), which is predicted to be damaging by SIFT (Figure 1, Table 1). Compared to alanine, the two extra methyl groups on valine result in a bulkier side chain. The variant c.1631A>G affects a completely conserved tyrosine residue within a YWTD-repeat containing domain, which is common to all LRP family members (Figure 1). This variant leads to a p.Tyr544Cys substitution which is predicted to be probably damaging by PolyPhen (Table 1). Tyrosine has a large and hydrophobic ring form, while cysteine is a small and rather reactive amino acid. Both variants c.4445C>T and c.4721G>T changed highly conserved residues in the cytoplasmic domain of LRP6 (Figure 1). Variant c.4721G>T was predicted to be damaging by both PolyPhen and SIFT and c.4445C>T was predicted to be benign by PolyPhen and tolerated by SIFT (Table 1).
Figure 1.
Rare, novel LRP6 SNVs identified in human spina bifida cases. (A) A partial alignment of human LRP6 with seven other orthologous sequences. The LRP6 variants found in NTD patients affect conserved residues (indicated by arrows). Accession numbers: Homo sapiens LRP6 (human), NP_002327.2; Danio rerio Lrp6 (zebrafish), NP_001128156.1; Xenopus tropicalis Lrp6 (frog), NP_001079233.1; Mus musculus Lrp6 (mouse), NP_032540.2; Canis lupus familiaris LRP6 (dog), XP_534886.2; Pan troglodytes LRP6 (chimpanzee), XP_001152103.1; Gallus gallus Lrp6 (chicken), XP_417286.3; Macaca mulatta LRP6 (Rhesus monkey), NP_001244648.1. (B). A schematic diagram of LRP6 showing the approximate locations of the 4 variants, p.Ala3Val, p.Tyr544Cys, p.Pro1482Leu and p.Arg1574Leu.
Co-Immunoprecipitation
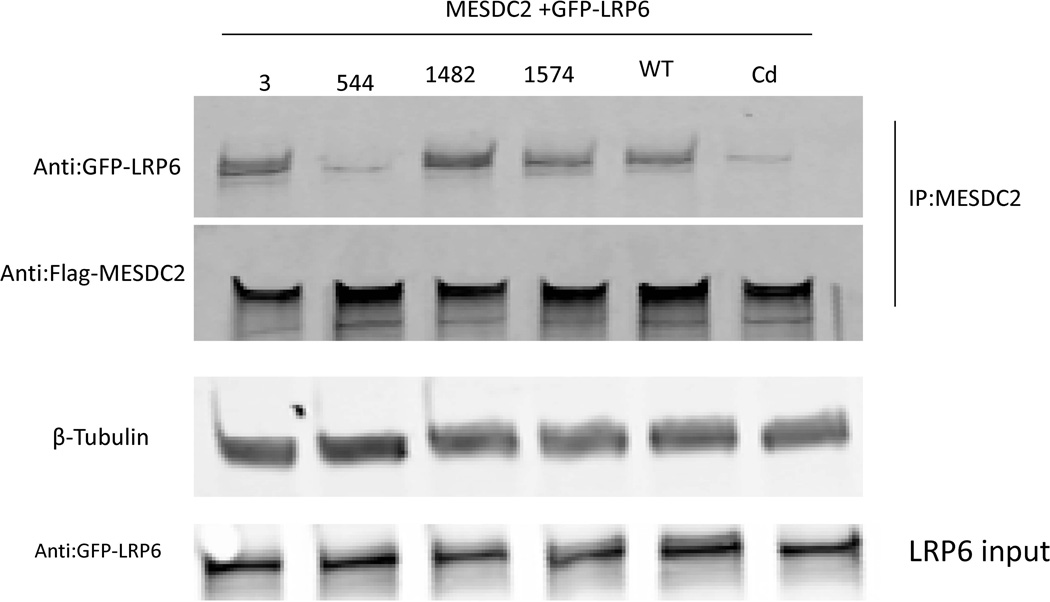
The physical interaction between the Lrp6 mutated constructs and their chaperone protein MESD was detected using co-immunoprecipitation (Co-IP). It has already been observed that LRP6 binds its MESD chaperone to correctly insert itself into the plasma membrane, whereas the Cd mutant does not interact with MESD [Gray, et al., 2013]. Here, we demonstrate that the Lrp6 p.Tyr544Cys mutant behaves in a manner similar to the established Cd mutant. Figure 2 shows that all of the Lrp6 variants found in cases normally bind MESD, except the p.Tyr544Cys point mutation.
Figure 2.
Co-Immunoprecipitation illustrates the effect of novel mutations on the physical interaction of LRP6 with Mesdc2. Following Flag immunoprecipitation, blotting with anti-GFP confirmed physical interaction between Lrp6 wildtype and Mesdc2. This interaction was decreased in the p.Tyr544Cys variant as well as the Cd point mutation. β-Tubulin was taken as an internal control.
Cellular Localization
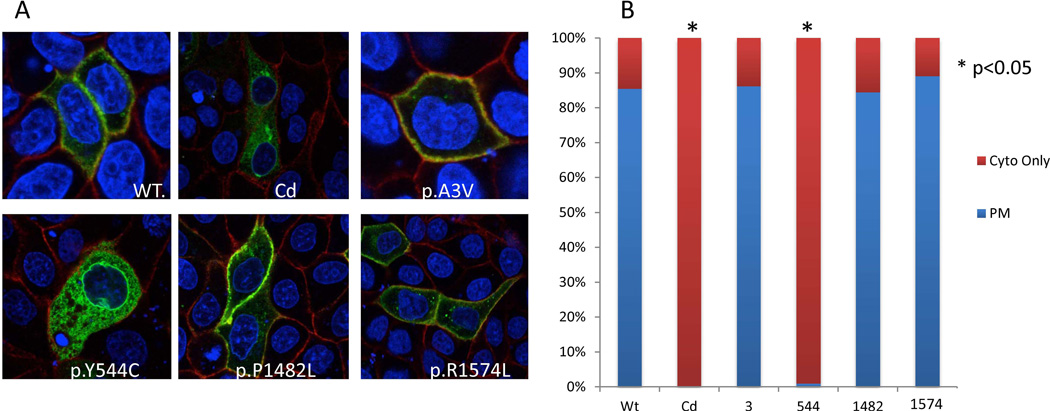
The physical interaction between the Lrp6 protein and MESD assists Lrp6 in correctly inserting into the plasma membrane. The localization of the mutant forms of Lrp6 was therefore investigated in MDCK II cells. The Lrp6 p.Tyr544Cys mutant followed the same pattern of mislocalization as the Lrp6 Cd mutant, as the lost interaction with MESD would predict. Figure 3A shows representative images of the cellular localization of these mutant proteins that are quantified in Figure 3B.
Figure 3.
Subcellular localization of LRP6 variants. (A) Protein subcellular localization in MDCK II cells. Blue color indicates DAPI stain of the nucleus, red color indicates SCRIB stain for plasma membrane, and green color indicates GFP from the LRP6-GFP fusion plasmid constructs. (B) Quantitative analysis of protein localization for GFP-Lrp6 wildtype and novel variants. Fields of cells were imaged randomly and at least 100 GFP positive cells were counted per construct for each of the triplicates. Cells were classified as either GFP only in the cytoplasm or GFP predominantly in the plasma membrane. Asterisks represent a significant (p<0.05) departure from the wildtype construct using a chi-squared analysis.
Assessment of Canonical and Non-Canonical Wnt Signaling
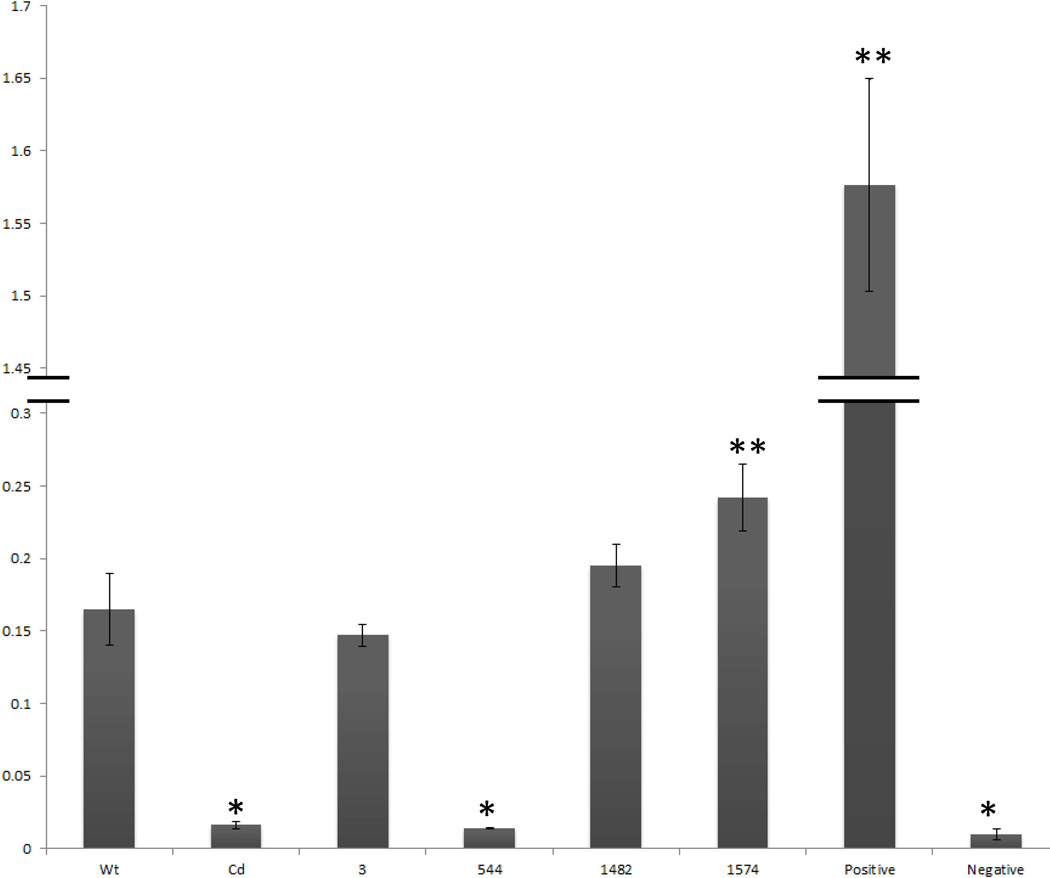
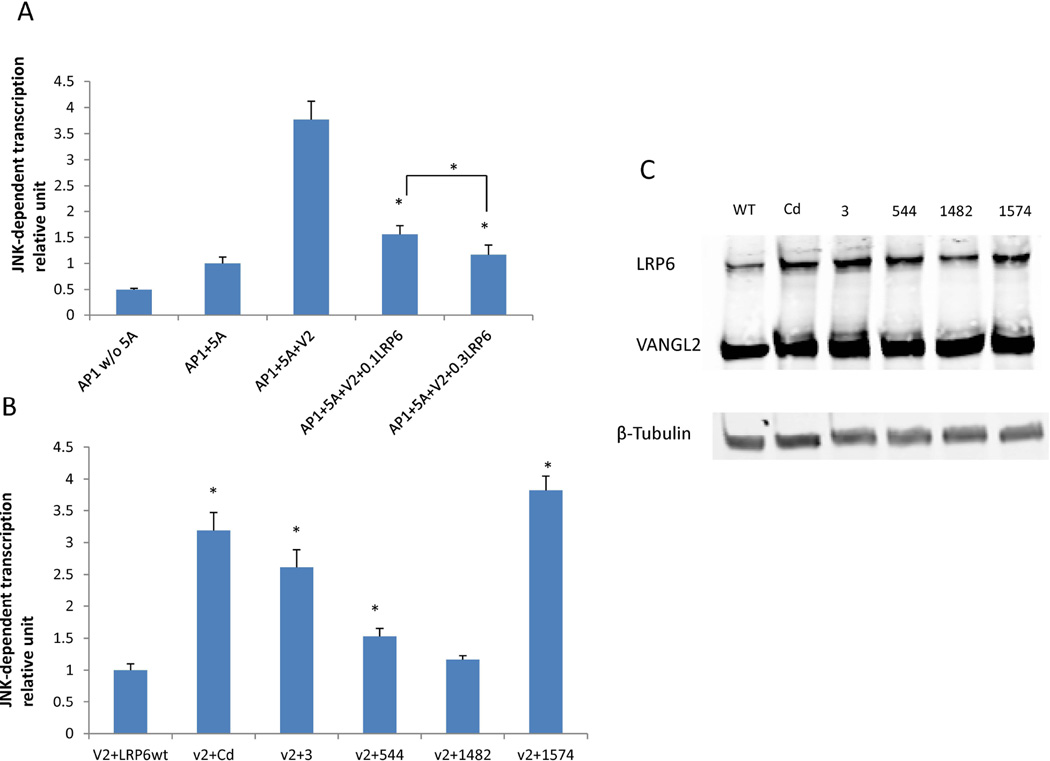
Using a Topflash reporter gene, canonical Wnt signaling was measured in HEK293T cells transfected with the different LRP6 mutants being examined. Previous reports showed that the Lrp6 Cd construct produced attenuated Wnt signaling compared to a wildtype Lrp6 construct [Gray, et al., 2013; Allache, et al., 2014]. Figure 4 demonstrates that the point mutation at p.Tyr544Cys in the LRP6 gene of human spina bifida cases mimics the decrease seen in Wnt signaling in the Cd point mutation. Interestingly, the p.Arg1574Leu mutant is a hyperactive canonical Wnt signaling variant, with a significant increase over the wildtype plasmid. Using an AP1 reporter gene, non-canonical PCP signaling, stimulated by the presence of Wnt5a, was measured in HEK293T cells transfected with the different Lrp6 variants and VANGL2. Figure 5 demonstrates that the wildtype Lrp6 protein decreases the PCP signaling in cells from a positive control in a dose dependent fashion. In the presence of the VANGL2 plasmid, all but one (p.Pro1482Leu) of the NTD-specific Lrp6 mutant constructs significantly increased PCP signaling compared to the wildtype Lrp6 construct (Figure 5B).
Figure 4.
LRP6 variant effects on canonical Wnt signaling pathway. TopFlash Reporter shows that both the Cd variant and the Y544C variant to the Lrp6 gene decrease canonical Wnt/β Catenin signaling. The p.R1524L variant shows a significant upregulation in Wnt/β Catenin signaling compared to the wildtype plasmid. Single asterisks indicate a significant (p<0.05) down regulation in Wnt signaling (two-tailed t-test), whereas double asterisks indicate a significant (P<0.05) up regulation in Wnt signaling (two-tailed t-test)
Figure 5.
LRP6 mutant effects non-canonical Wnt signaling pathway. (A) JNK-AP1 activity was detected in the presence of Wnt5a. Increasing concentrations of Lrp6 significantly inhibited AP1 activity in the presence of Wnt5a and VANGL2. (B) In the presence of VANGL2 and Wnt5a conditioned medium, all tested LRP6 mutants, except p.P1482L, inhibited AP1 activity significantly less when compared with the wildtype LRP6 construct. (C) A representative of western blot analysis showed that LRP6 and VANGL2 constructs expressed at a comparable level. (*p<0.05, two-tailed t-test).
DISCUSSION
Mutations in the Lrp6 gene lead to NTDs in three different mouse models, the Lrp6 knockout mouse, the Lrp6 Cd and the Lrp6 Ringleschwanz (rs) point mutations in mouse [Carter, et al., 2005; Kokubu, et al., 2004; Pinson, et al., 2000], and have been associated with human NTDs [Allache, et al., 2014]. Herein we present four rare LRP6 SNVs that were found in infants with spina bifida, but that were absent in matched controls. Three (p.Ala3Val, p.Tyr544Cys and p.Pro1482Leu) of the four rare variants were found in ExAC exome variant browser with very low frequency. Two of them (p.Ala3Val and p.Tyr544Cys) showed significant difference between NTD cohort and public controls also affected Lrp6 function as demonstrated by in vitro assays. The p.Pro1482Leu which had no difference between NTD cohort and public controls did not affect Lrp6 protein function in both in silico prediction and in vitro functional assays. The p.Tyr544Cys variant was of particular interest because a highly conserved tyrosine is replaced by cysteine within the second YWTD-repeat domain, a motif characteristic of all low density lipoprotein like receptor family members [Johnson, et al., 2004]. Only the p.Tyr544Cys variant was close to the Cd in the second beta-barrel domain, suggesting that p.Tyr544Cys alone may follow the same mechanism of dysregulation as the Cd allele. Another Lrp6 allele, rs, is a hypomorphic point mutation seen in the third beta-propeller domain of the protein [Kokubu, et al., 2004], which also abrogates binding to the MESD chaperone protein [Kubota, et al., 2008], and decreases canonical Wnt signaling, similarly to the Cd variant [Kokubu, et al., 2004]. Data presented here show that the p.Tyr544Cys variant, but not the other three rare SNPs, mimics the previously described Lrp6 Cd and rs variants [Gray, et al., 2013; Kubota, et al., 2008], by failing to properly translocate and insert into the plasma membrane, associated with a significantly abrogated level of canonical Wnt signaling and PCP signaling.
The YWTD-EGF domain pairing that is found in LDL like receptors allows for the proteins to pass through the membrane a single time and form the four β -propeller structures that provide extracellular binding surfaces [Jeon, et al., 2001]. Mutations that compromise the integrity of the key amino acid sequences for these folds are predicted to be damaging to the functionality of the protein by either not allowing for proper binding of partner proteins or simply not allowing the protein to fold correctly in the membrane. The Cd variant of Lrp6, and the recently described novel variants in the gene that have been shown to alter functionality of the Lrp6 protein, are located in the YWTD domains, like the p.Tyr544Cys variant described here [Allache, et al., 2014]. Previous studies provide evidence that the second and third YWTD-repeat containing β-propeller domains of LRP5/6 are necessary for the binding of the chaperone protein, MESD [Carter, et al., 2005] [Chen, et al., 2011; Kubota, et al., 2008]. The work presented herein supports this hypothesis. The p.Tyr544Cys variant directly affects the signature YWTD domain in the second β-propeller by mutating the conserved tyrosine to cysteine, disrupting the binding of MESD to LRP6.
Not only does this work emphasize the importance of a properly placed Wnt signaling receptor for efficient canonical Wnt signal transduction, it further informs the recently proposed hypothesis that Lrp6, traditionally only associated with canonical Wnt signaling, may have an important role in the PCP pathway as well. It has recently been shown that LRP6 complexes with Disheveled-associated activator of morphogeneisis 1 (DAAM1) to influence non-canonical Wnt signaling, presumably by stabilizing the guanine exchange factor (GEF) activity of the DAAM1 complex and increasing levels of active RhoA-GTP [Gray, et al., 2013]. Other recent reports also emphasize the idea of Lrp6-dependent reciprocal canonical and non-canonical Wnt signaling. In a cohort of NTD patients, Allache and colleagues demonstrated that three SNPs in the LRP6 gene decreased canonical Wnt signaling and in return, also increased the flux through the PCP pathway [Allache, et al., 2014]. Here we show that in the presence of VANGL2, three of four SNPs increased PCP signaling compared with wildtype Lrp6. The p.Pro1482Leu variant, which was predicted to be tolerated by SIFT and benign by PolyPhen, mimics the response of the wildtype protein. This substitution of a small hydrophobic amino acid for a different small hydrophobic amino acid falls in the cystoplasmic tail portion of the LRP6 protein. It is unsurprising that this SNV behaves no differently than the wildtype Lrp6 protein.
Interestingly, even though three of the variants found here increased PCP signaling significantly over wildtype, only one (p.Tyr544Cys) of them decreased canonical Wnt signaling and one (p.Arg1574Leu) increased canonical Wnt signaling, another one (p.Ala3Val) had no effect on canonical Wnt signaling. The p.Arg1574Leu mutant increased beta-catenin dependent signaling over the wildtype protein. This mutation changes a charged amino acid in the cytoplasmic portion of the protein to a hydrophobic amino acid. This dramatic change may increase the affinity of the intercellular binding partners of the Lrp6 protein and increase the downstream effects. The cytoplasmic tail is a target for kinases specific to several PPPSPXS motifs in the Lrp6 intracellular domain (ICD) [Piao, et al., 2008] whose phosphorylation governs the interaction between LRP6 and its cytosolic binding partners including GSK3 and Axin [Zeng, et al., 2008]. The p.Arg1574Leu mutation changes a PPPTP-R-S sequence to mutant PPPTP-L-S motif. The change from a highly conserved, charged amino acid to a small hydrophobic residue here may influence phosphorylation of the nearby threonine or serine, and so may increase Wnt/B-Catenin signaling by stabilizing complex formation. The stability of this complex formation for all of the mutations presented here could be a focus of future studies that could help more mechanistically explain the phenomena observed in this initial study. The p.Ala3Val mutation changed a conserved residue in the signal peptide region of LRP6. This variant was seen to increase PCP signaling but, it did not change canonical Wnt signaling. It has previously been reported that the N-terminal of the LRP6 protein is not required for β-catenin dependent Wnt signaling to occur [Brennan, et al., 2004; Tamai, et al., 2004], therefore it is expected that there is no significant change in canonical Wnt signaling for this varaiant. The dysregulation that is seen in PCP signaling with this variant however, underscores the putative participation of LRP6 in PCP signaling and neurulation as well. In any case, the fact that all of the Lrp6 mutant proteins thus far examined in connection with NTD in mouse and man alter flux through the Wnt/PCP pathway while reducing (p.Tyr544Cys), [Allache, et al., 2014; Gray, et al., 2013], leaving unchanged (p.Ala3Val) or enhancing (p.Arg1547Leu) the canonical pathway, suggests that PCP signaling is the Wnt function most critical to successful neurulation.
The p.Tyr544Cys mutant and the Cd mutant both exhibit the expected reciprocal signaling previously described for other variants. Allache and colleagues observed [Allache, et al., 2014] that in the presence of VANGL2 or Dishevelled 3, wildtype LRP6 inhibits PCP signaling in a dose-dependent manner, and a similar phenomenon was also observed in our study (Figure 5A). Three previously identified NTD-specific SNPs were reported to increase PCP signaling in the presence of Dishevelled 3. Here, we found that all three of the identified NTD-specific LRP6 mutants increased PCP signaling in the presence of VANGL2 compared to co-transfected wildtype LRP6. The underlying mechanism for this disruption is unclear, and there may be several. One possible explanation is that VANGL2 plays a role in helping the PCP proteins to assemble in a signaling complex in the cytosol. This complex has been previously suggested as one potential reason increased PCP signaling is seen in Cd mutants [Gray, et al., 2013]. Wild type Lrp6 was seen to diminish PCP signaling in the presence of VANGL2. Potentially, the mutations to the Lrp6 protein make Lrp6 less efficient at inhibiting the VANGL2 and Wnt5a stimulated PCP signaling by making a more stable protein assembly in the cytoplasm.
Despite the existence of three genetically modified mouse models of the Lrp6 gene that are susceptible to NTDs, and a total of over 240 mouse models of NTDs [Harris and Juriloff, 2010], it has proven difficult to relate these mouse models to variants in human populations. Interestingly, the p.Tyr544Cys variant of the LRP6 gene, described here, is found to behave similarly to the Cd and rs variants in mice. The p.Tyr544Cys Cd-likeprotein fails to bind the chaperone protein MESD, which results in an inappropriate localization of the Lrp6 receptor protein and a decrease in canonical Wnt signaling. This discovery relates the folate-responsive Cd mouse model to a relevant variant found in a human NTD population, and underscores its translational importance. In addition, of interest, two new LRP6 SNVs in humans are shown to appropriately bind MESD, localize to the membrane and transduce canonical Wnt signals, but still disregulate PCP signal transduction, suggesting yet another LRP6-dependent interaction between canonical and non-canonical Wnt signaling pathways that is independent of MESD binding. The potential role(s) of LRP6 in PCP signaling expands the importance of this receptor during the proper closure of the neural tube. The novel genetic findings presented here open new avenues for investigation of the etiology and underlying mechanisms of NTDs, their interaction with diet and complex genetics in human patients. For example, future inquiries might target the presence/absence of such variants in parents of infants with spina bifida or in mothers who used or did not use folic acid supplements. One major limitation of this study is that other family members were not studied. Therefore, we could not determine the causation of these variants. In future study, NTD-trios, NTD-tetras or NTD-pedigrees would be collected and studied to overcome this limitation.
ACKNOWLEDGMENTS
We thank the California Department of Public Health, Maternal Child and Adolescent Health Division for providing data. We thank Dr. Bernadette Holdener of Stony Brook University, New York, USA for providing us Mesdc2-flag plasmid. We also appreciate the outstanding confocal microscopy support provided by Dr. Yue Li of the Dell Pediatric Research Institute. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the California Department of Public Health or the National Institutes of Health.
Grant Sponsor: This work was supported by National Institutes of Health [P01HD067244 to MER and RHF, R01ES021006 to RHF].
REFERENCES
- Allache R, Lachance S, Guyot MC, De Marco P, Merello E, Justice MJ, Capra V, Kibar Z. Novel mutations in Lrp6 orthologs in mouse and human neural tube defects affect a highly dosage-sensitive Wnt non-canonical planar cell polarity pathway. Hum Mol Genet. 2014;23(7):1687–1699. doi: 10.1093/hmg/ddt558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan K, Gonzalez-Sancho JM, Castelo-Soccio LA, Howe LR, Brown AM. Truncated mutants of the putative Wnt receptor LRP6/Arrow can stabilize beta-catenin independently of Frizzled proteins. Oncogene. 2004;23(28):4873–4884. doi: 10.1038/sj.onc.1207642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M, Chen X, Slowinska B, Minnerath S, Glickstein S, Shi L, Campagne F, Weinstein H, Ross ME. Crooked tail (Cd) model of human folate-responsive neural tube defects is mutated in Wnt coreceptor lipoprotein receptor-related protein 6. Proc Natl Acad Sci U S A. 2005;102(36):12843–12848. doi: 10.1073/pnas.0501963102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M, Ulrich S, Oofuji Y, Williams DA, Ross ME. Crooked tail (Cd) models human folate-responsive neural tube defects. Hum Mol Genet. 1999;8(12):2199–2204. doi: 10.1093/hmg/8.12.2199. [DOI] [PubMed] [Google Scholar]
- Chen J, Liu CC, Li Q, Nowak C, Bu G, Wang J. Two structural and functional domains of MESD required for proper folding and trafficking of LRP5/6. Structure. 2011;19(3):313–323. doi: 10.1016/j.str.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croen LA, Shaw GM, Jensvold NG, Harris JA. Birth defects monitoring in California: a resource for epidemiological research. Paediatr Perinat Epidemiol. 1991;5(4):423–427. doi: 10.1111/j.1365-3016.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- Gray JD, Kholmanskikh S, Castaldo BS, Hansler A, Chung H, Klotz B, Singh S, Brown AM, Ross ME. LRP6 exerts non-canonical effects on Wnt signaling during neural tube closure. Hum Mol Genet. 2013;22(21):4267–4281. doi: 10.1093/hmg/ddt277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MJ, Juriloff DM. An update to the list of mouse mutants with neural tube closure defects and advances toward a complete genetic perspective of neural tube closure. Birth Defects Res A Clin Mol Teratol. 2010;88(8):653–669. doi: 10.1002/bdra.20676. [DOI] [PubMed] [Google Scholar]
- Herz J, Marschang P. Coaxing the LDL receptor family into the fold. Cell. 2003;112(3):289–292. doi: 10.1016/s0092-8674(03)00073-4. [DOI] [PubMed] [Google Scholar]
- Hsieh JC, Lee L, Zhang L, Wefer S, Brown K, DeRossi C, Wines ME, Rosenquist T, Holdener BC. Mesd encodes an LRP5/6 chaperone essential for specification of mouse embryonic polarity. Cell. 2003;112(3):355–367. doi: 10.1016/s0092-8674(03)00045-x. [DOI] [PubMed] [Google Scholar]
- Jeon H, Meng W, Takagi J, Eck MJ, Springer TA, Blacklow SC. Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair. Nat Struct Biol. 2001;8(6):499–504. doi: 10.1038/88556. [DOI] [PubMed] [Google Scholar]
- Johnson ML, Harnish K, Nusse R, Van Hul W. LRP5 and Wnt signaling: a union made for bone. J Bone Miner Res. 2004;19(11):1749–1757. doi: 10.1359/JBMR.040816. [DOI] [PubMed] [Google Scholar]
- Kokubu C, Heinzmann U, Kokubu T, Sakai N, Kubota T, Kawai M, Wahl MB, Galceran J, Grosschedl R, Ozono K, et al. Skeletal defects in ringelschwanz mutant mice reveal that Lrp6 is required for proper somitogenesis and osteogenesis. Development. 2004;131(21):5469–5480. doi: 10.1242/dev.01405. [DOI] [PubMed] [Google Scholar]
- Kolligs FT, Hu G, Dang CV, Fearon ER. Neoplastic transformation of RK3E by mutant beta-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol Cell Biol. 1999;19(8):5696–5706. doi: 10.1128/mcb.19.8.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Michigami T, Sakaguchi N, Kokubu C, Suzuki A, Namba N, Sakai N, Nakajima S, Imai K, Ozono K. Lrp6 hypomorphic mutation affects bone mass through bone resorption in mice and impairs interaction with Mesd. J Bone Miner Res. 2008;23(10):1661–1671. doi: 10.1359/jbmr.080512. [DOI] [PubMed] [Google Scholar]
- Lei Y, Zhu H, Duhon C, Yang W, Ross ME, Shaw GM, Finnell RH. Mutations in planar cell polarity gene SCRIB are associated with spina bifida. PLoS One. 2013;8(7):e69262. doi: 10.1371/journal.pone.0069262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Zhu H, Yang W, Ross ME, Shaw GM, Finnell RH. Identification of novel CELSR1 mutations in spina bifida. PLoS One. 2014;9(3):e92207. doi: 10.1371/journal.pone.0092207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell LE. Epidemiology of neural tube defects. Am J Med Genet C Semin Med Genet. 2005;135C(1):88–94. doi: 10.1002/ajmg.c.30057. [DOI] [PubMed] [Google Scholar]
- Piao S, Lee SH, Kim H, Yum S, Stamos JL, Xu Y, Lee SJ, Lee J, Oh S, Han JK, et al. Direct inhibition of GSK3beta by the phosphorylated cytoplasmic domain of LRP6 in Wnt/beta-catenin signaling. PLoS One. 2008;3(12):e4046. doi: 10.1371/journal.pone.0004046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature. 2000;407(6803):535–538. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- Smithells RW, Sheppard S, Schorah CJ, Seller MJ, Nevin NC, Harris R, Read AP, Fielding DW. Possible prevention of neural-tube defects by periconceptional vitamin supplementation. Lancet. 1980;1(8164):339–340. doi: 10.1016/s0140-6736(80)90886-7. [DOI] [PubMed] [Google Scholar]
- Tamai K, Zeng X, Liu C, Zhang X, Harada Y, Chang Z, He X. A mechanism for Wnt coreceptor activation. Mol Cell. 2004;13(1):149–156. doi: 10.1016/s1097-2765(03)00484-2. [DOI] [PubMed] [Google Scholar]
- Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol. 2003;13(8):680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- Wallingford JB, Niswander LA, Shaw GM, Finnell RH. The continuing challenge of understanding, preventing, and treating neural tube defects. Science. 2013;339(6123):1222002. doi: 10.1126/science.1222002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hamblet NS, Mark S, Dickinson ME, Brinkman BC, Segil N, Fraser SE, Chen P, Wallingford JB, Wynshaw-Boris A. Dishevelled genes mediate a conserved mammalian PCP pathway to regulate convergent extension during neurulation. Development. 2006;133(9):1767–1778. doi: 10.1242/dev.02347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Huang H, Tamai K, Zhang X, Harada Y, Yokota C, Almeida K, Wang J, Doble B, Woodgett J, et al. Initiation of Wnt signaling: control of Wnt coreceptor Lrp6 phosphorylation/activation via frizzled, dishevelled and axin functions. Development. 2008;135(2):367–375. doi: 10.1242/dev.013540. [DOI] [PMC free article] [PubMed] [Google Scholar]